Abstract
Agrobacterium tumefaciens and Agrobacterium rhizogenes are related pathogens that cause crown gall and hairy root diseases, which result from integration and expression of bacterial genes in the plant genome. Single-stranded DNA (T strands) and virulence proteins are translocated into plant cells by a type IV secretion system. VirD2 nicks a specific DNA sequence, attaches to the 5′ end, and pilots the DNA into plant cells. A. tumefaciens translocates single-stranded DNA-binding protein VirE2 into plant cells where it likely binds T strands and may aid in targeting them into the nucleus. Although some A. rhizogenes strains lack VirE2, they transfer T strands efficiently due to the GALLS gene, which complements an A. tumefaciens virE2 mutant for tumor formation. Unlike VirE2, full-length GALLS (GALLS-FL) contains ATP-binding and helicase motifs similar to those in TraA, a strand transferase involved in conjugation. GALLS-FL and VirE2 contain nuclear localization signals (NLS) and secretion signals. Mutations in any of these domains abolish the ability of the GALLS gene to substitute for virE2. Here, we show that the GALLS gene encodes two proteins from one open reading frame: GALLS-FL and a protein comprised of the C-terminal domain, which initiates at an internal in-frame start codon. On some hosts, both GALLS proteins were required to substitute for VirE2. GALLS-FL tagged with yellow fluorescent protein localized to the nucleus of tobacco cells in an NLS-dependent manner. In plant cells, the GALLS proteins interacted with themselves, VirD2, and each other. VirD2 interacted with GALLS-FL and localized inside the nucleus, where its predicted helicase activity may pull T strands into the nucleus.
Agrobacterium rhizogenes causes hairy root disease in which adventitious roots proliferate from infected plant tissue. Pathogenesis results when transformed plant cells express rol (root loci) genes transferred from the root-inducing (Ri) plasmid (45). In contrast, Agrobacterium tumefaciens causes unorganized growth of infected plant cells. Oncogenes transferred from the tumor-inducing (Ti) plasmid into plant cells encode proteins involved in synthesis of the plant growth hormones auxin (iaaM and iaaH) and cytokinin (ipt), which results in formation of crown galls (50).
Regions of the Ri and Ti plasmids that are transferred to plant cells (T-DNA) are delimited by border sequences (30, 43). T-DNA transfer initiates when border sequences are nicked by VirD2 and VirD1 (46). VirD2, which contains a secretion signal (42), attaches to the 5′ end of the nicked strand (14, 44, 47) and is transported into plant cells along with attached T-strand DNA (37). Transport requires a type IV secretion system that includes 11 virB-encoded proteins (4) and VirD4 (29). VirD2 contains a nuclear localization signal (NLS) and interacts with host proteins involved in nuclear import, including importin α proteins (1, 18, 34, 39).
Ri and Ti plasmids share many similarities, including nearly identical organization of the vir operons (25). One exception is the absence of virE1 and virE2 from the Ri plasmid (and the genome) in some strains of A. rhizogenes (15, 25). The single-stranded DNA-binding (SSB) protein VirE2 and its secretory chaperone VirE1 are critical for pathogenesis by A. tumefaciens (9, 38, 49). VirE2 is required only in plant cells; transgenic plants that produce VirE2 are fully susceptible to A. tumefaciens virE2 mutants (7). Inside plant cells, VirE2 protects T strands from nuclease attack (32, 48) and may promote their nuclear import (13, 32, 51). The genome of A. rhizogenes 1724 lacks virE1 and virE2 but still transfers T strands efficiently due to the presence of the GALLS gene on the Ri plasmid (15). The GALLS gene can complement an A. tumefaciens virE2 mutant, and the GALLS gene is essential for virulence in A. rhizogenes strains that lack virE1 and virE2 (15).
Although full-length GALLS (GALLS-FL) protein can substitute for VirE2 function, these proteins lack obvious similarities in their amino acid sequences. The closest known relatives of GALLS-FL are helicases and proteins involved in conjugative transfer of plasmids. The amino terminus of GALLS-FL resembles plasmid-encoded TraA (strand transferase) from A. tumefaciens and Sinorhizobium meliloti (12). This portion of GALLS-FL contains ATP-binding motifs (Walker boxes A and B) and a third motif found in members of a helicase/replicase superfamily (Fig. 1) (12, 17), but VirE2 lacks these motifs. Changes in each motif abolished the ability of GALLS-FL to substitute for VirE2 even though the mutant proteins were stable in A. tumefaciens (16).
FIG. 1.
Domains in the GALLS proteins. Boxes indicate the locations of the ATP binding sites (Walker A and B), helicase motif III, NLS, GALLS repeats 1 to 3, and type IV secretion signal. Restriction sites relevant to gene manipulation are shown. The GALLS gene encodes GALLS-FL and GALLS-CT, which results from translation initiated at an in-frame start codon (Met 808). The bottom boxes depict tryptic peptides of GALLS-CT detected by MS as indicated on the figure.
The GALLS proteins (16) and VirE2 (35, 40, 41) contain C-terminal signals for translocation into plant cells mediated by the VirB/D4 type IV secretion system (Fig. 1). VirE2 contains two NLSs that may aid in targeting VirE2-bound T strands to plant nuclei (7, 51), whereas GALLS-FL contains a single bipartite NLS (Fig. 1) (15), which is important for its ability to substitute for VirE2 (16). This indicates that GALLS-FL performs a critical function inside the nucleus or at the nuclear membrane.
In this study, we show that the GALLS gene encodes the 1,769-amino-acid GALLS-FL and a C-terminal domain protein (GALLS-CT) of 962 amino acids. Translation of GALLS-CT initiates at an internal in-frame start codon (Met 808), which is required for production of GALLS-CT. On some hosts, both GALLS proteins were required to substitute for VirE2, but on others GALLS-FL was sufficient. Most of GALLS-CT consists of nearly identical 266-residue sequences repeated three times (Fig. 1). Mutant GALLS-FL and GALLS-CT proteins that contain a single copy of this 266-residue sequence are unable to substitute for VirE2 effectively although the proteins remain stable (15). We also show that GALLS-FL expressed in tobacco protoplasts localized inside the nucleus, whereas GALL-CT expressed separately remained in the cytoplasm. However, when the two GALLS proteins were expressed together, these proteins interacted with each other and accumulated inside the nucleus. VirD2 interacted with GALLS-FL and localized inside the nucleus, suggesting that VirD2 may recruit GALLS-FL to the 5′ end of the T strand inside the nucleus. If the T strand-VirD2-GALLS complex is anchored to host proteins inside the nucleus, the predicted helicase activity of GALLS-FL may pull T strands into the nucleus as it translocates along the DNA strand, thereby obviating the need for VirE2 to facilitate nuclear import of T strands.
MATERIALS AND METHODS
Bacterial transformation and virulence assays.
Plasmids containing mutant or wild-type GALLS genes were transformed into A. tumefaciens mx358 (virE2::Tn3-lacZ) (36). A. tumefaciens strains were tested for virulence on carrot root disks, Kalanchoe daigremontiana leaves, and Arabidopsis thaliana root explants as described previously (16, 28). The basal surfaces of 10 to 18 carrot root slices were inoculated with overnight cultures of each bacterial strain (10 μl/slice); five replicates were performed. Leaves of K. daigremontiana were wounded with a sterile toothpick and inoculated with A. tumefaciens cultured on AB-glucose agar (3 g KH2PO4, 1 g NaH2PO4, 1 g NH4Cl, 300 mg MgSO4·7H2O, 150 mg KCl, 10 mg CaCl2, 2.5 mg FeSO4·7H2O, 5 g glucose, 15 g agar [per liter]) containing appropriate antibiotics. Wounds on each leaf were inoculated with A. tumefaciens mx358 harboring vector plasmid (2 wounds/leaf), the wild-type GALLS gene (2 wounds/leaf), or the GALLS M808I M815V gene (8 wounds/leaf); six replicates were performed. Root segments from four A. thaliana plants (ecotype Ws-2) were pooled and randomized for each of 6 to 8 replicates prior to inoculation with three different concentrations of A. tumefaciens (108, 107, and 106 CFU/ml). Each bacterial strain was inoculated onto 100 to 150 root segments per replicate.
Fusion of a six-histidine affinity tag to GALLS-FL.
The coding sequence of the GALLS-FL gene was fused to a start codon and six histidine codons located downstream of the trc promoter and ribosome binding site in the expression vector pTrc99 (Pharmacia). All constructions (here and in subsequent sections) were confirmed by DNA sequencing. The resulting plasmid was integrated by Cre-mediated site-specific recombination (21) into the loxP site of a vector derived from pCGN5927, which encodes resistance to gentamicin and has a Ri plasmid origin of replication (24). The resulting cointegrate, pLH404, can replicate in A. tumefaciens and encodes His6-tagged GALLS-FL under the control of the isopropyl β-d-thiogalactoside (IPTG)-inducible trc promoter.
A gene encoding His6-tagged GALLS-FL was constructed as follows. Four oligonucleotides (His A to His D) (Table 1) comprising codons 3 to 28 of the GALLS gene, which includes BglII site 1 (Fig. 1), were annealed and inserted into the NcoI and KpnI sites of pTrc99 to create pLH378. Annealed oligonucleotides (GALLS 3′ sense/antisense) (Table 1) containing the last eight codons of the GALLS gene, including the SacII site (Fig. 1), were inserted into the KpnI and BamHI sites of pLH378 to create pLH381. The sequences of the cloned oligonucleotides were confirmed. To reconstruct the GALLS gene, a BglII restriction fragment containing the central region of the GALLS gene (Fig. 1) was inserted into the BglII site of pLH381 to create pLH387, and then the coding sequence between EcoRI site 2 and the SacII site (Fig. 1) was inserted into the corresponding sites in pLH387 to create pLH388. Annealed oligonucleotides containing a loxP site (Lox F and R) (Table 1) were inserted into the BamHI site of pLH388 to create pLH401. The entire pLH401 plasmid was integrated by Cre-mediated site-specific recombination into the loxP site of pDM13, creating pLH404. To construct pDM13, we inserted annealed oligonucleotides that comprise a loxP sequence into the KpnI site of pCGN5927.
TABLE 1.
Annealed oligonucleotides, PCR primers, and probes
| Primer (strand) | Plasmid | Sequence (5′ to 3′)a |
|---|---|---|
| His A (sense) | pLH378 | cATGGCACATCACCATCACCATCACACCGACGACATTGTAATGTCCGATCCCGGAATG |
| His B (sense) | pLH378 | GCTGCTGTTGACACGTCTGTCCCTATGCGCTTCCAGACAGATCTGgtac |
| His C (antisense) | pLH378 | cAGATCTGTCTGGAAGCGCATAGGGACAGACGTGTCAACAGCAGCCATTCCGGGATCGGACA |
| His D (sense) | pLH378 | TACAATGTCGTCGGTGTGATGGTGATGGTGATGTGC |
| GALLS 3′ (sense) | pLH381 | CGCGGGGATGGACGTGGACTCTAAg |
| GALLS 3′ (antisense) | pLH381 | gatccTTAGAGTCCACGTCCATCCCCGCGGtac |
| Lox F | pLH401 | gatccataacttcgtataatgtatgctatacgaagttatggtaccg |
| Lox R | pLH401 | gatccggtaccataacttcgtatagcatacattatacgaagttatg |
| Pst #2 kill (sense) | pLH407 | GGCAGCGAGCGTCATTTTTCGCAGACTTCCATTGTGGAAGCCTTGCA |
| Pst #2 kill (antisense) | pLH407 | AGGCTTCCACAATGGAAGTCTGCGAAAAATGACGCTCGCTGCCTGCA |
| Eco #2 (sense) | pJNM10 | CAACCGGAATTCTGCGGGTCG |
| M808I M815V (antisense) | pJNM10 | CTCGCTGCCTGCAGAGTCAGCGCCTTGGACTTTCTGGTCATTCGAAGCAATATTTTTTTCTCC |
| +1 frameshift (antisense) | pLH433 | CTCGCTGCCTGCAGAGTCAGCGCCTTGCATTTTCTGGTCATTCGAAGCCATATTTTTTTCTCCTCGAGTCTTTGG |
| Kozak/start (sense) | pDM6 | cgcggatccgccgccaccATGCCAACCGACGACATTGTAATGTCCG |
| 240′-217′ (antisense) | pDM6 | tggaattcCAGGTCGATTTCTTGTACCGGCCG |
| Hind-Asc (sense/antisense) | pJP1 | agctgggcgcgccc |
| SXXB (sense) | pDM16 | GGGGATGGACGTGGACTCtctagactcgagg |
| BXXS (antisense) | pDM16 | gatccctcgagtctagaGAGTCCACGTCCATCCCCGC |
| Xba YFP 5′ (sense) | pDM18 | cgcagatctagaATGGTGAGCAAGGGCGAGGAGCTG |
| Bam YFP 3′ (antisense) | pDM18 | cgcggatccTCACTTGTACAGCTCGTCCATGCCGAG |
| GALLS-CT 5′ (sense) | pDM21 | cgctctagagccgccaccATGGCTTCGAATGACCAGAAAATGCAAGGCG |
| GALLS-CT 3′ (antisense) | pDM21 | tctagaTTAGAGTCCACGTCCATCCCCGCGGACGCG |
| nYFPXba (sense) | pLH425 | aacagctatgaccatgtctagaATGGTGAGCAAGGGCG |
| nYFPBam (antisense) | pLH425 | gtaaaacgacgagtggatccTCAGTCCTCGATGTTGTGGCG |
| pLH437 | ||
| cYFPXba (sense) | pLH426 | aacagcttgaccatgtctagaGCCGACAAGCAGAAGAACG |
| cYFPBam (antisense) | pLH426 | gtaaaacgacggccagtggatccTCACTTGTACAGCTCGTC |
| pLH438 | ||
| nYFPXho (sense) | pLH437 | aacagctatgaccatgctcgagATGGTGAGCAAGGGCG |
| cYFPXho (sense) | pLH438 | aacagctatgaccatgctcgagGCCGACAAGCAGAAGAACG |
| SXEB (sense) | pLH435 | GGGGATGGACtcgagaattcg |
| BEXS (antisense) | pLH435 | gatcccgaattctcgaGTCCACGTCCATCCCCGC |
| Sense 1-11 (sense) | probe | cgaggggaataggagtggcaagcATGCCAACCGACGACATTGTAATGTCCGATCCCG |
| Anti 1-11 (antisense) | probe | CGGGATCGGACATTACAATGTCGTCGGTTGGCATgcttgccactcctattcccctcg |
| Sense 808-818 (sense) | probe | ccaaagactcaggagaaaaaaatATGGCTTCGAATGACCAGAAAATGCAAGGCGCTG |
| Anti 808-818 (antisense) | probe | CAGCGCCTTGCATTTTCTGGTCATTCGAAGCCATatttttttctcctgagtctttgg |
Nucleotides that differ from the wild-type sequence are shown in bold type, as are the Kozak sequence and His6 tag. Lowercase letters indicate sequences outside the coding sequence. Restriction sites are italicized. Unpaired ends of annealed oligonucleotides are underlined.
Construction of mutations in the GALLS gene.
A plasmid containing the GALLS gene with M808I and M815V mutations was constructed as follows. Plasmid pLH389 (16) contains the coding sequence and promoter of the wild-type GALLS gene in a derivative of pBluescript SK(−) (Stratagene) with a loxP site inserted into its KpnI site. The NcoI fragment of the GALLS gene, which contains PstI site 3 (Fig. 1), was removed from this plasmid by NcoI digestion and ligation to form pLH400. PstI site 2 (Fig. 1) was destroyed by inserting annealed oligonucleotides with PstI-compatible ends (Table 1, Pst 2 kill sense/antisense) into PstI-cut pLH400 to create pLH407; these oligonucleotides did not alter the encoded amino acids. The region of the GALLS gene from EcoRI site 2 through PstI site 1 (Fig. 1) was amplified by PCR using Pfu DNA polymerase. The antisense primer included the M808I and M815V mutations and PstI site 1 (Table 1, M808I M815V antisense and Eco 2 sense). This amplicon was inserted into pCR2.1 (Invitrogen) by topoisomerase-mediated ligation to create pJNM10, and the cloned amplicon was sequenced. The amplicon was excised from pJNM10 with EcoRI and PstI and inserted into pLH407 digested with these enzymes, forming pJNM13. The EcoRI fragment containing the 5′ end of the GALLS gene coding sequence (Fig. 1) was inserted in the proper orientation into pJNM13, creating pLH415. The NcoI fragment near the 3′ end of the GALLS gene (Fig. 1) was inserted in the correct orientation into the NcoI site of pLH415 to yield pLH416, which contains the entire GALLS gene with the M808I and M815V mutations. pLH416, which contains a loxP site, was integrated by Cre-mediated site-specific recombination into the loxP site of pDM13 to create pLH417.
To construct a plasmid containing the GALLS gene with a +1 frameshift in codon 803, we amplified the region of the GALLS gene from EcoRI site 2 through PstI site 1 by PCR using an antisense primer containing the +1 frameshift mutation (an extra C base) and PstI site 1 (Table 1, +1 frameshift antisense and Eco 2 sense).This amplicon was inserted into pCR2.1 to create pJNM11. The sequenced amplicon was excised from pJNM11 with EcoRI and PstI and inserted into pLH407, forming pLH427. The GALLS-FL gene was reconstructed by inserting the appropriate NcoI and EcoRI fragments, and this plasmid was integrated into pDM13 by Cre-mediated recombination to yield pLH434.
Fusion of the GALLS-FL gene to the CaMV35S promoter and YFP.
Plasmid pCGN8059 contains an enhanced CaMV35S promoter joined to an HSP70 5′ untranslated region (UTR) followed by unique restriction sites and a nopaline synthase 3′ untranslated region (24). We fused these regulatory elements to the coding sequence of the GALLS-FL gene and replaced the stop codon with XbaI and BamHI sites, creating pDM16, as follows. We inserted a self-complementary oligonucleotide containing an AscI site (Table 1, Hind-Asc) into the HindIII site of pCGN8059 to create pJP1. The first 240 bp of the coding sequence of the GALLS gene was amplified by PCR using primers that added a BamHI site and Kozak sequence adjacent to the start codon and an EcoRI site to the opposite end of the amplicon (Table 1, Kozak/start sense and 240′-217′ antisense). This amplicon was digested with BamHI and EcoRI and inserted into pJP1 at the BglII and EcoRI sites, creating pDM6; the sequence of the cloned amplicon was confirmed. A restriction fragment extending from BglII site 1 to EcoRI site 2 (Fig. 1) of the coding sequence of the GALLS gene was inserted into the BglII and EcoRI sites of pDM6 to yield pDM9. A restriction fragment extending from EcoRI site 2 to the BamHI site of the GALLS gene (Fig. 1) was inserted into the EcoRI and BamHI sites of pDM9 to produce pDM10. The stop codon of the coding sequence of the GALLS gene was replaced with an XbaI site by inserting annealed oligonucleotides (Table 1, SXXB and BXXS) into the SacII and BamHI sites near the 3′ end of the GALLS gene in pDM10 to create pDM16. The yellow fluorescent protein (YFP) coding sequence was amplified by PCR using primers that fused XbaI and BamHI sites to the 5′ and 3′ ends. This amplicon was inserted into the XbaI and BamHI sites of pDM16, fusing the YFP coding sequence to the 3′ end of GALLS to create pDM18. We digested pDM18 with BglII to remove the region that contains the NLS, and we replaced it with the corresponding BglII fragment from pLH379, from which the NLS was deleted, creating pDM19.
Fusion of the GALLS-CT gene to the CaMV35S promoter.
The coding sequence of the GALLS-CT gene and stop codon (codons 808 to 1770) were amplified by PCR using primers that added XbaI sites to each end and a Kozak sequence immediately upstream of the start codon (Table 1, GALLS-CT 5′ and 3′). The amplicon was inserted into pCR4.0 by topoisomerase-mediated ligation, excised with XbaI, and inserted into the XbaI site of pCGN8059, which lies between the CaMV35S promoter/HSP70 leader and the nopaline synthase polyadenylation signal (24); the resulting plasmid (pDM26) was used to construct fusions for bimolecular fluorescence complementation (BiFC).
Construction of fusions for BiFC.
The coding sequences of the GALLS-FL (in pDM16) and GALLS-CT (in pDM26) genes expressed from the CaMV35S promoter were fused to sequences encoding the N-terminal or C-terminal fragment of YFP (nYFP or cYFP, respectively). Amplicons encoding nYFP (codons 1 to 174) or cYFP (codons 155 to 240, including the stop codon) were generated by PCR using primers that added sites for XbaI upstream and BamHI downstream of the coding sequences (Table 1, nYFPXba with nYFPBam and cYFPXba with cYFPBam). These amplicons were inserted into the XbaI and BamHI sites of pDM16 to create pLH425, which expresses a GALLS-FL::nYFP fusion protein, and pLH426, which expresses GALLS-FL::cYFP. The coding sequence of the GALLS-CT gene expressed from the CaMV35S promoter (in pDM26) was also fused to sequences encoding nYFP and cYFP. The last four codons of the GALLS-CT gene were replaced with an XhoI site by inserting annealed oligonucleotides (Table 1, SXEB and BEXS) into the SacII and BamHI sites in pDM26 to create pLH435. Amplicons encoding nYFP and cYFP were produced by PCR using primers that added sites for XhoI upstream and BamHI downstream (nYFPXho with nYFPBam and cYFPXho with cYFPBam) (Table 1). These amplicons were inserted into the XhoI and BamHI sites of pLH435, creating plasmids that express GALLS-CT::nYFP (pLH437) and GALLS-CT::cYFP (pLH438).
BiFC fusions to another secreted virulence protein (VirD2) and a host protein involved in nuclear targeting (importin α4) were prepared as follows. The virD2 gene from pTiA6 was cloned as an EcoRI-SacII fragment into the BiFC vector pSAT6-nEYFP-C1 (which contains an N-terminal fragment of the enhanced YFP) (6). A cDNA (lacking the translation stop codon) of the A. thaliana importin α4 (AtImpa-4) gene was cloned into pSAT6-cEYFP-N1 (6) as an NcoI-XmaI fragment.
Immunoblot analysis.
To compare levels of GALLS proteins in bacterial cells, A. tumefaciens mx358 cells harboring mutant or wild-type GALLS genes were cultured as described previously (16). Harvested cells were lysed in a French pressure cell, and soluble proteins were prepared as described previously (16). Samples containing 18 μg of protein were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) through 4 to 20% gradient gels (Bio-Rad), and proteins were transferred to a polyvinylidene difluoride membrane using a Bio-Rad miniProtean II electroblotting cell. Immunoblots were probed as described previously (16) with GALLS-specific antibodies from rabbits immunized with GALLS-FL protein expressed in Escherichia coli; antibodies were diluted 1:2,500.
Protein purification and mass spectrometry.
GALLS-CT protein was purified by SDS-PAGE and excised from a Coomassie-stained gel. The protein was eluted from the gel and digested with trypsin at the mass spectrometry (MS) facility at the University of Texas Medical Branch, Galveston, TX. The peptides were separated by high-performance liquid chromatography and analyzed by matrix-assisted laser desorption ionization—time of flight (MALDI-TOF) MS.
RNA extraction and hybridization.
To compare levels of mRNA encoded by the GALLS gene at start codons 1 and 808, we cultured A. tumefaciens mx358 harboring a wild-type GALLS gene in plasmid pLH338 (15) as described previously (16) and extracted total nucleic acids from 30-ml cultures using an SDS lysis-phenol extraction method (31). Agarose gel electrophoresis revealed sharp rRNA bands, indicating good preservation of RNA during extraction (data not shown). Total nucleic acids from A. tumefaciens WR5004 served as a negative control; this derivative of WR3095 (30) lacks the GALLS gene but contains the virE2::Tn3-lacZ mutation from A. tumefaciens mx358 (36). A plasmid (pLH337) (15) containing the GALLS gene was linearized by digestion with BamHI, denatured by boiling, and used as a standard to normalize hybridization signals. Samples containing 6 μg of total nucleic acids from A. tumefaciens or 100 ng of denatured pLH337 DNA in 10× SSC (pH 7; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) were spotted in triplicate onto 15-mm squares of Gene Screen Plus hybridization transfer membrane (Perkin Elmer) and irradiated with 1,200 μJ/m2 of UV light in a Stratalinker 1800 instrument (Stratagene). DNA oligonucleotides (Invitrogen) corresponding to the sense (Sense 1-11 and Sense 808-818) and antisense (Anti 1-11 and Anti 808-818) strands of the GALLS gene (Table 1) were labeled in reaction mixtures containing 10 pmol of DNA, 10 pmol of [γ-32P]ATP (7,000 Ci/mmol; 2 mCi/12 μl) (MP Biomedicals) and 10 units of T4 polynucleotide kinase (Invitrogen). Labeled DNA was precipitated with ethanol to remove unincorporated ATP. Irradiated filters were incubated at 42°C in hybridization solution (1 M NaCl, 10% dextran sulfate, 1% SDS, 50% formamide, 0.1 ml/ml calf thymus DNA) (31) without probe for 2 h prior to addition of 5 pmol of 32P-labeled probe. Incubations were continued overnight at 42°C followed by three washes each in 2× SSC-1% SDS at 65°C and 0.1× SSC-1% SDS at 42°C (31).
Bound probe was quantified by liquid scintillation counting in a Beckman LS6500. Hybridizations included filters (in triplicate) with no sample or with nucleic acids from A. tumefaciens WR5004, which lacks the GALLS gene. Radioactivity detected on these filters was 1.3 to 10% of that retained on filters with 100 ng of denatured pLH337 DNA. The average signal on the blank filters was subtracted from that on filters with pLH337 DNA, and the signal on filters containing nucleic acids from WR5004 was subtracted from that on filters with nucleic acids from mx358(pLH338). To normalize the signals from each probe, the mean signal (minus background) present on filters containing nucleic acids from mx358(pLH338) was divided by the corresponding value for filters containing denatured pLH337 DNA. For each of three biological replicates, normalized values for the Anti 1-11 probe were divided by the corresponding figure for the Anti 808-818 probe, and the ratios for all three replicates were averaged.
Tobacco protoplast preparation and microscopy.
Tobacco BY-2 protoplasts were prepared and coelectroporated with 20 μg of each BiFC construction as described previously (27). In some instances, the cells were stained with 20 ng/ml Hoechst 33242 (Molecular Probes, Eugene, OR) in phosphate-buffered saline-0.4 M mannitol buffer, and fluorescence was visualized using a Nikon Eclipse E600 fluorescence microscope or a Zeiss LSM510 Meta confocal microscope.
RESULTS
The GALLS gene encodes two proteins.
To investigate proteins encoded by the GALLS gene, we extracted soluble proteins from an A. tumefaciens virE2 mutant (mx358) expressing the GALLS gene from its native promoter (15). Immunoblot analysis with GALLS-specific antibodies revealed protein bands with estimated molecular weights of 193,000 (GALLS-FL) and 115,000 (GALLS-CT) (Fig. 2A). These proteins were absent from vector-only controls (Fig. 2A). GALLS-CT protein was significantly more abundant than GALLS-FL. To facilitate purification of GALLS-FL, we added an N-terminal six-histidine affinity tag. A. tumefaciens mx358 expressing His6-GALLS-FL and GALLS-CT was fully virulent on carrot (data not shown), indicating that histidine-tagged GALLS-FL is fully active. We fractionated soluble proteins from these cells using a Ni-nitrilotriacetic acid resin. Most of the GALLS-CT protein eluted in the flowthrough fraction, as expected due to the absence of the histidine tag (Fig. 2B). The Ni column retained His6-GALLS-FL and GALLS-CT, which eluted with imidazole (Fig. 2B). The retention of untagged GALLS-CT on the Ni column suggests that GALLS-CT interacts with His6-GALLS-FL.
FIG. 2.
Immunoblot analysis of proteins encoded by the GALLS gene. Blots were probed with GALLS-specific antibodies. FL, GALLS-FL; His-FL, His6-tagged GALLS-FL; CT, GALLS-CT. Numbered bars indicate the size (in thousands) of molecular weight standards. (A) Soluble proteins from A. tumefaciens containing vector plasmid (left lane) or the wild-type GALLS gene (right lane). (B) Soluble proteins were extracted from A. tumefaciens cells harboring the His6-GALLS-FL gene and fractionated on Ni-nitrilotriacetic acid resin; bound proteins were eluted with imidazole. Proteins from the flowthrough (flow), wash, and first and second eluates are shown. (C) Soluble proteins from A. tumefaciens containing the vector plasmid (lane 1), wild-type GALLS gene (lane 2), the +1 frameshift mutation in codon 803 of the GALLS gene (lane 3, +1 FS), or the GALLS gene with the Met808Ile Met815Val mutations (lane 4, MIMV). (D) GALLS proteins expressed in A. tumefaciens cells containing the GALLS gene fused to the Trc promoter and 5′ UTR from pTrc99 in the absence (−) or presence (+) of IPTG.
The GALLS gene contains an in-frame methionine (Met) codon (808) that may be the start codon for translation of GALLS-CT. This putative alternative start codon lies 20 codons upstream of the first GALLS repeat (Fig. 1) and is preceded by a strong ribosome binding site (RBS [AGGAG]) 8 bp upstream and a favored A at −3 (20). The RBS, Met codon 808, and the A at −3 are conserved in three known GALLS genes from pRi1724, pRiA4, and pRi2659 (accession numbers NP_066636, BAB47245, and ABW33585) (23, 25). Initiation of translation at Met 808 would yield a protein with a calculated molecular weight of 104,697, which is close to the molecular weight estimated by SDS-PAGE (Fig. 2A). A nearby methionine codon (815) lacks an RBS and is not conserved in pRiA4, which encodes a valine at the corresponding position (accession number BAB47245). The fact that Met 808 is conserved and is preceded by a conserved RBS suggests that it is the start codon for GALLS-CT.
To test whether translation of GALLS-CT initiates at Met codon 808 or 815, we converted the residue to Ile or Val, respectively. Only the double mutation was constructed to eliminate both potential start sites. This double mutation abolished production of GALLS-CT (Fig. 2C), as expected if translation of GALLS-CT initiates at one of these codons. A nonpolar +1 frameshift in codon 803, upstream of the RBS, suggests that translation of GALLS-CT begins at Met 808. This mutation did not affect production of GALLS-CT (Fig. 2C), as predicted if translation of GALLS-CT begins at Met 808. GALLS-CT cannot result from proteolysis of GALLS-FL because the frameshift truncates GALLS-FL three amino acids after Met 808.
We used MS to confirm the identity of GALLS-CT because the N terminus was blocked to Edman degradation. We purified GALLS-CT by SDS-PAGE, excised it from Coomassie-stained gels, and digested it with trypsin. High-performance liquid chromatography separation of the resulting peptides and MALDI-TOF MS analysis confirmed that all peptides were derived from the three GALLS repeats and the GALLS-CT (Fig. 1). Five of these peptides could be derived from each of the three repeats, whereas another nine peptides occur in repeats 1 and 3 but not in repeat 2 (Fig. 1). Twelve peptides are unique, with seven derived from repeat 2, one from repeat 3, and four from the region between repeat 3 and the secretion signal (Fig. 1). Although repeat 1 did not yield unique peptides due to its similarity to repeat 3, we can infer that repeat 1 is present in GALLS-CT based on the estimated molecular weight of GALLS-CT (Fig. 2A). The mass spectrum did not include any peptides derived from the region upstream of the GALLS repeats. The MS data support our hypothesis that the GALLS-CT protein results from initiation at Met 808. Translation of GALLS-CT initiated at this codon would yield an N-terminal tryptic peptide containing seven amino acids; peptides containing seven or fewer residues were not detected by the MALDI-TOF MS procedure due to interference by matrix ions.
Both GALLS proteins are translated from the same mRNA.
We fused the coding sequence of the GALLS gene to the trc promoter and 5′ UTR of pTrc99 in an attempt to increase expression of GALLS-FL protein. However, this construction (pLH404) did not increase the level of GALLS-FL protein relative to GALLS-CT (Fig. 2D), even though pTrc99 is designed to express proteins at high levels. Thus, replacing the native promoter and 5′ UTR of the GALLS gene did not increase expression of GALLS-FL. This construction also allowed us to ask whether the GALLS coding sequence contains a second promoter upstream of the start of GALLS-CT. Expression of GALLS-FL and high levels of GALLS-CT from the trc promoter required IPTG (Fig. 2D), strongly suggesting that one promoter controls expression of both proteins. Low levels of GALLS-CT produced in the absence of IPTG may result from incomplete repression of the trc promoter, despite the presence of the lacI gene in pTrc99.
Transcription of the GALLS gene.
We characterized transcription through the GALLS gene to understand better its regulation and to seek an explanation for the different levels of proteins encoded by the GALLS gene. Conceivably, low levels of GALLS-FL relative to GALLS-CT might result from preferential degradation of the 5′ end of the GALLS gene-encoded mRNA. Alternatively, an antisense RNA complementary to the region of the mRNA encoding the start of GALLS-FL might reduce translation of GALLS-FL protein. To test these possibilities, we probed both translation start regions of the GALLS gene-encoded mRNA (expressed from the native promoter) with 32P-labeled oligonucleotides corresponding to the sense and antisense strands of the GALLS gene. Each 57-nucleotide probe contained 23 bases (including the RBS) immediately upstream of the start codons (at position 1 or 808) and the first 34 bases of the coding sequence or its complement. Both antisense oligonucleotides (Anti 1-11 and Anti 808-818) (Table 1) yielded strong signals when hybridized to mRNA encoded by the GALLS gene [from A. tumefaciens mx358(pLH338)] or denatured plasmid DNA (pLH337) containing the GALLS gene. These probes did not hybridize to mRNA from A. tumefaciens WR5004, which lacks the GALLS gene, or to double-stranded pLH337 DNA. For each probe, the average signal bound to GALLS mRNA was normalized to that bound to denatured pLH337 DNA. These normalized values were used to determine the ratio of the signals bound to the 5′ end (FL) and central region (CT) of the mRNA. Three separate preparations of mRNA (biological replicates) yielded FL/CT ratios of 1.45 ± 0.27, 0.88 ± 0.16, and 0.79 ± 0.21, for an average ratio of 1.04 ± 0.37. Immunoblot analysis of total cellular protein from the cultures used to produce the GALLS mRNA for this experiment showed typically high levels of GALLS-CT protein relative to GALLS-FL, as expected (data not shown). Thus, mRNA transcribed from the native promoter of the GALLS gene produced similar signals when annealed to labeled oligonucleotides complementary to the translation start regions of GALLS-FL and GALLS-CT, indicating that instability of the 5′ end of the mRNA is not responsible for the low levels of GALLS-FL protein relative to GALLS-CT.
In contrast, both sense oligonucleotides (Sense 1-11 and Sense 808-818) (Table 1) yielded weak signals when hybridized to GALLS gene-encoded mRNA because antisense transcription through the GALLS gene is minimal (15). In a single replicate, the Sense 1-11 probe gave only 8.7% of the signal produced by the Anti 1-11 probe; similarly, the Sense 808-818 probe yielded 16% of the signal generated by the Anti 808-818 probe. These direct measurements confirmed our earlier observations with sense and antisense lacZ fusions, which showed weak transcription in the antisense direction (15). Because relatively little antisense RNA is present at either translation start site, antisense RNA probably does not affect translation of either GALLS protein.
Importance of GALLS-CT to plant transformation.
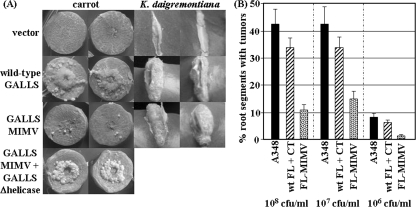
The M808I M815V double mutation eliminated production of GALLS-CT (Fig. 2C), but these mutations did not affect complementation of a virE2 mutant A. tumefaciens strain in virulence assays on K. daigremontiana (Fig. 3A), confirming that GALLS-FL M808I M815V remained active. In contrast, this mutation severely reduced (but did not abolish) tumorigenesis on carrot (Fig. 3A) and A. thaliana (Fig. 3B). Full virulence was restored on carrot roots coinfected with two A. tumefaciens virE2 mutants expressing (i) GALLS-FL M808I M815V and (ii) wild-type GALLS-CT and inactive GALLS-FL with a deletion of helicase motif III (GALLS-FLΔhelicase III) (16) (Fig. 3A), indicating that both GALLS-FL and GALLS-CT are required for full virulence on carrot. Thus, GALLS-FL alone is sufficient for full virulence on K. daigremontiana, but GALLS-CT is also important on carrot and A. thaliana.
FIG. 3.
Virulence of strains expressing wild-type or mutant GALLS genes. (A) K. daigremontiana leaves and the basal surfaces of carrot root slices were inoculated with the A. tumefaciens mx358 virE2 mutant strain harboring vector plasmid (top row), the wild-type GALLS gene (second row), or the mutated GALLS gene (M808I M815V [MIMV]) inoculated alone (third row) or coinoculated with a virE2 mutant harboring the nonpathogenic GALLSΔhelicase III mutant (16) (carrots only). (B) A. thaliana root explants were inoculated with wild-type A. tumefaciens A348 (black bars) or the A. tumefaciens mx358 virE2 mutant strain expressing both wild-type GALLS-FL and GALLS-CT (wt FL + CT) or only GALLS-FL (FL-MIMV). Inoculum size is indicated as CFU/ml.
Localization and interaction of GALLS-FL and GALLS-CT proteins in plant cells.
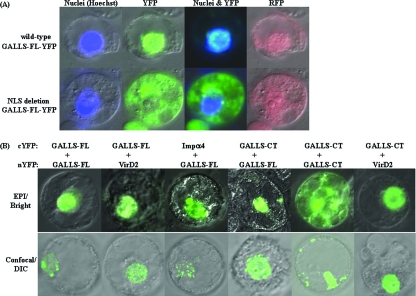
GALLS-FL contains an NLS (Fig. 1), which targets a GALLS-FL::YFP fusion protein to the nucleus of transiently transformed tobacco protoplasts (Fig. 4A). The NLS was required for nuclear localization; GALLS-FL::YFP with a deletion of the GALLS NLS (GALLS-FLΔNLS::YFP) was found throughout the cytoplasm but appeared excluded from the nucleus (Fig. 4A). Some GALLS-FLΔNLS::YFP protein may associate with the nucleus, which is consistent with our observation that deletion of the NLS severely reduced but did not abolish tumorigenesis (16).
FIG. 4.
Localization of GALLS proteins expressed in plant cells. (A) Tobacco protoplasts were electroporated with plasmids expressing GALLS-FL::YFP or GALLS-FLΔNLS::YFP from a CaMV35S promoter. A plasmid expressing RFP was included to mark transformed cells; although RFP is distributed throughout transformed cells, higher amounts concentrate in the nucleus. Nuclei were stained with Hoechst dye (left). Cells were examined by epifluorescence microscopy using filters for YFP or RFP, as indicated. Column 3 shows Hoechst (Nuclei) and YFP signals superimposed. (B) Epifluorescence and confocal microscopy of GALLS protein interactions. Tobacco cells were electroporated with plasmids expressing proteins fused to complementary nYFP and cYFP. BiFC was detected by epifluorescence microscopy (top row) and confocal microscopy with the focal plane through the nucleus (bottom row). YFP fluorescence detected by epifluorescence microscopy (EPI) is superimposed on bright-field images. Confocal images of YFP fluorescence are superimposed on differential interphase contrast (DIC) images.
Plant-encoded importin α proteins interact with many proteins that are targeted to the nucleus. VirD2, the T-strand pilot protein, is translocated from A. tumefaciens into plant cells. VirD2 interacts with A. thaliana importin α1 in Saccharomyces cerevisiae (1) and importin α1, -2, -3, -4, -7, and -9 in plant cells (2). A. thaliana importin α4 (AtImpa-4) is essential for A. tumefaciens-mediated transformation on root explants, whereas importin α1, -2, and -3 are not (2). GALLS-FL is also translocated to plant cells and targeted to the nucleus (Fig. 4A), leading us to ask whether GALLS-FL interacts with importin α4. BiFC is a powerful method to visualize the subcelluar localization of protein interactions in plant cells (6). To test for interactions, two proteins are fused to complementary nonfluorescent fragments of YFP. Interaction between the proteins may bring nYFP and cYFP together so that fluorescence is reconstituted. We fused the nYFP fragment to the C terminus of GALLS-FL and fused cYFP to importin α4. Plasmids expressing these fusion proteins were coelectroporated into tobacco protoplasts along with a plasmid encoding free monomeric red fluorescent protein (RFP) as a positive control for transformation (data not shown). Half-YFP fusions to cDNAs derived from the petunia isoeugenol synthase 1 (IGS1) and coniferyl alcohol acyltransferase (CFAT) genes (10, 19) were used as negative controls. These fusion proteins did not interact with GALLS fused to the complementary half of YFP (data not shown). Cells were examined by epifluorescence and confocal microscopy (Fig. 4B). Confocal images show optical sections with the focal plane through the nucleus superimposed on differential interphase contrast images. Epifluorescence images show that GALLS-FL interacted with AtImpa-4 and localized at the nucleus, and confocal images confirmed that this complex accumulated inside the nucleus (Fig. 4B).
GALLS-FL interacts with itself and GALLS-CT in plant cells.
We fused nYFP and cYFP fragments to the C-termini of GALLS-FL and GALLS-CT. Pairs of plasmids expressing these fusion proteins were coelectroporated into tobacco protoplasts, and protein interactions were visualized by epifluorescence and confocal microscopy. GALLS-FL interacted with itself and localized inside the nucleus (Fig. 4B). GALLS-CT lacks an NLS, and epifluorescence images showed the GALLS-CT self-interaction throughout the cytoplasm (Fig. 4B). Confocal images confirmed that this self-interaction is cytoplasmic but not evenly distributed (Fig. 4B). In contrast, GALLS-FL interacted with GALLS-CT and localized primarily inside the nucleus (Fig. 4B). Thus, interaction of GALLS-FL with GALLS-CT did not interfere with nuclear localization of GALLS-FL.
GALLS proteins interact with the VirD2 pilot protein in plant cells.
Protein interactions often direct DNA-binding proteins to sites where they are needed (5, 26). Mobilization proteins (e.g., the GALLS-related proteins TraA and MobA) often contain separate domains with oriT nicking and helicase activities. VirD2 has T-DNA border-specific nicking activity, but it does not exhibit helicase activity, whereas GALLS-FL has a predicted TraA-like helicase domain, but it lacks a nicking domain. VirD2 and GALLS-FL are both translocated to plant cells, targeted to the nucleus, and required for T-DNA transfer. Because VirD2 and GALLS contain domains that are often present in a single protein, we asked whether they interact in plant cells. Confocal and epifluorescence micrographs show that both GALLS-FL::cYFP and GALLS-CT::cYFP interacted with VirD2::nYFP, and these complexes localized inside the nucleus of tobacco protoplasts (Fig. 4B). The C-terminal domain of GALLS was sufficient to mediate the interaction with VirD2, and this interaction did not prevent nuclear localization of VirD2. Thus, VirD2 may recruit GALLS-FL to the 5′ end of T strands inside the nucleus of host cells.
DISCUSSION
Our data show that the GALLS gene encodes two proteins, GALLS-FL and GALLS-CT, and that translation of GALLS-CT initiates at an internal in-frame start codon. Other genes that encode translational restart proteins include mobA of RSF1010 (33), the primases of plasmids ColI (sog) (3) and R16 (pri) (8), the phage T7 gene 4 primase/helicase (11), and the cisA nickase/helicase of φX174 (22). RSF1010 mobA produces full-length MobA (relaxase/primase) and RepB′ (primase), which is identical to the C-terminal portion of MobA; this sequence also encodes MobB, another relaxase subunit, from a different internal reading frame (33). These genes share intriguing similarities with the GALLS gene, including their involvement in plasmid conjugation (mobA, sog, and pri) and the enzymatic activities they encode (cisA and gene 4 helicases). The translational restart proteins encoded by sog, pri, and cisA are more abundant than the corresponding full-length proteins (3, 8, 22), as is the case for the proteins encoded by the GALLS gene (Fig. 2A). Although the functions of these restart proteins are unknown, their abundance suggests a structural role rather than an enzymatic activity (3).
The low abundance of GALLS-FL may result from the high incidence of rarely used leucine codons upstream of codon 808 in the coding sequence of the GALLS gene. Leucine codons (UUA and CUA), which are used rarely, occur eight times in the GALLS-FL coding sequence but only twice in the GALLS-CT portion. Lower pools for tRNAs that recognize UUA and CUA codons may limit translation of GALLS-FL. We have ruled out differences in the mRNA sequences upstream of the start codons as a possible explanation for the different levels of the proteins encoded by the GALLS gene. Both start codons are preceded by identical RBSs (AGGAG) and a favored A at −3. Fusion of the coding sequence of the GALLS gene to the trc promoter and 5′ UTR of pTrc99 did not increase the level of GALLS-FL protein relative to GALLS-CT even though this plasmid is designed to express proteins at high levels. Expression of both GALLS proteins from the trc promoter required IPTG, strongly suggesting that they are expressed from one promoter. We also eliminated instability of the 5′ end of the mRNA as a possible explanation because mRNA levels are similar at both translation start regions. In addition, we did not detect antisense RNA at levels sufficient to interfere with translation of the mRNA. Thus, reduced translation of GALLS-FL due to codon bias or instability of GALLS-FL protein may account for its low abundance although we have no evidence for either.
GALLS-CT contains a protein interaction domain that promotes self-interaction and binding to GALLS-FL and VirD2. Because GALLS-CT is much more abundant than GALLS-FL in bacterial cells, it may bind GALLS-FL and prevent premature interaction of GALLS-FL with VirD2 prior to export from A. tumefaciens. Similarly, GALLS-CT may prevent self-aggregation of GALLS-FL in bacterial cells. Both GALLS proteins contain identical type IV secretion signals and probably are translocated into plant cells with equal efficiency, thereby maintaining an excess of GALLS-CT relative to GALLS-FL in the cytoplasm of the plant cell. However, GALLS-CT is excluded from the nucleus unless it interacts with either VirD2 or GALLS-FL. Therefore, inside the nucleus, the level of GALLS-CT may be lower than in the cytoplasm. Once inside the nucleus, GALLS-CT may be displaced, allowing GALLS-FL and VirD2 to assemble into a complex at the 5′ end of the T strand, or all three proteins may form a multisubunit complex. GALLS-CT also may modulate the predicted helicase activity of GALLS-FL, or it may anchor the DNA-protein complex to host proteins in the nucleus. Alternatively, high levels of GALLS-CT may saturate proteases that would otherwise degrade GALLS-FL.
GALLS-FL protein compensates for the absence of VirE2, apparently without duplicating its activities. VirE2 is an abundant SSB protein required in stoichiometric amounts to coat and protect T strands (32, 48) and perhaps promote their nuclear import (13, 32, 48, 51). In contrast, GALLS-FL is likely a low-abundance enzyme that may mobilize T strands into the nucleus using its predicted ATP-dependent strand transferase/helicase activity (16). GALLS-FL requires its NLS to function (16) and localizes inside the nucleus of host cells (Fig. 4), suggesting that it provides an alternative means to transport T strands into the nucleus. GALLS-FL and VirD2 can interact when they are coexpressed in plant cells, and this complex localizes inside the nucleus. Thus, during T-strand transfer, GALLS-FL may be anchored to VirD2 at the leading (5′) end of the T strand. GALLS-FL has a predicted helicase domain and may translocate along T strands in a 5′-to-3′ direction, disrupting secondary structures that may form in T strands in the absence of VirE2 SSB. If the helicase remains in a fixed position, translocation along DNA would cause the DNA to move. Thus, GALLS-FL may pull T strands into the nucleus, obviating the need for VirE2 to mediate nuclear import of T strands. Extensive degradation of T strands observed in plant cells infected with an A. tumefaciens virE2 mutant strain (32, 48) may occur because progress of T strands into the nucleus is stalled in plant cells lacking both VirE2 and GALLS-FL. Efficient nuclear import of T strands in the presence of either VirE2 or GALLS-FL may minimize opportunities for nuclease attack in the cytoplasm.
The GALLS gene illustrates the evolution of a novel type IV secretion system-effector protein combination that may confer the ability to mobilize bacterial DNA into the nucleus of eukaryotic cells. To compensate for loss of virE1 and virE2, A. rhizogenes appropriated an unusual conjugation gene (the GALLS gene) to restore its ability to deliver T-DNA to plant cells. The GALLS gene adjoins conjugation (tra) genes ∼60 kb away from the nearest vir gene. Thus, a promiscuous gene transfer system capable of delivering DNA to eukaryotic cells apparently evolved from a type IV secretion system and a bacterial conjugation system.
Acknowledgments
This work was supported by grants from the National Science Foundation (MCB-0344939 and IOS-0724067 to W.R. and MCB-0418709 to S.B.G.); Hank McNett received a summer fellowship from the Howard Hughes Medical Institute.
We thank Annette Vergunst, INSERM, UFR Medecine, Nimes Cedex, for critical reading of the manuscript; Jason Neal-McKinney and Deborah Moyer (OSU) for building several plasmids; Chris Brown (OSU) for assistance with statistical analysis; Anthony Qualley and Natalia Dudareva (Purdue) for providing petunia CFAT and IGS1 cDNAs; Joerg Spantzel (Purdue) for conducting the A. thaliana transformation assays; and Lin-Yun Kuang and Mei-Jane Fang, Academia Sinica, Taipei, Taiwan, for help with plant cell transformation and confocal microscopy.
Footnotes
Published ahead of print on 24 October 2008.
REFERENCES
- 1.Ballas, N., and V. Citovsky. 1997. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci. USA 9410723-10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhattacharjee, S., L. Y. Lee, H. Oltmanns, H. Cao, Veena, J. Cuperus, and S. B. Gelvin. 2008. IMPa-4, an Arabidopsis importin α isoform, is preferentially involved in Agrobacterium-mediated plant transformation. Plant Cell. doi: 10.1105/tpc.108.060467. [DOI] [PMC free article] [PubMed]
- 3.Boulnois, G. J., B. M. Wilkins, and E. Lanka. 1982. Overlapping genes at the DNA primase locus of the large plasmid ColI. Nucleic Acids Res. 10855-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie, P. J. 1997. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J. Bacteriol. 1793085-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Churchill, J. J., D. G. Anderson, and S. C. Kowalczykowski. 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of chi, resulting in constitutive recombination activation. Genes Dev. 13901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Citovsky, V., L. Y. Lee, S. Vyas, E. Glick, M. H. Chen, A. Vainstein, Y. Gafni, S. B. Gelvin, and T. Tzfira. 2006. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 3621120-1131. [DOI] [PubMed] [Google Scholar]
- 7.Citovsky, V., J. Zupan, D. Warnick, and P. Zambryski. 1992. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 2561802-1805. [DOI] [PubMed] [Google Scholar]
- 8.Dalrymple, B. P., and P. H. Williams. 1984. Analysis of the R16 primase gene. Plasmid 12206-210. [DOI] [PubMed] [Google Scholar]
- 9.Deng, W., L. Chen, W. T. Peng, X. Liang, S. Sekiguchi, M. P. Gordon, L. Comai, and E. W. Nester. 1999. VirE1 is a specific molecular chaperone for the exported single-stranded-DNA-binding protein VirE2 in Agrobacterium. Mol. Microbiol. 311795-1807. [DOI] [PubMed] [Google Scholar]
- 10.Dexter, R., A. Qualley, C. M. Kish, C. Je Ma, T. Koeduka, D. A. Nagegowda, N. Dudareva, E. Pichersky, and D. Clark. 2007. Characterization of a petunia acetyltransferase involved in the biosynthesis of the floral volatile isoeugenol. Plant J. 49265-275. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, J. J., and F. W. Studier. 1981. Nucleotide sequence from the genetic left end of bacteriophage T7 DNA to the beginning of gene 4. J. Mol. Biol. 148303-330. [DOI] [PubMed] [Google Scholar]
- 12.Farrand, S. K., I. Hwang, and D. M. Cook. 1996. The tra region of the nopaline-type Ti plasmid is a chimera with elements related to the transfer systems of RSF1010, RP4, and F. J. Bacteriol. 1784233-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelvin, S. B. 1998. Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J. Bacteriol. 1804300-4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrera-Estrella, A., Z. Chen, M. Van Montagu, and K. Wang. 1988. VirD proteins of Agrobacterium tumefaciens are required for the formation of a covalent DNA-protein complex at the 5′ terminus of T-strand molecules. EMBO J. 74055-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges, L. D., J. Cuperus, and W. Ream. 2004. Agrobacterium rhizogenes GALLS protein substitutes for Agrobacterium tumefaciens single-stranded DNA-binding protein VirE2. J. Bacteriol. 1863065-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodges, L. D., A. C. Vergunst, J. Neal-McKinney, A. den Dulk-Ras, D. M. Moyer, P. J. J. Hooykaas, and W. Ream. 2006. Agrobacterium rhizogenes GALLS protein contains domains for ATP binding, nuclear localization, and type IV secretion. J. Bacteriol. 1888222-8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgman, T. C. 1988. A new superfamily of replicative proteins. Nature 33322-23. [DOI] [PubMed] [Google Scholar]
- 18.Howard, E. A., J. R. Zupan, V. Citovsky, and P. C. Zambryski. 1992. The VirD2 protein of Agrobacterium tumefaciens contains a C-terminal bipartite nuclear localization signal: implications for nuclear uptake of DNA in plant cells. Cell 68109-118. [DOI] [PubMed] [Google Scholar]
- 19.Koeduka, T., E. Fridman, D. R. Gang, D. G. Vassao, B. L. Jackson, C. M. Kish, I. Orlova, S. M. Spassova, N. G. Lewis, J. P. Noel, T. J. Baiga, N. Dudareva, and E. Pichersky. 2006. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 10310128-10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak, M. 1989. The scanning model for translation: an update. J. Cell Biol. 108229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, E. C., D. Yu, J. M. de Velasco, L. Tessarollo, D. A. Swing, D. L. Court, N. A. Jenkins, and N. G. Copeland. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 7356-65. [DOI] [PubMed] [Google Scholar]
- 22.Linney, E., and M. Hayashi. 1974. Intragenic regulation of the synthesis of phiX174 gene A proteins. Nature 249345-348. [DOI] [PubMed] [Google Scholar]
- 23.Mankin, S. L., D. S. Hill, P. M. Olhoft, E. Toren, A. R. Wenck, L. Nea, L. Xing, J. A. Brown, H. Fu, L. Ireland, H. Jia, H. Hillebrand, T. Jones, and H. S. Song. 2007. Disarming and sequencing of Agrobacterium rhizogenes strain K599 (NCPPB2659) plasmid pRi2659. In Vitro Cell. Dev. Biol. 43521-535. [Google Scholar]
- 24.McBride, K. E., and K. Summerfelt. 1990. Improved binary vectors for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 14269-276. [DOI] [PubMed] [Google Scholar]
- 25.Moriguchi, K., Y. Maeda, M. Satou, N. S. Hardayani, M. Kataoka, N. Tanaka, and K. Yoshida. 2001. The complete nucleotide sequence of a plant root-inducing (Ri) plasmid indicates its chimeric structure and evolutionary relationship between tumor-inducing (Ti) and symbiotic (Sym) plasmids in Rhizobiaceae. J. Mol. Biol. 307771-784. [DOI] [PubMed] [Google Scholar]
- 26.Morimatsu, K., and S. C. Kowalczykowski. 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: a universal step of recombinational repair. Mol. Cell 111337-1347. [DOI] [PubMed] [Google Scholar]
- 27.Mysore, K. S., B. Bassuner, X. Deng, N. S. Darbinian, A. Motchoulski, W. Ream, and S. B. Gelvin. 1998. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant-Microbe Interact. 11668-683. [DOI] [PubMed] [Google Scholar]
- 28.Nam, J., A. G. Matthysse, and S. B. Gelvin. 1997. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9317-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okamoto, S., A. Toyoda-Yamamoto, K. Ito, I. Takebe, and Y. Machida. 1991. Localization and orientation of the VirD4 protein of Agrobacterium tumefaciens in the cell membrane. Mol. Gen. Genet. 22824-32. [DOI] [PubMed] [Google Scholar]
- 30.Peralta, E. G., and L. W. Ream. 1985. T-DNA border sequences required for crown gall tumorigenesis. Proc. Natl. Acad. Sci. USA 825112-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ream, W., and K. G. Field. 1999. Molecular biology techniques: an intensive laboratory course. Academic Press, San Diego, CA.
- 32.Rossi, L., B. Hohn, and B. Tinland. 1996. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 93126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholz, P., V. Haring, B. Wittmann-Liebold, K. Ashman, M. Bagdasarian, and E. Scherzinger. 1989. Complete nucleotide sequence and gene organization of the broad-host-range plasmid RSF1010. Gene 75271-288. [DOI] [PubMed] [Google Scholar]
- 34.Shurvinton, C. E., L. Hodges, and W. Ream. 1992. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. USA 8911837-11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simone, M., C. A. McCullen, L. E. Stahl, and A. N. Binns. 2001. The carboxy-terminus of VirE2 from Agrobacterium tumefaciens is required for its transport to host cells by the virB-encoded type IV transport system. Mol. Microbiol. 411283-1293. [DOI] [PubMed] [Google Scholar]
- 36.Stachel, S. E., and E. W. Nester. 1986. The genetic and transcriptional organization of the vir region of the A6 Ti plasmid of Agrobacterium tumefaciens. EMBO J. 51445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stachel, S. E., B. Timmerman, and P. Zambryski. 1986. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature 322706-712. [Google Scholar]
- 38.Sundberg, C. D., and W. Ream. 1999. The Agrobacterium tumefaciens chaperone-like protein, VirE1, interacts with VirE2 at domains required for single-stranded DNA binding and cooperative interaction. J. Bacteriol. 1816850-6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tinland, B., Z. Koukolikova-Nicola, M. N. Hall, and B. Hohn. 1992. The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals. Proc. Natl. Acad. Sci. USA 897442-7446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vergunst, A. C., B. Schrammeijer, A. den Dulk-Ras, C. M. T. de Vlaam, T. J. G. Regensburg-Tuink, and P. J. J. Hooykaas. 2000. VirB/D4-dependent protein translocation from Agrobacterium into plant cells. Science 290979-982. [DOI] [PubMed] [Google Scholar]
- 41.Vergunst, A. C., M. C. M. van Lier, A. den Dulk-Ras, and P. J. J. Hooykaas. 2003. Recognition of the Agrobacterium tumefaciens VirE2 translocation signal by the VirB/D4 transport system does not require VirE1. Plant Physiol. 1331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vergunst, A. C., M. C. M. van Lier, A. den Dulk-Ras, T. A. G. Stuve, A. Ouwehand, and P. J. J. Hooykaas. 2005. Positive charge is an important feature of the C-terminal transport signal of the VirB/D4-translocated proteins of Agrobacterium. Proc. Natl. Acad. Sci. USA 102832-837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, K., L. Herrera-Estrella, M. Van Montagu, and P. Zambryski. 1984. Right 25 bp terminus sequence of the nopaline T-DNA is essential for and determines direction of DNA transfer from Agrobacterium to the plant genome. Cell 38455-462. [DOI] [PubMed] [Google Scholar]
- 44.Ward, E. R., and W. M. Barnes. 1988. VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242927-930. [Google Scholar]
- 45.White, F. F., B. H. Taylor, G. A. Huffman, M. P. Gordon, and E. W. Nester. 1985. Molecular and genetic analysis of the transferred DNA regions of the root-inducing plasmid of Agrobacterium rhizogenes. J. Bacteriol. 16433-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanofsky, M. F., S. G. Porter, C. Young, L. M. Albright, M. P. Gordon, and E. W. Nester. 1986. The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47471-477. [DOI] [PubMed] [Google Scholar]
- 47.Young, C., and E. W. Nester. 1988. Association of the VirD2 protein with the 5′ end of T strands in Agrobacterium tumefaciens. J. Bacteriol. 1703367-3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yusibov, V. M., T. R. Steck, V. Gupta, and S. B. Gelvin. 1994. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci. USA 912994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhou, X. R., and P. J. Christie. 1999. Mutagenesis of the Agrobacterium VirE2 single-stranded DNA-binding protein identifies regions required for self-association and interaction with VirE1 and a permissive site for hybrid protein construction. J. Bacteriol. 1814342-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu, J., P. M. Oger, B. Schrammeijer, P. J. J. Hooykaas, S. K. Farrand, and S. C. Winans. 2000. The bases of crown gall tumorigenesis. J. Bacteriol. 1823885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zupan, J. R., V. Citovsky, and P. Zambryski. 1996. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl. Acad. Sci. USA 932392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]