Abstract
The genome of Geobacter sulfurreducens contains three genes whose sequences are quite similar to sequences encoding known members of an RNA nucleotidyltransferase superfamily that includes tRNA nucleotidyltransferases and poly(A) polymerases. Reverse transcription-PCR using G. sulfurreducens total RNA demonstrated that the genes encoding these three proteins are transcribed. These genes, encoding proteins designated NTSFI, NTSFII, and NTSFIII, were cloned and overexpressed in Escherichia coli. The corresponding enzymes were purified and assayed biochemically, resulting in identification of NTSFI as a poly(A) polymerase, NTSFII as a C-adding tRNA nucleotidyltransferase, and NTSFIII as an A-adding tRNA nucleotidyltransferase. Analysis of G. sulfurreducens rRNAs and mRNAs revealed the presence of heteropolymeric RNA 3′ tails. This is the first characterization of a bacterial system that expresses separate C- and A-adding tRNA nucleotidyltransferases and a poly(A) polymerase.
RNA nucleotidyltransferases play an important role in the maturation, processing, and degradation of RNAs. tRNA nucleotidyltransferases (TNTs) and poly(A) polymerases (PAPs) are both members of a superfamily of nucleotidyltransferases (13, 27). TNTs add C and A residues to the 3′ ends of immature or damaged tRNA molecules and are found in bacteria, archaea, and eukaryotes (24). PAPs, which are present in both eukaryotes and bacteria, polyadenylate RNA 3′ ends (27). With regard to the distribution of RNA nucleotidyltransferases in bacteria, with only a few exceptions, the genomes of the alpha- and epsilonproteobacteria and of the low- and high-G+C-content gram-positive bacteria appear to encode only one member of this superfamily, presumably a CCA-adding TNT (1). Several of the so-called deeply branching bacterial species, including Aquifex aeolicus and Deinococcus radiodurans, and some cyanobacteria contain two RNA nucleotidyltransferases, which have been shown to function as separate C- and A-adding TNTs (22, 23). Interestingly, at least one low-G+C-content gram-positive species, Bacillus halodurans, has also been shown to contain separate C- and A-adding TNTs (1). The beta- and gammaproteobacteria, such as Escherichia coli, also contain two RNA nucleotidyltransferases, a CCA-adding TNT and a PAP, PAP I, that adds 3′ tails to the ends of RNA molecules in cells of these organisms (8, 11, 18, 19). mRNAs and 16S and 23S rRNAs are polyadenylated in E. coli and presumably in other beta- and gammaproteobacteria, and polyadenylation facilitates the exonucleolytic degradation of these RNAs by RNase II and polynucleotide phosphorylase (9, 16, 18, 19).
The genomes of several deltaproteobacteria also encode two members of the RNA nucleotidyltransferase superfamily (NTSF), and the genomes of a few deltaproteobacteria appear to encode three NTSF enzymes (1). In particular, the genomes of Geobacter sulfurreducens, Geobacter metallireducens, Pelobacter carbinolicus, and Desulfotalea psychrophila all contain three open reading frames with significant homology to open reading frames encoding other members of the superfamily of nucleotidyltransferases. We describe here biochemical characterization of the three NTSF enzymes of the dissimilatory metal- and sulfur-reducing bacterium G. sulfurreducens (6). This report is the first characterization of NTSF enzymes from a bacterial species with three RNA nucleotidyltransferases.
MATERIALS AND METHODS
Bacterial strains and genomic DNA.
G. sulfurreducens ATCC 51573 genomic DNA was obtained from the American Type Culture Collection. Frozen late-log-phase cells of G. sulfurreducens PCA were generously provided by Lori Hayes (laboratory of Derek Lovley) of the University of Massachusetts.
Sequences, alignment, and phylogenetic analysis.
Sequences were retrieved from the microbial genomes database at www.ncbi.nlm.nih.gov. Sequences were aligned using ClustalX, and neighbor-joining phylogenetic trees were constructed using PHYLIP 3.6b (12) as described previously (1) and in the supplemental material.
RT-PCR analysis.
Reverse transcription (RT)-PCR was carried out essentially as described by Bralley et al. (1). For studies of expression of the enzymes designated NTSFI, NTSFII, and NTSFIII, 5 μg of total RNA was reverse transcribed with random hexamer primers. PCR was then performed using primers specific for each of the NTSF enzymes using the Expand high-fidelity PCR system (Roche). PCR primers were designed so that they were complementary to nonhomologous regions of the three proteins to avoid detecting the transcript of one gene with more than one set of primers. The gene-specific primers were GeoBac I F1, GeoBac I R1, GeoBac II F1, GeoBac II R1, GeoBac III F1, and GeoBac III R1 (see Table S1 in the supplemental material).
Cloning and overexpression of the NTSF genes.
PCR primers corresponding to the 5′ and 3′ ends of the three genes were designed and used in the PCR as described previously (1). Primers were designed to incorporate BamHI sites at the 5′ and 3′ ends of each of the NTSF genes, and the PCR products were cloned into pET11A for overexpression in E. coli BL21(DE3)/pLysS Rosetta cells. Overexpressed proteins were purified as described in the supplemental material.
Preparation of tRNAAsp-C and tRNAAsp-CC.
Plasmid pmBsDCCA encodes E. coli tRNAAsp (15). Plasmid DNA (27 μg) was digested with either BbsI or FokI. The digested DNAs were isolated by phenol extraction and ethanol precipitation and used as templates for transcription with T7 RNA polymerase. The reaction mixtures (300 μl) contained 5× reaction buffer, 9 μg DNA template, 10 mM dithiothreitol, 300 U 5 PRIME Stop RNase inhibitor (Fisher), 2 mM ATP, 2 mM CTP, 2 mM GTP, 2 mM UTP, and 270 U T7 RNA polymerase (Promega). The reaction mixtures were incubated for 4 h at 37°C, brought to 1.5 mM with CaCl2, and RNase-free DNase (15 U; Promega) and additional RNase inhibitor (45 U) were added. Then the mixtures were incubated for 30 min at 37°C, DNase and RNase inhibitor were added again as described above, and the mixtures were incubated for 30 min. RNAs were collected by phenol extraction and two ethanol precipitations. Transcription of the BbsI-digested template produced tRNAAsp-CC, while transcription of the FokI-digested template produced tRNAAsp-C.
Enzyme assays.
Generally, PAP and TNT assays were performed using 60-μl reaction mixtures as described previously (1, 2, 5). The PAP assay reaction mixtures contained [α-32P]ATP, while the TNT assay mixtures contained [α-32P]ATP or [α-32P]CTP (3,000 Ci/mmol; GE Health Sciences). To examine the products of the TNT and PAP reactions, large-scale mixtures (60 to 240 μl) were incubated as described above and extracted with phenol-chloroform. Labeled RNAs were precipitated with ethanol. RNAs were examined by electrophoresis on 5 or 10% denaturing polyacrylamide gels along with Ambion Century-Plus size markers which had been end labeled with [32P]pCp and T4 RNA ligase. The amounts of enzyme (per 60-μl reaction mixture) used to generate the data shown in Fig. 3 were as follows: PAP I (New England Biolabs), 5 U; NTSFI, 5. 9 μg; NTSFII, 7.5 μg; NTSFIII, 7.1 μg; and SCO3896, 8.6 μg.
FIG. 3.
Autoradiogram of denaturing polyacrylamide gels of the reaction products from assays of E. coli PAP I (lane 3), G. sulfurreducens (G. sulf.) NTSFI (lane 4), S. coelicolor (S. coel.) SCO3896 with CTP and tRNAAsp-C as substrates (lane 5), SCO3896 with ATP and tRNAAsp-CC as substrates (lane 8), NTSFII with CTP and tRNAAsp-C as substrates (lane 6), NTSFII with ATP and tRNAAsp-CC as substrates (lane 7), NTSFIII with ATP and tRNAAsp-CC as substrates (lane 9), and NTSFIII with CTP and tRNAAsp-C as substrates (lane 10). Lanes 1 to 4 are lanes from a 5% polyacrylamide gel, and lanes 5 to 11 are lanes from a 10% polyacrylamide gel. Lanes 1 and 11 contained end-labeled size markers (STDS). For comparison, lane 2 shows the migration position of the end-labeled rpsO-pnp transcript, but it should be noted that unlabeled transcript was used in the polyadenylation assays. The rpsO-pnp transcript migrated more rapidly than expected, presumably due to the presence of some secondary structure in the transcript even in the presence of urea (20).
Determination of poly(A) tail lengths, cDNA cloning, and sequencing.
Poly(A) tails were isolated and characterized as described previously by end labeling total RNA with [32P]pCp and then digesting the RNA with RNases A and T1 (1, 4, 5). Products of this digestion were displayed on a 12% denaturing polyacrylamide gel. RT of G. sulfurreducens total RNA was carried out using primer AD20 containing an oligo(dT17) sequence that could anneal to poly(A) tails. For cDNA amplification by PCR, gene-specific forward primers and a reverse primer complementary to the primer used for RT were used. The sequences of the AD20 primers have been described previously (3). The gene-specific primers used in these experiments were 23SF, 16S1, NTSFI F3, and NTSFII F2 (see Table S1 in the supplemental material).
In an alternative method for cDNA preparation, a DNA adaptor, GSUENDR2 (see Table S1 in the supplemental material) was attached to the 3′ ends of G. sulfurreducens total RNA using RNA ligase. The reaction mixtures (20 μl) contained 25 μg of RNA, 0.4 nmol of adaptor, 10% dimethyl sulfoxide, 30 U of 5 PRIME Stop RNase inhibitor, 10 U of T4 RNA ligase (Ambion), and 5× RNA ligase buffer (Ambion). The reaction mixtures were incubated overnight at 4°C, and the products were purified by phenol extraction and ethanol precipitation. The RNA products were then reverse transcribed, and cDNA amplification was performed as described previously using GSURT (see Table S1 in the supplemental material) and the gene-specific primers indicated above (3). Amplification products were cloned using a TOPO-TA cloning kit (Invitrogen) and then sequenced.
RESULTS AND DISCUSSION
Phylogenetic analysis of the Geobacter NTSF protein sequences.
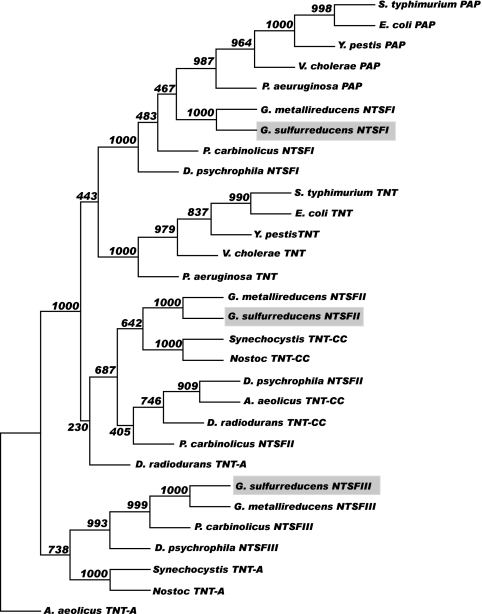
We initially subjected the three Geobacter NTSF protein sequences, arbitrarily designated NTSFI (annotated GSU3250 in the G. sulfurreducens genome database), NTSFII (GSU2184), and NTSFIII (GSU1581), to phylogenetic analysis and compared them with RNA nucleotidyltransferases from other bacteria to which functions have been assigned. Figure 1 shows an unrooted neighbor-joining phylogenetic tree constructed using PHYLIP (12). Representative NTSF enzymes from species with separate C- and A-adding TNTs belonging to the beta-and gammaproteobacteria and from other deltaproteobacteria with three NTSF genes were compared in this analysis. Protein sequences were aligned using ClustalX, and the alignments were refined using TuneClustal as described previously (1). The results of this analysis show that G. sulfurreducens NTSFI, along with the corresponding NTSF enzymes from the other deltaproteobacteria that were examined, occurs in a clade that contains the PAPs from the beta- and gammaproteobacteria. G. sulfurreducens NTSFII and the corresponding proteins from the other deltaproteobacteria occur in the clade that contains the C-adding enzymes from the species with separate C- and A-adding TNTs, while NTSFIII occurs in the clade containing the A-adding TNTs (Fig. 1).
FIG. 1.
Neighbor-joining phylogenetic tree for the bacterial NTSF enzymes. The tree was constructed using PHYLIP. Bootstrap scores are shown at the nodes. The branch lengths do not represent evolutionary distances. The species represented are E. coli, Salmonella enterica serovar Typhimurium, Vibrio cholerae, Yersinia pestis, Pseudomonas aeruginosa, D. radiodurans, A. aeolicus, Nostoc spp., Synechocystis spp., G. sulfurreducens, G. metallireducens, P. carbinolicus, and D. psychrophila.
G. sulfurreducens NTSF genes are transcribed.
We next determined whether the three NTSF genes were transcribed in G. sulfurreducens. RNA was isolated from frozen cells, and RT-PCR was performed using random primers for RT and primers specific for each NTSF enzyme for PCR. The primers utilized for PCR were expected to yield 1,160-, 1,183-, and 1,250-bp amplification products for NTSFI, NTSFII, and NTSFIII, respectively. An agarose gel of the PCR products (Fig. 2A) revealed bands of the expected size in each case and also showed that the PCR templates were generated by RT rather than by contaminating DNA in the RNA preparations. Thus, the genes encoding NTSFI, NTSFII, and NTSFIII are transcribed in G. sulfurreducens.
FIG. 2.
(A) RT-PCR analysis showing the expression of the genes encoding G. sulfurreducens NTSF enzymes. Lane 1, size standards (the arrow indicates the position of the 1-kb standard); lanes 2, 4, and 6, RT-PCR products obtained with primers designed to identify NTSFI, NTSFII, and NTSFIII, respectively; lanes 3, 5, and 7, PCR products obtained with the same primers but without RT. (B) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel of purified NTSFI (1 μg) (lane 2), NTSFII (2.5 μg) (lane 3), and NTSFIII (2.5 μg) (lane 4). Lane 1 contained size standards having molecular weights ranging from 20,000 to 106,000. The arrow indicates the position of the 50,000-Mr standard.
Cloning and overexpression of the G. sulfurreducens NTSF enzymes.
PCR products corresponding to the coding sequences of NTSFI, NTSFII, and NTSFIII were prepared using primers that incorporated BamHI sites at the ends of each gene (NTSFI, NTSFII, and NTSFIII forward and reverse primers [see Table S1 in the supplemental material]) and G. sulfurreducens chromosomal DNA as the template. The products were then cloned into the BamHI site of the overexpression vector pET11A, and the proteins were overexpressed in E. coli BL21(DE3)/pLysS Rosetta cells. We note that the overexpressed proteins contained a 13-amino-acid N-terminal tag upstream of each coding sequence, contributed by the cloning vector. These amino acids were derived from the sequence between the NdeI and BamHI sites in pET11A. Inclusion of this tag was necessary in order to express the proteins in E. coli. Attempts to express the NTSF enzymes from PCR products cloned into pET11A as NdeI/BamHI fragments lacking the upstream tag were unsuccessful. We speculate that the additional amino acids facilitated the translation of the mRNAs for the G. sulfurreducens NTSF enzymes by E. coli ribosomes. Overexpressed proteins were purified by a combination of anion-exchange chromatography and hydrophobic interaction chromatography (NTSFI and NTSFIII) or by ammonium sulfate fractionation, size exclusion chromatography, and hydrophobic interaction chromatography (NTSFII) as described in the supplemental material. Figure 2B shows that nearly homogeneous protein was obtained for each NTSF enzyme. The predicted molecular weights of the NTSF enzymes, based on the genomic sequences, are 50,633 for NTSFI, 45,586 for NTSFII, and 98,394 for NTSFIII.
Functional analysis of the G. sulfurreducens NTSF enzymes.
The phylogenetic analysis suggested that G. sulfurreducens NTSFI functioned as a PAP, NTSFII functioned as a C-adding TNT, and NTSFIII functioned as an A-adding TNT. To confirm these assignments, the activity of NTSFI was tested using reaction mixtures optimized for RNA polyadenylation with a model mRNA, the 5650 transcript derived from the rpsO-pnp operon of Streptomyces coelicolor, as a substrate (10). Positive control reaction mixtures for polyadenylation contained authentic E. coli PAP I. For the TNT reactions, an E. coli tRNAAsp lacking either the 3′-terminal CA residues or the terminal A residue and prepared by transcription of plasmid pmBsDCCA (15) as described in Materials and Methods was used as the substrate. NTSFII and NTSFIII were examined for C- and A-adding TNT activity, and the S. coelicolor SCO3896 gene product, which has been shown to be a TNT (2, 21), was used as a positive control for C and A addition to tRNA 3′ ends. Products from these reactions were separated by polyacrylamide gel electrophoresis on 5 or 10% denaturing gels, followed by autoradiography of the gels, and the results are shown in Fig. 3. Lane 2 shows the migration position of the rpsO-pnp transcript used in these experiments (note that the unlabeled transcript was used as a substrate in the PAP assays), lane 3 shows the products obtained by using the polyadenylation reaction mixture containing authentic E. coli PAP I, and lane 4 shows the products obtained by using the reaction mixture containing G. sulfurreducens NTSFI. Essentially identical patterns were observed in each case, showing the higher-molecular-weight products expected from addition of A residues to the 3′ end of the mRNA substrate. Lanes 5 and 8 show the products obtained by using TNT reaction mixtures containing SCO3896 with CTP and tRNAAsp-C and ATP and tRNAAsp-CC as substrates, respectively. A single band reflecting the addition of C or A to the tRNAAsp substrate was observed. Lanes 6 and 7 show the products obtained by using similar reaction mixtures containing G. sulfurreducens NTSFII. It is apparent that the product obtained by using the reaction mixture containing CTP and tRNAAsp-C as substrates (lane 6) migrated in the same fashion as the corresponding product obtained by using the reaction mixture containing SCO3896 (lane 5), while only a faint band was observed with ATP and tRNAAsp-CC (lane 7). Similarly, lane 9 shows that the product of the reaction catalyzed by NTSFIII with ATP as the substrate and tRNAAsp-CC had a mobility identical to that observed for the product obtained by using the reaction mixture containing SCO3896 (lane 8), while NTSFIII was inactive with CTP and tRNAAsp-C as substrates (lane 10). These results confirmed the identity of G. sulfurreducens NTSFII as a C-adding TNT and the identity of NTSFIII as an A-adding TNT. NTSFII and NTSFIII were inactive with the 5650 transcript as the substrate (data not shown).
Characterization of the G. sulfurreducens RNA 3′ tails.
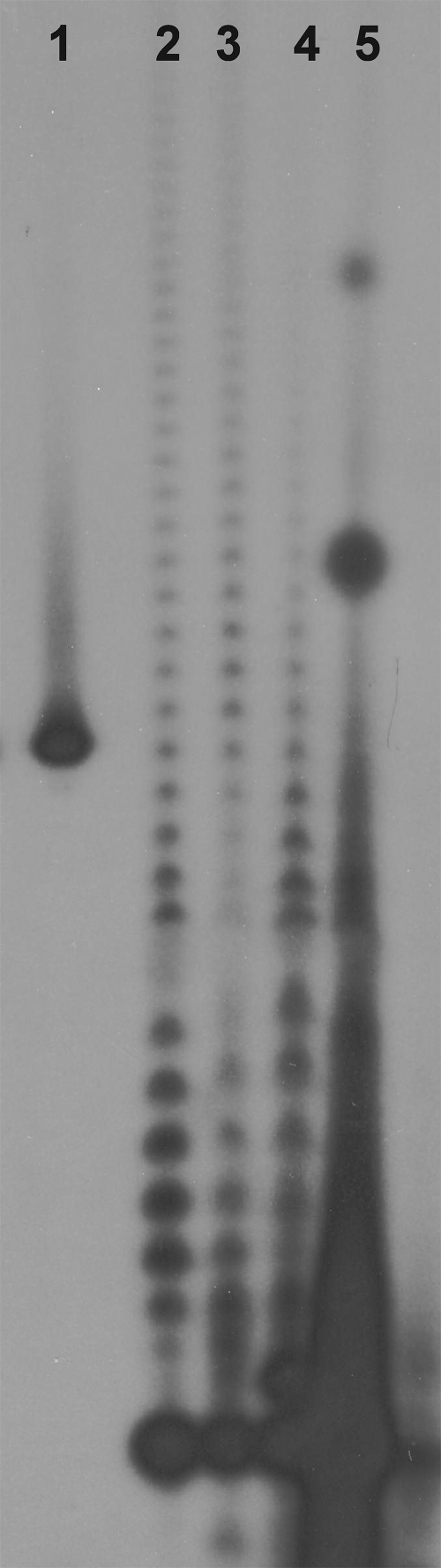
Given that G. sulfurreducens contains a functional PAP, it was of interest to confirm that G. sulfurreducens RNAs possess poly(A) tails. Total RNA from G. sulfurreducens cells was end labeled with [32P]pCp and digested with a combination of RNases A and T1 as described previously (1, 4, 5). This digestion procedure cleaves all phosphodiester bonds except those following A residues, leaving the poly(A) tails from the 3′ ends of the labeled RNA intact. Products of this digestion were displayed on a 12% denaturing polyacrylamide gel, as shown in Fig. 4. Lane 1 shows the migration of a size marker, oligo(dT18). Lane 2 shows the migration of tails associated with RNAs from E. coli, lane 3 shows the migration of tails from RNAs of Bacillus subtilis, lane 4 shows the migration of tails from B. halodurans RNA, and lane 5 shows the migration of tails from G. sulfurreducens RNA. Lane 5 contains prominent bands for tails that are ca. 20 and 30 residues long, but most of the poly(A) tails associated with the G. sulfurreducens RNAs are relatively short (15 residues or less). The pattern obtained for G. sulfurreducens is similar to that observed for organisms with heteropolymeric 3′ tails, including Streptomyces species (2, 4, 5).
FIG. 4.
Measurement of the lengths of oligonucleotide and poly(A) stretches found in the 3′ tails of RNAs from E. coli DH5α (lane 2), B. subtilis (lane 3), B. halodurans C-125 (lane 4). and G. sulfurreducens (lane 5). Lane 1 contained an oligo(dT18) size standard. Labeled 3′ tails were fractionated on a 12% denaturing polyacrylamide gel.
To characterize the G. sulfurreducens 3′ tails further, cDNA cloning was performed as described previously (1, 3, 7) and in Materials and Methods, using total RNA as the template for RT. PCR was performed using gene-specific primers designed from the sequences of the 16S and 23S rRNA genes and the genes encoding NTSFI and NTSFII (see Table S1 in the supplemental material). Results of this analysis are shown in Fig. 5. cDNA cloning revealed the presence of 3′ tails associated with decay intermediates (or premature transcription termination products) of 16S and 23S rRNAs and with intermediates from the mRNA populations corresponding to NTSFI and NTSFII. Tails of various lengths were observed for the rRNA and mRNA populations and ranged from a single residue attached to putative decay intermediates of 16S and 23S rRNAs to a 253-nucleotide tail attached at position 2618 of 23S rRNA. It is noteworthy that two 23S cDNAs and one 16S cDNA representing putative decay intermediates that did not possess a 3′ tail (designated “adaptor attachment site” [Fig. 5]) were isolated. Rather, these products terminated at presumed cleavage or processing sites in the transcripts in question and may have been isolated prior to posttranscriptional tail addition. Of particular interest is the observation that, as suggested by the results shown in Fig. 4, the 3′ tails associated with the Geobacter RNAs are heteropolymeric, containing G, C, and U residues as well as A residues. In E. coli, Streptomyces, some cyanobacteria, and spinach chloroplasts, polynucleotide phosphorylase is the enzyme responsible for the addition of G, C, and U residues to RNA 3′ tails (2, 14, 17, 21, 25). This may also be the case in G. sulfurreducens, which, like all other bacteria except mycoplasmas, contains a gene for polynucleotide phosphorylase. It should be noted that, at least in vitro, G. sulfurreducens NTSFI utilized CTP and UTP (but not GTP) as substrates in reactions performed under PAP assay conditions (data not shown). E. coli PAP I also utilizes other nucleoside triphosphates as substrates in vitro, although only A residues appear to be added to RNA 3′ ends in vivo (26). It is possible that, unlike the situation in E. coli, G. sulfurreducens NTSFI utilizes all three nucleoside triphosphates in vivo. This would explain our failure to observe tails composed of only A residues in the cDNA cloning experiments whose results are shown in Fig. 5. Alternatively, a combination of NTSFI and polynucleotide phosphorylase activities may be responsible for the synthesis of 3′ tails in G. sulfurreducens. It is also possible that examination of other G. sulfurreducens transcripts will reveal the presence of tails composed of only A residues.
FIG. 5.
Positions and compositions of G. sulfurreducens RNA 3′ tails. The position of each tail was determined from the mature 5′ end of rRNA gene or the translation start site of the NTSF enzyme. The number of the last nucleotide of the mature rRNA or translation stop codon is indicated at the top right. The figure is not drawn to scale.
In summary, we showed that unlike the alpha-, beta-, gamma- and epsilonproteobacteria, the low- and high-G+C-content gram-positive bacteria, and many other bacterial genera, at least some deltaproteobacteria encode separate C- and A-adding TNTs and a PAP. We demonstrated that the genes encoding all three of these enzymes are transcribed in G. sulfurreducens, and although we did not determine whether the transcripts were translated, we were able to detect all three enzymatic activities in crude G. sulfurreducens extracts (data not shown). We speculate that the CCA-adding TNT activity observed in the beta- and gammaproteobacteria evolved via mutations in the genes encoding either the C- or A-adding TNTs of an ancestral species and that subsequent mutations resulted in the conversion of the CCA-adding TNT to a PAP. The apparent absence of a PAP I-like activity in the so-called deeply branching bacterial species, such as Aquifex, Thermotoga, and Deinococcus species, and the cyanobacteria (1, 22, 23) is consistent with the hypothesis that polyadenylation of RNA 3′ tails by a PAP I-like enzyme evolved subsequent to the evolution of TNTs. The G. sulfurreducens PAP gene and the PAP I genes of the beta- and gammaproteobacteria may have arisen via independent evolutionary events. The G. sulfurreducens PAP gene and the E. coli pcnB (PAP I) gene are ca. 53% identical. It is also possible, therefore, that the G. sulfurreducens gene arose by horizontal gene transfer from a beta- or gammaproteobacterium.
Acknowledgments
We thank Lori Hayes and Derek Lovley for providing G. sulfurreducens cells and Norman Pace for providing pmBsDCCA.
This work was supported in part by a grant from the Emory University Research Committee.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bralley, P., S. A. Chang, and G. H. Jones. 2005. A phylogeny of bacterial nucleotidyltransferases: Bacillus halodurans contains two tRNA nucleotidyltransferases. J. Bacteriol. 1875927-5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bralley, P., B. Gust, S. A. Chang, K. F. Chater, and G. H. Jones. 2006. RNA 3′-tail synthesis in Streptomyces: in vitro and in vivo activities of RNase PH, the SCO3896 gene product and PNPase. Microbiology 152627-636. [DOI] [PubMed] [Google Scholar]
- 3.Bralley, P., and G. H. Jones. 2002. cDNA cloning confirms the polyadenylation of RNA decay intermediates in Streptomyces coelicolor. Microbiology 1481421-1425. [DOI] [PubMed] [Google Scholar]
- 4.Bralley, P., and G. H. Jones. 2003. Overexpression of the polynucleotide phosphorylase gene (pnp) of Streptomyces antibioticus affects mRNA stability and poly(A) tail length but not ppGpp levels. Microbiology 1492173-2182. [DOI] [PubMed] [Google Scholar]
- 5.Bralley, P., and G. H. Jones. 2001. Poly(A) polymerase activity and RNA polyadenylation in Streptomyces coelicolor A3(2). Mol. Microbiol. 401155-1164. [DOI] [PubMed] [Google Scholar]
- 6.Caccavo, F., Jr., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 603752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campos-Guillén, J., P. Bralley, G. H. Jones, D. H. Bechhofer, and G. Almeda-Alvarez. 2005. Addition of poly(A) and heteropolymeric 3′ ends in Bacillus subtilis wild-type and polynucleotide phosphorylase-deficient strains. J. Bacteriol. 1874698-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao, G. J., and N. Sarkar. 1992. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc. Natl. Acad. Sci. USA 8910380-10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpousis, A. J., N. F. Vanzo, and L. C. Raynal. 1999. mRNA degradation: a tale of poly(A) and multiprotein machines. Trends Genet. 1524-28. [DOI] [PubMed] [Google Scholar]
- 10.Chang, S. A., M. Cozad, G. A. Mackie, and G. H. Jones. 2008. Kinetics of polynucleotide phosphorylase: comparison of enzymes from Streptomyces and Escherichia coli and effects of nucleoside diphosphates. J. Bacteriol. 19098-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cudny, H., J. R. Lupski, G. N. Godson, and M. Deutscher. 1986. Cloning, sequencing, and species relatedness of the Escherichia coli cca gene encoding the enzyme tRNA nucleotidyltransferase. J. Biol. Chem. 2616444-6449. [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 13.Holm, L., and C. Sander. 1995. DNA polymerase beta belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 20345-347. [DOI] [PubMed] [Google Scholar]
- 14.Mohanty, B. K., and S. R. Kushner. 2000. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. USA 9711966-11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh, B. K., and N. R. Pace. 1994. Interaction of the 3′-end of tRNA with ribonuclease P RNA. Nucleic Acids Res. 224087-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23353-370. [DOI] [PubMed] [Google Scholar]
- 17.Rott, R., G. Zipor, V. Portnoy, V. Liveanu, and G. Schuster. 2003. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from E. coli. J. Biol. Chem. 27815771-15777. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar, N. 1996. Polyadenylation of mRNA in bacteria. Microbiology 1423125-3133. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar, N. 1997. Polyadenylation of mRNA in prokaryotes. Annu. Rev. Biochem. 66173-197. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki, N., M. Izawa, Y. Sugahara, T. Tanaka, M. Watahiki, K. Ozawa, E. Ohara, H. Funaki, Y. Yoneda, S. Matsuura, M. Muramatsu, Y. Okazaki, and Y. Hayashizaki. 1998. Identification of stable RNA hairpins causing band compression in transcriptional sequencing and their elimination by use of inosine triphosphate. Gene 22217-23. [DOI] [PubMed] [Google Scholar]
- 21.Sohlberg, B., J. Huang, and S. N. Cohen. 2003. The Streptomyces coelicolor polynucleotide phosphorylase homologue, and not the putative poly(A) polymerase, can polyadenylate RNA. J. Bacteriol. 1857273-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tomita, K., and A. M. Weiner. 2002. Closely related CC- and A-adding enzymes collaborate to construct and repair the 3′-terminal CCA of tRNA in Synechocystsis sp. and Deinococcus radiodurans. J. Biol. Chem. 27748192-48198. [DOI] [PubMed] [Google Scholar]
- 23.Tomita, K., and A. M. Weiner. 2001. Collaboration between CC- and A-adding enzymes to build and repair the 3′-terminal CCA of tRNA in Aquifex aeolicus. Science 2941334-1336. [DOI] [PubMed] [Google Scholar]
- 24.Weiner, A. M. 2004. tRNA maturation: RNA polymerization without a nucleic acid template. Curr. Biol. 14R883-R885. [DOI] [PubMed] [Google Scholar]
- 25.Yehudai-Resheff, S., M. Hirsh, and G. Schuster. 2001. Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 215408-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yehudai-Resheff, S., and G. Schuster. 2000. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 281139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yue, D., N. Maizels, and A. M. Weiner. 1996. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyl transferase superfamily:characterization of the hyperthermophile Sulfolobus shibatae. RNA 2895-908. [PMC free article] [PubMed] [Google Scholar]