Abstract
The MtrR transcriptional-regulatory protein is known to repress transcription of the mtrCDE operon, which encodes a multidrug efflux pump possessed by Neisseria gonorrhoeae that is important in the ability of gonococci to resist certain hydrophobic antibiotics, detergents, dyes, and host-derived antimicrobials. In order to determine whether MtrR can exert regulatory action on other gonococcal genes, we performed a whole-genome microarray analysis using total RNA extracted from actively growing broth cultures of isogenic MtrR-positive and MtrR-negative gonococci. We determined that, at a minimum, 69 genes are directly or indirectly subject to MtrR control, with 47 being MtrR repressed and 22 being MtrR activated. rpoH, which encodes the general stress response sigma factor RpoH (sigma 32), was found by DNA-binding studies to be directly repressed by MtrR, as it was found to bind to a DNA sequence upstream of rpoH that included sites within the rpoH promoter. MtrR also repressed the expression of certain RpoH-regulated genes, but this regulation was likely indirect and a reflection of MtrR control of rpoH expression. Inducible expression of MtrR was found to repress rpoH expression and to increase gonococcal susceptibility to hydrogen peroxide (H2O2) and an antibiotic (erythromycin) recognized by the MtrC-MtrD-MtrE efflux pump system. We propose that, apart from its ability to control the expression of the mtrCDE-encoded efflux pump operon and, as a consequence, levels of gonococcal resistance to host antimicrobials (e.g., antimicrobial peptides) recognized by the efflux pump, the ability of MtrR to regulate the expression levels of rpoH and RpoH-regulated genes also modulates levels of gonococcal susceptibility to H2O2.
Neisseria gonorrhoeae is a gram-negative strict human pathogen and the causative agent of the sexually transmitted disease gonorrhea. Worldwide, over 60 million cases of gonorrhea occur each year (7, 33). Although the incidence of disease in the United States has decreased since the 1970s, there has since been an increase in antibiotic-resistant strains reported in the United States and elsewhere (25, 33). Indeed, the prevalence of gonococci resistant to penicillin, tetracycline, and/or fluoroquinolones became so significant over the past 25 years that in 2007 the Centers for Disease Control and Prevention added N. gonorrhoeae to the list of “superbugs”.
The capacity of gonococci and other bacteria to employ drug efflux pumps to recognize and export antibiotics has attracted considerable attention, as these pumps can impact the efficacy of antibiotic therapy of infections (18, 19, 25, 30, 34, 35). Moreover, since certain efflux pumps recognize host-derived antimicrobial agents, such as antimicrobial peptides (24), there is increasing suspicion that they provide bacteria with a mechanism to escape innate host defenses (13, 21). The gonococcal MtrC-MtrD-MtrE efflux pump can export a variety of antibiotics (17, 28), including penicillin (30) and macrolides (35), and antimicrobial peptides, such as human LL-37 (24).
The mtrCDE operon is negatively regulated by the MtrR repressor (10, 16, 23), which is encoded by a gene (mtrR) immediately upstream of, but transcriptionally divergent from, mtrCDE (19). Two homodimers of MtrR bind to a DNA sequence within the mtrCDE promoter (12). Point mutations in the MtrR-binding site (16), a single-base-pair deletion within a 13-bp inverted-repeat sequence in the mtrR promoter (10), or missense mutations that cause radical amino acid replacements within the helix-turn-helix motif of MtrR all can result in derepression of mtrCDE expression (23). Derepression of mtrCDE results in decreased antimicrobial susceptibility of gonococci (10, 11, 23) and increased fitness of gonococci in an experimental murine lower genital tract infection model (31).
Consequently, it is important to define the regulatory properties of MtrR. DNA-binding proteins such as MtrR that negatively regulate bacterial efflux pump genes have been considered to be “local” gene regulators, although there is increasing evidence that they can directly or indirectly influence the expression of other genes (6, 15). In order to define the genes in gonococci regulated by MtrR, we employed a microarray analysis of total RNA extracted from mid-logarithmic-phase broth cultures of isogenic MtrR-positive and MtrR-negative gonococci. Through this analysis, we found that MtrR can directly or indirectly regulate 69 genes, which are distributed throughout the genome. We identified 47 MtrR-repressed genes (including the previously described mtrCDE operon) and 22 MtrR-activated genes. An MtrR-repressed gene of particular interest, which was the subject of this investigation, was rpoH. rpoH encodes the alternative stress response sigma factor RpoH (sigma 32), which appears to be essential for gonococcal viability even under normal growth conditions (3, 8). Earlier studies showed that rpoH expression is increased during exposure of gonococci to certain stress conditions, such as growth at elevated temperatures (8), and during bacterial contact with cultured epithelial cells (2, 3). Moreover, certain RpoH-regulated genes are differentially expressed during exposure of gonococci to H2O2 (29). Thus, apart from its regulation of mtrCDE and levels of gonococcal resistance to hydrophobic antimicrobial agents (20, 21, 24, 30), MtrR regulation of rpoH expression may have significance for gonococcal survival in vivo, particularly at sites rich in neutrophils producing large quantities of H2O2.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and culture conditions.
The bacterial strains used in this study are listed in Table 1. Escherichia coli strain TOP10 (Invitrogen, Carlsbad, CA) or strain DH5α mcr was used in all cloning experiments. E. coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar plates at 37°C. N. gonorrhoeae strain FA19 was used as the primary gonococcal strain (22). N. gonorrhoeae strains were grown on gonococcal medium base (GCB) agar (Difco Laboratories, Detroit, MI) containing glucose and iron supplements (22) at 37°C under 3.8% (vol/vol) CO2 or in GCB broth with supplements and sodium bicarbonate as described previously (22). The plasmids and oligonucleotide primers used in this investigation are listed in Tables 1 and 2. All chemicals were purchased from Sigma Biochemical (St. Louis, MO).
TABLE 1.
Gonococcal strains and plasmids used
| Strain or plasmid | Relevant genotype or remarks | Source or reference |
|---|---|---|
| Strains | ||
| FA19 | Wild type | P. F. Sparling |
| JF1 | Like FA19 but ΔmtrR | 5 |
| JF6 | Like JF1 but mtrR+a | This study |
| JF7 | Like JF1 but mtrR+brpoH-lacZ | This study |
| JF8 | Like FA19 but rpoH-lacZ | This study |
| JF9 | Like JF1 but rpoH-lacZ | This study |
| JF10 | Like JF6 but rpoH-lacZ | This study |
| JF11 | Like FA19 but grpE-lacZ | This study |
| JF12 | Like JF1 but grpE-lacZ | This study |
| JF13 | Like JF6 but grpE-lacZ | This study |
| JF14 | Like FA19 but rpoH1-lacZc | This study |
| JF15 | Like JF1 but rpoH1-lacZc | This study |
| JF16 | Like FA19 but mtrR-lacZ | This study |
| Plasmids | ||
| pLES94 | Cloning vector containing promoterless lacZ for insertion of gonococcal genes between proA and proB (27) | V. Clark |
| pGCC3 | NICS vector (25) used for insertion of mtrR between lctP and aspC | H. Seifert |
| pGCC4 | Same as pGCC3 with IPTG-inducible lacZ promoter | H. Seifert |
| pJF1 | Like pGCC3 but mtrR+ | This study |
| pJF2 | Like pGCC4 but mtrR+ | This study |
mtrR from pJF1 is under the control of its own promoter in this strain.
mtrR from pJF2 is under the control of the lacZ promoter in this strain.
This fusion lacks the 172-bp rpoH leader sequence.
TABLE 2.
Oligonucleotides used
| Oligonucleotide primer name | Sequence (5′→3′) |
|---|---|
| 5′PrpoH | GGGATCCGGCGAAACGCCCTATATGAA |
| 3′PrpoHB | GGGATCCCGATTCATTTGGGCATTTCCTTT |
| 5′ΔPrpoH | GGACAGGATGAGTTGTTTGG |
| 3′ΔPrpoH | TATCGGCGGTTGTAAACCTGATAGCTCAATTC |
| 3′PrpoHB | GGGATCCCGATTCATTTGGGCATTTCCTTT |
| 5′ΔPrpoH | GGACAGGATGAGTTGTTTGG |
| 3′ΔPrpoH | TATCGGCGGTTGTAAACCTGATAGCTCAATTC |
| 5′mtrR | GGTTAATTAACCGCCCTCGTCAAACCGA |
| 3′mtrR | GGGTTTAAACTTATTTCCGGCGCAGGCAG |
| mtrC1 | CGGGATCCCGAGCCATTATTTATCCTATCTGTC |
| pmtrR | GGATCCTCTCATAATGGCGTTTTCGTTTCG |
| 5′PgrpE | GGGGATCCACACGGTTTGGTGCAAAAAAC |
| 3′PgrpE | GGGGATCCGGCTCATATCGTATCCCTCAAA |
RNA isolation.
RNA was isolated from 50-ml GCB broth cultures of strains FA19 and JF1 (the same as FA19 but with mtrR deleted [ΔmtrR] [5]) grown to mid-log phase using a hot-phenol method as previously described (4). Samples were then DNase treated on column (Qiagen DNase kit), and the RNA was recovered using the Qiagen RNeasy Mini kit. RNA was quantified by NanoDrop1000 (NanoDrop Technolgies), and the RNA integrity was analyzed on an Agilent bioanalyzer (Agilent Technologies). The RNA samples were stored at −80°C until further use.
Microarray design.
A custom multipathogen Affymetrix GeneChip, MPAUT1a520274F, was utilized for the microarray experiments in the present study. Probe pairs were designed for each gene, consisting of 11 probe pairs (22 features) with a perfect match and a single-nucleotide mismatch. The N. gonorrhoeae genome probe set (1,925 genes) was represented on the array.
cDNA labeling and hybridization.
Sample preparation and hybridization were done at the University of Iowa DNA Facility (http://dna-9.int-med.uiowa.edu/) according to the Affymetrix Genechip Expression Analysis technical manual prokaryotic protocol. Briefly, biotinylated cDNA was generated from 10 μg of total RNA and fragmented prior to hybridization to the custom chip at 45°C for 16 h. The arrays were stained using streptavidin-phycoerythrin (ProkGE-WS2v3_450 protocol; Affymetrix, Inc.), and the scanned images (Affymetrix GeneChip Scanner 3000) were processed using Genechip Operating Software version 1.4 (Affymetix, Inc.). Hybridizations were done on three separate biological replicates of each isolate.
Array analysis.
Data files generated by Genechip Operating Software (Affymetix, Inc.) were imported into GeneSpring GX 7.3.1 (Agilent Technologies) and normalized per chip by dividing each measurement by the 50th percentile of all the measurements in that sample and per gene by dividing each gene by the median of its measurements across all samples. The cross-gene error model was operating based on replicates. The normalized data from all samples were filtered on genes flagged as present or marginal, with the resulting gene list used for further gene expression analysis and clustering. Change was expressed as the ratio of gene expression of the wild-type FA19 over the same gene expression for the mutant. Genes with a differential expression of ≥1.5-fold and a P value of 0.05 were selected.
Construction of mtrR-complemented strains, inducible expression of mtrR, detection of MtrR, and antimicrobial susceptibility in induced cultures.
The construction of pJF1, which contains the wild-type mtrR coding sequence and promoter sequence from strain FA19 cloned into pGCC3 (27), and strain JF6 (like JF1 but with this mtrR sequence, including its promoter, located between aspC and lctP) has been described by Folster and Shafer (5). In order to engineer an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible copy of mtrR, the mtrR coding sequence with its ribosome binding site was PCR amplified from chromosomal DNA prepared from wild-type strain FA19 using primers 5′mtrR and 3′mtrR (Table 2). The resulting DNA fragment was inserted into the PmeI and PacI sites of pGCC4, which contains the lacZ promoter (27) (kindly provided by A. Jerse and H. Seifert) to produce pJF2, and the correct orientation and nucleotide sequence were confirmed by DNA sequencing. pJF2 was digested with ClaI, and the fragment containing the gonococcal lctP gene, mtrR, ermC (an erythromycin [Ery] resistance cassette), and aspC was purified and used to transform gonococcal strain FA19 ΔmtrR. Transformations were performed as previously described (9). Transformants were selected on GCB agar containing 1 μg/ml of Ery, and the resulting strain was named JF7 (like JF1, but mtrR+).
For inducible expression of mtrR, strain JF7 was grown on GCB agar as described above. The overnight growth was then used to inoculate 50 ml of GCB broth, and the culture was grown at 37°C with shaking (200 rpm) until the optical density at 600 nm reached 0.2, at which time it was split into two equal samples. One sample received IPTG (1 mM), while the other received an equal volume of sterile distilled water. These cultures were grown for 1 h, and samples were then harvested by centrifugation. MtrR was detected in solubilized cell extracts by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western immunoblotting as described previously (6). MtrR was detected using a rabbit anti-MtrR antiserum (1:5,000 dilution) prepared by Protein Tech Group, Inc. (Chicago, IL), with purified MtrR fused to maltose-binding protein (MBP) (15) as the antigen and was visualized on the blot using goat anti-rabbit immunoglobulin G coupled to alkaline phosphatase (1:5,000 dilution). IPTG-induced and control samples were also evaluated for their susceptibility to Ery (0.5 μg/ml) by an efficiency-of-plating assay described previously (22), using agar plates with or without 1 mM IPTG. H2O2 susceptibility was determined using a disk diffusion assay containing filter disks (1 cm) presoaked in 3% (vol/vol) H2O2 that were placed on freshly inoculated GCB agar plates that contained or lacked 1 mM IPTG.
Construction and analysis of rpoH-lacZ, grpE-lacZ, and mtrR-lacZ fusions.
Translational lacZ fusions were constructed as previously described (5, 6). In brief, the promoter sequence of rpoH was amplified by PCR from strain FA19 using primers 5′PrpoH and 3′PrpoHB (Table 2). The resulting DNA fragment was inserted into the BamHI site of pLES94 (26), and the recombinant plasmid was introduced into E. coli DH5α mcr by transformation. The correct orientation of the insertion was confirmed by PCR analysis and DNA sequencing. The plasmid was used to transform strains FA19, JF1, and JF6 to allow insertion into the chromosomal proAB gene, and transformants were selected on GCB agar containing 1 μg/ml of chloramphenicol. Representative transformants obtained with the three recipient strains were termed JF8, JF9, and JF10, respectively (Table 1). Strains bearing a grpE-lacZ fusion were prepared essentially as described above, but the oligonucleotide primers 5′PgrpE and 3′PgrpE (Table 2) were used to PCR amplify a 300-bp fragment encompassing the upstream sequence of grpE and the first two codons. The strains bearing the grpE-lacZ fusion were termed JF11 (FA19 grpE-lacZ), JF12 (FA19 ΔmtrR grpE-lacZ), and JF13 (FA19 ΔmtrR mtrR+ grpE-lacZ), respectively (Table 1). For construction of an mtrR-lacZ fusion in strain FA19, oligonucleotide primers mtrC1 and pmtrR (Table 2) were used to PCR amplify mtrR; the strain bearing the mtrR-lacZ fusion was termed JF16 (FA19 mtrR-lacZ). β-Galactosidase (β-Gal) assays were conducted as previously described (5, 6).
Deletion of the rpoH extended leader sequence.
PCR mutagenesis was used to delete the 172-bp rpoH leader sequence and was performed using the overlapping primers 5′ΔPrpoH and 3′ΔPrpoH (Table 2). First, two fragments were amplified by PCR from FA19 chromosomal DNA using the primer set pairs 5′PrpoH/3′ΔPrpoH and 3′PrpoHB/5′ΔPrpoH. The resultant fragments were purified after agarose gel electrophoresis using the Qiaquick purification kit (Qiagen Inc., Valencia, CA), and they then served as both primers and templates for a second PCR. After eight reaction cycles, primers 5′PrpoH and 3′PrpoHB were added to the PCR mixture, and amplification continued for an additional 25 cycles. The resulting DNA fragment containing the deletion was purified and served as the template for the last PCR using primers 5′PrpoH and 3′PrpoHB. The resulting DNA fragment was inserted into the BamHI site of pLES94, resulting in the rpoHΔ-lacZ construct. The recombinant plasmid was introduced into DH5α mcr by transformation. Correct insertion and orientation were confirmed by PCR analysis, and DNA-sequencing analysis confirmed the mutation of the MtrR-binding site (data not presented). The plasmid was used to transform strains FA19 and JF1 to allow insertion into the chromosomal proAB gene. Transformants were selected on GCB agar containing 1 μg/ml of chloramphenicol; representative transformants obtained with recipient strains FA19 and JF1 were termed JF14 and JF15, respectively (Table 1).
EMSA and DNase I protection.
Electrophoretic mobility shift assays (EMSAs) using purified MBP fused to MtrR were performed as previously described (5, 6, 15). In brief, the 184-bp promoter region of rpoH lacking the leader sequence was amplified by PCR using 5′PrpoH and 3′PrpoHB from strain JF14 (Table 1). The 184-bp nonspecific probe was PCR amplified from FA19 chromosomal DNA using primers 5′ΔPrpoH and 3′PrpoHB (Table 2). The resulting PCR products were end labeled with [γ-32P]dATP using T4 polynucleotide kinase (New England Biolabs, Beverly, MA). The labeled DNA fragments (10 ng) were incubated with purified MtrR in 30 μl of reaction buffer [10 mM Tris-HCl (pH 7.5), 0.5 mM dithiothreitol, 0.5 mM EDTA, 4% (vol/vol) glycerol, 1 mM MgCl2, 50 mM NaCl, poly(dI-dC) (0.05 μg/ml), salmon sperm (200 ng/ml)] at 4°C for 20 min. Samples were subjected to electrophoresis on a 6% (wt/vol) polyacrylamide gel at 4°C, followed by autoradiography.
DNase I protection assays were performed as previously described (16). Target DNA sequences for DNase I footprints were generated by PCR using the oligonucleotide primers 5′PrpoH and 3′PrpoHB (Table 2) to generate a 184-bp promoter region of rpoH lacking the leader sequence from strain JF14. Target DNA was labeled at the 5′ end of one strand as described for EMSA. Purified MBP-MtrR protein was allowed to bind in the binding buffer as in the EMSA before CaCl2 and MgCl2 were added to final concentrations of 2.5 mM and 5 mM, respectively. Five units of DNase I (Promega, Madison, WI) were then added to the reaction mixture, which was incubated at room temperature for 1 min. Digestion was stopped by the addition of NaCl to 250 mM, and the reaction mixture was extracted with phenol-chloroform-isoamyl alcohol (25:24:1) before precipitation with 100% ethanol for 30 min at −20°C. The pellet was washed four times with 70% (vol/vol) ethanol, vacuum dried, and resuspended in gel loading buffer, which consisted of 0.1 M NaOH-formamide (1:2 [vol/vol]), 0.1% (vol/vol) xylene cyanol, and 0.1% (vol/vol) bromophenol blue. Regions protected from DNase I digestion were resolved on a 6% denaturing polyacrylamide gel, which was dried and exposed to X-ray film for autoradiography.
Microarray data expression number.
Gene expression data for all microarray experiments can be retrieved from the Gene Expression Omnibus (GEO) database at NCBI (http://www.ncbi.nih.gov/geo/) (accession number GSE12686).
RESULTS AND DISCUSSION
Identification of MtrR-regulated genes in gonococci.
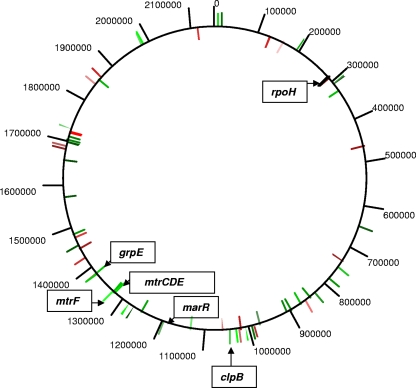
To identify MtrR-regulated genes in N. gonorrhoeae, a microarray analysis was performed that permitted a comparison of gonococcal transcript levels in total RNA prepared from mid-logarithmic-phase cultures of wild-type strain FA19 and its isogenic mtrR deletion mutant, strain JF1 (Table 1). Using an expression difference of 1.5-fold (P ≤ 0.05), analysis of the microarray results allowed us to establish an MtrR regulon that at a minimum consists of 47 MtrR-repressed and 22 MtrR-activated genes (Table 3); these MtrR-regulated genes were distributed throughout the genome (Fig. 1). Four previously identified MtrR-repressed genes (mtrC, mtrD, mtrE, and mtrF) were confirmed as being MtrR repressed by the array, which validated the array as a means to identify MtrR-regulated genes. Based on the annotation of the FA1090 genome sequence (GenBank accession no. AE004969), the majority of MtrR-regulated genes were predicted to encode hypothetical proteins, but several genes encoding proteins with known or putative functions were identified (Table 3). In particular, we noted that expression of rpoH, which encodes the stress-related sigma factor RpoH, and some RpoH-regulated genes (grpE, clpB, and marR [8, 29]) were increased in strain JF1 relative to the MtrR-positive parental strain FA19, suggesting that they are MtrR repressed. Since RpoH expression in gonococci is increased during exposure to certain environmental stresses (8) when gonococci bind to cervical epithelial cells (2, 3) and is essential for viability (3, 8), we studied the ability of MtrR to regulate rpoH expression in N. gonorrhoeae.
TABLE 3.
MtrR-regulated genesa in N. gonorrhoeae
| Gene | Common name | Change (fold) | Functional classification |
|---|---|---|---|
| MtrR repressed | |||
| NGO0006 | leuS | 1.59 | Leucyl-tRNA synthetase |
| NGO0018 | NGO0018 | 1.65 | Unknown |
| NGO0195 | potH | 1.56 | Putative ABC-type polyamine transporter |
| NGO0288 | rpoH | 1.58 | Alternative sigma factor |
| NGO0302 | NGO0302 | 1.98 | Unknown |
| NGO0308 | bioA | 1.59 | Putative aminotransferase |
| NGO0309 | bioD | 1.53 | Dithiobiotin synthetase |
| NGO0658 | pdxH | 1.51 | Pyrodoxamine 5′-phosphate oxidase |
| NGO0756 | NGO0756 | 1.53 | Unknown |
| NGO0795 | brfB | 1.98 | Bacterioferrritin B |
| NGO0846 | umpA | 1.54 | Putative prolipoprotein diacylglycerol transferase |
| NGO0863 | NGO0863 | 1.71 | Unknown |
| NGO0891 | NGO0891 | 1.92 | Unknown |
| NGO0916 | accB | 1.58 | Dihydrolipoamide acetyltransferase |
| NGO0927 | NGO0927 | 1.51 | Unknown |
| NGO1002 | NGO1002 | 1.54 | Unknown (putative phage associated) |
| NGO1029 | fumC | 1.57 | Fumarate hydratase |
| NGO1046 | clpB | 2.10 | Heat shock chaperone |
| NGO1069 | NGO1069 | 1.63 | Unknown |
| NGO1084 | NGO1084 | 1.75 | Unknown |
| NGO1189 | hsp33 | 2.24 | Heat shock chaperonin |
| NGO1244 | NGO1244 | 1.52 | MarRfamily regulator |
| NGO1293 | NGO1293 | 2.11 | Unknown |
| NGO1294 | lrp | 1.77 | AsnC family transcriptional regulator |
| NGO1314 | NGO1314 | 1.80 | Putative protease |
| NGO1325 | gcvP | 2.35 | Glycine dehydrogenase |
| NGO1363 | mtrE | 2.20 | Mtr efflux pump protein component |
| NGO1364 | mtrD | 2.40 | Mtr efflux pump protein component |
| NGO1365 | mtrC | 1.78 | Mtrefflux pump protein component |
| NGO1368 | mtrF | 2.21 | Mtrefflux pump protein component |
| NGO1418 | nqrF | 1.74 | Subunit F, NADH-quinone reductase |
| NGO1422 | grpE | 1.83 | Heat shock protein |
| NGO1494 | potF | 1.58 | Putative periplasmic polyamine binding protein |
| NGO1520 | NGO1520 | 1.55 | Unknown |
| NGO1600 | glnA | 1.63 | Glutamine synthetase |
| NGO1699 | NGO1699 | 1.56 | Unknown |
| NGO1738 | nuoM | 1.52 | NADH dehydrogenase subunit M |
| NGO1741 | nuoK | 1.80 | NADH dehydrogenase kappa subunit |
| NGO1742 | nuoJ | 1.56 | NADH dehydrogenase I chain J |
| NGO1749 | nuoC | 1.57 | NADH dehydrogenase subunit C |
| NGO1751 | nuoA | 1.78 | Putative NADH dehydrogenase I chain A |
| NGO1765 | pglA | 1.63 | Glycosyltransferase |
| NGO1890 | gltS | 1.66 | Putative glutamate permease |
| NGO2011 | glnM | 1.55 | Putative ABC-type polyamine transporter |
| NGO2013 | glnQ | 1.75 | Putative ATP-binding protein |
| NGO2014 | cjaA | 2.00 | Amino acid periplasmic binding protein |
| MtrR activated | |||
| NGO0113 | rng | 1.57 | Putative RNase G/cytoplasmic axial filament protein |
| NGO0150 | NGO0150 | 1.61 | Unknown |
| NGO0290 | NGO0290 | 1.57 | Unknown |
| NGO0481 | NGO0481 | 1.85 | Unknown (putative phage associated) |
| NGO0739 | NGO0739 | 1.62 | Putative DNA helicase |
| NGO0876 | NGO0876 | 1.83 | Unknown |
| NGO1024 | NGO1024 | 1.57 | Unknown |
| NGO1059 | NGO1059 | 1.77 | Unknown |
| NGO1062 | NGO1062 | 1.71 | Unknown |
| NGO1097 | NGO1097 | 1.70 | Unknown (phage associated) |
| NGO1420 | abpE | 1.65 | Thiamine biosynthesis |
| NGO1460 | NGO1460 | 1.72 | Unknown |
| NGO1488 | NGO1488 | 1.53 | Unknown |
| NGO1511 | NGO1511 | 1.67 | Unknown |
| NGO1722 | recQ | 1.67 | Recombination |
| NGO1728 | NGO1728 | 1.60 | Unknown |
| NGO1758 | glnE | 7.04 | Glutamine synthetase adenylyltransferase |
| NGO1759 | NGO1759 | 148.70 | Unknown |
| NGO1760 | NGO1760 | 1.73 | Unknown |
| NGO1874 | NGO1874 | 1.58 | Unknown |
| NGO1897 | rfbB | 1.73 | dDTP-d-glucose-4,6 dehydratase |
| NGO2135 | NGO2135 | 1.63 | Putative transglycosylase |
Genes of interest to this investigation are highlighted in boldface.
FIG. 1.
Chromosomal-map positions of MtrR-regulated genes. The positions of MtrR-regulated genes in strain FA19 that were identified by the microarray analysis (Table 3) are shown on the circular map of the gonococcal chromosome (strain FA1090). MtrR-activated genes are shown in red, while MtrR-repressed genes are shown in green.
Expression of rpoH and certain RpoH-regulated genes is repressed by MtrR.
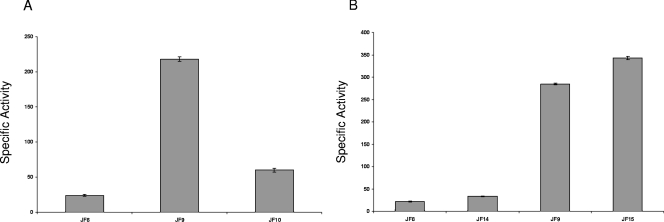
To study the capacity of MtrR to regulate the expression of rpoH, a translational lacZ reporter fusion system was engineered. This translational fusion consisted of 365 bp of DNA sequence upstream of rpoH, which included the promoter region (14), the first two codons of rpoH (Fig. 2), and a promoterless lacZ gene. This construct was transformed into isogenic strains FA19 and JF1 to create the transformant strains JF8 (FA19 rpoH-lacZ) and JF9 (FA19 ΔmtrR rpoH-lacZ), respectively, resulting in a single copy of the rpoH promoter fused translationally to lacZ within the proAB chromosomal locus. β-Gal activity in cell lysates from these strains indicated that expression of rpoH increased more than eightfold in the MtrR-negative strain JF9 compared to the MtrR-positive strain JF8 (FA19 rpoH-lacZ) (Fig. 3A). To confirm that these results were due to deletion of mtrR and not a polar effect on other genes, strain JF9 was complemented with the wild-type mtrR gene from strain FA19, which was inserted at a secondary site within the gonococcal chromosome (between the lctP and aspC genes) to create strain JF10 (FA19 ΔmtrR mtrR+ rpoH-lacZ) (Table 1). Using this strain, we found that complementation of the mtrR deletion in JF9 (FA19 ΔmtrR rpoH-lacZ) with the wild-type mtrR gene restored rpoH repression to a level similar to that of JF8 (like FA19, but rpoH-lacZ) (Fig. 3A).
FIG. 2.
Nucleotide sequence upstream of rpoH and identification of the MtrR-binding site. The nucleotide sequence of the DNA upstream of rpoH is shown with the 172-bp leader sequence indicated by the dashed lines; the first two codons encoding methionine (M) and asparagine (N) are also shown in boldface. The position of the transcriptional start point (8) is shown by the boldface A nucleotide downstream of the −10 sequence. The −10 and −35 sequences of the rpoH promoter identified previously (8) are shown with the −10 and −35 hexamers and are highlighted by a single line under the sequence. The 4-bp ribosome binding site (RBS) sequence is shown by an underline. The MtrR-binding site identified by DNase I protection (Fig. 6) is shown by the wide bars above the coding strand and below the noncoding strand. The boxed region shows a 15-nucleotide sequence on the coding strand (5′-GGC[-]CGTTTACATACA-3′) that is 73.3% identical (the identical nucleotides are boldface in above sequence) to the MtrR-binding sequence (5′-GGCACGTTAGCACATA-3′) upstream of mtrCDE on the noncoding strand (12); the hyphen in the sequence above represents a single-nucleotide gap in the rpoH sequence compared to the mtrCDE bound by MtrR.
FIG. 3.
MtrR regulation of rpoH expression. (A) Levels of β-Gal activity (specific activity expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per mg of protein) in strains JF8 (like FA19, but rpoH-lacZ), JF9 (like JF1, but rpoH-lacZ), and JF10 (like JF6 [Table 1], but rpoH-lacZ). Complementation of the mtrR deletion mutation resulted in reduced rpoH expression. The results are shown as average values (± standard deviations) from three independent assays. The difference in β-Gal activities between strains JF8 and JF9 was significant (P < 0.001), as was that between JF9 and JF10 (P < 0.001). (B) MtrR repression of rpoH was independent of the 172-bp leader sequence upstream of rpoH, as shown by similar levels of β-Gal production between isogenic pairs (MtrR-positive strains JF8 and JF14 and MtrR-negative strains JF9 and JF15) with (strains JF8 and JF9) or without (JF14 and JF15) the 172-bp leader sequence. The differences in β-Gal activities between strains JF8 and JF14 and between JF9 and JF15 were not statistically significant.
The sequence of the gonococcal rpoH promoter, as well as the transcriptional start point and ribosome binding site, were defined previously (8) (Fig. 2). This region contains an extended 172-bp RNA leader sequence, which was previously observed to be involved in rpoH repression during growth under normal conditions (14). To determine if the leader sequence was involved in MtrR repression of rpoH, an internal deletion was engineered within the rpoH-lacZ construct and transformed into strains FA19 and JF1 (FA19 ΔmtrR) to create strains JF14 (FA19 rpoH-lacZ) and JF15 (FA19 ΔmtrR rpoH-lacZ) (Table 1), respectively, and expression was measured. We found that rpoH-lacZ expression from the wild-type promoter and the promoter with the leader deleted did not differ in either the MtrR-positive (JF14) or MtrR-negative (JF15) background (Fig. 3B), indicating that MtrR-mediated repression of rpoH is independent of the leader sequence.
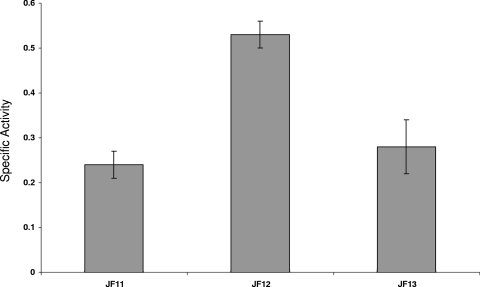
Gunesekere et al. (8) identified a panel of RpoH-regulated genes in gonococci, and Stohl et al. (29) showed that some of these were upregulated during bacterial exposure to H2O2. Since our microarray analysis (Table 3) suggested that a subset of these RpoH-regulated genes (clpB, grpE, and marR) is also subject to MtrR regulation, we examined the ability of MtrR to control the expression of RpoH-regulated genes in gonococci and used grpE for this purpose. We examined its expression in isogenic MtrR-positive and MtrR-negative gonococci utilizing a grpE-lacZ construct introduced into strains FA19, JF1 (FA19 ΔmtrR), and JF6 (FA19 ΔmtrR mtrR+); the transformed strains were termed JF11, JF12, and JF13, respectively (Table 1). The results indicated that expression of grpE (Fig. 4) was elevated in the absence of MtrR (see strain JF12) and restored to near-wild-type levels (strain JF11) by complementation with the intact mtrR gene (strain JF13).
FIG. 4.
MtrR regulation of the RpoH-regulated grpE gene. Shown are the β-Gal-specific activities expressed in grpE-lacZ fusion strains JF11 (MtrR positive) and JF12 (MtrR negative) and the mtrR+ complemented strain JF13. The results are shown as average values (± standard deviations) from three independent assays. The differences between strains JF11 and JF12 and between JF12 and JF13 were significant (P < 0.001 and 0.003, respectively).
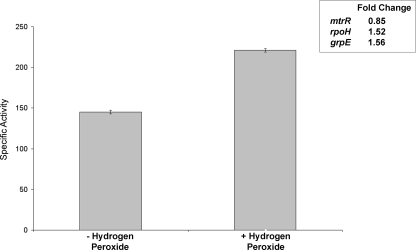
As expression of RpoH-regulated grpE is induced during exposure of gonococci to low levels of H2O2 (29), we next evaluated whether expression of mtrR, rpoH, and grpE was altered when gonococci were exposed to the oxidizing agent. Using the lacZ fusion strains JF8 (FA19 rpoH-lacZ), JF11 (FA19 grpE-lacZ), and JF16 (FA19 mtrR-lacZ) (Table 1), we monitored the expression of rpoH, grpE, and mtrR during a brief (15-min) exposure of gonococci to 1.5 mM H2O2; this concentration of H2O2 did not reduce viability (data not presented). As is shown in Fig. 5, we found that under these conditions, expression of both rpoH and grpE was increased significantly (P < 0.0001 and 0.002, respectively), while mtrR expression was slightly but significantly (P = 0.049) dampened. In order to determine if H2O2 induction of rpoH expression would be enhanced in the absence of MtrR, we next compared its expression when strains JF8 (FA19 rpoH-lacZ) and JF9 (like JF8, but ΔmtrR) were exposed to 1.0 mM H2O2 for 15 min. Although rpoH expression was nearly 15-fold greater in strain JF9 than in JF8, the changes in expression after H2O2 exposure were nearly identical (data not presented). We propose that although MtrR can control constitutive levels of rpoH and grpE exposure, it is not necessarily involved in their H2O2-inducible expression. Alternatively, under the experimental conditions employed, rpoH expression in JF9 reached maximal levels, and induction above the observed level was not possible or detectable.
FIG. 5.
H2O2 induction of rpoH expression. Shown in the graph are the β-Gal-specific activities in rpoH-lacZ fusion strain JF8 after a 15-min exposure to 1 mM H2O2. The results are shown as average values (± standard deviations) from three independent assays; the difference was significant (P < 0.00001). The inset shows the changes in expression for mtrR, rpoH, and grpE when fusion strains were exposed to H2O2.
MtrR directly binds to the rpoH promoter region.
Initial EMSA experiments showed that MtrR bound to the DNA sequence upstream of rpoH encompassing its promoter region in a specific, concentration-dependent manner and did not require the 172-bp leader sequence (data not presented). In order to determine if MtrR directly or indirectly regulates the RpoH-regulated grpE gene, an EMSA was performed using MtrR and a 305-bp probe containing the upstream DNA (including a putative promoter element) of the grpE coding sequence. Unlike its binding to the DNA sequence upstream of mtrC and rpoH, MtrR failed to bind the grpE target DNA sequence (data not presented). Accordingly, we believe that the ability of MtrR to regulate grpE is indirect and due to its capacity to repress rpoH.
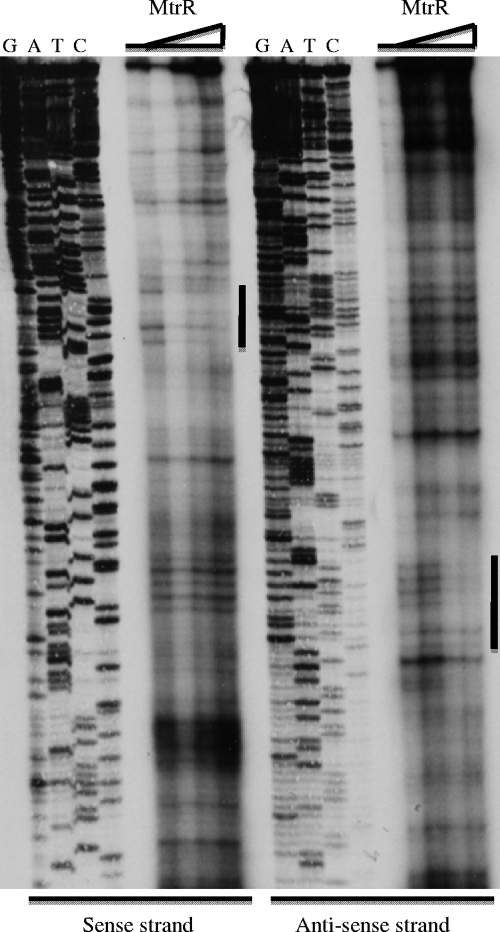
In order to identify the MtrR-binding site within the rpoH promoter region, a DNase I protection assay was performed. The rpoH promoter probe described above was radiolabeled separately at its 5′ and 3′ ends and utilized in DNase I protection assays that employed increasing amounts (0 to 20 μg) of MtrR (Fig. 6). With increasing amounts of MtrR, an area of protection of seventeen nucleotides on the sense strand (Fig. 2) could be identified as being involved in binding MtrR. A second area of protection of sixteen nucleotides was observed on the antisense strand (Fig. 2), which overlapped the area observed on the sense strand. The region of protection indicates that MtrR binds to the rpoH promoter at an area that overlaps the −35 hexamer sequence of the rpoH promoter. Using the nucleotide sequence of the MtrR-binding site on the noncoding strand within the mtrCDE promoter (5′-GGCACGTTAGCACATA-3′) defined by Hoffman et al. (12), we were able to identify a region of similarity (73.3% identity) (Fig. 2) within the rpoH promoter region, which was also found by DNase I protection to bind MtrR.
FIG. 6.
Identification of the MtrR-binding site within the rpoH promoter. The MtrR-binding site (see the nucleotide sequences in Fig. 2 and its legend) was identified by DNase I protection using increasing amounts of MtrR (0, 5, 10, and 20 μg) in assays that employed sense and antisense probes. The site of protection on each strand is shown by the vertical bar adjacent to the protected region. The nucleotide-sequencing reaction for each strand is shown (G, A, T, C) adjacent to the lanes with the protection reactions.
Modulation of MtrR expression and levels of antimicrobial resistance.
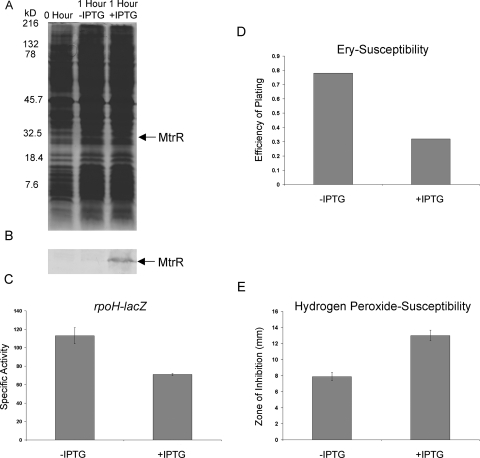
In order to further connect MtrR expression with levels of rpoH expression and to assess the biologic consequence of MtrR gene control in gonococci, we next inserted an IPTG-inducible copy of the wild-type mtrR gene from strain FA19 into its transformant strain JF9 (FA19 ΔmtrR rpoH-lacZ) to create strain JF7 (FA19 ΔmtrR mtrR+ rpoH-lacZ). Strain JF7 was grown in GCB broth to mid-log phase and split into two samples, one receiving 1 mM IPTG and one left untreated. After 1 h of induction, MtrR was clearly produced by the IPTG-treated culture (Fig. 7B), and rpoH expression was decreased by nearly twofold (Fig. 7C) compared to the untreated sample. In order to determine whether induction of mtrR expression resulted in biological changes of gonococci, we next examined the susceptibility of the IPTG-treated and control cultures to Ery and H2O2. Ery susceptibility analysis was performed because MtrR repression of the mtrCDE-encoded efflux pump increases gonococcal susceptibility to this antibiotic and other hydrophobic antimicrobial agents (10, 11, 19). In support of this, we found that inducible expression of mtrR resulted in increased susceptibility of gonococci to Ery (Fig. 7D). H2O2 assays were performed because loss of RpoH production was found to enhance the susceptibility of Brucella melitensis (1) to the oxidizing agent. Since we (unpublished observations) and others (3, 8, 14) have been unsuccessful in obtaining an rpoH null mutant, we hypothesized that under conditions where rpoH expression is dampened due to the repressive action of MtrR, gonococcal susceptibility to H2O2 would increase. Indeed, we found that inducible expression of mtrR increased gonococcal susceptibility to H2O2 (Fig. 7E).
FIG. 7.
Inducible production of MtrR represses rpoH expression and modulates antimicrobial susceptibility levels in gonococci. (A and B) Strain JF7 was grown in the presence or absence of 1 mM IPTG as described in Materials and Methods, and cell lysates before induction (0 Hour) or after 1 h of incubation in the absence (−) or presence (+) of IPTG were solubilized and subjected to SDS-PAGE using a 12.5% (wt/vol) SDS-PAGE gel with the separated proteins stained by Coomassie brilliant blue (A); the positions of the molecular mass markers are shown on the left of the gel, and the approximate positions of MtrR as determined by immunoblotting with detection using anti-MtrR antiserum (B) are shown. (C) Specific activity values for β-Gal levels in the control (−IPTG) and induced (+IPTG) cultures. The results are shown as average values (± standard deviations) from three independent assays. The difference was significant (P = 0.0012). (D) The efficiencies of plating of the control and IPTG-induced cultures on GCB agar with or without 0.5 μg/ml of Ery are shown. (E) The H2O2 susceptibilities of the control and IPTG-induced cultures were assessed by a disk diffusion assay, and the difference in growth inhibition between the cultures was significant (P < 0.001).
We believe that the MtrR regulon defined in this study represents the minimum number of gonococcal genes under MtrR control, as other genes (farR, ponA, and pilMNOPQ) known (6, 15) to be controlled by MtrR were not identified as being regulated by ≥1.5-fold in the microarray data set. In this respect, the MtrR-repressed farR (15) and pilM (6) genes were measured as being repressed by 1.44- and 1.40-fold, respectively, in the microarray data set, while the MtrA-activated ponA gene was measured to be 1.24-fold activated. Other MtrR-controlled genes might be uncovered under different growth conditions or by exposure of gonococci to stress conditions or antimicrobial agents.
Although the microarray results indicated that expression of rpoH was modestly repressed by MtrR, the level of repression (1.58-fold) was only slightly less than those of the mtrCDE genes (1.7- to 2.2-fold). Since loss of MtrR repression of mtrCDE in strain JF1 (FA19 ΔmtrR) can have important consequences for certain gonococcal properties (levels of antimicrobial susceptibility and in vivo fitness [31]), we elected to use rpoH as a model gene outside of the mtrCDE locus to study the global regulatory properties of MtrR. The combined translational fusion data that examined rpoH expression in the presence or absence of MtrR, especially those involving the inducible production of MtrR (Fig. 7), the increased expression of three RpoH-activated genes (clpB, marR, and grpE) in the MtrR-deficient strain JF1, and the ability of MtrR to bind within the rpoH promoter (Fig. 6), led us to conclude that MtrR acts as a direct repressor of rpoH. As EMSA experiments failed to show MtrR binding to grpE, we also conclude that the ability of MtrR to control the expression of this RpoH-regulated gene is likely indirect and is most easily explained by the ability of MtrR to dampen rpoH expression.
What is the biological significance of MtrR regulation of rpoH? We considered the possibility that because rpoH expression can enhance bacterial resistance to killing by H2O2 (1) and because certain genes under RpoH control are upregulated in gonococci during bacterial exposure to H2O2 (29), MtrR expression in gonococci would impact the levels of such resistance. The results presented here support this hypothesis. Moreover, based on these results, we suggest that MtrR is a general regulator of levels of gonococcal susceptibility to antimicrobial agents of the innate host defense, particularly the nonoxidative and oxidative killing systems of neutrophils. Thus, by repressing mtrCDE expression, MtrR can enhance gonococcal susceptibility to LL-37, which is produced by neutrophils, epithelial cells, and certain organs (e.g., testis) (24) and is a mediator of nonoxidative killing of bacteria. As MtrR production also increases bacterial susceptibility to H2O2, its control of gene expression, along with other resistance systems, may be important in the ability of gonococci to survive oxidative killing by neutrophils. We do not yet know if the observed decrease in H2O2 susceptibility in mtrR mutants directly or indirectly involves RpoH, but the ability of MtrR to control rpoH expression suggests that this regulatory scheme is important in the general stress response. We also noted that hsp33 expression is also subject to MtrR repression (2.24-fold) (Table 3). This is of interest because Hsp33 has been implicated in bacterial responses to heat and peroxide (32). Unlike, grpE, MtrR regulation of hsp33 may be direct, as a potential MtrR-binding site consisting of 31 bp is positioned from nucleotides −110 to −141 (data not presented); this putative binding site is 61.3% identical to the 31-bp region upstream of mtrCDE shown by Lucas et al. (16) to bind MtrR. Taking these data together, we propose that naturally occurring mtrR mutants (22, 23, 34, 35) may have an advantage during infection, based on their increased resistance to LL-37 (24) and H2O2 (Fig. 7). This linkage of bacterial resistance to distinct mediators of innate host defense through MtrR may help to explain the recent observation of Warner et al. (31) that mtrR mutants have a competitive advantage over otherwise wild-type gonococci in a female mouse lower genital tract infection model. The potential roles of other transcriptional regulators similar to MtrR in modulating bacterial resistance to innate host defense mechanisms should be considered.
Against this background, it should be asked why mtrR mutants are not the dominant strain in the community. At first glance, it seems to be a disadvantage for gonococci to have maintained mtrR, as its presence would downregulate the expression of genes needed for resistance to antimicrobials. We propose that because MtrR can also transcriptionally activate certain genes (Table 3) involved in metabolism (e.g., abpE, glnE, and rfbB), the need to resist host-derived antimicrobials through constitutive MtrR-repressible systems may be essential only at those sites rich in these agents. For most strains and infections, the need to have expression of MtrR-activated genes is more critical and overrides any advantage afforded by mtrR mutations, particularly since the mtrCDE efflux operon can be transcriptionally induced by MtrA in the presence of sublethal levels of antimicrobials (20), even in the presence of a functional MtrR.
Supplementary Material
Acknowledgments
We thank L. Pucko for help in manuscript preparation and submission, N. Kamal and Y. Zalucki for critical reading of the paper, M. Apicella for facilitating access to the microarray slides, and the staff at the University of Iowa DNA Facility for sample preparation and hybridizations.
This work was supported by NIH grants AI-21150 (W.M.S.), RR-015564 (J. Iandolo and D.W.D.), and RR016478 (F. Waxman and D.W.D.) and by a VA Merit Award to W.M.S. W.M.S. was supported by a Senior Research Career Scientist Award from the Department of Veterans Affairs Medical Research Service.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Delory, M., R. Hallez, J.-J. Letesson, and X. De Bolle. 2006. An RpoH-like heat shock sigma factor is involved in stress response and virulence in Brucella melitensis 16M. J. Bacteriol. 1887707-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Du, Y., and C. G. Arvidson. 2006. RpoH mediates the expression of some, but not all, genes induced in Neisseria gonorrhoeae adherent to epithelial cells. Infect. Immun. 742767-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Du, Y., J. Lenz, and C. G. Arvidson. 2005. Global gene expression and the role of sigma factors in Neisseria gonorrhoeae in interactions with epithelial cells. Infect. Immun. 734834-4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ducey, T. F., M. B. Carson, J. Orvis, A. P. Stintzi, and D. W. Dyer. 2005. Identification of the iron-responsive genes of Neisseria gonorrhoeae in microarray analysis in defined medium. J. Bacteriol. 1874865-4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folster, J. P., and W. M. Shafer. 2005. Regulation of mtrF expression in Neisseria gonorrhoeae and its role in high-level antimicrobial resistance. J. Bacteriol. 1873713-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Folster, J. P., V. Dhulipala, R. A. Nicholas, and W. M. Shafer. 2007. Differential regulation of ponA and pilMNOPQ expression by the MtrR transcriptional regulatory protein in Neisseria gonorrhoeae. J. Bacteriol. 1894569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerbase, A. C., J. T. Rowley, D. H. Heymann, S. F. Berkley, and P. Piot. 1998. Global prevalence and incidence estimates of selected curable STDs. Sex. Transm. Infect. 74(Suppl.):S12-S16. [PubMed] [Google Scholar]
- 8.Gunesekere, I. C., C. M. Kahler, D. R. Powell, L. A. Snyder, N. J. Saunders, J. I. Rood, and J. K. Davies. 2006. Comparison of the RpoH-dependent regulon and stress response in Neisseria gonorrhoeae. J. Bacteriol. 1884769-4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunn, J. S., and D. C. Stein. 1996. Use of a non-selective transformation technique to construct a multiply restriction/modification-deficient mutant of Neisseria gonorrhoeae. Mol. Gen. Genet. 251509-517. [DOI] [PubMed] [Google Scholar]
- 10.Hagman, K. E., and W. M. Shafer. 1995. Transcriptional control of the mtr effux system of Neisseria gonorrhoeae. J. Bacteriol. 1774162-4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hagman, K. E., C. E. Lucas, J. T. Balthazar, L. Snyder, M. Nilles, R. C. Judd, and W. M. Shafer. 1997. The MtrD protein of Neisseria gonorrhoeae is a member of the resistance/nodulation/division protein family constituting part of an efflux pump. Microbiology 1432117-2125. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman, K. M., W. M. Shafer, and R. G. Brennan. 2005. Characterization of the multiple transferable resistance repressor, MtrR. J. Bacteriol. 1875008-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jerse, A. E., N. D. Sharma, A. N. Bodner, L. A. Snyder, and W. M. Shafer. 2003. A gonococcal efflux pump system enhances bacterial survival in a female mouse model of genital tract infection. Infect. Immun. 715576-5582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laskos, L., C. S. Ryan, J. A. M. Fyfe, and J. K. Davies. 2004. The RpoH-mediated stress response in Neisseria gonorrhoeae is regulated at the level of activity. J. Bacteriol. 1868443-8452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, E.-H., C. Rouquette-Loughlin, J. P. Folster, and W. M. Shafer. 2003. FarR regulates the farAB-encoded efflux pump in Neisseria gonorrhoeae via an MtrR regulatory mechanism. J. Bacteriol. 1857145-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas, C. E., J. T. Balthazar, and W. M. Shafer. 1997. The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J. Bacteriol. 1794123-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maness, M. J., and P. F. Sparling. 1973. Multiple antibiotic resistance due to a single mutation in Neisseria gonorrhoeae. J. Infect. Dis. 128321-330. [DOI] [PubMed] [Google Scholar]
- 18.Nikaido, H. 1998. The role of the outer membrane and efflux pumps in the resistance of Gram-negative bacteria: can we improve drug access? Drug Resist. Updat. 193-98. [DOI] [PubMed] [Google Scholar]
- 19.Pan, W., and B. G. Spratt. 1994. Regulation of the permeability of the gonococcal cell envelope. Mol. Microbiol. 11769-775. [DOI] [PubMed] [Google Scholar]
- 20.Rouquette, C., J. B. Harmon, and W. M. Shafer. 1999. Induction of the mtrCDE-encoded efflux pump system of Neisseria gonorrhoeae requires MtrA, an AraC-like protein. Mol. Microbiol. 33651-658. [DOI] [PubMed] [Google Scholar]
- 21.Rouquette-Loughlin, C., W. L. Veal, E.-H. Lee, L. Zarantonelli, J. T. Balthazar, and W. M. Shafer. 2001. Antimicrobial efflux systems possessed by Neisseria gonorrhoeae and Neisseria meningitidis viewed as virulence factors, p. 187-200. In I. T. Paulsen and K. Lewis (ed.), Multidrug efflux. Horizon Scientific Press, Wymonham, United Kingdom.
- 22.Shafer, W. M., L. F. Guymon, I. Lind, and P. F. Sparling. 1984. Identification of an envelope mutation (env-10) resulting in increased antibiotic susceptibility in a clinical isolate of Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 25767-769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shafer, W. M., J. T. Balthazar, K. E. Hagman, and S. A. Morse. 1995. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology 141907-911. [DOI] [PubMed] [Google Scholar]
- 24.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. USA 951829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shafer, W. M., and J. P. Folster. 2006. Towards an understanding of chromosomally mediated resistance in Neisseria gonorrhoeae: evidence for a porin-efflux pump collaboration. J. Bacteriol. 1882297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silver, L. E., and V. L. Clark. 1995. Construction of a translational lacZ fusion system to study gene regulation in Neisseria gonorrhoeae. Gene 166101-104. [DOI] [PubMed] [Google Scholar]
- 27.Skaar, E. P., M. P. Lazio, and H. S. Seifert. 2002. Roles of the recJ and recN genes in homologous recombination and DNA repair pathways of Neisseria gonorrhoeae. J. Bacteriol. 184919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sparling, P. F., F. A. Sarubbi, and E. Blackman. 1975. Inheritance of low-level resistance to penicillin, tetracycline and chloramphenicol in Neisseria gonorrhoeae. J. Bacteriol. 124740-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stohl, E. A., A. K. Criss, and H. S. Seifert. 2005. The transcriptosome response of Neisseria gonorrhoeae to hydrogen peroxide reveals genes with previously uncharacterized roles in oxidative damage protection. Mol. Microbiol. 58520-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Veal, W. L., R. A. Nicholas, and W. M. Shafer. 2002. Overexpression of the MtrC-MtrD-MtrE efflux pump due to an mtrR mutation is required for the chromosomally mediated penicillin resistance in Neisseria gonorrhoeae. J. Bacteriol. 1845619-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warner, D. M., J. P. Folster, W. M. Shafer, and A. E. Jerse. 2007. Regulation of the MtrC-MtrD-MtrE efflux-pump system modulates the in vivo fitness of Neisseria gonorrhoeae. J. Infect. Dis. 1961804-1812. [DOI] [PubMed] [Google Scholar]
- 32.Winter, J., K. Linke, A. Jatzek, and U. Jakob. 2005. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol. Cell 17381-392. [DOI] [PubMed] [Google Scholar]
- 33.Workowski, K. A., S. M. Berman, and J. M. Douglas. 2008. Emerging antimicrobial resistance in Neisseria gonorrhoeae: urgent need to strengthen prevention strategies. Ann. Intern. Med. 148606-613. [DOI] [PubMed] [Google Scholar]
- 34.Xia, M., W. L. H. Whittington, W. M. Shafer, and K. K. Holmes. 2000. Gonorrhea among men who have sex with men: outbreak caused by a single genotype of erythromycin-resistant Neisseria gonorrhoeae with a single base pair deletion in the mtrR promoter region. J. Infect. Dis. 1812080-2082. [DOI] [PubMed] [Google Scholar]
- 35.Zarantonelli, L., G. Borthagary, E.-H. Lee, and W. M. Shafer. 1999. Decreased azithromycin susceptibility of Neisseria gonorrhoeae due to mtrR mutations. Antimicrob. Agents Chemother. 432468-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.