Abstract
The mechanisms by which chromosomes condense and segregate during developmentally regulated cell division are of interest for Streptomyces coelicolor, a sporulating, filamentous bacterium with a large, linear genome. These processes coordinately occur as many septa synchronously form in syncytial aerial hyphae such that prespore compartments accurately receive chromosome copies. Our genetic approach analyzed mutants for ftsK, smc, and parB. DNA motor protein FtsK/SpoIIIE coordinates chromosome segregation with septum closure in rod-shaped bacteria. SMC (structural maintenance of chromosomes) participates in condensation and organization of the nucleoid. ParB/Spo0J partitions the origin of replication using a nucleoprotein complex, assembled at a centromere-like sequence. Consistent with previous work, we show that an ftsK-null mutant produces anucleate spores at the same frequency as the wild-type strain (0.8%). We report that the smc and ftsK deletion-insertion mutants (ftsK′ truncation allele) have developmental segregation defects (7% and 15% anucleate spores, respectively). By use of these latter mutants, viable double and triple mutants were isolated in all combinations with a previously described parB-null mutant (12% anucleate spores). parB and smc were in separate segregation pathways; the loss of both exacerbates the segregation defect (24% anucleate spores). For a triple mutant, deletion of the region encoding the FtsK motor domain and one transmembrane segment partially alleviates the segregation defect of the smc parB mutant (10% anucleate spores). Considerable redundancy must exist in this filamentous organism because segregation of some genomic material occurs 90% of the time during development in the absence of three functions with only a fourfold loss of spore viability. Furthermore, we report that scpA and scpAB mutants (encoding SMC-associated proteins) have spore nucleoid organization defects. Finally, FtsK-enhanced green fluorescent protein (EGFP) localized as bands or foci between incipient nucleoids, while SMC-EGFP foci were not uniformly positioned along aerial hyphae, nor were they associated with every condensing nucleoid.
Streptomyces coelicolor is a filamentous sporulating soil saprophyte and it is the best-characterized model organism for an important group of bacteria known for their elaborate life cycle and their production of biologically active compounds. During the culmination of their life cycle, streptomycetes make chains of unicellular, metabolically quiescent reproductive cells that subsequently separate for dispersion and long-term survival in the environment (13). The genome of S. coelicolor is a large 8.7-Mb linear molecule with a centrally located oriC region, ending with long terminal inverted repeats (TIRs) (3). An aging surface-grown Streptomyces colony contains two basic zones of filamentous cells with different cytological properties (15). The vegetative mycelium contains branching syncytial hyphae, where the genome copies are not segregated or condensed into nucleoids and septation is infrequent and unevenly spaced. In contrast, the aerial mycelium contains mostly unbranched syncytial aerial hyphae, and within a given hypha, uniformly spaced septation synchronously occurs to form a chain of spores, each with a highly condensed nucleoid. Coincident with the synchronous septation, dozens of dispersed copies of the genome are accurately partitioned and condensed in the forming unicellular spores. It is conceivable that these synchronous processes coordinating the accurate separation of multiple copies of a linear genome may require a special or augmented system of segregation and condensation.
Condensation and segregation of the genome is a subject of great interest in microbial cell biology. Most of the known information has come from the study of bacterial cells that undergo binary fission (14, 51). These bacterial models typically have a single circular chromosome that is compacted into a nucleoid, and genome segregation occurs simultaneously with DNA replication. In rod-shaped bacteria, three well-characterized systems dovetail to ensure that chromosomes are equally and faithfully partitioned to daughter cells. FtsK (SpoIIIE), SMC (structural maintenance of chromosomes; functional homologue, MukB in Escherichia coli), and ParAB (Soj-Spo0J) have been identified as key players. First, FtsK/SpoIIIE homologues are well-studied sequence-directed DNA motor proteins that localize at the invaginating septa and clear each genome copy to the appropriate side of the septum during vegetative growth and sporulation (references 2, 6, 23, and 42 and references therein). The homologues typically have an N-terminal domain with membrane-spanning segments responsible for septal localization separated by a linker from a more highly conserved C-terminal motor protein domain (34). Second, SMC proteins are widely distributed from bacteria to mammals and participate in higher levels of chromatin organization, acting either as condensins or cohesins (references 18, 22, 38, and 50 and references therein). DNA may be pulled into the cell by the action of these proteins from a single site or a limited number of sites. The molecular details of how these large hinged proteins function in association with non-SMC modulators (ScpAB/MukEF) to compact the chromosome into higher-order structures are being determined (8, 9, 21). Third, chromosomes from a wide range of bacteria use a partition system related to ones described for certain low-copy-number plasmids. Two trans-acting factors, ParA (Soj) ATPase and ParB (Spo0J), and cis-acting parS (“centromere”) sites in the origin-proximal region are utilized (references 5, 12, and 51 and references therein). Spo0J (ParB) binds to the parS sites to form a large nucleoprotein complex that can spread beyond the origin region (7). ParA is required for the proper placement of the ParB-parS nucleoprotein complex in the cell. For unicellular bacteria, individual inactivation of any of these three systems results in a severe segregation phenotype, and combining them typically leads to synthetic lethal phenotypes.
How copies of the linear genome are synchronously partitioned during sporulation in S. coelicolor is beginning to be appreciated. If utilized, the physical landmarks in the apical aerial hypha, the tip (pole) and the septum at the base of the long apical cell, can be separated by 40 to 50 μm or more. DNA segregation occurs after active rounds of DNA replication populate the predivisional apical cell with several dozen dispersed copies of the genome (44). After DNA replication ceases, septa synchronously form over these unsegregated copies of the genome (37, 46, 54). ParB binds to about 20 parS sites densely packed around oriC in the chromosome, forming a nucleoprotein complex that helps to accurately partition the chromosome during division (24, 28). ParB-enhanced green fluorescent protein (EGFP) localizes as regularly spaced foci assembled along aerial hyphae before DNA segregation and septation begin, suggesting that the origin region of the chromosome is accurately positioned between where adjacent septa form (25). ParA assembles into helical structures along aerial hyphae to facilitate the organization of evenly spaced ParB-oriC complexes (26). It was anticipated that the FtsK motor protein would be instrumental in ensuring that the genomic DNA trapped by invaginating septa would be properly partitioned; however, strains containing a complete deletion of ftsK resulted in a mild segregation phenotype (1, 53). SffA, another FtsK/SpoIIIE-like protein, localizes to sporulation septa and may function in chromosomal DNA organization during differentiation (1). Septal localization of SffA requires a small protein called SmeA, which itself appears to have a more pleiotropic role in spore maturation. Thus, progress has been made, but there are still substantial gaps in our understanding of how the linear genome is segregated and condensed.
In this study, we evaluated the functions of ftsK and smc in S. coelicolor by constructing ftsK and smc deletion-insertion mutant strains. In contrast to what was found in two recent studies, the ftsK allele reported here results in a C-terminal truncation, removing the DNA motor domain of FtsK and a membrane-spanning segment. Each mutation studied here was found to affect the accuracy of genome segregation into spores. We then extended our analysis using a previously published parB mutation. We report that all double and triple mutants are viable. Surprisingly, the majority of the spores produced by a triple mutant still receive genomic material with a modest fourfold decrease in viability, suggesting that there are additional genes responsible for proper developmental genome segregation and condensation. In addition, we show that mutations in the genes encoding SMC-associated proteins ScpA and ScpB mainly affect the nucleoid morphology in prespore compartments rather than impairing segregation. Finally, we determined the subcellular localization of FtsK and SMC in aerial hyphae undergoing differentiation using EGFP translational fusions.
MATERIALS AND METHODS
Bacterial strains and media.
S. coelicolor A3(2) strains used in this study are listed in Table 1. YEME (liquid), R2YE (agar), MS (agar), and minimal medium (MM; agar with 0.5% glucose [wt/vol]) were used for growth of S. coelicolor at 30°C (27, 36). Standard procedures for protoplast preparation and transformation were used (27). Antibiotics used for S. coelicolor were as follows: apramycin at 25 μg ml−1, neomycin at 10 μg ml−1, hygromycin at 200 μg ml−1, kanamycin at 160 μg ml−1, and thiostrepton at 50 μg ml−1. The viability of the wild type and of certain mutant strains was determined using the average of at least two independent spore preparations (growth on MS) analyzed from duplicate serial dilutions by direct count using phase-contrast microscopy and viable count after 3 days of incubation on glucose MM.
TABLE 1.
S. coelicolor A3(2) strains used in this study
| Strain | Genotypea | Isolationb | Reference or source |
|---|---|---|---|
| 2709 | proA1 hisA1 argA1 cysD18 uraA1 strA1 SCP1− SCP2− | 27 | |
| HJ2 | Δsmc::hyg | M145 × pHJ2 | This study |
| J2537 | ΔparB::acc(3)IV | 28 | |
| J2540 | ΔparB::acc(3)IV/parB+ | J2537::pIJ6539 | 28 |
| JM148 | ΔftsK::aphI (aphI inserted in same orientation as ftsK′) | M145 × pJM148 | This study |
| JM152 | ΔftsK::aphI (aphI inserted in opposite orientation from ftsK′) | M145 × pJM152 | This study |
| M145 | Prototroph SCP1− SCP2− | 27 | |
| RMD1 | ftsK-egfp acc(3)IV | M145 × 7C7ftsK-egfp | This study |
| RMD2 | smc-egfp acc(3)IV | M145 × 7A1smc-egfp | This study |
| RMD3 | Δsmc::hyg ΔftsK::aphI | HJ2 × JM148 | This study |
| RMD4 | Δsmc::hyg ΔparB::acc(3)IV | HJ2 × J2537 | This study |
| RMD5 | ΔparB::acc(3)IV ΔftsK::aphI | J2537 × JM148 | This study |
| RMD6 | Δsmc::hyg ΔparB::acc(3)IV ΔftsK::aphI | RMD3 × J2537 | This study |
| RMD7 | Δsmc::hyg/smc+ | HJ2/pHW35 | This study |
| RMD8 | ΔftsK::aphI/ftsK+ | JM148/pRMD6 | This study |
| RMD9 | Δsmc::hyg ΔftsK::aphI/ftsK+ | RMD3/pRMD6 | This study |
| RMD10 | Δsmc::hyg ΔftsK::aphI/smc+ | RMD3/pHW35 | This study |
| RMD11 | Δsmc::hyg ΔparB::acc(3)IV/smc+ | RMD4/pHW35 | This study |
| RMD12 | Δsmc::hyg ΔparB::acc(3)IV/parB+ | J2540 × HJ2 | This study |
| RMD13 | ΔparB::acc(3)IV ΔftsK::aphI/ftsK+ | RMD5/pRMD6 | This study |
| RMD14 | ΔparB::acc(3)IV ΔftsK::aphI/parB+ | J2540 × RMD5 | This study |
| RMD15 | Δsmc::hyg ΔparB::acc(3)IV ΔftsK::aphI/ftsK+ | RMD6/pRMD6 | This study |
| RMD16 | Δsmc::hyg ΔparB::acc(3)IV ΔftsK::aphI/smc+ | RMD6/pHW35 | This study |
| RMD17 | ΔSCO1768::acc(3)IV | M145 × pRMD7 | This study |
| RMD18 | ΔscpA::acc(3)IV | M145 × pRMD8 | This study |
| RMD19 | ΔscpAB::acc(3)IV | M145 × pRMD9 | This study |
| RMD20 | Δsmc::hyg ΔscpA::acc(3)IV | HJ2 × RMD18 | This study |
| RMD21 | Δsmc::hyg ΔscpAB::acc(3)IV | HJ2 × RMD19 | This study |
| RMD22 | ΔscpA::acc(3)IV/scpAB+ | RMD18/pRMD10 | This study |
| RMD23 | ΔscpAB::acc(3)IV/scpAB+ | RMD19/pRMD10 | This study |
| RMD24 | ΔftsK::acc(3)IV (null, deletion begins upstream of 5′ end) | M145 × pAEB232 | This study |
All mutants are derivatives of prototrophic strain M145.
Strains were constructed by protoplast transformation: the recipient strain is listed first and the source of donor DNA second (e.g., HJ2 × JM148 indicates that HJ2 was transformed with DNA prepared from JM148), or the recipient strain is listed first and the mutagenic plasmid second (e.g., M145 × pHJ2).
E. coli strain TG1 was used for standard plasmid manipulation (45). E. coli strain BW25113 containing λ RED plasmid pIJ790 (19) was used for in vivo construction of egfp fusions or deletion of scpAB. E. coli strain ET12567 (dam dcm hsdM) (31) was used to prepare unmodified plasmid and cosmid DNA to circumvent the restriction system of S. coelicolor. Final concentrations of antibiotics in LB medium used for E. coli were as follows: for ampicillin, 100 μg ml−1; for carbenicillin, 100 μg ml−1; for apramycin, 100 μg ml−1; for chloramphenicol, 25 μg ml−1; and for kanamycin, 50 μg ml−1.
Plasmids and general DNA techniques.
Plasmids used in this study are listed in Table 2. S. coelicolor total DNA preparations were obtained using the Wizard genomic DNA purification kit (Promega). DNA restriction and modifying enzymes were used according to the manufacturer's recommendation (New England BioLabs). Taq (Promega) and Pfx (Invitrogen) DNA polymerases were used according to the manufacturer's instructions. Redirect technology (19) was used for λ RED-mediated recombination using mutagenic linear DNA cassettes in E. coli.
TABLE 2.
Plasmids and cosmids used in this study
| Plasmid | Description | Reference or source |
|---|---|---|
| SC7A1 | Cosmid source of smc | 43 |
| SC7C7 | Cosmid source of ftsK | 43 |
| SCI51 | Cosmid source of scpAB | 43 |
| H24parB-egfp | egfp inserted in frame 3′ of parB in cosmid SCH24; acc(3)IV aphII | 25 |
| 7A1smc-egfp | egfp inserted in frame 3′ of smc in cosmid SC7A1; acc(3)IV aphII | This study |
| 7C7ftsK-egfp | egfp inserted in frame 3′ of ftsK in cosmid SC7C7; acc(3)IV aphII | This study |
| pAEB228 | 4.2-kb PvuII fragment from SC7C7 containing ftsK cloned into EcoRV-digested pBluescript (ftsK; same orientation as lacZ′) | This study |
| pAEB232 | 2.5-kb MluI-SphI deletion of pJR143 [ΔftsK::acc(3)IV; same orientation as ftsK]; aadA | This study |
| pAH137 | Source of neomycin resistance gene aphI | A. Hausler, unpublished data |
| pBluescriptII SK(+) | Standard cloning vector | Stratagene |
| pIJ773 | Source of acc(3)IV for in vivo recombination | 19 |
| pIJ6539 | 5.3-kb EcoRI fragment containing parB; tsr (parB complementation plasmid) | 28 |
| pKC1053 | Source of hygromycin resistance gene hyg | 29 |
| pHJ2 | 3.2-kb SalI-PstI deletion of pHW25 (Δsmc::hyg; same orientation as smc); acc(3)IV | This study |
| pHW24 | 5.4-kb XhoI fragment from SC7A1 cloned into pBluescript (smc; opposite orientation from lacZ′) | This study |
| pHW25 | 5.4-kb XhoI fragment from SC7A1 cloned into pBluescript (smc; same orientation as lacZ′) | This study |
| pHW35 | 24-kb BamHI-EcoRI fragment from pJRM10 cloned into pHW24 (smc complementation plasmid) | This study |
| pJR143 | 6.1-kb BamHI-PstI fragment of SC7C7 containing ftsK cloned into pOJ260 (ftsK; same orientation as lacZ′); acc(3)IV | This study |
| pJR148 | 1.8-kb SphI fragment deletion of pJR143 (ΔftsK::aphI; same orientation as ftsK′); acc(3)IV | This study |
| pJR152 | 1.8-kb SphI fragment deletion of pJR143 (ΔftsK::aphI; opposite orientation from ftsK′); acc(3)IV | This study |
| pJRM10 | Bifunctional cloning vector, low copy number in S. coelicolor; bla tsr | 35 |
| pOJ260 | pUC-like plasmid containing oriT and acc(3)IV in place of bla | 4 |
| pOJ427 | Source of apramycin resistance gene acc(3)IV for in vitro recombination | B. Schoner, unpublished data |
| pRMD6 | 24-kb EcoRI-HindIII fragment from pJRM10 cloned into pAEB228 (ftsK complementation plasmid) | This study |
| pRMD7 | Cosmid SCI51 containing ΔSCO1768::acc(3)IV by in vivo recombination; aphII | This study |
| pRMD8 | Cosmid SCI51 containing ΔscpA::acc(3)IV by in vivo recombination; aphII | This study |
| pRMD9 | Cosmid SCI51 containing ΔscpAB::acc(3)IV by in vivo recombination; aphII | This study |
| pRMD10 | 4.8-kb XbaI fragment from SCI51 containing scpAB cloned into pBluescript (scpAB; opposite orientation from lacZ′) | This study |
| pRMD11 | 24-kb HindIII-SpeI fragment from pJRM10 cloned into pRMD10 (scpAB complementation plasmid) | This study |
Isolation of ftsK insertion-deletion mutant strains.
Insertion-deletion mutations for ftsK of S. coelicolor were constructed in E. coli by use of traditional in vitro recombinant DNA technology. In plasmids pJR148 and pJR152 (Table 2), 1.8 kb of the 3′ end of the ftsK gene, encoding the DNA motor domain and one membrane-spanning segment, was replaced by a neomycin resistance marker (aphI), inserted in either orientation. The insertion-deletion mutations in pJR148 and pJR152 were introduced into the chromosome by transformation, selecting for neomycin resistance and screening for apramycin sensitivity. Two independent representative strains were named JM148 and JM152, respectively. These strains could express a C-terminally truncated version of FtsK containing the first 203 amino acids (out of 929). In plasmid pAEB232 (Table 2), a 2.5-kb deletion, beginning 57 bases upstream of the translation start site and ending at the same position as described above, results in the entire 5′ end of the ftsK gene being replaced by an apramycin resistance marker [acc(3)IV]. The insertion-deletion mutation in pAEB232 was introduced into the S. coelicolor chromosome by transformation, selecting for apramycin resistance and screening for spectinomycin sensitivity. The resulting strain RMD24 should not express FtsK.
Isolation of smc deletion-insertion mutant strains.
Insertion-deletion mutations in smc of S. coelicolor were constructed in E. coli by use of traditional in vitro recombinant DNA technology. Initially, we isolated a Δsmc::acc(3)IV mutant (strain HW1) to determine that the smc gene was dispensable (55). We isolated another smc-null strain, containing Δsmc::hyg, for compatibility with other mutation markers used in this study. In plasmid pHJ2 (Table 2), the majority of smc was replaced by a hygromycin resistance gene (hyg). By use of pHJ2, the Δsmc::hyg mutation was introduced into the S. coelicolor chromosome by transformation, selecting for hygromycin resistance and screening for apramycin sensitivity. One representative strain was named HJ2.
Characterization of a parB mutant strain.
Strain J2537 [ΔparB::acc(3)IV] strain was a gift from Keith Chater (28). By Southern blot hybridization analysis, we determined that the apramycin resistance gene was in the same orientation as parB.
Isolation of scpA and scpAB mutants.
Insertion-deletion mutations in SCO1768, scpA, and scpB of S. coelicolor were constructed in vivo in E. coli by use of λ RED-mediated recombination and cosmid SCI51. In cosmid derivatives pRMD7, pRMD8, and pRMD9, the majority of the SCO1768 (encoding a putative pseudouridine synthase), scpA, and scpAB genes, respectively, were replaced by a cassette obtained from pIJ773, containing an apramycin resistance gene and oriT flanked by FRT sites (19). Oligonucleotides listed in Table 3 were used as primers in PCRs to add homology to the ends of the apramycin resistance gene cassette. Primers 60Sco1768 and 59Sco1768 were used for completely deleting SCO1768, oScpA60 and oScpA59 were used for deleting the middle 777 bases of the 792-base scpA, and oScpA60 and oScpB59 were used for deleting scpA and the first 426 bases of the 5′ end of the 672-base scpB. The insertion-deletion mutations in each of the cosmids were introduced into the chromosome by conjugation, selecting for apramycin resistance and screening for kanamycin sensitivity. Representative strains were named RMD17 [ΔSCO1768::acc(3)IV], RMD18 [ΔscpA::acc(3)IV], and RMD19 [ΔscpAB::acc(3)IV].
TABLE 3.
Oligonucleotides used in this study
| Oligonucleotide | Sequencea | Application |
|---|---|---|
| oSMCgfpF | CGGTGTGTCGAAGGTCATCAGCCAGCGGTTGCGTCAGCCCCTGCCGGGCCCGGAGCTG | Construction of smc-egfp |
| oSMCgfpR | GAAGTGGTTATGTGTTCAAGTCTTGAAGAACGAGGGTTCACATATGTAGGCTGGAGCTGCTTC | Construction of smc-egfp |
| oKgfpF | GGACGGCGTGCTCGCGGTGATCCGTGGGGAGTCTGAAGGGCTGCCGGGCCCGGAGCTG | Construction of ftsK-egfp |
| oKgfpR | CGGCGGATCGTCGGCGGAACCCTTCTCCTACCCGCCCCTACATATGTAGGCTGGAGCTGCTTC | Construction of ftsK-egfp |
| 60Sco1768 | CCGGGGACGCAGACGACAAGACGGAATTTTGATGCGAAGCATTCCG GGGATCCGTCGACC | Construction of ΔSCO1768::acc(3)IV |
| 59Sco1768 | TCGTCGGGGCAGGGGTTGCCCAGAGGCCGGTCTAGAGGTCTGTAGGCTGGAGCTGCTTC | Construction of ΔSCO1768::acc(3)IV |
| oScpA60 | CTCCGACGACCCGGACGACGGTGTCTTCAAGGTGCGGCTCATTCCGGGGATCCGTCGACC | Construction of ΔscpA::acc(3)IV |
| oScpA59 | GTGGCCTCCTCGGCCTCCGTGATCCGCTCACTCACGCCTTTGTAGGCTGGAGCTGCTTC | Construction of ΔscpA::acc(3)IV |
| oScpB59 | TGCCCGCCTCCTCGACCAGACCCCGCTGCAGGAGGGTGCGTGTAGGCTGGAGCTGCTTC | Construction of ΔscpAB::acc(3)IV |
Start codons and reverse complements of stop codons are in boldface.
Isolation of double and triple mutant strains.
Double and triple mutant strains were constructed using protoplasts prepared from one mutant strain and transforming with chromosomal DNA prepared from a second mutant strain (Table 1). All double and triple mutant strains were readily isolated by selection using the antibiotic resistance gene marking each insertion-deletion mutation.
Southern blot hybridization analysis.
Each mutation in single, double, and triple deletion-insertion mutant strains was confirmed by Southern blot hybridization analysis. Descriptions of probes used and data obtained are located in the supplementary material. Hybridization reactions were performed at 65°C using buffer that contained 5% sodium dodecyl sulfate (52). Nylon membranes (Hybond-N; Amersham) were used as solid support, and probes were nonisotopically labeled before immunological detection (digoxigenin DNA labeling and detection kit; Boehringer Mannheim).
Construction of plasmids for genetic complementation.
Genetic complementation plasmids were derived from the self-transmissible, low-copy-number, bifunctional plasmid pJRM10 (Table 2). ftsK+ is the only complete reading frame on the DNA insert of plasmid pRMD6. smc+ is the only complete reading frame on the DNA insert of plasmid pHW35. The DNA insert of scpAB complementation plasmid pRMD11 contains five complete genes: SCO1772, SCO1771 (hypothetical gene), scpAB+, and the downstream gene SCO1768 (putative pseudouridine synthase). The insert in pRMD11 ends 1 base downstream of SCO1768. These plasmids were introduced into the multiple auxotroph 2709 by transformation, selecting for thiostrepton resistance. The plasmids were mated from the 2709 derivative into the desired mutant strains, selecting for thiostrepton resistance and prototrophy. Transconjugants were tested for the presence of the antibiotic resistance genes marking the chromosomal mutations.
Strain J2540 (ΔparB/parB+) and derivatives isolated here had parB+ integrated by single homologous crossover recombination using plasmid pIJ6539 (28).
Construction of strains expressing EGFP fusion proteins.
Genes expressing EGFP fusions were constructed in one step by in vivo recombination. Cosmid H24parB-egfp (25) was used as the source of an egfp acc(3)IV oriT cassette to make ftsK-egfp and smc-egfp fusions. PCR was performed using H24parB-egfp as the template with specific oligonucleotides which anneal to the linker sequence at the 5′ end of egfp and downstream of the 3′ end of acc(3)IV and introduce 40 bases of homology to the 3′ ends of smc or ftsK, as well as removing the stop codons (Table 3). PCR products were introduced into E. coli strains expressing λ RED recombination functions and containing the cognate cosmid with either ftsK (SC7C7) or smc (SC7A1). Apramycin-resistant transformants were selected creating 7C7ftsK-egfp and 7A1smc-egfp. The gene fusion junction and egfp sequences were verified. The proline- and glycine-rich 5-amino-acid linker peptide (LPGPE) was similar to that of H24parB-egfp (25). Fusion-containing cosmids were conjugated from E. coli into strain M145, selecting for apramycin resistance and kanamycin sensitivity. In these transconjugants (RMD1 and RMD2), the fusion genes (ftsK-egfp and smc-egfp, respectively) are the only copies of ftsK or smc.
Microscopy.
Strains were characterized by confocal laser scanning microscopy using a Leica SP2 microscope equipped with a phase-contrast 100× oil objective (numerical aperture, 1.40) and 488- and 543-nm lasers. Autofluorescence seen in wide-field microscopy is not an issue with confocal microscopy with appropriately adjusted excitation light. There was no background autofluorescence in the wild-type strain under the conditions used. Standardized amounts of spores (CFU) were inoculated on MS agar adjacent to sterile coverslips that were embedded at a 45° angle in the agar medium and incubated for the indicated lengths of time. Coverslips were removed and cell material was fixed at room temperature for 10 min using a phosphate-buffered saline solution containing 4.375% glutaraldehyde and 0.028% paraformaldehyde. The coverslips were washed twice with phosphate-buffered saline, allowed to air dry, and mounted in 50% glycerol containing 0.1% propidium iodide, a total nucleic acid stain. Images were processed to adjust the brightness and contrast using Adobe Photoshop and assembled in Adobe Illustrator.
Computer analysis.
Predicted gene and protein products were obtained from the S. coelicolor genome database StrepDB (http://streptomyces.org.uk/).
RESULTS
Identification of ftsK and smc homologues in S. coelicolor.
FtsK (SpoIIIE) is a DNA motor protein that is involved in the process of chromosome partitioning during cell division or spore formation (30, 57, 59). The ftsK (SCO5750) gene is 2,790 bp and is predicted to encode a 929-amino-acid protein, FtsKSc (Fig. 1A). Complementing our study, recently published articles also reported genetic analysis of SCO5750 (1, 53). Transmembrane prediction program analyses for FtsKSc are consistent with the models containing four transmembrane domains proposed for SpoIIIEBs (56) and FtsKEc (11). The N terminus is required for targeting FtsK to the division plane (53).
FIG. 1.
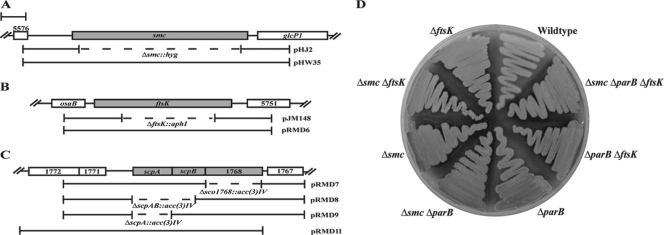
Physical maps of regions of chromosomal DNA containing smc, ftsK, or scpAB. In each case, the flanking genes are in the same orientation as the genes of interest. Bar: 1 kb. (A) The smc region of the chromosome. SCO5576 is predicted to encode an acyl phosphatase and glcP1 is involved in glucose uptake. For mutagenic plasmid pHJ2, the region deleted and replaced with a hygromycin resistance gene is shown (dashed line). The insert in complementation plasmid pHW35 is shown (bracketed line). (B) The ftsK region of the chromosome. osaB is part of a two-component regulator involved in osmoadaptation and SCO5751 is predicted to encode a hypothetical membrane protein. For mutagenic plasmid pJR148, the region of ftsK deleted and replaced with a neomycin resistance gene is shown (dashed line). The insert in complementation plasmid pRMD6 is shown (bracketed line). (C) The scpAB region of the chromosome. SCO1768, SCO1771, and SCO1767 are predicted to encode a pseudouridine synthase, a hypothetical protein found in actinomycetes, and a possible DNA hydrolase, respectively. For mutagenic cosmids pRMD7, -8, and -9, respectively, the regions of SCO1768, scpA, and scpAB deleted and replaced with an apramycin resistance gene are shown (dashed lines). The insert in complementation plasmid pRMD11 is shown (bracketed line); the DNA insert ends 1 base after SCO1768. (D) Single, double, and triple mutant strains macroscopically resemble the wild type. Strains were grown on MS agar for 4 days at 30°C. Mutant strains formed robust, gray-pigmented, aerial mycelia with no obvious growth or developmental delays. Strains M145 (wild type), J2537 (ΔparB), JM148 (ΔftsK), HJ2 (Δsmc), RMD3 (Δsmc ΔftsK), RMD4 (Δsmc ΔparB), RMD5 (ΔparB ΔftsK), and RMD6 (Δsmc ΔparB ΔftsK) are shown.
SMC proteins are extremely well conserved throughout prokaryotes, archaea, and eukaryotes; they are involved in chromosome maintenance and structure (21a, 28b, 50a). The smc gene of S. coelicolor (SCO5577) is 3,561 bp in length and is predicted to encode a 1,186-amino-acid protein (Fig. 1B). SMC proteins consist of N- and C-terminal DNA-binding domains connected by two coiled-coil regions joined by a hinge. The highest similarities to homologues were in the N- and C-terminal ATPase domains (54% and 46% identical to SMC of Bacillus subtilis, respectively). The functional analogue of SMC is MukB in E. coli.
ftsK and smc are dispensable for growth and viability of S. coelicolor.
Plasmids pJR148 and pJR152 were constructed in vitro and contain a deletion-insertion mutation for ftsK marked with a neomycin resistance gene (ΔftsK::aphI), with aphI cloned in each orientation. The altered ftsK genes (ftsK′) can express a C-terminally truncated protein of 203 amino acids (FtsK′). Mainly, the motor domain was lost as the truncation is in a loop between the final two predicted transmembrane segments. Plasmid pHJ2 contains a deletion-insertion mutation for smc marked with a hygromycin resistance gene (Δsmc::hyg). By use of these plasmids, the mutations were introduced into the chromosome by transformation. Marker replacement strains were readily isolated, indicating that these genes are not essential for growth or viability. Macroscopically, these strains looked very similar to the wild-type strain. They grew well and formed a robust, gray, aerial mycelium (Fig. 1D). As noticed in two recently published papers, strains containing ΔftsK mutations showed increased colony heterogeneity upon plating (1, 53). The ΔftsK strains reported here also had increased colony heterogeneity, as did the Δsmc strain.
These new strains were further characterized using confocal laser scanning microscopy. The DNA segregation phenotype associated with each mutant strain was determined by staining the DNA with propidium iodide. DNA segregation did not appear to be affected in vegetative hyphae of ftsK and smc mutants (data not shown). Figure 2 illustrates that spores formed in aerial hyphae in the single mutants (Fig. 2B to D) were similar to the wild type in shape and size (Fig. 2A) and had detectable defects in DNA partitioning (Table 4). The wild-type strain produced aerial hyphae with 0.8% anucleate spores. Because of its similarity to eukaryotic condensins and cohesins, we anticipated a chromosome condensation defect in the Δsmc mutant, but no obvious phenotype was observed and nucleoids in mature spore compartments were no less condensed. The Δsmc mutant produced aerial hyphae with 7% anucleate spores, which indicated a defect in segregation. For the Δsmc mutant, we did not observe spores with trapped DNA inside the septa (guillotine effect) as was observed for the E. coli mukB mutant (39). Consistent with the gene product's role as a DNA motor protein, the ΔftsK truncation mutants, isolated with different orientations of aphI, behaved similarly and produced aerial hyphae with 15 to 17% anucleate spores (Table 4). Here, for certain hyphae, we did observe DNA trapped in septa and small spore compartments with less DNA than normal (data not shown). The observed segregation defect was surprising, because complete deletions of ftsK resulted in strains that produced spores with reduced DNA staining but no anucleate spores (1, 53). To rule out a difference in laboratory copies of the same wild type, we isolated a strain containing a complete deletion of ftsK [RMD24; ΔftsK::acc(3)IV] and observed that it produced anucleate spores at a level similar to that seen for the wild-type parent (Table 4), in agreement with the other recent studies. We concluded that expressing the truncated version, FtsK′, was responsible for anucleate spore production in the ΔftsK::aphI strains (JM148 and JM152). For comparison, we analyzed the previously published ΔparB mutant (28). Analysis of the ΔparB strain under our growth conditions indicated that it produced aerial hyphae with 12% anucleate spores (Table 4), the same level as was originally reported. Thus, loss of parB or the portion of ftsK encoding the DNA motor domain and a membrane-spanning segment had a similar effect on the fidelity of simultaneous DNA segregation into each prespore compartment. The effect of losing smc was more modest, i.e., about half that observed for the loss of parB or the truncation of ftsK. Completely removing ftsK had no effect on anucleate spore production.
FIG. 2.
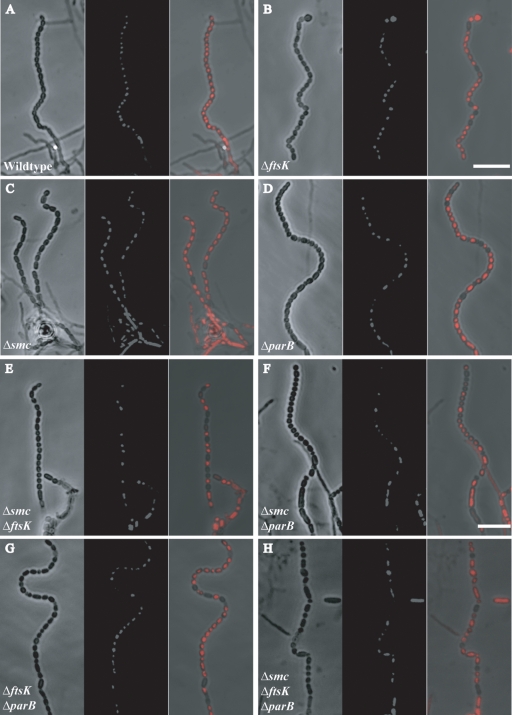
Confocal microscope images showing sporulation and DNA segregation phenotypes of the wild type and of single, double, and triple mutant strains. Four-day-old cultures grown on MS agar were sampled on coverslips, fixed, and stained with propidium iodide (red). In each panel, phenotypes of representative aerial hyphae are shown as phase-contrast and fluorescent image pairs, followed by a merged image. (A) M145 (wild type). (B) JM148 (ΔftsK). (C) HJ2 (Δsmc). (D) J2537 (ΔparB). (E) RMD3 (Δsmc ΔftsK). (F) RMD4 (Δsmc ΔparB). (G) RMD5 (ΔparB ΔftsK). (H) RMD6 (Δsmc ΔparB ΔftsK). Bar: 7 μm.
TABLE 4.
Quantitative analysis of genome segregation defects during spore formation
| Strain or mutation | % Anucleate sporesa | Total no. of spores |
|---|---|---|
| Wild type | 0.8 | 2,233 |
| ΔftsK::acc(3)IV (null) | 0.8 | 1,876 |
| ΔftsK::aphI (aphI in same orientation as ftsK′)b | 15 | 3,469 |
| ΔftsK::aphI (aphI in opposite orientation from ftsK′) | 17 | 1,463 |
| Δsmc::hyg | 7 | 4,122 |
| ΔparB::acc(3)IV | 12 | 1,783 |
| Δsmc::hyg ΔftsK::aphI | 13 | 2,389 |
| Δsmc::hyg ΔparB::acc(3)IV | 24 | 3,663 |
| ΔparB::acc(3)IV ΔftsK::aphI | 13 | 3,230 |
| Δsmc::hyg ΔparB::acc(3)IV ΔftsK::aphI | 10 | 1,503 |
| Δsmc::hyg/smc+ | 3 | 2,033 |
| ΔftsK::aphI/ftsK+ | 5 | 2,180 |
| Δsmc::hyg ΔftsK::aphI/ftsK+ | 7 | 2,008 |
| Δsmc::hyg ΔftsK::aphI/smc+ | 11 | 1,667 |
| Δsmc::hyg ΔparB::acc(3)IV/smc+ | 14 | 1,809 |
| Δsmc::hyg ΔparB::acc(3)IV/parB+ | 6 | 1,807 |
| ΔparB::acc(3)IV ΔftsK::aphI/ftsK+ | 16 | 2,009 |
| ΔparB::acc(3)IV ΔftsK::aphI/parB+ | 18 | 2,125 |
| Δsmc::hyg ΔparB::acc(3)IV ΔftsK::aphI/ftsK+ | 27 | 2,075 |
| Δsmc::hyg ΔparB::acc(3)IV ΔftsK::aphI/smc+ | 11 | 1,712 |
One hundred random spore chains were quantified and the percentages of anucleate spores were determined. All spores were considered regardless of the compartment size.
Double and triple mutants contained this allele of ftsK′.
Genetic complementation experiments were completed to confirm that the segregation phenotypes observed were due to the truncation of ftsK or the loss of smc and not to an unlinked mutation. Wild-type copies of each gene were cloned into the low-copy-number vector pJRM10 (SCP2* derivative; one or two copies/chromosome). The DNA inserts contained only one complete gene, ftsK or smc (Fig. 1A and B). The phenotypes of the complemented ΔftsK and Δsmc mutants were analyzed (Table 4). The ΔftsK/ftsK+ strain produced aerial hyphae with 5% anucleate spores. Thus, about two-thirds of the segregation phenotype associated with the ftsK truncation mutation was complemented. The Δsmc/smc+ strain produced aerial hyphae that produced 3% anucleate spores, indicating that 50% of the phenotype associated with the mutation was complemented. Our complementation data suggest that the observed defects in segregation resulted, at least in part, from the introduced ftsK and smc insertion-deletion mutations (see additional complementation results below). At least one promoter, active in aerial hyphae, is present on the DNA inserts used for genetic complementation. However, proper temporal and spatial expression may require that the genes be located in their native chromosomal locations.
Δsmc ΔftsK, Δsmc ΔparB, and ΔftsK ΔparB double mutants have more-severe developmental defects.
For the construction of double mutants, we used ΔftsK mutant JM148 (aphI inserted in the same orientation as ftsK′) because the two mutants with either orientation of the marker had the same phenotype (Table 4). The Δsmc ΔftsK, Δsmc ΔparB, and ΔftsK ΔparB double mutants were constructed by transformation using DNA prepared from one mutant strain and using a second mutant as the recipient (Table 1). The double mutant strains were readily isolated, which indicated that the three double mutants were not synthetic lethal combinations. Macroscopically, these double mutants had a wild-type appearance without any obvious growth delay and formed robust gray aerial mycelia (Fig. 1D), although they all displayed colony heterogeneity relative to what was seen for the wild-type strain. Thus, the double mutants did not even have a synthetic sick phenotype.
DNA segregation did not appear to be affected in vegetative hyphae of the double mutants (data not shown). Observations by phase-contrast microscopy revealed aerial hyphae containing spores that were heterogeneous in size and shape compared to those of the single mutants or the wild type. In addition, Fig. 2E to G illustrates that spore chains in the double mutants had defects in developmental DNA partitioning, and the segregation defect in each strain was quantified (Table 4). The Δsmc ΔftsK mutant produced aerial hyphae containing 13% anucleate spores, similar to those of the ΔftsK strain (Fig. 2E). Therefore, additive effects of the mutations were not seen when combining the loss of a condensation gene (7%) with the segregation gene (15%). Interestingly, the ΔftsK ΔparB mutant also was only 13% anucleate, which again indicated that this strain did not have segregation defects additive to those observed for the ΔftsK (15%) and ΔparB (13%) single mutant strains (Fig. 2G and Table 4). It seems that without the function of the two DNA-partitioning and segregation genes, S. coelicolor still can disseminate its DNA just as well as the most defective single mutant. In contrast, the Δsmc ΔparB mutant produced aerial hyphae containing 24% anucleate spores (Fig. 2F), slightly more than the additive effect (19%) of each individual gene loss (Table 4). Commensurate with the rise in the segregation defect, the Δsmc ΔparB mutant had a more defective developmental phenotype than the other double mutants. Aerial hyphae of the Δsmc ΔparB mutant contained more spore compartments with altered shapes and sizes. This observation, along with the single mutant data, suggested that there was a phenotypic effect when both a partitioning and a condensation gene in S. coelicolor were deleted, indicating genetic overlap or interaction.
In order to verify that the phenotypes in the double mutants resulted from the introduced mutations, genetic complementation analyses were completed with the low-copy-number plasmids bearing smc or ftsK or an integrated copy of parB (Table 4). Complementation data for the double mutant strains suggest that the phenotypes were linked to the mutations introduced by transformation into the Δsmc mutant. The Δsmc ΔftsK/ftsK+ strain produced aerial hyphae containing spores that were 7% anucleate, which indicates the that ftsK truncation mutation was completely complemented. The Δsmc ΔparB/parB+ strain produced aerial hyphae that contained 6% anucleate spores, also indicating complete complementation. Finally, the ΔftsK ΔparB/ftsK+ strain produced aerial hyphae with 16% anucleate spores, which is similar to the 13% anucleate spores in aerial hyphae of the ΔparB strain. Complementation data suggest that the introduced second mutation was responsible for the observed phenotypes compared to the single mutant strains.
A Δsmc ΔparB ΔftsK triple mutant is viable and does not have as severe of a developmental segregation defect as a Δsmc ΔparB double mutant.
The Δsmc ΔparB ΔftsK triple mutant was easily isolated by transformation with DNA prepared from a ΔparB strain and using the Δsmc ΔftsK strain as the recipient. Macroscopically, the triple mutant showed no obvious growth delay and produced a robust gray aerial mycelium (Fig. 1D), but an elevation in colony heterogeneity was evident. Mutations in the three genes did not result in a synthetic lethal or synthetic sick phenotype.
Using phase-contrast microscopy, the spores of the Δsmc ΔparB ΔftsK strain appeared heterogeneous in shape and size, most like those of the Δsmc ΔparB strain (Fig. 2H). Segregation did not appear to be affected in vegetative hyphae of the triple mutant (data not shown). Surprisingly, determination of the segregation phenotype for the triple mutant indicated that only 10% of the spores were anucleate (Table 4). The number of anucleate spores in aerial hyphae for the Δsmc ΔparB ΔftsK strain was not the total sum of each single mutant combined (34%). It was not even as poor as the most severe phenotype of the double mutant strains (Δsmc ΔparB strain, 24%). Without all three of these genes, 90% of the spores still received some genomic material. This observation suggests that there must be considerable redundancy, and another unidentified system for developmental DNA segregation must function in this filamentous organism.
We performed genetic complementation analyses with the low-copy-number plasmids bearing smc or ftsK (Table 4). In particular, the complementation strain with the genotype Δsmc ΔparB ΔftsK/ftsK+ produced aerial hyphae with an increase in the number of anucleate spores to 27%, which was similar to the Δsmc ΔparB phenotype of 24% (Table 4). This verified that the introduced ΔftsK truncation mutation was responsible for partially alleviating the segregation defects in the Δsmc ΔparB strain. Therefore, introducing a functional copy of ftsK back into the triple mutant made the genome segregation phenotype worse, suggesting that these genes interact genetically.
The fact that only 10% of triple mutant spores were anucleate prompted us to test if the viability of the spores was more adversely affected and the percent anucleate values were misleading. The viability of spores plated on MM supplemented with glucose was compared for the most pertinent strains. Wild-type strain produced spore compartments that were 49% viable. Although the ΔftsK truncation and Δsmc ΔparB mutants have significant amounts of anucleate spores (15% and 24%, respectively), surprisingly, the viabilities of those produced were equal to or slightly higher than those of the wild type, respectively. In contrast, spores of the triple mutant were 12.5% viable, a level one-forth of that of the wild type. Although 90% of the spores in the triple mutant receive some genetic material, they are fourfold less fit than the wild type.
FtsK-EGFP localizes between nucleoids of aerial hyphae.
ftsK was replaced by ftsK-egfp in the chromosome of the wild-type strain. This strain (RMD1) produced aerial hyphae containing spores with normal size and shape, as judged by phase-contrast microscopy. DNA segregation defects were not observed, as judged by fluorescence microscopy (data not shown). We concluded that the FtsK-EGFP fusion protein was functional.
Localization of FtsK-EGFP was analyzed at several time points during the life cycle, and FtsK-EGFP foci were apparent at both early vegetative and early aerial hyphae stages of growth. FtsK-EGFP was localized as distinct foci at several places within young germ tubes growing out of spores (Fig. 3A). Distinct gaps in the propidium iodide staining were repeatedly observed, and FtsK-EGFP foci coincided with those gaps in staining. This result suggested that DNA segregation could be occurring during the outgrowth of newly germinated spores and that there was a role for FtsK in this process. However, gaps in the propidium iodide staining were observed for ftsK mutants as well, indicating that the gaps in staining were not solely caused by the action of FtsK (data not shown). Although we attempted to stain membranes and cross-walls with unsatisfactory results, we assume that at least some gaps in DNA result from vegetative cross-walls. In addition, FtsK-EGFP localized as foci or bands in reproductive aerial hyphae prior to the formation of sporulation septa (Fig. 3B). The ladder-like pattern in the aerial hyphae was similar to what was observed for FtsZ localization (17, 46). These data indicated that FtsK localized to evenly spaced positions, as expected if it were to have a role in correctly positioning the nucleoids into prespore compartments before the DNA becomes trapped by the invaginating septa. Similar FtsK localization during synchronous septation in aerial hyphae has been reported independently (1, 53). In addition, we also observed potentially helical structures that preceded the appearance of the evenly spaced ladder-like arrays, as reported by Wang et al. (53).
FIG. 3.
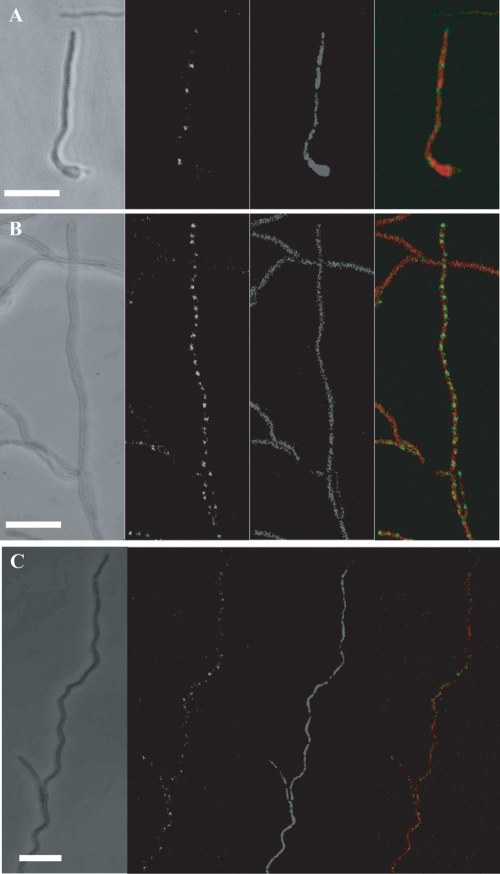
FtsK-EGFP and SMC-EGFP fusion protein localization in vegetative and reproductive hyphae. Coverslips were inoculated and hyphae grown for 1 to 3 days on MS agar, fixed, and stained with propidium iodide. In each panel, representative hyphae are shown as phase-contrast and fluorescent images as observed by scanning laser confocal microscopy. From left to right, panels contain phase-contrast, EGFP, and DNA images and a merge of the two fluorescent images. (A) FtsK-EGFP localized in a vegetative filament newly emerging from a spore (lower right; 1 day of growth). (B) FtsK-EGFP localized in a regular pattern in a predivisional aerial filament just prior to prespore formation (long, central filament oriented with long axis of panel; 2 days of growth). (C) The SMC-EGFP fusion protein had an irregular punctuate pattern of localization in a predivisional aerial filament (2 to 3 days of growth). SMC-EGFP foci are not uniformly distributed, nor are they associated with every nucleoid. Bar: 7 μm.
SMC-EGFP is localized in a punctuate pattern in aerial hyphae.
smc was replaced by smc-egfp in the chromosome of the wild-type strain. This strain (RMD2) produced aerial hyphae containing spores with normal size and shape, as judged by phase-contrast microscopy. DNA segregation defects were not observed, as judged by fluorescence microscopy (data not shown). We concluded that the SMC-EGFP fusion protein was functional.
We anticipated that SMC might bind to every chromosome, helping to condense the DNA into nucleoids during genome segregation into prespore compartments. However, SMC-EGFP foci were not uniformly positioned and seemed to localize in a punctate pattern only in predivisional aerial hyphae. The foci were not observed to be associated with every nucleoid, nor with any particular region of the aerial filament (Fig. 3C). SMC-EGFP signal was not observed at any other time in the life cycle. Similar observations were made using immunofluorescence microscopy and an SMC-specific antibody (28a), indicating that the SMC-EGFP fusion did reflect the behavior of native SMC.
scpA and scpB mutants cause DNA condensation defects.
SMC functions in association with non-SMC proteins called ScpA and ScpB (segregation and condensation protein) (32, 47). These proteins (MukEF) may regulate the SMC (MukB) association with DNA (33, 41). scpA (SCO1770) is 795 bp and is predicted to encode a 264-amino-acid protein and scpB (SCO1769) is 672 bp and is predicted to encode a 223-amino-acid protein (Fig. 1C). ScpA and ScpB are approximately 27% and 32% identical, respectively, to the B. subtilis homologues. The genes encoding ScpA and ScpB are adjacent and are likely to be cotranscribed with SCO1768, predicted to encode a pseudouridine synthase (Fig. 1C). Plasmids were constructed containing deletion-insertion mutations for SCO1768, scpA, and scpAB marked with an apramycin resistance cassette (Table 2). These mutations were introduced individually into the chromosome by conjugation, and marker replacement strains were readily isolated (Table 1), indicating that these genes are not essential for growth or viability. These strains produced normal gray, aerial mycelia characteristic of the wild-type strain (data not shown).
Deletion of rluD, coding for one of the many pseudouridine synthases in E. coli, causes a growth rate defect (20). Because scpB and SCO1768 overlap by 1 base and mutations in scpA or scpB were likely to have polar effects on the expression of SCO1768, we isolated a SCO1768 mutant to determine its phenotype in S. coelicolor. The introduced mutation for SCO1768 did not have an obvious effect on the growth rate, nor did it have a DNA segregation or condensation phenotype (data not shown). We conclude that the phenotypic effects observed for the ΔscpA or ΔscpAB mutants resulted from the deletion of those genes.
We anticipated that the scpA or scpAB mutants might have a segregation phenotype similar to that observed for the smc mutant. However, the observed frequency of anucleate spores was 1% (Table 5), insignificantly higher than that of the wild type (0.8%) and less than that of the smc deletion-insertion mutant (7%) (Table 4). Instead, a substantial fraction of nucleoids of the ΔscpA and ΔscpAB strains had an altered morphology in sporulating aerial hyphae (Fig. 4). Certain nucleoids did not follow the contours of the spore compartment and contain two lobes of DNA, a phenotype we have called “bilobed.” In late aerial filaments, 15% of spores in the ΔscpA strain and 26% of spores in the ΔscpAB strain were bilobed (Table 5). In contrast, bilobed spores were not observed in the wild-type strain. Although the isolated deletion of scpA might be polar on the expression of scpB, deleting both genes resulted in a higher frequency of bilobed spores, indicating that some scpB must be expressed in the scpA mutant. Genetic complementation experiments indicated that the observed nucleoid morphology defect was the result of the introduced deletion-insertion mutation for scpA or scpAB. Introduction of pRMD11, containing scpAB+ and SCO1768, substantially reduced the occurrence of bilobed spores in the mutants (Table 5).
TABLE 5.
Quantitative analysis of scp mutant genome segregation and nucleoid morphology phenotypes during spore formation
| Strain or mutation | % Anucleate sporesa | % Bilobed sporesa | Total no. of spores |
|---|---|---|---|
| Wild type | 0.8 | 0 | 2,233 |
| ΔscpA::acc(3)IV | 1 | 15 | 1,797 |
| ΔscpAB::acc(3)IV | 1 | 26 | 1,787 |
| ΔscpA::acc(3)IV/scpAB+ | 1 | 2 | 1,790 |
| ΔscpAB::acc(3)IV/scpAB+ | 1 | 4 | 1,626 |
| Δsmc::hyg ΔscpA::acc(3)IV | 4 | 14 | 3,589 |
| Δsmc::hyg ΔscpAB::acc(3)IV | 3 | 17 | 3,148 |
One hundred random spore chains were quantified and the percentages of anucleate and bilobed spores were determined. All spores were considered regardless of the compartment size.
FIG. 4.
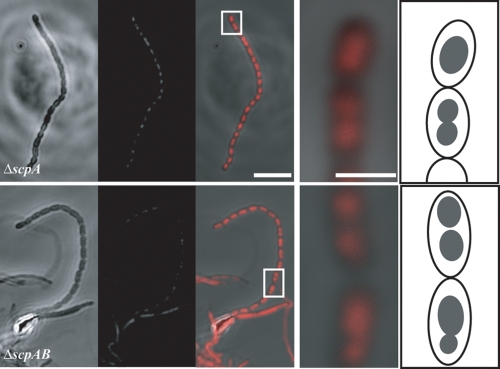
The scpA and scpAB mutants have a bilobed nucleoid phenotype. Phase-contrast and fluorescent images of the same fields were taken by scanning laser confocal microscopy. Coverslips were inoculated and grown for 1 to 3 days on MS agar, fixed, and stained with propidium iodide. In each panel, phenotypes of representative aerial hyphae are shown as phase-contrast and fluorescent image pairs, followed by a merged image. For the merged images, the areas in the white boxes are enlarged and a cartoon interpretation is provided. (A) Images of ΔscpA (RMD18) showing spores with DNA appearing bilobed, unlike wild-type spores. (B) Images of ΔscpAB spores (RMD19) also appearing to have a bilobed DNA pattern. Bar: 7 μm.
Because ScpA and ScpB are thought to regulate the interaction of SMC with DNA, we were interested in learning the phenotypes obtained when combining scpA or scpAB mutations with smc (Table 1). The data suggest that deleting smc in the scpA or scpAB mutant backgrounds combines their effects. While bilobed spores were not observed for the Δsmc mutant, aerial hyphae of the Δsmc ΔscpA double mutant produced 4% anucleate spores, and an additional 14% had a bilobed nucleoid morphology (Table 5). Aerial hyphae of the Δsmc ΔscpAB triple mutant produced 3% anucleate spores, and an additional 17% had a bilobed nucleoid morphology. An approximately similar number of spore compartments were affected in the triple mutant compared to the scpAB double mutant; however, the percentage of bilobed spores decreased and the percentage of anucleate spores increased. The data are consistent with an interpretation that ScpA and/or ScpB has functions in addition to regulating SMC activity.
DISCUSSION
smc and chromosome partitioning.
In this study, we described the isolation of an smc mutant. The main defect observed was in segregation, not in DNA condensation. The magnitude of the segregation defect in the smc mutant was modest, resulting in 7% anucleate prespore compartments, less than 10-fold more than that of the wild-type parent. The assembly of SMC condensation complexes presumably functions to compact and help pull the linear genome into the space available in prespore compartments. Consistent with the expected function and genetic defect, SMC-EGFP foci form in irregular patterns along aerial hyphae at a stage prior to when synchronous septation occurred and nucleoids were condensed. These results may suggest that nucleoid condensation in adjacent compartments of aerial hyphae is nonsynchronous. Alternatively, SMC-EGFP may not aggregate to discrete foci that can be visualized easily. SMC-EGFP foci were not observed in vegetative hyphae or in prespore compartments (i.e., postseptation). This could indicate that expression is developmentally regulated and SMC is present in a low concentration in vegetative hyphae, where DNA is not condensed, or early aerial hyphae, before DNA segregation occurs. Further regulation may be at the level of activity. SMC may only transiently and dynamically localize at points in the life cycle, such as when particular DNA sequences must be condensed in the maturing spore compartments. Similar results for the segregation phenotype of an smc mutant and SMC localization have been obtained independently (28a).
ftsK and chromosome partitioning.
In this study, we also described the isolation of mutants with different deletion-insertion mutations for ftsK. Consistent with two recent studies (1, 53), genome segregation during spore formation was not as appreciably affected as anticipated with an ftsK null mutation [ΔftsK::acc(3)IV]. Spores with reduced DNA content were readily apparent, but anucleate spores were observed at the same level as for the wild type. Other ftsK mutants described here (ΔftsK::aphI mutants) harbor a mutant allele (ftsK′) lacking the region encoding the DNA motor domain but still have the potential to produce a truncated protein of 203 native amino acids containing several N-terminal membrane-spanning segments (three out of four segments). In contrast, the truncation of ftsK resulted in a more deleterious segregation phenotype producing 15% anucleate spores, ∼20-fold higher than that of the wild type. Interestingly, despite the obvious increase in missegregation, the spores produced did not suffer a dramatic loss of fitness and were as viable as the wild type when germination and growth were tested on MM with glucose (46% versus 49%, respectively).
Except for segregation in the mutant with a truncated ftsK gene, everything else in growth and development—aerial mycelium formation, septation, septum spacing, spore maturation (compartment rounding up), and spore pigment production—appeared to be normal. In addition, FtsZ-EGFP localized correctly in the frsk mutant, indicating that the C-terminal motor domain is not required for, and the N-terminal fragment did not interfere with, Z ring formation (data not shown). Thus, FtsK′ does not appear to be pleiotropically poisoning a central process such as the general secretion/membrane insertion pathway. A segregation phenotype is what would be predicted based on previous work with other organisms, and that is in fact the phenotype observed. Because the phenotype of ftsK′ is different from the loss of the entire gene, we think that the results are consistent with the interpretation that the FtsK′ fragment (truncated cleanly between the third and fourth predicted transmembrane helices) associates with the components with which FtsK may normally interact. We predict that FtsK′ is where it is supposed to be but that it is unable to transport DNA. We conclude that the FtsK motor domain is important for proper genome segregation during prespore formation and that the N-terminal 203-amino-acid FtsK′ must interact with some component during prespore formation. The expression of FtsK′ did not result in a dominant-negative phenotype, as the genome segregation defect could be complemented by expression of ftsK+ from a low-copy-number plasmid (Table 4). In speculation, this may indicate that the first three transmembrane segments of the FtsK/SpoIIIE family are important for the majority of normal function and that the final transmembrane segment is responsible for positioning the DNA motor domain on the correct side of the membrane.
Consistent with the expected function of moving DNA trapped by invaginating septa, FtsK-EGFP foci and bands were observed to form in regular ladder-like patterns along aerial hyphae at a stage when synchronous septation occurred and the nucleoids would become condensed. Our data support similar observations from two other recent studies (1, 53). In addition, FtsK-EGFP foci were observed in germ tubes during spore outgrowth, suggesting that FtsK might function in the vegetative hyphae, presumably moving DNA trapped when forming the widely spaced vegetative cross-walls.
Genetic interactions between segregation and condensation genes.
We combined our mutations for smc and truncated ftsK (ftsK′) with each other and that of parB. ParB is responsible for organizing the origin proximal-region of the linear chromosomes into nucleoprotein complexes that are synchronously positioned at regularly spaced distances to help ensure that copies of the genome are captured within each prespore compartment (25, 26). A parB mutant produces 13% anucleate spores, ∼20-fold higher than the wild type (Table 4) (28). Combining ftsK′ with smc or parB mutations did not give an additive effect, and the production of anucleate spores was approximately 13% for both double mutants, similar to the worst single mutant. In contrast, smc and parB are in two pathways, as combining the two mutations led to 24% anucleate spores, approximately the sum of the phenotypes for each single mutant. Despite the obvious increase in missegregation, the spores produced by an smc parB mutant did not suffer a dramatic loss of fitness and were slightly more viable than the wild type when germination and growth were tested on MM with glucose.
The most interesting result was observed when we isolated an smc parB ftsK′ triple mutant. We anticipated that removing ftsK+ would exacerbate the segregation phenotype. Instead, introducing the ftsK′ mutation into the smc parB background improved the ability to segregate genomic material into prespore compartments (24% anucleate for the double mutant versus 10% for the triple mutant). We confirmed the alteration in the segregation phenotype resulted from the introduced ftsK′ mutation. Genetic complementation of ftsK′ (smc parB ftsK′/ftsK+) resulted in a phenotype similar to that for the smc parB double mutant (27% anucleate spores). Our data indicate that there must be considerable redundancy and more genes that can provide backup function in segregation when smc, parB, and ftsK are inactive, or we are left to conclude that unassisted postreplication diffusion of nucleoids accounts for the even distribution of genomes during development. In the absence of the three well-characterized genes involved in bacterial genome segregation, normally resulting in lethal or synthetic lethal phenotypes when combined in rod-shaped bacteria, an impressive 90% of the spore compartments receive at least some genomic material. A portion of those genomes in spore compartments must be complete or nearly complete copies, as the triple mutant spores are approximately 25% as viable as those produced by the wild-type parent when tested on MM with glucose. Certainly, some genomes must be guillotined by the septa and account for the loss of viability.
When normal genome segregation is disturbed by the loss of ParB and SMC, the FtsK DNA motor domain may clear trapped genomes to the “wrong” side of invaginating septa too quickly, resulting in 24% anucleate compartments. When FtsK′ is expressed, a less efficient backup system(s) may function in the smc parB mutant. The less efficient redundant system is able to eventually segregate genomes into a larger proportion of prespore compartments when the strong FtsK motor is not rapidly clearing DNA from invaginating septa. A better understanding of segregation will require additional analysis. One obvious candidate might be the recently described FtsK-like protein SffA, although the functions of SffA and FtsK did not appear to overlap (1). Due to the considerable redundancy demonstrated here, the contributions of SffA may not be apparent until they are tested in smc parB or smc parB ftsK′ mutants. Related to the subtle phenotype of an sffA mutant (1), infrequent spores with double the DNA intensity were not observed adjacent to anucleate spores in any of the mutants described here. Likewise, another ftsK-like gene (SCO4508) could be a candidate and was tested for a role in segregation (53), but its function may not be apparent until combined with the other genes tested here. Due to the lack of compatible markers, we have yet to test the effect of ftsK-null allele when combined with smc parB. Finally, we anticipate that additional components influencing segregation might be cytoskeletal proteins MreB or Mbl (9a, 16, 49) or genes under the regulation of SsgC (40).
Genetic instability of the linear genome.
A recent study with an ftsK-null mutant reported that portions of the right end of the chromosome were frequently lost in some colonies during routine propagation of the mutant strain (53). The interpretation of that study was that loss of FtsK resulted in an increase in genome instability. We and Ausmees et al. (1) also observed an increase of colony heterogeneity with ftsK mutants. However, we note that an increase in genome instability resulted in most of our mutants. We believe that increased genome instability is not specific for the loss of FtsK sweeping the arms of the linear chromosome from being strangled and lost but rather is a property of DNA segregation mutants in general. To test if the segregation mutants in this study suffered unanticipated genetic loss, we subjected all of our strains to Southern blot hybridization analyses using the inserts of two nonoverlapping cosmids as probes to see if there were any observable differences in the cmlR (SCO7526) region near the right end of the chromosome. The strains from this study did not have observable deletions or rearrangements (data not shown).
Nucleoid morphology defects in a scpAB-null mutant.
We anticipated that mutants for genes encoding non-SMC segregation and condensation proteins ScpA and ScpB would have a phenotype similar to the smc mutant but perhaps more modest (32). These proteins were thought to interact with SMC to influence the interaction of SMC with DNA and are required to form higher-order structures involved in genome condensation. However, the phenotype of the smc mutant is distinct from those of the scpA and scpAB mutants. The loss of scpA or scpAB mainly affected the morphology of the condensed nucleoid and did not give rise to a significant segregation defect. Isolation of smc scpA and smc scpAB mutants indicated that the phenotypes of the double and triple mutants were additive. These mutants have both a segregation phenotype (smc) and a nucleoid morphology phenotype (scpAB). We speculate that in addition to the normal interaction with SMC, the nucleoid morphology perturbation might be the result of ScpA and ScpB interacting with additional proteins required for and regulating processes associated with condensation and genome organization. The SMC-ScpA-ScpB complex was shown to have a role in DNA repair and gene regulation in B. subtilis (10). Those authors proposed that these roles are evolutionarily conserved and our data may agree.
Finally, the bilobed morphology of the nucleoids observed for the scpA and scpAB mutants could offer insight into the way the genome is stored in the quiescent spore. The genome in streptomycetes is a linear DNA molecule with TIRs. For vegetative hyphae, evidence suggests that the TIRs are held in association near each other (58), and perhaps the ends are also normally in association in the spore. In the scpAB mutants, the TIRs may no longer be closely associated, revealing two independently folding domains of the nucleoid. Alternatively, a DNA replication checkpoint may have failed and the mutant prespore compartments might undergo an additional round of DNA replication. Other possibilities exist and we will be exploring the problem in future studies.
Supplementary Material
Acknowledgments
We thank Agnieszka Kois and Jolanta Zakrzewska-Czerwińska for communicating their work to us prior to publication. We thank Aimee Belanger for constructing plasmids pAEB228 and pAEB232, Dagmara Jakimowicz for H24parB-egfp, Keith Chater for parB mutant strains, Bertolt Gust for technical advice, and Patrick Viollier for helpful discussions.
This work was supported, in part, by grant GM56915 from the National Institutes of Health (to J.R.M.).
Footnotes
Published ahead of print on 31 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ausmees, N., H. Wahlstedt, S. Bagchi, M. A. Elliot, M. J. Buttner, and K. Flardh. 2007. SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Mol. Microbiol. 651458-1473. [DOI] [PubMed] [Google Scholar]
- 2.Barre, F. X. 2007. FtsK and SpoIIIE: the tale of the conserved tails. Mol. Microbiol. 661051-1055. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Rajandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417141-147. [DOI] [PubMed] [Google Scholar]
- 4.Bierman, M., R. Logan, K. O'Brien, E. T. Seno, R. N. Rao, and B. E. Schoner. 1992. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 11643-49. [DOI] [PubMed] [Google Scholar]
- 5.Bignell, C., and C. M. Thomas. 2001. The bacterial ParA-ParB partitioning proteins. J. Biotechnol. 911-34. [DOI] [PubMed] [Google Scholar]
- 6.Bigot, S., V. Sivanathan, C. Possoz, F. X. Barre, and F. Cornet. 2007. FtsK, a literate chromosome segregation machine. Mol. Microbiol. 641434-1441. [DOI] [PubMed] [Google Scholar]
- 7.Breier, A. M., and A. D. Grossman. 2007. Whole-genome analysis of the chromosome partitioning and sporulation protein Spo0J (ParB) reveals spreading and origin-distal sites on the Bacillus subtilis chromosome. Mol. Microbiol. 64703-718. [DOI] [PubMed] [Google Scholar]
- 8.Chen, N., A. A. Zinchenko, Y. Yoshikawa, S. Araki, S. Adachi, M. Yamazoe, S. Hiraga, and K. Yoshikawa. 2008. ATP-induced shrinkage of DNA with MukB protein and MukBEF complex of Escherichia coli. J. Bacteriol. 1903731-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cui, Y., Z. M. Petrushenko, and V. V. Rybenkov. 2008. MukB acts as a macromolecular clamp in DNA condensation. Nat. Struct. Mol. Biol. 15411-418. [DOI] [PubMed] [Google Scholar]
- 9a.Defeu Soufo, H. J., and P. L. Graumann. 2003. Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr. Biol. 131916-1920. [DOI] [PubMed] [Google Scholar]
- 10.Dervyn, E., M. F. Noirot-Gros, P. Mervelet, S. McGovern, S. D. Ehrlich, P. Polard, and P. Noirot. 2004. The bacterial condensin/cohesin-like protein complex acts in DNA repair and regulation of gene expression. Mol. Microbiol. 511629-1640. [DOI] [PubMed] [Google Scholar]
- 11.Dorazi, R., and S. J. Dewar. 2000. Membrane topology of the N-terminus of the Escherichia coli FtsK division protein. FEBS Lett. 47813-18. [DOI] [PubMed] [Google Scholar]
- 12.Ebersbach, G., and K. Gerdes. 2005. Plasmid segregation mechanisms. Annu. Rev. Genet. 39453-479. [DOI] [PubMed] [Google Scholar]
- 13.Elliot, M. A., M. J. Buttner, and J. R. Nodwell. 2008. Multicellular development in Streptomyces, p. 419-438. In D. E. Whitworth (ed.), Myxobacteria: multicellularity and differentiation. ASM Press, Washington, DC.
- 14.Errington, J., H. Murray, and L. J. Wu. 2005. Diversity and redundancy in bacterial chromosome segregation mechanisms. Philos. Trans. R. Soc. Lond. B 360497-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flärdh, K. 2003. Growth polarity and cell division in Streptomyces. Curr. Opin. Microbiol. 6564-571. [DOI] [PubMed] [Google Scholar]
- 16.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120329-341. [DOI] [PubMed] [Google Scholar]
- 17.Grantcharova, N., U. Lustig, and K. Flärdh. 2005. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2). J. Bacteriol. 1873227-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graumann, P. L. 2001. SMC proteins in bacteria: condensation motors for chromosome segregation? Biochimie 8353-59. [DOI] [PubMed] [Google Scholar]
- 19.Gust, B., G. Chandra, D. Jakimowicz, T. Yuqing, C. J. Bruton, and K. F. Chater. 2004. Lambda red-mediated genetic manipulation of antibiotic-producing Streptomyces. Adv. Appl. Microbiol. 54107-128. [DOI] [PubMed] [Google Scholar]
- 20.Gutgsell, N. S., M. Del Campo, S. Raychaudhuri, and J. Ofengand. 2001. A second function for pseudouridine synthases: a point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 7990-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirano, M., and T. Hirano. 2006. Opening closed arms: long-distance activation of SMC ATPase by hinge-DNA interactions. Mol. Cell 21175-186. [DOI] [PubMed] [Google Scholar]
- 21a.Hirano, T. 1999. SMC-mediated chromosome mechanics: a conserved scheme from bacteria to vertebrates? Genes Dev. 1311-19. [DOI] [PubMed] [Google Scholar]
- 22.Hirano, T. 2006. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 7311-322. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, L. M., K. S. Makarova, E. V. Koonin, and L. Aravind. 2004. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 325260-5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakimowicz, D., K. Chater, and J. Zakrzewska-Czerwińska. 2002. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Mol. Microbiol. 451365-1377. [DOI] [PubMed] [Google Scholar]
- 25.Jakimowicz, D., B. Gust, J. Zakrzewska-Czerwińska, and K. F. Chater. 2005. Developmental-stage-specific assembly of ParB complexes in Streptomyces coelicolor hyphae. J. Bacteriol. 1873572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jakimowicz, D., P. Zydek, A. Kois, J. Zakrzewska-Czerwińska, and K. F. Chater. 2007. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Mol. Microbiol. 65625-641. [DOI] [PubMed] [Google Scholar]
- 27.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 28.Kim, H.-J., M. J. Calcutt, F. J. Schmidt, and K. F. Chater. 2000. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-like parAB locus. J. Bacteriol. 1821313-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Kois, A., M. Świątek, D. Jakimowicz, and J. Zakrzewska-Czerwińska. 2009. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. J. Bacteriol. 191310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28b.Koshland, D., and A. Strunnikov. 1996. Mitotic chromosome condensation. Annu. Rev. Cell Dev Biol. 12305-333. [DOI] [PubMed] [Google Scholar]
- 29.Kuhstoss, S., and R. N. Rao. 1991. A thiostrepton-inducible expression vector for use in Streptomyces spp. Gene 10397-99. [DOI] [PubMed] [Google Scholar]
- 30.Liu, G., G. C. Draper, and W. D. Donachie. 1998. FtsK is a bifunctional protein involved in cell division and chromosome localization in Escherichia coli. Mol. Microbiol. 29893-903. [DOI] [PubMed] [Google Scholar]
- 31.MacNeil, D. J., K. M. Gewain, C. L. Ruby, G. Dezeny, P. H. Gibbons, and T. MacNeil. 1992. Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 11161-68. [DOI] [PubMed] [Google Scholar]
- 32.Mascarenhas, J., J. Soppa, A. V. Strunnikov, and P. L. Graumann. 2002. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 213108-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mascarenhas, J., A. V. Volkov, C. Rinn, J. Schiener, R. Guckenberger, and P. L. Graumann. 2005. Dynamic assembly, localization and proteolysis of the Bacillus subtilis SMC complex. BMC Cell Biol. 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey, T. H., C. P. Mercogliano, J. Yates, D. J. Sherratt, and J. Löwe. 2006. Double-stranded DNA translocation: structure and mechanism of hexameric FtsK. Mol. Cell 23457-469. [DOI] [PubMed] [Google Scholar]
- 35.McCormick, J. R., and R. Losick. 1996. Cell division gene ftsQ is required for efficient sporulation but not growth and viability in Streptomyces coelicolor A3(2). J. Bacteriol. 1785295-5301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCormick, J. R., E. P. Su, A. Driks, and R. Losick. 1994. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Mol. Microbiol. 14243-254. [DOI] [PubMed] [Google Scholar]
- 37.Miguélez, E. M., B. Rueda, C. Hardisson, and M. B. Manzanal. 1998. Nucleoid partitioning and the later stages of sporulation septum synthesis are closely associated events in the sporulating hyphae of Streptomyces carpinensis. FEMS Microbiol. Lett. 15959-62. [Google Scholar]
- 38.Nasmyth, K., and C. H. Haering. 2005. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 74595-648. [DOI] [PubMed] [Google Scholar]
- 39.Niki, H., A. Jaffe, R. Imamura, T. Ogura, and S. Hiraga. 1991. The new gene mukB codes for a 177 kD protein with coiled-coil domains involved in chromosome partitioning of E. coli. EMBO J. 10183-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Noens, E. E., V. Mersinias, B. A. Traag, C. P. Smith, H. K. Koerten, and G. P. van Wezel. 2005. SsgA-like proteins determine the fate of peptidoglycan during sporulation of Streptomyces coelicolor. Mol. Microbiol. 58929-944. [DOI] [PubMed] [Google Scholar]
- 41.Petrushenko, Z. M., C. H. Lai, and V. V. Rybenkov. 2006. Antagonistic interactions of kleisins and DNA with bacterial condensin MukB. J. Biol. Chem. 28134208-34217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ptacin, J. L., M. Nollmann, E. C. Becker, N. R. Cozzarelli, K. Pogliano, and C. Bustamante. 2008. Sequence-directed DNA export guides chromosome translocation during sporulation in Bacillus subtilis. Nat. Struct. Mol. Biol. 15485-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 2177-96. [DOI] [PubMed] [Google Scholar]
- 44.Ruban-Ośmiałowska, B., D. Jakimowicz, A. Smulczyk-Krawczyszyn, K. F. Chater, and J. Zakrzewska-Czerwińska. 2006. Replisome localization in vegetative and aerial hyphae of Streptomyces coelicolor. J. Bacteriol. 1887311-7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 46.Schwedock, J., J. R. McCormick, E. R. Angert, J. R. Nodwell, and R. Losick. 1997. Assembly of the cell division protein FtsZ into ladder-like structures in the aerial hyphae of Streptomyces coelicolor. Mol. Microbiol. 25847-858. [DOI] [PubMed] [Google Scholar]
- 47.Soppa, J., K. Kobayashi, M. F. Noirot-Gros, D. Oesterhelt, S. D. Ehrlich, E. Dervyn, N. Ogasawara, and S. Moriya. 2002. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 4559-71. [DOI] [PubMed] [Google Scholar]
- 48.Reference deleted.
- 49.Srivastava, P., G. Demarre, T. S. Karpova, J. McNally, and D. K. Chattoraj. 2007. Changes in nucleoid morphology and origin localization upon inhibition or alteration of the actin homolog, MreB, of Vibrio cholerae. J. Bacteriol. 1897450-7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Strunnikov, A. V. 2006. SMC complexes in bacterial chromosome condensation and segregation. Plasmid 55135-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Strunnikov, A. V. 1998. SMC proteins and chromosome structure. Trends Cell Biol. 8454-459. [DOI] [PubMed] [Google Scholar]
- 51.Thanbichler, M., and L. Shapiro. 2008. Getting organized—how bacterial cells move proteins and DNA. Nat. Rev. Microbiol. 628-40. [DOI] [PubMed] [Google Scholar]
- 52.Virca, G. D., W. Northemann, B. R. Sheils, G. Widera, and S. Broome. 1990. Simplified Northern blot hybridization using 5% sodium dodecyl sulfate. BioTechniques 8370-371. [PubMed] [Google Scholar]
- 53.Wang, L., Y. Yu, X. He, X. Zhou, Z. Deng, K. F. Chater, and M. Tao. 2007. Role of an FtsK-like protein in genetic stability in Streptomyces coelicolor A3(2). J. Bacteriol. 1892310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wildermuth, H., and D. A. Hopwood. 1970. Septation during sporulation in Streptomyces coelicolor. J. Gen. Microbiol. 6051-59. [DOI] [PubMed] [Google Scholar]
- 55.Wildschutte, H. 2000. Analysis of smc and in vivo transposon mutagenesis in Streptomyces coelicolor. MS thesis. Duquesne University, Pittsburgh, PA.
- 56.Wu, L. J., and J. Erington. 1997. Septal localization of the SpoIIIE chromosome partitioning protein in Bacillus subtilis. EMBO J. 162161-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, L. J., and J. Errington. 1994. Bacillus subtilis spoIIIE protein required for DNA segregation during asymmetric cell division. Science 264572-575. [DOI] [PubMed] [Google Scholar]
- 58.Yang, M. C., and R. Losick. 2001. Cytological evidence for association of the ends of the linear chromosome in Streptomyces coelicolor. J. Bacteriol. 1835180-5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu, X.-C., E. K. Weihe, and W. Margolin. 1998. Role of the C terminus of FtsK in Escherichia coli chromosome segregation. J. Bacteriol. 1806424-6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.