Abstract
One of the features of the periodontal pathogen Porphyromonas gingivalis is the presence of complex iron acquisition systems that include an hmuYRSTUV locus. HmuY and HmuR are hemin binding proteins required for P. gingivalis growth. Previous studies have demonstrated that expression of the hmu locus is regulated in response to environmental changes, such as growth phases. However, the mechanisms involved in hmu gene regulation are poorly understood. Here we report that a novel transcriptional activator, PG1237, is required for the expression of humY and humR, but not other iron acquisition-related genes, such as fetB and tlr, which also encode hemin binding proteins. Real-time reverse transcription-PCR analysis revealed that a mutation in the pg1237 gene decreased expression of hmuY and hmuR 149- and 25-fold, respectively, compared to that observed in the wild-type strain. In addition, differential expression of hmuY, hmuR, and the pg1237 gene was found to be quorum-sensing dependent, such that higher expression levels of these genes were observed when P. gingivalis was grown at a lower cell density, such as that seen during the early exponential growth phase. This work demonstrates the involvement of a novel transcriptional activator, PG1237, in expression of the hmu operon in a cell density-dependent fashion.
Ability to uptake iron/hemin plays an important role in bacterial growth, survival, and virulence (15). Porphyromonas gingivalis is considered a periodontopathogen, associated with several forms of periodontitis (20, 21). The organism grows in the periodontal packets of patients with destructive periodontitis, an environment bathed by gingival crevicular fluid which exudes from vessels of the microcirculation. Proteins in gingival crevicular fluid, such as transferrin and hemoglobin, are an important source of iron for the organism. Based on sequence analysis of the P. gingivalis W83 genome, at least 50 genes have been identified as being possibly involved in iron/hemin acquisition (13). Several specific iron/hemin acquisition systems have been identified in P. gingivalis. For example, P. gingivalis produces a number of proteases which are involved in binding and degrading hemoglobin (3, 9, 22). Iron/hemin is acquired by P. gingivalis surface proteins, including the outer membrane hemin binding proteins, HmuR and Tlr (18, 19). In addition, the FetB (iron hemin transport) protein may function to assimilate hemin (15). Iron/hemin is then transported from the cell surface to the cytoplasm by P. gingivalis ABC transport systems and permeases (19).
Recent studies have shown that the hmuR gene belongs to an hmu locus that is composed of six genes, hmuYRSTUV, which are cotranscribed (10). It has been suggested that HmuY and HmuR function as outer membrane hemin receptors, while the HmuSTUV proteins are involved in hemin processing and transport across the cell inner membrane. Interestingly, Northern blot analysis has indicated a differential expression of the hmuYRSTUV genes within the hmu operon (10). The expression level of hmuY is at least sevenfold higher than that of hmuR, and it has been suggested that secondary structures in hmu mRNA are likely responsible for differential expression of these two genes (10).
Expression of genes encoding iron uptake systems in bacteria often is regulated by the level of iron in their environments. It is reported that under hemin-limited conditions, P. gingivalis elevates expression of hmuY and hmuR (10, 15, 16, 18). However, the mechanism of regulation of the expression of iron uptake genes is not well established for P. gingivalis. The P. gingivalis ferric uptake regulator (fur) gene with homology to the fur gene from gram-negative bacteria has been identified (15). The P. gingivalis Fur protein appears to complement the functional activity of the Escherichia coli Fur protein and a Fur binding consensus sequence (Fur box) is located upstream of the hmuY start codon, suggesting that the Fur protein may regulate hmuY expression (18). Recently, LuxS, an AI-2 synthase, has been shown to play a role in the transcriptional regulation of the P. gingivalis iron/hemin acquisition mechanism (2, 8). In these studies, AI-2 was found to negatively regulate expression of hmuR, but positively regulate tlr expression.
In the study reported here, we examined the role of the PG1237 transcriptional regulator, which tightly controls the transcriptional level of the hmu operon. Using mutagenic analysis, we demonstrated that PG1237 specifically activates expression of hmu genes but not other iron acquisition-related genes. Expression of the pg1237 gene appears to be modulated by cell density. In addition, we demonstrated that the PG1237 transcriptional regulator binds directly to the promoter region upstream of hmuY.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains used in this study are listed in Table 1. P. gingivalis strains were grown from frozen stocks in Trypticase soy broth (TSB) or on TSB blood agar plates, supplemented with yeast extract (1 mg/ml), hemin (5 μg/ml), and menadione (1 μg/ml), at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). E. coli strains were grown in Luria-Bertani broth at 37°C. Antibiotics were used when appropriate, at the following concentrations: gentamicin (100 μg/ml), erythromycin (10 μg/ml), ampicillin (50 μg/ml), and kanamycin (50 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| P. gingivalis | ||
| 33277 | Type strain from ATCC | Lab collection |
| 1237E | P. gingivalis mutant with the pg1237 gene inactivated by insertion of an ermF-ermAM cassette; Emr | This study |
| Plasmids | ||
| PCRII-TOPO | Linearized plasmid with single 3′ deoxyribosylthymine residues; Kmr Amr | Invitrogen |
| pTOPO1237 | PCRII-TOPO plasmid carrying a pg1237 gene | This study |
| pET30b | Circle plasmid carrying an N-terminal His tag/thrombin/enterokinase configuration plus an optional C-terminal His tag sequence | Novagen |
| pET30b-1237 | pET30b plasmid carrying a pg1237 gene in the expression region | This study |
Kmr, Emr, and Amr indicate resistance to kanamycin, erythromycin, and ampicillin, respectively.
RNA isolation and quantitative PCR.
P. gingivalis strains were grown anaerobically in 5 ml of TSB. Bacteria were harvested by centrifugation at 10,000 rpm and homogenized in Trizol reagent (Invitrogen, Carlsbad, CA). The RNA in the supernatant was then purified using an RNeasy mini spin column (Qiagen, Valencia, CA). RNA samples were digested on the column with RNase-free DNase. Total RNA was tested using an Agilent 2100 bioanalyzer to ensure the quality of the samples. Real-time RT-PCR analysis was performed using the QuantiTect Sybr green RT-PCR kit (Qiagen) on the iCycler MyiQ real-time PCR detection system (Bio-Rad Laboratories, Inc.) according to the manufacturer's instructions. Primers were designed using Primer3 software and are listed in Table 2. Amplification reactions consisted of an RT cycle at 50°C for 30 min, an initial activation at 95°C for 15 min, and 40 cycles of 94°C for 15 s, 58°C for 30 s, and 72°C for 30 s. The melting curve profile was analyzed to verify a single peak for each sample, which indicates primer specificity. The expression levels of the investigated genes for the test sample were determined relative to the untreated calibrator sample by using the comparative cycle threshold (ΔΔCT) method. The ΔΔCT values were calculated by subtracting the average CT value of the test sample from the average CT value of the calibrator sample and were then used to calculate the ratio of the two by assuming 100% amplification efficiency. By loading the same amount of total RNA for any comparable samples, the ΔΔCT represents the difference in gene expression levels between the samples.
TABLE 2.
Oligonucleotide primers
| Gene | Primer | Primer sequence (5′-3′)a | Application |
|---|---|---|---|
| pg1237 | r1237F-EcoRV | CAGTCGATATCCATGAATATCATCACACTCAAGG | Full-length pg1237 open reading frame amplification |
| r1237R-HindIII | CAGTCAAGCTTTTAGTGGGCGATTTCGTCGA | ||
| pg1237(1)F | GCATACGAGAAGTCTGTGAGGA | Construction of pg1237 mutation | |
| pg1237(1)R-erm | GATGTTGCAAATACCGATGAGCAATCGGCGTAGTGAGCAAAC | ||
| pg1237(2)F-erm | CCTCTAGAGTCGACCTGCAGCGAAGAGAGCGGAGAAGAAC | ||
| pg1237(2)R | GAAACGGTCAAGCAGTGGAT | ||
| pg1237-164F | CCACGCCACAGTAGAGGAAT | Real-time PCR | |
| pg1237-164R | GCTCTTCGGCAATCTCTTTG | ||
| ermF-ermAM | ErmF | GCTCATCGGTATTTGCAACA | ermF-ermAM amplification |
| ErmR | CTGCAGGTCGACTCTAGAGG | ||
| pg1551 (hmuY) | pg1551-149F | GGCTACTACCGTTCCGACAG | Real-time PCR |
| pg1551-149R | ATCCCTGTGCGTTCTTCTTG | ||
| TSR1 | TTCGGGTGTGGAGGGTTGGT | Identification of transcriptional start site | |
| TSR2 | CGGCTCGTCTTTCTTCTTCC | ||
| RTR4 | CTGTCGGAACGGTAGTAGCC | ||
| RTF1 | CCTACAAATTGGGATTGCTC | ||
| RTF2 | CCTATTAAAAGTAAGGTCAG | ||
| RTF3 | AAATGTTACCGACTATAAGA | ||
| Prom-F | Bio-ACCTATGTATCGAGGGCTTT | Generating biotin-labeled hmuY promoter region for EMSA | |
| Prom-R1 | CTGACCTTACTTTTAATAGG | ||
| Prom-R2 | CACAGAGTGCGGAGAAAATG | ||
| pg1552 (hmuR) | pg1552-134F | GCGACGGACAGAAATACGAT | Real-time PCR |
| pg1552-134R | GCCTGCAACATTCAGTTCCT | ||
| pg1737 | pg1737-131F | ATGAATCCGATCCGCCACCAC | Real-time PCR |
| pg1737-131R | GCCTCCCATCCCAAAGCACT | ||
| tlr | 5′tlr-454 | CCTGCGGGAACGGACAATATC | Real-time PCR |
| 3′tlr-630 | GCTACCGCCGAAGAGAGAAAC | ||
| fetB (ihtB) | 5′ihtB-636 | TATTGCCGAACTGAAAGAAACC | Real-time PCR |
| 3′ihtB-805 | TGCCATTGTCCAGCTTGTC | ||
| fimA | fimA88F | CGGAACGAATAACCCAGAGA | Real-time PCR |
| fimA88R | CTGACCAACGAGAACCCACT | ||
| fimAProm-F | CGACGCTATATGCAAGACAA | Generating biotin-labeled fimA promoter region for EMSA | |
| fimAProm-R | Bio-TGTAACGGGTTCTGCCTCGT | ||
| mfa | mfa121F | CAGATGGGTTGTTGCTCA | Real-time PCR |
| mfa121R | ATAGAAAGTGCTGCTGGTAG |
The underlined sequences indicate either restriction enzyme sites or sequences corresponding to the sequences of ermF-ermAM gene.
Construction of the pg1237 mutants.
An insertional pg1237 mutant was generated by using ligation-independent cloning of PCR-mediated mutagenesis (1, 25). A 2.1-kb ermF-ermAM cassette was introduced into the pg1237 gene by three steps of PCR to yield a pg1237-erm-pg1237 DNA fragment as described previously (23). The fragment was then introduced into P. gingivalis 33277 by electroporation (24). The pg1237-deficient mutant was generated via a double crossover event that replaces the pg1237 gene with the pg1237-erm-pg1237 DNA fragment in the 33277 chromosome. The mutants were selected on TSB plates containing erythromycin (5 μg/ml). The insertional mutation was confirmed by PCR analysis, and the mutants were designated P. gingivalis 1237E.
5′ RACE analysis of humY transcripts.
The transcriptional start site of humY was determined by using the FirstChoice RNA ligase-mediated rapid amplification of cDNA ends (RACE) kit (Ambion, Austin, TX) as described previously (24). Briefly, after the P. gingivalis cells were harvested by centrifugation, total RNA was purified using an RNeasy mini kit according to the manufacturer's instructions (Qiagen, Inc., Valencia, CA). A 45-base 5′ RNA adapter oligonucleotide was ligated to the 5′ end of the total RNA (10 μg) using T4 RNA ligase. RT was performed by using Moloney murine leukemia virus reverse transcriptase with the specific reverse primer HmuTSR1 of hmuY. Nested PCR was performed by first using 5′ RACE outer primer HmuTSR1 to amplify 5′ adapter-linked cDNA molecules of hmuY. Inner PCR was then conducted with 5′ RACE inner primer that anneals to the adapter oligonucleotide and HmuTSR2, and with the PCR product generated from the outer primers as templates. Five microliters of each PCR product was analyzed by 1.5% agarose gel electrophoresis. The PCR fragments of the inner PCR product were extracted and cloned into a pCRII-TOPO vector (Invitrogen) and sequenced by using an ABI capillary sequencer (Perkin-Elmer).
Production of PG1237 recombinant protein.
The DNA fragment encoding PG1237 was amplified by PCR with primers r1237F-EcoRV and r1237R-HindIII, which produced a 600-bp PCR product (Table 2). The PCR products were then cloned into pCRII-TOPO (Invitrogen, Carlsbad, CA). The recombinant PG1237 (rPG1237) was expressed in E. coli by using a pET protein expression system (Novagen, Madison, WI). The DNA fragment of pg1237 was subcloned into the pET-30b downstream of a histidine tag. rPG1237 was expressed in E. coli BL21(DE3) cells carrying the pET-30b/pg1237 plasmid in the presence of IPTG (isopropyl-β-d-thiogalactopyranoside) and kanamycin. His-tagged rPG1237 was purified with Ni2+-charged His-bind resin (Novagen, Madison, WI). The His tag on the recombinant protein was cleaved with enterokinase and removed by His-bind resin. Enterokinase was then removed by using Ekapture agarose.
EMSA.
Electrophoretic mobility shift assays (EMSA) were performed using the LightShift chemiluminescent EMSA kit (Pierce, Rockford, IL) as described previously (24). Biotin-labeled DNA fragments were generated by using 5′ biotin-incorporated primers (Table 2). The binding of rPG1237 to DNA was carried out in a 20-μl reaction mixture containing 20 fmol biotin-labeled DNA, 10 mM Tris (pH 7.5), 50 mM KCl, 1 mM dithiothreitol, 10 ng/μl poly(dI-dC), 2% glycerol, 0.05% NP-40, and 2 mM MgCl with various amounts of purified rPG1237 protein (0, 2.5, 5, and 10 μg) at room temperature for 30 min. Samples were then loaded and run into a 5% nondenaturing polyacrylamide gel in 0.5 × Tris-borate-EDTA buffer. The DNA and protein complexes were then transferred to a positively charged nylon membrane (380 mA, 30 min). The biotin end-labeled DNA was detected using the streptavidin-horseradish peroxidase conjugate and the chemiluminescent substrate. Each EMSA was repeated at least three times.
RESULTS
PG1237 serves as a transcriptional activator of hmu genes.
It is well documented that expression of hmu genes is under tight control in P. gingivalis (8, 10, 16). However, the mechanism responsible for this regulation of hmu gene expression is unknown. We postulated that a transcriptional element, either an activator or repressor, may be involved in the mediation of hmu gene expression. We initially investigated several genes which encode transcriptional factors, such as FimR, a transcriptional activator of fimA, and OxyR, a transcriptional activator of the sod gene (4, 14, 23). The fimR mutant or the oxyR mutant was examined for expression levels of hmuY and hmuR genes. The results from this study indicated that FimR and OxyR, two well-known transcriptional factors, are not involved in the expression of hmu genes (data not shown).
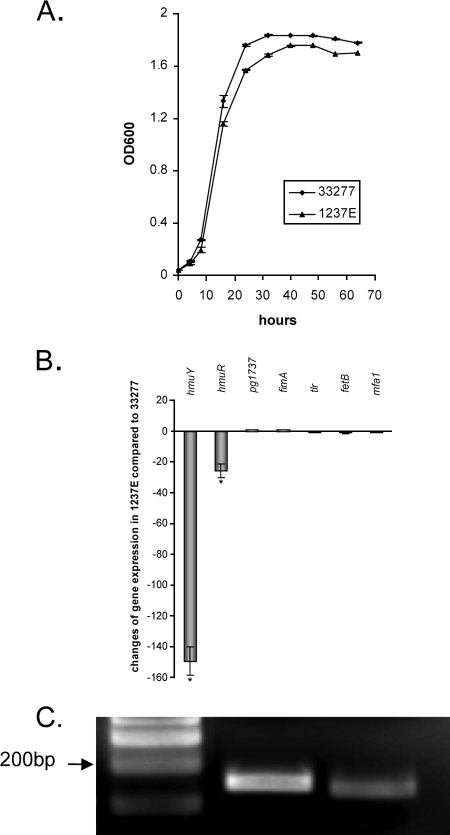
To test our hypothesis, we also constructed a mutant carrying an insertional mutation in the pg1237 gene by replacing the coding sequence of pg1237 with an erythromycin resistance gene cassette. pg1237 encodes a protein annotated in the genome of P. gingivalis as a transcriptional regulator with an unknown target (13). Comparison of the growth curves of P. gingivalis 33277 and the pg1237 mutant (1237E) indicated that the growth rate of the mutant was slightly lower than that of the wild-type strain (Fig. 1A). We then examined the functionality of the PG1237 protein in hmu gene regulation by comparing the expression level of hmu genes in the pg1237 mutant (1237E) with that in wild-type strain 33277. The expression level of hmu genes in early log phase cultures of the bacteria was determined using real-time RT-PCR analysis. As shown in Fig. 1B, significant differential expressions of hmuY and hmuR genes were detected for P. gingivalis wild-type strain 33277 and the pg1237 mutant. Expression of hmuY was downregulated approximately 150-fold, while expression of hmuR was decreased 25-fold in P. gingivalis 1237E compared to expression in wild-type strain 33277. In contrast, expression of glk (pg1737), a gene encoding glucokinase, was consistent between the wild type and the mutant strains. The different regulatory efficiencies of the PG1237 protein with hmuY and hmuR may be due to the different transcriptional rates of these two genes. A previous study by Lewis et al. showed that the hmuY transcript is more prevalent than the hmuR transcript in P. gingivalis W83, which may result from the secondary structures within hmu mRNA (10). In agreement with the earlier result, differential expression in the hmu locus was also observed for the hmuY and hmuR genes of P. gingivalis 33277 (Fig. 1C).
FIG. 1.
Differential expression of hmu genes in the pg1237 mutant (P. gingivalis 1237E) and wild-type 33277 strain. (A) Comparison of the growth curves of P. gingivalis strains. Cells were grown in TSB media. Shown in the curves are the means of four samples, with error bars representing standard errors of the means. One-milliliter aliquots were taken, and the OD600 was measured over a period of 70 h. (B) Gene expression in 33277 or in the pg1237 mutant was determined using real time RT-PCR analysis. Change in expression levels was calculated by the ΔΔCT method, where ΔΔCT = CT(33277) − CT(1237E) and ratio = 2−ΔΔCT. The expression levels of the pg1737, fimA, and mfa1 genes were used as controls. Standard errors are indicated (n = 3). Bars with the asterisks are significant (P < 0.05; t test and Bonferroni's test). (C) Differential expression of hmuY and hmuR in P. gingivalis 33277. Expression levels of hmuR and hmuY were determined by using RT-PCR. Lane 1, molecular size standards; lane 2, RT-PCR product of hmuY; lane 3, RT-PCR product of hmuR.
Expression of two other well-known genes involved in hemin acquisition, tlr and fetB, also was examined with P. gingivalis 1237E. These genes appear to not be regulated by the PG1237 protein (Fig. 1B). Moreover, the expression of genes fimA and mfa1, which encode long and short fimbrial proteins, key virulence factors of P. gingivalis, was not influenced by the deletion of the pg1237 gene. These data suggest that expression of the hmu genes is tightly controlled by PG1237, a transcriptional regulator that selectively activates the hmu promoter.
Binding of PG1237 protein to the promoter region of hmu genes.
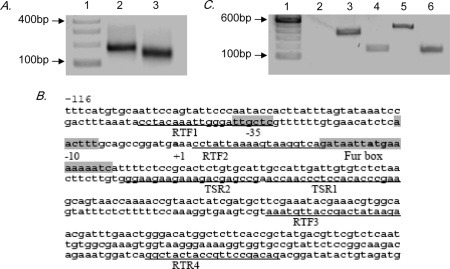
In order to investigate the interaction of PG1237 and the promoter region of the hmu locus, we first determined the transcriptional start site of hmu genes using 5′ RACE-PCR. The RACE experiment was performed with the hmuY-specific reverse primers HmuTSR1 and HmuTSR2 located 64 to 105 bp downstream of the potential start codon (Fig. 2B). Nucleotide sequence analysis of the RACE-PCR product (Fig. 2A) indicated that the transcription start site (TSS) is located 29 bases upstream of the hmuY open reading frame. Based on the consensus promoter sequences of P. gingivalis that were defined by Jackson et al. (7), the potential −10 and −35 sequences, 5′-CAACTT and 5′-GGATTG, are found to be centered at nucleotide position −14/15 and −39/40, respectively. The TSS for hmuY is not located at the same position found by Lewis et al. (10), who suggested that the TSS of hmuY in P. gingivalis W83 was positioned 292 bp upstream of the hmuY start codon. The basis for this strain-dependent location of the TSS is not known, but the data below emphasize the functional relevance of the TSS for hmuY identified in P. gingivalis 33277.
FIG. 2.
Mapping the promoter elements of hmuY of P. gingivalis 33277. (A) cDNA of hmuY mRNA was generated by 5′ RACE-PCR. The RACE-PCR products were visualized on a 1.5% agarose gel. Lane 1, molecular size standards; lane 2, outer 5′ RACE-PCR product; lane 3, inner 5′ RACE-PCR product. (B) DNA sequence of the hmuY promoter region. The transcriptional start site A (+1) established by 5′ RACE-PCR and the potential start codon, ATG, are bolded. The primers used for RACE -PCR are underlined. The potential Fur box, −10, and −35 regions are shaded. (C) RT-PCR analysis of hmuY mRNA. Lane 1, 100-bp ladder marker. Lanes 2, 3, and 4 are RT-PCR products of 33277. Lane 2, RT-PCR with primers RTF1 (from −55 to −36) and RTR4 (from +347 to +366); lane 3, RT-PCR undertaken with primers RTF2 (from +4 to +24) and RTR4 (from +347 to +366); lane 4, RT-PCR RTF3 (from +215 to +234) and RTR4 (from +347 to +366). Lanes 5 and 6 are RT-PCR products of W83. Lane 5, RT-PCR with primers RTF1 (from −55 to −36) and RTR4 (from +347 to +366); lane 6, RT-PCR RTF3 (from +215 to +234) and RTR4 (from +347 to +366).
To verify the result of RACE, we performed RT-PCR using three sets of primers. As shown in Fig. 2C, the PCR products were generated either with primers RTF2 (corresponding to positions +4 to +24) and RTR4 (+347 to +366) or with primers RTF3 (corresponding to +215 to +234) and RTR4 (+347 to +366). There was no PCR product generated from RT-PCR using the primers RTF1 (corresponding to −55 to −36), which corresponds to the DNA sequences upstream from the TSS identified here, or with RTR4 (+347 to +366). In contrast, an RT-PCR product was detected when P. gingivalis W83 genomic DNA was used as a template (Fig. 2C). These data suggest that the TSS of hmuY vary between P. gingivalis strains.
To determine if the transcriptional regulator PG1237 directly interacts with the promoter region of hmuY, we performed an EMSA. The promoter regions of hmuY, positioned from +23 to −150 (without a Fur box sequence) and from +60 to −150 (with a Fur box sequence), were generated by PCR with the 5′ biotin-labeled primers (Table 2). The promoter region (positioned from −22 to −190) of fimA (17), a gene-encoding component of the long fimbriae of P. gingivalis, was used as a control. rPG1237 was expressed in a pET expression system and purified from E. coli. As shown in Fig. 3, the DNA fragments of the hmuR promoter regions were shifted in the presence of the rPG1237. As the concentration of rPG1237 was increased, the retarded protein-DNA complex became more evident, with a parallel loss of uncomplexed hmuY promoter DNA. Moreover, there was no significant difference in the binding affinities of PG1237 for the hmuY promoter region with or without the Fur binding sequence. These data suggest that the potential Fur binding sequence of hmuY is not critical for interaction between PG1237 and the hmuY promoter. Several control incubations increase our confidence in the interpretation of our findings, including that the DNA binding activity of rPG1237 was blocked in the presence of excess cold DNA probes, and that no DNA shift was detected when rPG1237 was incubated with the promoter region of fimA.
FIG. 3.
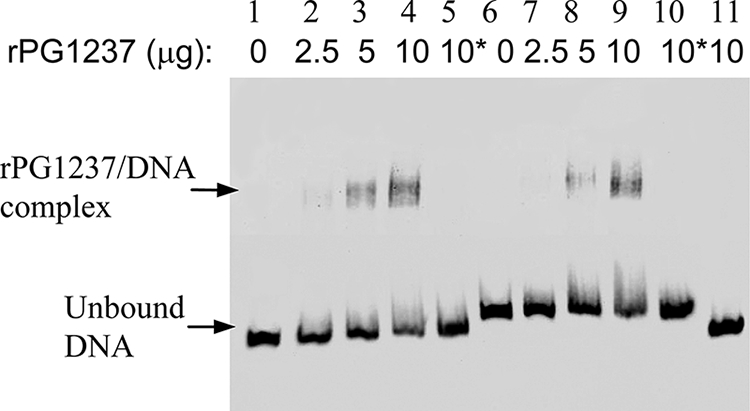
Interaction of rPG1237 with the promoter region of hmuY. EMSA were performed in the presence or absence of rPG1237 and 20 fmol biotin-labeled DNA (lanes 1 to 5, hmuY promoter region including Fur box; lanes 6 to 10, hmuY promoter region without Fur box; lane 11, fimA promoter region). Increasing amounts of rPG1237 were used in the assays. Asterisks indicate that 200-fold excess amounts of each specific competitor oligonucleotide were added to the reaction mixture with the labeled probe.
Expression of hmu genes is modulated in P. gingivalis in response to cell density.
Since expression of genes involved in iron uptake systems is known to be selectively regulated by the iron concentration in the microenvironment (11), several previous studies focused on the regulation of hmu expression in hemin-limited conditions (10, 16). These studies showed an upregulation of hmuY and hmuR expression in P. gingivalis grown under iron-depleted or -limited conditions. To examine if other environmental cues, such as growth phase, are involved in expression regulation of hmu genes, we analyzed and compared the mRNA levels of hmuY, hmuR, and the pg1237 gene in P. gingivalis 33277 strains grown in TSB for 8 (early log phase), 16 (mid-log phase), and 24 (late log phase) h, respectively. RNAs were extracted from the bacterial cells collected from the different time points, and expression levels of hmuY, hmuR, and pg1237 were determined using real time RT-PCR. As shown in Fig. 4A, hmuY, hmuR, and pg1237 were differentially expressed in P. gingivalis 33277 grown to different phases. The highest expression levels of hmuY and hmuR were observed in early log phase, while the expression levels of these genes were decreased as much as 92% in mid-log phase and 98% in late log phase compared to that in early log phase. A parallel expression pattern of hmu genes was found in the expression of pg1237 in P. gingivalis 33277 in response to growth phase. Expression of pg1237 was induced at the early log phase to a level comparable to that of expression of hmu genes (Fig. 4A) and then decreased in mid- and late log phases. However, the expression pattern of hmuY was completely different in the pg1237 mutant. Disruption of the pg1237 gene in P. gingivalis led to hmuY expression that was limited and independent of the growth phase of the bacteria (Fig. 4B). These results suggest that the transcriptional activator PG1237 is required for expression regulation of hmu genes in P. gingivalis in response to growth phase.
FIG. 4.
Differential expression of humY, hmuR, and pg1237 in P. gingivalis 33277 in different growth phases. (A) Differential expression of humY, hmuR, and pg1237 in P. gingivalis 33277 during different growth phases. The expression levels of each gene were determined by real time RT-PCR and then normalized to the expression level at 8 h (early log phase) as 100 units. (B) Comparison of the expression levels of hmuY in P. gingivalis 33277 and P. gingivalis 1237E (the pg1237 mutant). The expression levels were determined by real-time RT-PCR and normalized to the expression level in P. gingivalis grown for 8 h as 100 units. The results presented are the averages of four independent experiments.
Several factors may be involved in this early growth phase upregulation of hmuY and hmuR expression, such as low cell density, higher levels of nutrients, and lower levels of metabolic wastes. To investigate whether cell density is responsible for upregulation of hmu gene expression, we collected P. gingivalis 33277 cells grown in the early log phase by centrifugation and resuspended them in a one-fourth TSB medium containing three-fourths phosphate-buffered saline to reach a final cell density of 5 × 108 (1×) and 5 × 109 (10×)/ml, respectively. After 4 h of incubation at 37°C in an anaerobic chamber, RNA was extracted from each cell mixture, and the expression levels of hmuY, hmuR, and pg1237 were determined using real-time RT-PCR. All three genes were downregulated as cell density increased (Fig. 5). However, the other iron acquisition-related genes, fetB and tlr, were not regulated in P. gingivalis in response to different cell densities. Taken together, these data suggest that cell density likely contributes to the differential expression of hmuY, hmuR, and pg1237 in P. gingivalis grown at different phases.
FIG. 5.
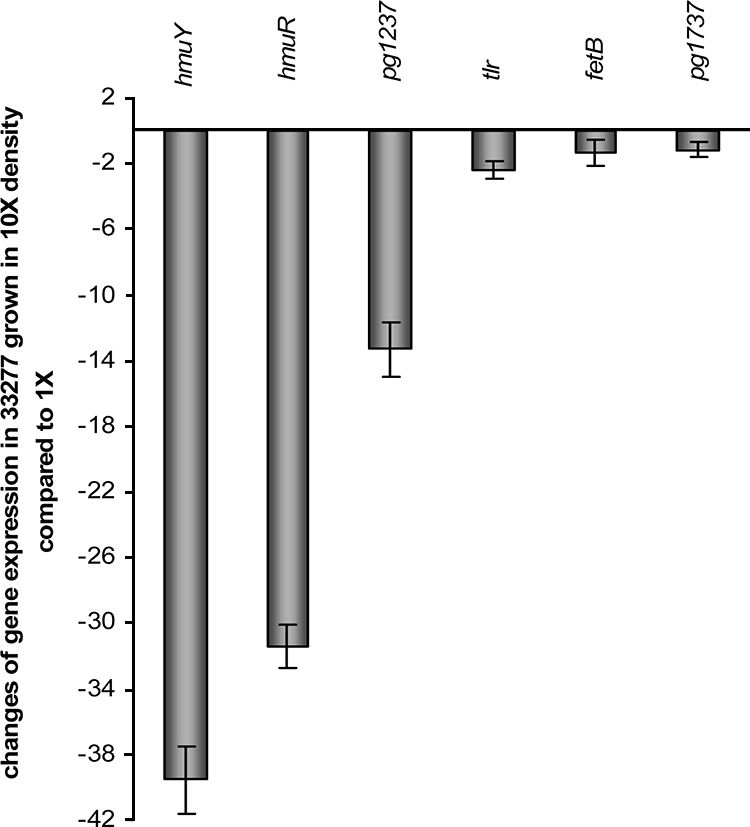
Impact of cell density on hmuY, hmuR, tlr, fetB, and pg1237 expression. P. gingivalis 33277 cells were grown to early log phase and resuspended in 4× diluted TSB media to reach a final cell density of 5 × 108 (1×) or 5 × 109 (10×) per ml, respectively. Expression of iron-related genes in 33277 in response to different cell densities was determined by using real-time RT-PCR. The change in expression levels was calculated by the ΔΔCT method, where ΔΔCT = CT(1× cell density) − CT(10× cell density) and ratio = 2−ΔΔCT. The expression of pg1737, tlr, and fetB was used as a control.
Expression of hmu genes is not upregulated in iron-limited conditions.
While cell density has been shown to be responsible for hmu gene regulation, we hypothesized that upregulation of hmu expression in P. gingivalis grown under hemin-limited or iron-depleted conditions might be a secondary regulation due to changes in cell density. It has also been shown that P. gingivalis grows at a significantly lower rate in the absence of iron/hemin (12, 15). We examined and compared expression levels of hmuY in P. gingivalis in the presence or absence of hemin for bacteria grown at similar cell densities. P. gingivalis 33277 was grown in TSB supplemented with hemin (5 μg/ml) for 8 or 24 h to reach a cell density of 2 × 108/ml (optical density at 600 nm [OD600] = 0.2) or 1.7 × 109/ml (OD600 = 1.7), respectively. The P. gingivalis cells were also cultured in the TSB medium without supplementary hemin. Under these conditions, P. gingivalis exhibited a little lower growth rate compared to bacteria grown with hemin for the first 24 h (Fig. 6), which may be due to the iron storage ability of P. gingivalis. However, the growth rate during the second passage of P. gingivalis 33277 in TSB without hemin was decreased, and cell density reached only to 2 × 108/ml (OD600 = 0.2) after 24 h of growth. Expression of hmuY and pg1237 was determined for P. gingivalis 33277 grown under these conditions using real-time RT-PCR. As shown in Table 3, the relative levels of hmuY and pg1237 mRNAs were negatively correlated with cell density. Expression levels of hmuY and pg1237 were repressed when P. gingivalis multiplied to high cell density, regardless of the presence or absence of hemin. Yet again, hmuY expression was induced at a lower cell density for P. gingivalis grown in the absence of hemin for the second 16 h or in the presence of hemin for 8 h. Our data indicate that hemin is required for P. gingivalis growth, but it does not serve as a signal in the regulatory pathway of hmu genes.
FIG. 6.
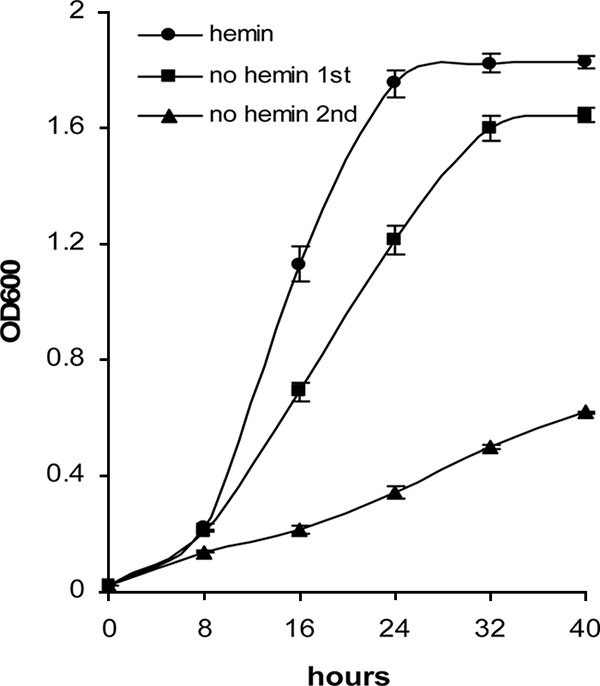
Comparison of the growth curves of P. gingivalis 33277 grown with or without hemin. Cells were grown in TSB media. Shown in the curves are the means of four samples, with error bars representing standard errors of the means. One-milliliter aliquots were taken, and the OD600 was measured over a period of 40 h.
TABLE 3.
Expression levels of hmuY mRNA in P. gingivali 33277 are not altered by the absence or presence of hemin when compared under similar cell density conditions
| Growth condition of 33277 | Growth period (h) | OD600 | mRNA levela:
|
||
|---|---|---|---|---|---|
| hmuY | pg1237 | pg1737 | |||
| With hemin | 24 | 1.76 ± 0.05 | 1.00 | 1.00 | 1.00 |
| 8 | 0.22 ± 0.01 | 166.57 ± 3.60 | 5.18 ± 0.11 | 0.80 ± 0.01 | |
| Without hemin | First 32 | 1.60 ± 0.04 | 1.17 ± 0.10 | 1.58 ± 0.03 | 1.62 ± 0.21 |
| Second 16 | 0.21 ± 0.01 | 172.45 ± 5.67 | 5.35 ± 0.10 | 0.95 ± 0.06 | |
Expression levels of hmuY, pg1237, and pg1737 mRNA were determined by using real-time RT-PCR. The mRNA levels of each gene in P. gingivalis 33277 grown under different conditions are indicated relative to the expression level in the presence of hemin for 24 h as 1 unit. The results shown are the means and standard deviations from three independent experiments.
DISCUSSION
Iron utilization systems are extremely complicated in P. gingivalis. Dozens of proteins are implicated in hemin processing, binding, and transporting functions (15). Both the HmuY and HmuR proteins exhibit the ability to bind hemin in vitro (10, 12). A site-directed mutagenesis analysis demonstrated that histidine 95, histidine 434, and an NPDL motif are involved in binding hemin to HmuR (12). In their recent publication, Lewis et al. showed that the hmuY and hmuR genes belong to the hmuYRSTUV locus and are cotranscribed (10). Although much has been learned about the function and gene organization of the P. gingivalis hmu locus, regulation of the hmu genes is not well understood. It is well known that bacteria are able to sense their environments and respond to environmental signals by selectively expressing the genes required for their survival. Iron concentration is a common regulatory signal that usually elicits expression modulation of genes associated with iron acquisition. Previous studies have shown that P. gingivalis hmu genes are upregulated in iron-depleted or -limited conditions (10, 16), suggesting the presence of an iron-dependent repression of hmu genes in P. gingivalis. In the present study, we also observed an upregulation of hmuY expression in P. gingivalis grown in hemin-limited conditions. However, our data provide new evidence that under the hemin-limited conditions, P. gingivalis enhances hmuY expression in response to low cell density rather than to low iron concentrations. We demonstrated that expression of hmu genes is at the highest level in P. gingivalis grown in the early log phase when cell density is low. Expression levels are significantly decreased in the late log phase, when cell density is much higher. Moreover, expression of hmuY in P. gingivalis cells grown in the same cell densities is similar in the presence of hemin and in the absence of hemin. Therefore, it is highly likely that the upregulation of hmu genes observed in P. gingivalis grown in the iron-depleted or -limited conditions is due to the slower growth of the organism under these conditions.
Currently, details concerning transcriptional regulation of hmu gene expression are not well understood. DNA sequence analysis revealed a 19-bp putative Fur box located upstream of the hmuY start codon, suggesting that a ferric uptake regulator (Fur) might be involved in the regulation of hmuY (15). Fur protein is well studied in E. coli (6), where the Fur protein acts as a repressor in iron-dependent gene regulation. A putative P. gingivalis Fur protein has been identified (6); however, there is no direct evidence that the P. gingivalis Fur protein is involved in expression of hmu genes. Here, we presented strong evidence that PG1237, a 22.4-kDa regulatory protein, is a transcriptional activator of the hmu genes, but not of other iron-related genes. We also showed that PG1237 binds to the promoter region of the hmuY genes, indicating a direct involvement of PG1237 in hmuY regulation. Moreover, the Fur box appears to be not required for the interaction between PG1237 and the hmuY promoter. DNA sequence analysis shows that PG1237 belongs to the LuxR family. Similar to the expression regulated by other members of LuxR family, expression of the pg1237 gene is regulated in a cell density-dependent manner. Generally, LuxR proteins sense the autoinducers, acyl-homoserine lactones, which are synthesized by members of the LuxI protein family (5). However, LuxI and acyl-homoserine lactoneshave not been found in P. gingivalis, properties which are usually required in order to identify new members of the LuxR family. Interestingly, another cell density-dependent sensory system, LuxS-AI-2, appears to be also involved in the regulation of hemin and iron acquisition pathways in P. gingivalis. LuxS proteins function as a key synthase that generates an autoinducer, AI-2. Previous studies have shown that expression levels of Ton-linked hemin binding protein (Tlr) and the lysine-specific protease Kgp are reduced in a luxS mutant, whereas expression levels of some other iron acquisition-related genes, including hmuR, are upregulated in the mutant (2, 8). The regulatory pathways of PG1237 and LuxS are likely not related, since expression of pg1237 is not modulated in the luxS mutant (data not shown).
Taken together, the present findings demonstrate that the expression of hmu genes in P. gingivalis is controlled through a novel transcriptional regulatory pathway involving the transcriptional regulator PG1237. Since the maximal expression of the hmu genes mediated by PG1237 is cell density dependent, but not iron dependent, inhibition of PG1237 production or function may thus represent a new target for the development of antimicrobial therapeutic agents against the periodontal pathogen P. gingivalis.
Acknowledgments
This work was supported by Public Health Service grant DE014699 (HX) from the National Institute of Dental and Craniofacial Research.
Footnotes
Published ahead of print on 17 October 2008.
REFERENCES
- 1.Aslanidis, C., and P. J. de Jong. 1990. Ligation-independent cloning of PCR products (LIC-PCR). Nucleic Acids Res. 186069-6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 1833903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeCarlo, A. A., M. Paramaesvaran, P. L. Yun, C. Collyer, and N. Hunter. 1999. Porphyrin-mediated binding to hemoglobin by the HA2 domain of cysteine proteinases (gingipains) and hemagglutinins from the periodontal pathogen Porphyromonas gingivalis. J. Bacteriol. 1813784-3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diaz, P. I., N. Slakeski, E. C. Reynolds, R. Morona, A. H. Rogers, and P. E. Kolenbrander. 2006. Role of oxyR in the oral anaerobe Porphyromonas gingivalis. J. Bacteriol. 1882454-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50727-751. [DOI] [PubMed] [Google Scholar]
- 6.Grenier, D., V. Goulet, and D. Mayrand. 2001. The capacity of Porphyromonas gingivalis to multiply under iron-limiting conditions correlates with its pathogenicity in an animal model. J. Dent. Res. 801678-1682. [DOI] [PubMed] [Google Scholar]
- 7.Jackson, C. A., B. Hoffmann, N. Slakeski, S. Cleal, A. J. Hendtlass, and E. C. Reynolds. 2000. A consensus Porphyromonas gingivalis promoter sequence. FEMS Microbiol. Lett. 186133-138. [DOI] [PubMed] [Google Scholar]
- 8.James, C. E., Y. Hasegawa, Y. Park, V. Yeung, G. D. Tribble, M. Kuboniwa, D. R. Demuth, and R. J. Lamont. 2006. LuxS involvement in the regulation of genes coding for hemin and iron acquisition systems in Porphyromonas gingivalis. Infect. Immun. 743834-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis, J. P., J. A. Dawson, J. C. Hannis, D. Muddiman, and F. L. Macrina. 1999. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 1814905-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis, J. P., K. Plata, F. Yu, A. Rosato, and C. Anaya. 2006. Transcriptional organization, regulation and role of the Porphyromonas gingivalis W83 hmu haemin-uptake locus. Microbiology 1523367-3382. [DOI] [PubMed] [Google Scholar]
- 11.Litwin, C. M., and S. B. Calderwood. 1993. Role of iron in regulation of virulence genes. Clin. Microbiol. Rev. 6137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu, X., T. Olczak, H. C. Guo, D. W. Dixon, and C. A. Genco. 2006. Identification of amino acid residues involved in heme binding and hemoprotein utilization in the Porphyromonas gingivalis heme receptor HmuR. Infect. Immun. 741222-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 1855591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishikawa, K., F. Yoshimura, and M. J. Duncan. 2004. A regulation cascade controls expression of Porphyromonas gingivalis fimbriae via the FimR response regulator. Mol. Microbiol. 54546-560. [DOI] [PubMed] [Google Scholar]
- 15.Olczak, T., W. Simpson, X. Liu, and C. A. Genco. 2005. Iron and heme utilization in Porphyromonas gingivalis. FEMS Microbiol. Rev. 29119-144. [DOI] [PubMed] [Google Scholar]
- 16.Olczak, T., A. Sroka, J. Potempa, and M. Olczak. 2008. Porphyromonas gingivalis HmuY and HmuR: further characterization of a novel mechanism of heme utilization. Arch. Microbiol. 189197-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, Y., H. Xie, and R. J. Lamont. 2007. Transcriptional organization of the Porphyromonas gingivalis fimA locus. FEMS Microbiol. Lett. 273103-108. [DOI] [PubMed] [Google Scholar]
- 18.Simpson, W., T. Olczak, and C. A. Genco. 2000. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 1825737-5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slakeski, N., S. G. Dashper, P. Cook, C. Poon, C. Moore, and E. C. Reynolds. 2000. A Porphyromonas gingivalis genetic locus encoding a heme transport system. Oral Microbiol. Immunol. 15388-392. [DOI] [PubMed] [Google Scholar]
- 20.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25134-144. [DOI] [PubMed] [Google Scholar]
- 21.Socransky, S. S., C. Smith, and A. D. Haffajee. 2002. Subgingival microbial profiles in refractory periodontal disease. J. Clin. Periodontol. 29260-268. [DOI] [PubMed] [Google Scholar]
- 22.Sroka, A., M. Sztukowska, J. Potempa, J. Travis, and C. A. Genco. 2001. Degradation of host heme proteins by lysine- and arginine-specific cysteine proteinases (gingipains) of Porphyromonas gingivalis. J. Bacteriol. 1835609-5616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu, J., X. Lin, and H. Xie. 2008. OxyR is involved in coordinate regulation of expression of fimA and sod genes in Porphyromonas gingivalis. FEMS Microbiol. Lett. 282188-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu, J., X. Lin, and H. Xie. 2007. Porphyromonas gingivalis short fimbriae are regulated by a FimS/FimR two-component system. FEMS Microbiol. Lett. 271214-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie, H., X. Lin, B. Y. Wang, J. Wu, and R. J. Lamont. 2007. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology 1533228-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]