Abstract
φRSΒ1 is a wide-host-range, T7-like bacteriophage that infects and efficiently lyses the phytopathogenic bacterium Ralstonia solanacearum. The φRSB1 genome comprises 43,079 bp of double-stranded DNA (61.7% G+C) with 325-bp terminal repeats and contains 47 open reading frames. Strong activity of tandem early promoters and wide specificity of phage promoters of φRSB1 were demonstrated.
The phytopathogenic gram-negative bacterium Ralstonia solanacearum causes bacterial wilt disease in many important crops (10). Recently, Yamada et al. (9, 12, 21) isolated and characterized various kinds of bacteriophages that specifically infect R. solanacearum strains belonging to different races and/or biovars. These phages may be useful as a tool not only for molecular biological studies of R. solanacearum pathogenicity but also for diagnosis and biocontrol of bacterial wilt. In this study, we report the genome and characteristic features of a new phage, φRSB1. φRSB1 was isolated from a soil sample from a tomato crop field and was selected for its ability to form large clear plaques on plate cultures of R. solanacearum strain M4S (for details of bacterial strains, see reference 21). Plaques formed on assay plates (21) were 1.0 to 1.5 cm in diameter. This phage has a wide host range and infected 13 of 15 strains tested, including strains of races 1, 3, and 4 and of biovars 3, 4, and N2. Under laboratory conditions (in standard one-step growth assays), host cells of R. solanacearum strains lyse after 2.5 to 3 h postinfection (p.i.) (with an eclipse period of 1.5 to 2 h), releasing approximately 30 to 60 PFU of new phage particles per cell (burst size) (data not shown). Electron microscopic observation of negatively stained phage particles revealed short-tailed icosahedral structures resembling those of the family Podoviridae. The particles consisted of a head of approximately 60 nm in diameter and a stubby tail of 20 nm in length (data not shown).
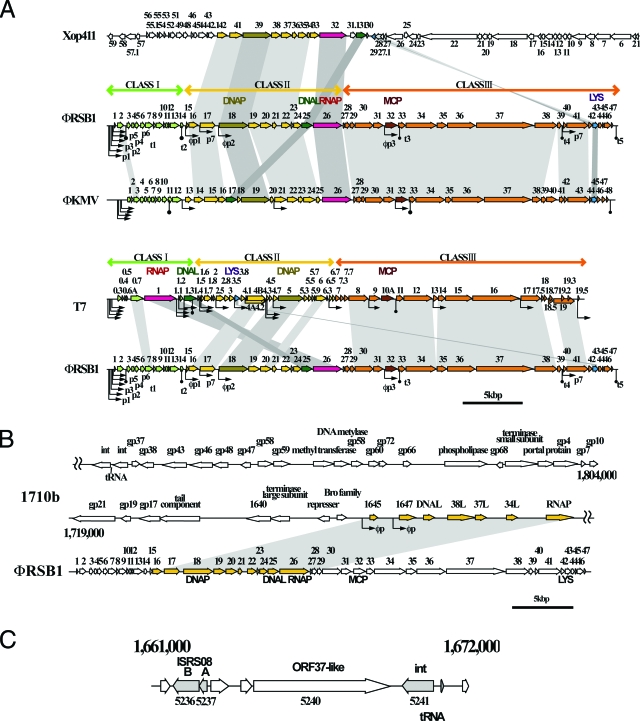
The φRSB1 genome is linear double-stranded DNA of approximately 43.0 kbp in size as determined by pulsed-field gel electrophoresis (data not shown). Since no oligomeric forms were formed after heat treatment, cohesive ends are absent. The sequence of the φRSB1 genome was determined using DNA purified from phage particles by shotgun sequencing and primer walking (9). Sequences were assembled into a circular contig of 42,754 bp, suggesting the presence of long terminal repeats. The precise sequence of the repeat was determined by direct sequencing of genomic DNA with outward-directed primers, located outside the possible terminal repeat region. The final sequence of the φRSB1 genome is 43,079 bp and includes direct terminal repeats of 325 bp. The φRSB1 genome size is comparable with that of Pseudomonas aeruginosa phage φKMV (42,519 bp; accession no. AJ505558) and slightly greater than those of coliphages T7 (39,937 bp; accession no. NC_001604) and T3 (38,208 bp; accession no. NC_003298). The size of the φRSB1 terminal repeats is less than that of φKMV (414 bp) but greater than that of T7 (160 bp) or T3 (231 bp). The G+C content of the genome is 61.7%. This value is lower than the G+C values of the large and small replicons of the R. solanacearum GMI1000 genome (67.04% and 66.86%, respectively) (18). Potential open reading frames (ORFs) consisting of more than approximately 50 codons and starting with ATG, GTG, or TTG were identified using the Orfinder and DNASIS programs. The presence of a Shine-Dalgarno ribosome-binding sequence preceding the initiation codon was taken into account for ORF prediction. Possible functions were assigned to ORFs by searching through databases using the BLAST, BLASTX, and BLASTP programs (1). Accordingly, a total of 47 potential ORFs oriented in the same direction were assigned on the genome (Fig. 1A; see the supplemental material). To find homologous sequences, nucleotide sequences from φRSB1 DNA were used to searched databases with the BLAST and BLASTX programs. Patchy or local homologies were detected in the genomic sequences of various phages, including Xanthomonas oryzae phages Xop411 (accession no. DQ777876) and Xp10 (accession no. AY299121) (22), Pseudomonas aeruginosa phages φKMV (accession no. AJ505558) (14) and LKD16 and LKA1 (6), Erwinia amylovora phage Era103 (accession no. EF160123), and Burkholderia cenocepacia phage BcepB1A (accession no. AY616033). All of these are members of the family Podoviridae. The genome of coliphage T7, the representative of T7-like viruses of the Podoviridae, generally consists of three functional gene clusters: one for early functions (class I), one for DNA metabolism (class II), and the other for structural proteins and virion assembly (class III) (8). These gene clusters are essentially conserved in the φRSB1 genome. Figure 1A shows putative ORFs identified on the φRSB1 genome compared with ORFs from other phages: Xanthomonas phage Xop411 (giving the highest local similarities), Pseudomonas phage φ KMV(showing marginal similarity but longest regions of similarity), and coliphage T7. The mosaic genetic relationship of φRSB1 indicates frequent recombinations on the φRSB1 ancestral genome during its evolution, in the way suggested for tailed phages and their prophages (2-5, 11).
FIG. 1.
Genetic organization and comparative analyses of the φRSB1 genome. (A) Comparison of the genomes of φRSB1 and T7-like phages. In each alignment, corresponding ORFs (horizontal arrows) are connected by shading. Three functional gene clusters, class I (green), class II (yellow), and class III (orange), are indicated above the φRSB1 and the T7 maps, and corresponding ORFs are colored. Putative bacterial promoters, phage promoters, and terminators of transcription are indicated under the φRSB1 map by p, φp, and t, respectively. Promoters and terminators are also shown in the φKMV and T7 maps. Xop411, Xanthomonas oryzae phage (44,520 bp, accession no. DQ777876); φKMV, Pseudomonas aeruginosa phage (42,519 bp, AJ50558); T7, coliphage T7 (39,937 bp, NC_00164). DNAP, DNA polymerase; MCP, major capsid protein; LYS, lysozyme. (B) The class II region of the φRSB1 genome (ORF16 to ORF26) often shows high homology with phage or prophage sequences of different phage groups. The φRSB1 region is aligned with the corresponding prophage sequence of Burkholderia pseudomallei 1710b (accession no. CP000124). (C) Region on the GMI1000 genome (positions 1661000 to 1672000) containing a large φRSB1 ORF37-like ORF (RSO5240), which encodes a putative transglycosylase protein. This GMI1000 region is flanked by two ORFs encoding ISRS08 transposase A and B on the left side and by integrase (RS05241) and arginine tRNA (AGA) on the right side.
One of the characteristic features found in the φRSB1 gene organization is that the predicted gene for RNA polymerase (RNAP) of φRSB1 (orf26) is not located in the early gene region (class I) but at the end of the class II region (Fig. 1A). Another exception is the gene for DNA ligase (DNAL); orf25, encoding the φRSB1 DNAL, is in front of the RNAP ORF (orf26), whereas the gene encoding T7 DNAL is downstream of the gene for RNAP at the end of the class I cluster (8). In Pseudomonas phages φKMV, LKD16, and LKA1, the DNAL gene is upstream of the gene for DNA polymerase in the class II gene cluster (Fig. 1A).
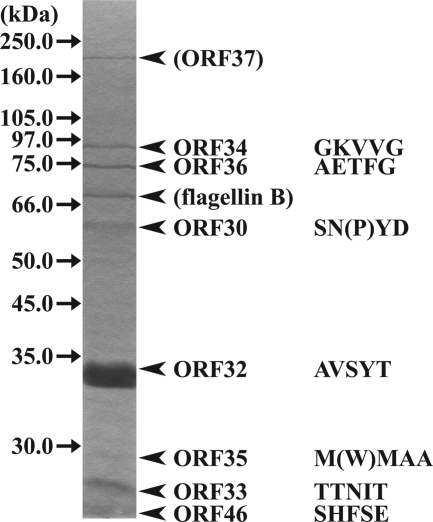
Similarly to the case for the T7 genome, structural proteins are predicted to be encoded in the class III gene cluster of the φRSB1 genome. Purified φRSB1 particles gave at least nine protein bands on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (Fig. 2). Each band was extracted from the gel and was subjected to N-terminal amino acid sequencing (19). The N-terminal sequence of each protein always started from the second amino acid residue of its corresponding ORF, except for ORF35, which included the first methionine (Fig. 2). In addition to known structural proteins, ORF35, ORF36, and ORF46 were identified as structural proteins. In this way, all predicted protein in the class III-structural region were identified, except for the scaffolding protein (ORF31) and a possible tail fiber protein, ORF38, which may be lost during purification of the phage particles. In the case of the largest structural protein (170 to 180 kDa), determination of the N-terminal sequence was unsuccessful using standard methods, possibly because of modification at the N terminus. However, it most likely corresponds to ORF37, as judged from its exceptionally large size (174 kDa); there is no other candidate for this size. ORF37 may encode a tail protein with a transglycosylase domain.
FIG. 2.
Identification of φRSB1 virion proteins. Proteins from purified φRSB1 particles were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% gel) and stained with Coomassie blue. The molecular size of each marker protein (Amersham full-range molecular weight markers and an LMW gel filtration calibration kit) is indicated on the left. The N-terminal amino acid sequence (five residues) determined for each φRSB1 protein band is shown on the right with its corresponding ORF. Amino acids in parentheses are obscure residues. Although the N-terminal sequence could not determined, the largest protein, approximately 175 to 180 kDa, is predicted to be ORF37, as there is no other candidate for this large size. A few small proteins were lost from the gel during electrophoresis.
T7-like phages are generally known as absolute lytic phages, with a few exceptions, such as integrase-coding phages, e.g., prophage 3 of Pseudomonas putida (17) and the cyanophage P-SSP7 (16). Sometimes nucleotide sequences related to T7-like phages are found in conjunction with other temperate phages such as λ-like phages that are integrated in various bacterial genomes (2-5, 11). BLAST and BLASTX database searches using the φRSB1 sequence revealed a significantly homologous region (at the nucleotide sequence level) in the genome of Burkholderia pseudomallei 1710b (accession no. CP000124). A matrix comparison plot showed that this homology is extended to a 20-kbp region (1710b positions 1740980 to 1761000) (data not shown). In the 1710b genome, this region is embedded in a large (85-kbp) prophage sequence (1710b positions 1719000 to 1804000), which is related to λ-like phages such as B. pseudomallei phage φ1026b (7) and B. thailandensis phage φE125 (20). The homologous region of the 1710b prophage contains eight ORFs encoding DNA primase, DNA helicase, DNAL, DNA polymerase, exonuclease, and RNAP, etc. These correspond to the class II genes of φRSB1, as shown in Fig. 1B. Interestingly, the putative φRSB1 promoters (see below) were also found in this 1710b region (positions 1743597 to 1743650 and 1744983 to 1745040) (Fig. 1B). Both Ralstonia and Burkholderia belong to the Betaproteobacteria and may share common bacteriophages (9). Database searches also showed that a 10-kbp genomic region of R. solanacearum GMI1000 (positions 1661000 to 1672000) contains a 1,589-amino-acid ORF (RS05240) showing significant similarity to φRSB1 ORF37, which encodes a putative transglycosylase-tail protein (see the supplemental material; E value, e−131). Amino acid sequence similarity extends to the entire region consisting of the N-terminal transglycosylase and C-terminal core or tail domains (1,606 amino acids in φRSB1 ORF37). This GMI1000 ORF is associated with two insertion sequence transposase sequences (ISRS08 transposases A and B; RS05237 and RS05236, respectively) on the left side. Immediately to the right of this ORF, there is an ORF (RS05241) for a putative integrase, which is closely associated with arginine tRNA (AGA), a possible att sequence (Fig. 1C). This structure indicates horizontal acquisition of this φRSB1 ORF by host cells, as well as some involvement of the phage integrase/att sequence and transposons in such an event. In the context of lysogenic conversion or introduction of a new fitness factor by phage in the pathogenic bacteria, the functions of φRSB1 ORF37 are interesting.
As shown in Fig. 3, several putative transcription promoters and terminators were identified in apparently noncoding regions (more than 100 bp long) in the φRSB1 genome. A typical prokaryotic promoter sequence (resembling E. coli σ70) was repeated five times (p1 to p5) in a left 1,000-bp region without ORFs (Fig. 1A and 3A). In addition, a few other putative sequences of the host σ70 promoter (p6 to p8) were detected in front of ORF1, -17, and -39 (Fig. 1A and 3A). There are possible ρ-independent terminator-like sequences (Fig. 3C). A terminator-like sequence (t2) present after ORF13 is located in the region that separates class I and class II genes (Fig. 1A). Another possible terminator (t3) is located immediately downstream of ORF32, encoding the major capsid protein. t4 is located in front of a putative promoter p8 for ORF40 and ORF41 (similar to the large subunit of a terminase). A final terminator (t5) was defined behind the last ORF47. The terminator positions of t2, t3, and t5 are consistent with those reported in Pseudomonas phages φKMV (14) and LKD16 and LKA1 (6) (Fig. 1A). Searching for core promoter-like sequences conserved in phages T3, T7, or SP6 in the φRSB1 intergenic regions could not find any significant ones. Instead, three sets of common sequence elements were found in front of ORF16, -18, and -32 (designated φp1, φp2, and φp3) (Fig. 1A). We found a set of sequence elements consisting of a GC-rich stretch and TTGT, TCTGG, and CGGGCAC motifs preceding an AG-rich Shine-Dalgarno sequence (Fig. 3B). The activity of transcriptional promoters of both host and phage types thus predicted on the φRSB1 genome was examined using a green fluorescent protein (GFP)-expressing single-copy plasmid, pRSS12 (13), where the lac promoter for GFP expression was replaced with a φRSB1 promoter sequence. When we tested bacterial σ70-type promoters p1 to p4, p1 to p5, and p1 to p6, which are located tandemly at the beginning of the class I gene cluster, transformed cells of R. solanacearum strains always showed strong GFP fluorescence. Fluorescence was 3 to 15 times greater than that of pRSS12 with a lac promoter. Results with strain MAFF301558 as the host are shown in Table 1. Increased GFP intensity from p1-p4 via p1-p5 to p1-p6 clearly demonstrates actual promoter activities of these φRSB1 early promoters. φp1 is located at the beginning of the class II gene cluster, after the possible terminator t2 and in front of ORF16, encoding possible DNA primase. φp3 is located upstream of ORF32, which encodes the major capsid protein. Both φp1 and φp3 also function as promoters for bacterial RNAP in φRSB1-uninfected R. solanacearum cells but show lower activity than p1-p6 (Table 1). The promoter activity was also examined in R. solanacearum cells after infection with φRSB1. As strain MAFF301558 was found to be a low-efficiency host, giving lower titers of phage progeny, the host was changed to strain M4S. After infection with φRSB1, GFP fluorescence intensity was retained at almost the same levels in cells containing the promoters p1 to p5 or p1 to p6, whereas cells with φp1 or φp3 showed increased GFP fluorescence after 30 min p.i. to 90 min p.i. (Table 1). At 120 min p.i. cell lysis began. These results indicate that φp1 and φp3 are functional in transcription by both bacterial and phage RNAPs. Bacterial σ70-type promoters are not shut down but continue to function after infection.
FIG. 3.
Predicted regulatory sequences found in the φRSB1 genome. (A) E. coli σ70-promoter-like sequences; (B) putative promoter sequences for φRSB1-encoded RNAP; (C) putative terminators.
TABLE 1.
Expression of GFP by φRSB1 promoters
| Promoter (position) | Relative intensity of GFP fluorescencea
|
|||||
|---|---|---|---|---|---|---|
| Strain MAFF301558b | Strain M4S at min p.i. with φRSB1:
|
|||||
| 0 | 30 | 60 | 90 | 120 | ||
| lac | 3.5 ± 0.2 (1.0) | 1.8 ± 0.1 | NDc | 2.0 ± 0.1 | ND | 2.0 ± 0.1 |
| p1-p4 (425-845) | 10.0 ± 0.3 (3.0) | ND | ND | ND | ND | ND |
| p1-p5 (425-921) | 14.3 ± 0.5 (4.1) | 4.2 ± 0.2 | 4.4 ± 0.2 | 6.0 ± 0.3 | 7.2 ± 0.3 | 9.0 ± 0.4 |
| p1-p6 (425-995) | 53.0 ± 0.9 (15.2) | 7.3 ± 0.3 | 8.4 ± 0.3 | 12.0 ± 0.4 | 11.7 ± 0.4 | 13.6 ± 0.5 |
| φp1 (6895-7009) | 11.0 ± 0.5 (3.2) | 6.7 ± 0.3 | 16.5 ± 0.5 | 19.0 ± 0.6 | 32.8 ± 0.8 | 15.1 ± 0.5 |
| φp3 (23515-23675) | 7.2 ± 0.3 (2.1) | 1.4 ± 0.1 | 4.1 ± 0.2 | 5.9 ± 0.2 | 11.4 ± 0.4 | 7.8 ± 0.3 |
The values are means ± standard errors for data from three independent experiments.
The ratio to the lac value (1.0) is in parentheses.
ND, not determined.
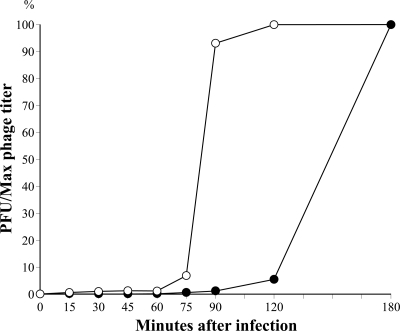
The occurrence of host σ70-type promoter sequences in the late gene clusters, class II and class III, and the low specificity of phage promoters further imply that expression of φRSB1 genes is highly dependent on the host RNAP. To determine whether host RNAP is involved in late stages of φRSB1 infection, rifampin was added to φRSB1-infected cultures at various times p.i., and the number of progeny phage was determined. The results are shown in Fig. 4. In samples that were incubated with rifampin, more than 90% of phage progeny was obtained when the drug was added at 90 min p.i. or later, and no or very few progeny phages were obtained when the drug was added at 75 min p.i. or earlier. These results indicate that a switch from host RNAP to φRSB1 RNAP occurs between 75 min p.i. and 90 min p.i. and that late stages of φRSB1 replication are independent of rifampin. The late genes can be transcribed by rifampin-resistant φRSB1 RNAP, at least in the presence of rifampin.
FIG. 4.
Late stages of φRSB1 development are resistant to rifampin. Cells of R. solanacearum M4S were infected with φRSB1 at a multiplicity of infection of 5. At the indicated times p.i., aliquots of the infected culture were withdrawn and incubated with 100 μg/ml rifampin for 2.5 h before CHCl3 treatment and determination of phage titers (PFU) (open circles). For a control, phage titers were also determined at the indicated times after CHCl3 addition without rifampin treatment (closed circles). The method is as described by Liao et al. (15).
Nucleotide sequence accession number.
The sequence data for φRSB1 genomic DNA have been deposited in the DDBJ database under accession no. AB451219.
Supplementary Material
Acknowledgments
This study was supported by the Industrial Technology Research Grant Program (04A09505) from the New Energy and Industrial Technology Development Organization (NEDO) of Japan.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brussow, H., C. Canchaya, and W.-D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canchaya, C., C. Proux, G. Fournous, A. Bruttin, and H. Brussow. 2003. Prophage genomics. Microbiol. Mol. Biol. Rev. 67238-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casjens, S. 2003. Prophages and bacterial genomics: what have we learned so far? Mol. Microbiol. 49277-300. [DOI] [PubMed] [Google Scholar]
- 5.Casjens, S. 2005. Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol. 8451-458. [DOI] [PubMed] [Google Scholar]
- 6.Ceyssens, P.-J., R. Lavigne, W. Mattheus, A. Chibeu, K. Hertveldt, J. Mast, J. Robben, and G. Volckaert. 2006. Genomic analysis of Pseudomonas aeruginosa phages LKD16 and LKA1: establishment of the φKMV subgroup within the T7 subgroup. J. Bacteriol. 1886924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeShazer, D. 2004. Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 1863938-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn, J. J., and F. W. Studier. 1983. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J. Mol. Biol. 166477-535. [DOI] [PubMed] [Google Scholar]
- 9.Fujiwara, A., T. Kawasaki, S. Usami, M. Fujie, and T. Yamada. 2008. Genomic characterization of Ralstonia solanacearum phage φRSA1 and its related prophage (φRSX) in strain GMI1000. J. Bacteriol. 190143-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayward, A. C. 1991. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu. Rev. Phytopathol. 2965-87. [DOI] [PubMed] [Google Scholar]
- 11.Hendrix, R. W., G. F. Hatfull, and M. C. M. Smith. 2003. Bacteriophages with tails: chasing their origins and evolution. Res. Microbiol. 154253-257. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki, T., S. Nagata, A. Fujiwara, H. Satsuma, M. Fujie, S. Usami, and T. Yamada. 2007. Genomic characterization of the filamentous integrative bacteriophage φRSS1 and φRSM1, which infect Ralstonia solanacearum. J. Bacteriol. 1895792-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawasaki, T., H. Satsuma, M. Fujie, S. Usami, and T. Yamada. 2007. Monitoring of phytopathogenic Ralstonia solanacearum cells using green fluorescent protein-expressing plasmid derived from bacteriophage φRSS1. J. Biosci. Bioeng. 104451-456. [DOI] [PubMed] [Google Scholar]
- 14.Lavigne, R., M. V. Burkal'tseva, J. Robben, N. N. Sykilinda, L. P. Kurochkina, B. Grymonprez, B. Jonckx, V. N. Krylov, V. V. Mesyanzhinov, and G. Volckaert. 2003. The genome of bacteriophage φKMV, a T7-like virus infecting Pseudomonas aeruginosa. Virology 31249-59. [DOI] [PubMed] [Google Scholar]
- 15.Liao, Y. D., J. Tu, and T. T. Kuo. 1987. Regulation of transcription of the Xp10 genome in bacteriophage-infected Xanthomonas campestris pv. oryzae. J. Virol. 611695-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindell, D., M. B. Sullivan, Z. I. Johnson, A. C. Tolonen, F. Rohwer, and S. W. Chisholm. 2004. Transfer of photosynthesis genes to and from Prochlorococcus viruses. Proc. Natl. Acad. Sci. USA 10111013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molineux, I. J. 1999. T7-like phages (Podoviridae), p. 1722-1729. In A. Granoff and R. Webster (ed.), Encyclopedia of virology. Academic Press, Ltd., London, United Kingdom.
- 18.Salanoubat, M., S. Genin, F. Artiguenave, J. Gouzy, S. Mangenot, M. Ariat, A. Billault, P. Brottier, J. C. Camus, L. Cattolico, M. Chandler, N. Choisene, S. Claudel-Renard, N. Cunnac, C. Gaspin, M. Lavie, A. Molsan, C. Robert, W. Saurin, T. Schlex, P. Siguier, P. Thebault, M. Whalen, P. Wincker, M. Levy, J. Weissenbach, and C. A. Boucher. 2002. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415497-502. [DOI] [PubMed] [Google Scholar]
- 19.Songsri, P., S. Hiramatsu, M. Fujie, and T. Yamada. 1997. Proteolytic processing of Chlorella virus CVK2 capsid proteins. Virology 227252-254. [DOI] [PubMed] [Google Scholar]
- 20.Woods, D. E., J. A. Jeddeloh, D. L. Fritz, and D. DeShazer. 2002. Burkholderia thailandensis E125 harbors a temperate bacteriophage specific for Burkholderia mallei. J. Bacteriol. 1844003-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamada, T., T. Kawasaki, S. Nagata, A. Fujiwara, S. Usami, and M. Fujie. 2007. Isolation and characterization of bacteriophages that infect the phytopathogen Ralstonia solanacearum. Microbiology 1532630-2639. [DOI] [PubMed] [Google Scholar]
- 22.Yuzenkova, J., S. Nechaev, J. Berlin, D. Rogulja, K. Kuznedelov, R. Inman, A. Mushegian, and K. Severinov. 2003. Genome of Xanthomonas oryzae bacteriophage XP10: an odd T-odd phage. J. Mol. Biol. 330735-748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.