Abstract
Although a great deal is known about the life cycle of bacteriophage P22, the mechanism of phage DNA transport into Salmonella is poorly understood. P22 DNA is initially ejected into the periplasmic space and subsequently transported into the host cytoplasm. Three phage-encoded proteins (gp16, gp20, and gp7) are coejected with the DNA. To test the hypothesis that one or more of these proteins mediate transport of the DNA across the cytoplasmic membrane, we purified gp16, gp20, and gp7 and analyzed their ability to associate with membranes and to facilitate DNA uptake into membrane vesicles in vitro. Membrane association experiments revealed that gp16 partitioned into the membrane fraction, while gp20 and gp7 remained in the soluble fraction. Moreover, the addition of gp16, but not gp7 or gp20, to liposomes preloaded with a fluorescent dye promoted release of the dye. Transport of 32P-labeled DNA into liposomes occurred only in the presence of gp16 and an artificially created membrane potential. Taken together, these results suggest that gp16 partitions into the cytoplasmic membrane and mediates the active transport of P22 DNA across the cytoplasmic membrane of Salmonella.
Phages T2 and T4 are commonly depicted in textbook images performing a “hypodermic syringe-like” mechanism to eject DNA from the phage into Escherichia coli (13, 14). This led to the idea that phage DNA is directly injected into the host cytoplasm by the contraction of the phage tail and driven by the release of pressure within the phage head (13). More recent evidence provided convincing evidence that this is not a general mechanism of phage DNA transport into the bacterial host (12, 32). The pressure inside the phage capsid due to DNA compression (7, 29) may promote the initial ejection of phage into the bacterial host, but the osmotic pressure within the bacterial cytoplasm exerts an opposing force that prevents complete transfer of phage DNA in vivo (13). For example, the initial 850 bp of phage T7 DNA enters the cell rapidly, but the remainder of the T7 genome is pulled into E. coli by RNA polymerase (9, 10, 13).
Phage P22 is a temperate, icosahedral, “lambdoid” bacteriophage that is commonly used for generalized transduction in Salmonella. P22 has a short, noncontractile tail that cannot penetrate both the outer and inner membranes of its host. The P22 gene 9 protein forms the hexameric tail spike that specifically recognizes the Salmonella O antigen. After reversible binding to the O antigen, the endorhamnosidase activity of the tail spike proteins cleaves the O-antigen subunits of the lipopolysaccharide until the proteins recognize an uncharacterized secondary receptor on the outer membrane of the host bacterium. Binding to the secondary receptor triggers release of the phage DNA, together with the phage-encoded ejection proteins (gp7, gp16, and gp20), into the periplasmic space of the host (17).
The phage-encoded ejection proteins are essential for the viability of the phage. One of the proposed functions of the ejection proteins is to protect the phage DNA from degradation by nucleases in the periplasmic space. In addition to protecting the DNA from degradation, the phage-encoded ejection proteins may target the DNA to the cytoplasmic membrane and facilitate transport into the cytoplasm (2, 17, 36).
As an initial step in transport of the phage DNA into the cytoplasm, the ejection proteins could target the phage DNA to the inner membrane of the host by binding to both the phage DNA and the cytoplasmic membrane. If the ejection proteins direct the translocation of the phage DNA across the cytoplasmic membrane of Salmonella, at least one of these proteins would be expected to interact with the lipid or a protein within the cytoplasmic membrane. Bioinformatic approaches have been used to determine if the three P22 ejection proteins have characteristics expected of membrane-associated proteins. The hydropathy plot of gp7 led Conlin et al. (5) to propose that the basic N terminus of gp7 might interact with DNA, while the hydrophobic C terminus might interact with the cytoplasmic membrane. In contrast, the sequence of gp20 is not homologous with any entries in the database, and the hydropathy plot does not reveal insights that suggest function (1). Umlauf and Dreiseikelmann reported an absence of bioinformatic evidence for α-helical membrane-spanning domains of gp16 (37). However, other reports suggest that gp16 may polymerize in vivo (36) and thus may associate with the membrane as amphipathic α-helices or multimeric β-strand pores like those of the porins (11).
This study provides direct evidence that gp16 associates with membrane lipids and mediates the transport of DNA by using the energy of the electrochemical gradient. The results suggest that P22-encoded proteins are necessary and sufficient for the active transport of DNA from the periplasmic space into the cytoplasm of the bacterial host.
MATERIALS AND METHODS
Plasmids and strains.
The pET46 enterokinase/ligation-independent cloning vector, NovaBlue cells, and E. coli BL21 expression host cells were purchased from Novagen (San Diego, CA). Plasmids with the appropriate inserts were maintained in NovaBlue E. coli cells {endA1 hsdR17 (rK-12− mK-12+) supE44 thi-1 recA1 gyrA96 relA1 lac F′ [proA+B+ lacIqlacZΔM15::Tn10] (Tetr)}.
Chemicals.
1,2-Dilauryl-sn-glycerol-3-phosphatidylcholine was obtained from Calbiochem. 3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), isopropyl-β-d-thiogalactopyranoside (IPTG), lysozyme, sucrose, Triton X-114, chloroform, ether, proteinase K, potassium chloride, and Triton X-100 were purchased from Fisher Scientific (Pittsburgh, PA). Calcein was purchased from Sigma (St. Louis, MO). Rabbit anti-six-His antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Horseradish peroxidase-conjugated goat anti-rabbit secondary antibody was purchased from Jackson ImmunoResearch (West Grove, PA). DNase I was purchased from Fermentas (Glen Burnie, MD). His-binding affinity column purification kits, enterokinase, and Benzonase were purchased from Novagen (San Diego, CA). The bicinchoninic acid protein assay was used to determine protein concentrations (Pierce Biotechnology, Inc., Rockford, IL).
Vector construction.
DNA sequences of genes coding for the P22 ejection proteins (gp7, gp16, and gp20) were obtained from GenBank. The P22 DNA template was extracted from concentrated P22 lysate as previously described (23). Primer sequences used were as follows: gene 7 forward, 5′-GACGACGACAAGATGAAAGGCGGTAAAGGTGGCGCAGATAAAAGC-3′; gene 7 reverse, 5′-GAGGAGAAGCCCGGTTTAAAACAACGAGCCAAGCAGACCAATACC-3′; gene 16 forward, 5′-GACGACGACAAGATGAAAGTTACCGCTAATGGCAAGACATTC-3′; gene 16 reverse, 5′-GAGGAGAAGCCCGGTCTACTGCCGGGTAGCTTCGTTAGCTAAAAG-3′; gene 20 forward, 5′-GACGACGACAAGATGGCTACGTGGCAGCAGGGCATTAATTCAGGT-3′; and gene 20 reverse, 5′-GAGGAGAAGCCCGGTTTATTCCACCGTGAATTTAATGCCAGATTT-3′. Truncated constructs of gp16 were constructed by amplifying shorter DNA derivatives of gene 16, using the same forward primer as that shown above, but with a different reverse primer for each construct. Reverse primers used were as follows: gp16Δ(531-609), 5′-GAGGAGAAGCCCGGTCTATCCCACTGGAGCGCTTCCCATTCTATT-3′; gp16Δ(476-609), 5′-GAGGAGAAGCCCGGTCTAACCAGTTTTTTCAGCAATCTTGCTGAT-3′; and gp16Δ(301-609), 5′-GAGGAGAAGCCCGGTCTACTGTGTGACAACATTTGCCGAATCAGA-3′.
The amplified sequences were cloned into the pET46 enterokinase/ligation-independent cloning expression vector from Novagen, placing the six-His tag in the N terminus of the expressed protein. The presence of the appropriate inserts was confirmed by PCR, restriction digests, and DNA sequencing. Expression vectors with the correct inserts were electroporated into the expression host cell line BL21 from Novagen and then plated on LB agar plates with 100 μg/ml ampicillin.
Protein expression and purification.
BL21 cells with each plasmid clone were inoculated into 100 ml of LB broth with 100 μg/ml ampicillin and incubated in a 37°C shaker until mid-exponential-phase growth. IPTG was added to a final concentration of 1 mM to induce protein expression, and the cultures were reincubated for an additional 3 h in a 30°C shaker. The cells were then centrifuged at 3,842 × g for 30 min at 4°C. The supernatant was removed, and the pellet was resuspended in 4 ml of lysis buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 10 mM imidazole, 10 mM CHAPS). Lysozyme was added to 0.5 mg/ml, and 100 units of Benzonase was also added to reduce the viscosity of the lysate. The lysate was incubated on ice for 30 min with occasional swirling and then sonicated three times using a Fisher Scientific model 100 ultrasonic dismembrator. The sonicated lysate was centrifuged at 11,000 × g for 30 min at 4°C. The supernatant was added to a column containing a 200-μl bed volume of nickel-charged His-binding resin, and samples were eluted at a flow rate of ca. 1 ml/min. The resin was washed three times with 1× binding buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 10 mM CHAPS, 20 mM imidazole) and twice with 1× wash buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 10 mM CHAPS, 60 mM imidazole) before the six-His-tagged ejection protein was eluted three times with 200 μl (each time) of 1× elution buffer (50 mM NaH2PO4, pH 8, 300 mM NaCl, 10 mM CHAPS, 1 M imidazole). The 1 M imidazole was removed from the eluted ejection protein by using a Centricon filter with a molecular mass cutoff of 5 kDa and by washing the ejection protein with 20 mM Tris buffer, pH 8-100 mM NaCl. The purified proteins were treated with enterokinase to remove the N-terminal six-His tag prior to using the purified proteins in the membrane disruption and DNA transport assays.
Membrane partitioning of ejection proteins.
The preparation of the Salmonella membrane extracts and the membrane partitioning assay were adapted from the work of Muro-Pastor et al. (26). Briefly, cells were grown to log phase in 500 ml of LB broth and then centrifuged for 30 min at 4°C and 3,800 × g, using a Sorvall GSA rotor. The pellet was resuspended in 20 ml of 0.1 M cacodylic buffer, pH 6.8, and then lysed twice using a French pressure cell at 12,000 lb/in2. The lysate was centrifuged for 10 min at 4°C and 7,800 × g, using a Sorvall SS-34 rotor, to pellet the cell debris. The supernatant was centrifuged at 228,000 × g for 3 h, using a Beckman Vti 65.2 class H rotor, to isolate the membrane fraction. The pellet containing the membrane vesicles was resuspended in 3 ml of 0.1 M cacodylic buffer with 5% glycerol. The membrane extract was passed through a 23-gauge syringe needle at least five times prior to performing the assay. A final concentration of 4 μM of the nickel-purified ejection proteins was mixed with 125 μl of the membrane extract in a total volume of 500 μl. MgCl2 was added to a final concentration of 10 mM. The mixture was separated by two-step sucrose gradient ultracentrifugation (25% and 65% sucrose layers) at 96,000 × g for 3 h, using a Ti44 rotor. After centrifugation, 20 μl of the soluble fraction found on top of the 25% layer and 20 μl of the membrane fraction found between the two sucrose layers were loaded on a 10.5% sodium dodecyl sulfate (SDS)-polyacrylamide gel. The proteins in the gel were transferred to a polyvinylidene difluoride (PVDF) membrane and probed with the rabbit anti-six-His antibody.
Western blotting.
The rabbit anti-six-His antibody was used at a 1:1,500 dilution. A goat anti-rabbit secondary antibody conjugated with horseradish peroxidase was used at a 1:10,000 dilution (31).
Triton X-114 phase separation of ejection proteins.
Triton X-114 phase separation of the ejection proteins was performed as described by Muro-Pastor et al. (26).
Liposome reconstitution.
Liposomes with large internal aqueous space or liposomes with encapsulated calcein fluorescent dye were created by reverse-phase evaporation, using the procedure described by Szoka and Papahadjopoulos (35). All preparations contained 66 μmol of total lipid per ml of aqueous phase. A thin film of phospholipid was formed on the wall of a round-bottomed flask by rotary evaporation of the chloroform at 42°C. The thin film of phospholipid was resuspended in 3 ml of diethyl ether, and then 1 ml of 20 mM Tris (pH 7.4)-100 mM NaCl was added to the phospholipid suspension to serve as the aqueous phase encapsulated within the liposomes. When encapsulation of fluorescent dye was desired, 1 ml of 40 mM calcein was added to the aqueous phase. Rotary evaporation of the diethyl ether allowed the thin layer of phospholipid to gradually form liposomes around the aqueous layer. Rotary evaporation was performed for approximately 2 h at room temperature until all traces of ether had been removed from the sample. The liposomes were then passed through a Sepharose 4B column to remove nonencapsulated material and residual organic solvent. To confirm that the liposomes were intact, an aliquot of the reconstituted liposome fraction was negatively stained and examined under a transmission electron microscope.
Liposomal partitioning assay.
A final concentration of 0.15 μM of the nickel-purified ejection proteins was mixed with 66 mM liposomes in a total volume of 150 μl. The mixture of liposomes and ejection proteins was rocked at room temperature for 30 min prior to ultracentrifugation at 227,640 × g for 40 min in a TLA-100.1 rotor. The supernatant was transferred to a microcentrifuge tube, and the pelleted liposomes were resuspended in 150 μl of 20 mM Tris, pH 7.4-100 mM NaCl. The soluble and liposomal fractions (20 μl of each) were heated for 5 min at 99°C in 4 μl of 6× SDS gel sample buffer and then loaded on a 12% SDS-polyacrylamide gel (31). The proteins were transferred to a PVDF membrane, and the membrane was probed with rabbit anti-six-His antibody to determine where the proteins partitioned.
Membrane leakage assay.
The ability of the ejection proteins to disrupt membranes was assayed as described by Zhu et al. (38) and Galloux et al. (8). Briefly, calcein was encapsulated inside liposomes. Molecular crowding inside liposomes inhibits calcein fluorescence, but fluorescence is enhanced upon release from the liposomes. A final concentration of 100 nM of the ejection proteins was added to 0.66 mM liposomes in a total volume of 100 μl. The samples were excited at 490 nm, and the relative fluorescence of each sample was followed for 3 min at an emission wavelength of 510 nm. Total dye release was completed by the addition of 20 μl of 1% Triton X-100. Background fluorescence from the liposomes alone was subtracted from the fluorescence reading of each sample. Bovine serum albumin (BSA) was used as the negative control, and ethanol was used as the positive control, resulting in complete disruption of the liposomes (38). The percent relative fluorescence for each sample was calculated as follows: percent relative fluorescence = (FP − FB) × 100/(FT − FB), where FP is the fluorescence intensity of the dye released by the protein, FT is the fluorescence intensity of the total dye released, and FB is the background fluorescence intensity.
DNA transport assay.
The transport of DNA across membranes was studied in vitro by following the uptake of 32P-labeled bla DNA into liposomes. DNA from the pUC19 bla gene was PCR amplified using the following forward and reverse primers: 5′-GACGACGACAAGATGAGTATTCAACATTTCCGTGTCGCCCTTATTCCC-3′ and 5′-GAGGAGAAGCCCGGTTTACCAATGCTTAATCAGTGAGGCACCTATCTC-3′. The PCR product was 5′ end labeled using T4 polynucleotide kinase (New England Biolabs) and 20 μCi of 6,000-Ci/mmol [γ-32P]ATP (30). A final concentration of 100 nM of ejection proteins was added to 1 nM of the radiolabeled DNA. Controls lacking ejection proteins were performed concurrently. The samples were incubated at 4°C for 1 h to allow the ejection proteins to potentially bind the DNA before adding 20 μl of 66 mM liposome to each sample. The samples were incubated at room temperature for at least 1 h, and then 1 unit of DNase I was added to each sample. The samples were incubated at 30°C for 1 h to degrade any DNA molecules that were not transported inside the liposomes, and then 2 μl of 25 mM EDTA was added to stop the DNase I reaction. The liposomes were centrifuged at 8,000 × g for 2 min and then washed with 20 mM Tris, pH 7.4. The pelleted liposomes were resuspended in 20 mM Tris buffer containing 60 μg of proteinase K and 2% Triton X-100. Samples were separated in a 5% native polyacrylamide gel, and the gel was vacuum dried for phosphorimaging.
Artificial membrane potential across liposomal membrane.
Liposomes (66 μmol of total lipid) were reconstituted in 1 ml of buffer solution containing 100 mM KCl, 20 mM Tris, pH 7. The buffer outside the liposomes was changed to 20 mM KCl, 20 mM Tris, pH 7, by carefully pouring the liposomes into a Bio-Rad Poly-Prep chromatography column (Hercules, CA) and equilibrating the column with a buffer containing 20 mM KCl. The column was capped, and the liposomes were resuspended in 1 ml of 20 mM KCl, 20 mM Tris, pH 7. Prior to performing the DNA transport assay, 1 μl of 100 mM valinomycin was added to the liposomes to equilibrate the potassium ion concentration inside and outside the liposomes. This created a net negative charge inside the liposomes, with approximately 100 mM Cl− in the interior and 20 mM Cl− in the exterior of the liposomes.
RESULTS
Do the P22 ejection proteins associate with the membrane?
If the ejection proteins facilitate transport of DNA across the cytoplasmic membrane, at least one of the ejection proteins must associate with the Salmonella cytoplasmic membrane. To test whether the ejection proteins alone or in combination partition into the cytoplasmic membrane, a membrane partitioning experiment was conducted. Purified ejection proteins or combinations of the proteins were added to membrane extracts purified from Salmonella serovar Typhimurium. The ejection proteins were allowed to equilibrate between the soluble and membrane fractions at room temperature for 30 min, and then the samples were separated in a sucrose step gradient. The material on top of the 25% sucrose contained the soluble fraction, and the material between the two layers contained the membrane fraction (26). The soluble and membrane fractions were separated in a 10.5% SDS-polyacrylamide gel, and the proteins were transferred to a PVDF membrane for Western blotting with rabbit anti-six-His. Both six-His-gp16 and the mixture of all three ejection proteins showed more six-His-gp16 protein in the membrane fraction than in the soluble fraction, indicating that six-His-gp16 partitions into the membrane fraction (Fig. 1A and B). In contrast, six-His-gp20 and six-His-gp7 remained primarily in the soluble fraction.
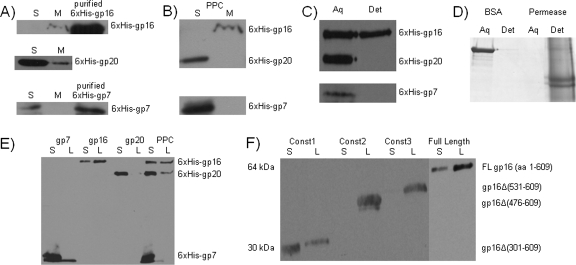
FIG. 1.
Membrane partitioning of ejection proteins. Membrane partitioning of individual ejection proteins (A) and the preformed protein complex (B) in crude membrane extract from Salmonella serovar Typhimurium. Proteins were detected by Western blotting with the rabbit anti-six-His antibody after transfer of the proteins to a PVDF membrane. S, soluble fraction; M, membrane fraction; PPC, preformed protein complex. (C) Partitioning of the preformed protein complex in Triton X-114 was detected by Western blotting with the rabbit anti-six-His antibody after transfer of the proteins to a PVDF membrane. Aq, aqueous phase; Det, detergent phase. (D) Controls demonstrating partitioning of BSA into the aqueous phase and the partitioning of the E. coli lactose permease (LacY) into the detergent phase. Proteins were detected by staining with Coomassie brilliant blue. (E) Liposomal partitioning of the ejection proteins. Proteins were detected by probing the PVDF membrane with the rabbit anti-six-His antibody. S, soluble fraction; L, liposomal fraction. (F) Domain of gp16 responsible for membrane association. Truncated constructs of gp16 were cloned and expressed in the E. coli expression host and purified using a nickel column. Each construct was subjected to the liposomal partitioning assay to determine if the construct would partition into the liposomal fraction or remain in the soluble fraction. The different constructs were detected on the PVDF membrane by using rabbit anti-six-His antibody.
The observed partitioning could be due to interactions of gp16 with integral membrane proteins or to hydrophobic interactions with membrane lipids. To distinguish between these possibilities, Triton X-114 was used in lieu of the membrane extract from Salmonella. Proteins mixed with Triton X-114 at 0°C form a monophasic solution. However, when this solution is incubated at 30°C, it separates into two phases, with an upper, aqueous phase and a lower, hydrophobic phase (3, 6, 28). Partitioning into the hydrophobic phase is characteristic of proteins that interact with membrane lipids. The purified gp16 partitioned into the hydrophobic Triton X-114 phase, while gp20 and gp7 partitioned into the aqueous phase (Fig. 1C). BSA was used as a control to confirm that soluble proteins partitioned into the aqueous phase, and the E. coli lactose permease (LacY) was used as a control to confirm that membrane proteins partitioned into the hydrophobic phase of Triton X-114 (Fig. 1D).
The membrane partitioning behavior of the ejection proteins was also confirmed by assaying their association with reconstituted liposomes. The liposomal partitioning assay confirmed the results obtained from the membrane partitioning assay and the partitioning of the ejection proteins in Triton X-114. gp16 was again found in the liposomal fraction, while gp20 and gp7 were found primarily in the soluble fraction (Fig. 1E). The overall behavior of the individual proteins in the liposomal partitioning assay was very similar to their behavior as part of the combined proteins, except that when the combined proteins were assayed, more gp16 protein was found in the soluble fraction and more gp20 was found in the liposomal fraction, possibly due to protein-protein interactions.
To determine the domain of gp16 required for membrane association, truncated constructs of gp16 were cloned into pET46 and expressed in E. coli BL21. All of the constructs of gp16 were truncated at the C-terminal end. The purified full-length (609 amino acids) and truncated constructs of gp16 were assayed for liposomal partitioning. gp16 derivatives truncated from residues 476 to 609 partitioned into the liposomal fraction (Fig. 1F), implying that the C terminus of gp16 is not required for membrane association. In contrast, derivatives that were truncated from residues 301 to 609 partitioned in the soluble fraction instead of the liposomal fraction, suggesting that the domain between amino acids 301 and 475 is critical for proper folding or membrane association of gp16.
Do the ejection proteins affect membrane integrity?
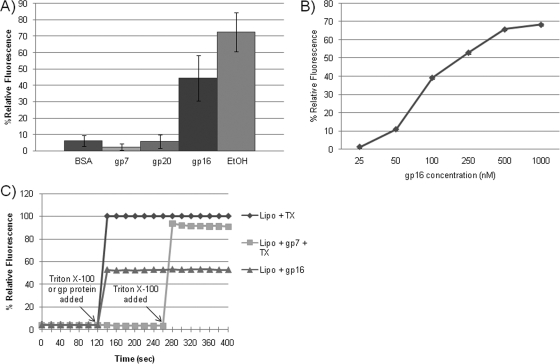
If the ejection proteins form membrane channels, then the proteins may disrupt liposomes. To test this possibility, we used liposomes containing the dye calcein. Molecular crowding inside liposomes limits the fluorescence of calcein, and thus, release of the dye from the liposomes results in increased fluorescence. Of the three ejection proteins, only gp16 promoted release of the fluorescent dye from inside the liposomes (Fig. 2A). The increase in fluorescence was concentration dependent (Fig. 2B) and was observed within seconds after the addition of gp16 to the liposomes (Fig. 2C). The addition of gp16 proteins did not lyse the liposomes, as evidenced by electron micrographs showing intact liposomes (data not shown). This suggests that gp16 forms membrane channels to allow the release of calcein from within the liposomes.
FIG. 2.
Membrane-disrupting activity of the ejection proteins. Samples were excited at 490 nm and were read at an emission wavelength of 510 nm. Ethanol was used as the positive control, inducing almost complete lysis of the liposomes, and BSA was used as the negative control. The percent relative fluorescence was determined by calculating the ratio of the relative fluorescence values of the different samples to the relative fluorescence exhibited with 0.2% Triton X-100. (A) The ejection proteins, BSA, and ethanol were added to liposomes with encapsulated fluorescent dye to determine their membrane-disrupting activity. The bar graph depicts the percent relative fluorescence at the 2-min time point. (B) Increasing concentrations of the gp16 protein were added to liposomes with encapsulated calcein dye to determine if dye leakage is concentration dependent. The graph reflects the percent relative fluorescence 2 min after the protein was added. (C) Triton X-100, gp16, and gp7 were added to the liposomes after 2 min of background fluorescence reading. After another 2 min, Triton X-100 was added to the liposomes with gp7 to demonstrate that the fluorescent dye was still inside the liposomes.
Do ejection proteins facilitate DNA transport?
The ability of the ejection proteins to facilitate DNA transport across membranes was investigated by following the uptake of 32P-labeled DNA into liposomes. After allowing time for DNA uptake, DNase I was added to degrade any extraliposomal DNA, allowing quantitation of DNA uptake into the liposomes. The added DNA was completely degraded by DNase I within 1 h at 30°C in the presence of 10 nM ejection proteins without membranes or in the presence of membranes without ejection proteins (data not shown).
No transport of DNA was observed in the absence of ejection proteins. Transport of radiolabeled DNA was optimal when gp16 was added (Fig. 3). Moreover, transport of the radiolabeled DNA occurred only in the presence of a membrane potential across the liposomal membrane. This suggests that the transport of phage P22 DNA across the cytoplasmic membrane of Salmonella is dependent on gp16 and the membrane potential of the host cell.
FIG. 3.
Transport of DNA inside liposomes is gp16 and membrane potential dependent. Ejection proteins were incubated with radiolabeled DNA prior to adding liposomes to the mixture. After 1 h at room temperature, DNase I was added to digest all untransported DNA. Liposomes were disrupted with Triton X-100 plus proteinase K, and samples were run in a 5% native polyacrylamide gel. The gel was vacuum dried and visualized with a phosphorimager. The artificial membrane potential was created by maintaining 100 mM KCl inside and 20 mM KCl outside the liposomes. The addition of valinomycin equilibrates the [K+] inside and outside the liposomes, generating a net negative charge inside the liposomes. Radiolabeled DNA was loaded in the first lane to mark the mobility of the DNA.
DISCUSSION
The process of phage infection requires translocation of the phage DNA across the cytoplasmic membrane of the host cell. For most phages, the mechanism whereby phage DNA is transported across the inner membrane of the bacterial host has remained elusive. The process of phage DNA translocation into the host cytoplasm may occur by different mechanisms in different types of phages. Susskind et al. (34) provided convincing evidence that P22 DNA is initially released into the periplasmic space and subsequently transported into the host cytoplasm. Our results indicate that for phage P22, the phage-encoded ejection protein gp16 is sufficient to catalyze the transport of DNA across the membrane in vitro. The membrane potential across the liposomal membrane is required to drive DNA transport into the liposome. Since DNA is a negatively charged molecule and the inside of the liposomes is also negatively charged, we expect that the DNA is associated with counterions upon transport into the liposomes.
P22 gp16 partitions into hydrophobic membranes. Deletion analysis indicated that a central segment of gp16 (amino acids 301 to 475) is critical for membrane association. Bioinformatic analysis reveals that this region has two potential amphipathic α-helices long enough to span the cytoplasmic membrane. Thomas and Prevelige (36) reported that gp16 self-polymerizes into large structures in a concentration-dependent manner. Hence, the amphipathic α-helices from multiple gp16 proteins may interact to form a channel to transport a large, hydrophilic DNA molecule.
The liposomal transport assay revealed that DNA transport by gp16 requires a membrane potential across the liposomal bilayer. In support of these in vitro results, uptake of P22 DNA in vivo is dependent on the membrane potential of the host (G. L. Perez and S. Maloy, submitted for publication). This agrees with previous studies showing that phage T4 and T7 infection in E. coli requires a minimum threshold membrane potential (19-21).
Although the addition of gp16 is necessary and sufficient for DNA transport in vitro, the other ejection proteins are essential during P22 infection (4, 17, 27). Previous studies indicate that gp7 and gp20 may be required to protect the phage DNA in the periplasmic space. In addition, they may be required during the recircularization of the phage DNA once the linear double-stranded DNA has entered the host cytoplasm (2).
With only a few exceptions, the mechanism of translocation of phage DNA into the bacterial host is poorly understood (18, 19, 22, 32, 33). However, it is clear that proteins coejected with DNA also facilitate the transport of DNA from other phage across the cytoplasmic membrane. The uptake of DNA from phage T7 is the best-understood example. Like that of P22, the T7 tail is too short to span the E. coli cell envelope, so a membrane channel is required to allow the phage DNA to enter the cytoplasm. Molineux et al. (18, 24, 25) proposed that virion proteins ejected into the cell functionally endow T7 with an extensible tail. The proteins ejected from T7 catalyze the localized degradation of the cell wall (24, 25), form a channel across the cell envelope, and facilitate the translocation of the leading end of the phage genome in an enzyme-catalyzed reaction (9, 10, 19, 33). Entry of the first 850 bp of T7 DNA is controlled by a phage-encoded protein ejected from the phage head at the time of infection (10, 19, 33). Elegant experiments by Molineux and colleagues demonstrated that the transport of phage T7 DNA across the cytoplasmic membrane of E. coli is enzyme driven, not dependent on the pressure within the phage capsid (26). After the initial 850 bp of T7 DNA enter the host cell, T7 DNA is pulled into the E. coli cytoplasm by RNA polymerases in a transcription-dependent manner.
Although phage-encoded proteins are required for transport of both P22 and T7 DNAs, the mechanisms are quite different. In contrast to the case for T7, the ejected proteins from P22 probably do not form an extensible tail. P22 gp16 can function in trans when Salmonella is coinfected with a gp16-negative P22 particle and a UV-inactivated P22 particle (15, 16). The difference in mechanism between P22 and T7 may be a consequence of differences in DNA packaging of these two phages. The transcription-dependent mechanism used by phage T7 relies on RNA polymerase binding sites located in the initial 850 bp of the T7 genome that enters the host cell. However, P22 DNA is inserted into the phage head by a headful packaging mechanism, and thus, the packaged DNA is circularly permuted. P22 is also an effective generalized transducing phage that can incorporate random fragments of the Salmonella host genome by a headful packaging mechanism. Because the initial ends of the DNA fragments have different RNA polymerase binding sites and because transport into liposomes can occur in vitro without additional enzymes, it is unlikely that P22 DNA is pulled into the host by a mechanism like that of T7.
In summary, these studies show that the P22-encoded protein gp16 and the host membrane potential are necessary and sufficient for DNA translocation across the membrane in vitro.
Acknowledgments
This work was supported by NIH/NIGMS MBRS grant 1R25GM8906-07 and by the generous support of the ARCS Foundation and a Biosite Fellowship.
We thank Anca Segall for advice and supplies and Keith Wright, who generously provided the purified E. coli lactose permease. Ian Booth provided valuable comments and suggestions on this work.
Footnotes
Published ahead of print on 31 October 2008.
REFERENCES
- 1.Adhikari, P., and P. B. Berget. 1993. Sequence of a DNA injection gene from Salmonella Typhimurium phage P22. Nucleic Acids Res. 211499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, N. R., and J. Roth. 1997. A Salmonella phage P22 mutant defective in abortive transduction. Genetics 14517-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bordier, C. 1981. Phase-separation of integral membrane proteins in Triton X-114 solution. J. Biol. Chem. 2561604-1607. [PubMed] [Google Scholar]
- 4.Botstein, D., C. H. Waddell, and J. King. 1973. Mechanism of head assembly and DNA encapsulation in Salmonella phage P22. I. Genes, proteins, structures and DNA maturation. J. Mol. Biol. 80669-695. [DOI] [PubMed] [Google Scholar]
- 5.Conlin, C. A., E. R. Vimr, and C. G. Miller. 1992. Oligopeptidase A is required for normal phage P22 development. J. Bacteriol. 1745869-5880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeSpicer, P. O., and S. R. Maloy. 1993. PutA protein, a membrane-associated flavin dehydrogenase, acts as a redox-dependent transcriptional regulator. Proc. Natl. Acad. Sci. USA 904295-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evilevitch, A., L. Lavelle, C. M. Knobler, E. Raspaud, and W. M. Gelbart. 2003. Osmotic pressure inhibition of DNA ejection from phage. Proc. Natl. Acad. Sci. USA 1009292-9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galloux, M., S. Libersou, N. Morellet, S. Bouaziz, B. Da Costa, M. Ouldali, J. Lepault, and B. Delmas. 2007. Infectious bursal disease virus, a non-enveloped virus, possesses a capsid-associated peptide that deforms and perforates biological membranes. J. Biol. Chem. 28220774-20784. [DOI] [PubMed] [Google Scholar]
- 9.Garcia, L. R., and I. J. Molineux. 1995. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 1774066-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia, L. R., and I. J. Molineux. 1996. Transcription-independent DNA translocation of bacteriophage T7 DNA into Escherichia coli. J. Bacteriol. 1786921-6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glaserwuttke, G., J. Keppner, and I. Rasched. 1989. Pore-forming properties of the adsorption protein of filamentous phage fd. Biochim. Biophys. Acta 985239-247. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg, E., L. Grinius, and L. Letellier. 1994. Recognition, attachment, and injection, p. 347-356. In J. D. Karam (ed.), Bacteriophage T4, 1st ed. ASM Press, Washington, DC.
- 13.Grayson, P., and I. J. Molineux. 2007. Is phage DNA ‘injected’ into cells—biologists and physicists can agree. Curr. Opin. Microbiol. 10401-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hershey, A. D., and M. Chase. 1952. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J. Gen. Physiol. 3639-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, B., and M. Levine. 1975. Bacteriophage P22 virion protein which performs an essential early function. 1. Analysis of 16− Ts mutants. J. Virol. 161536-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman, B., and M. Levine. 1975. Bacteriophage P22 virion protein which performs an essential early function. 2. Characterization of gene 16 function. J. Virol. 161547-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Israel, V. 1977. E proteins of bacteriophage P22. 1. Identification and ejection from wild-type and defective particles. J. Virol. 2391-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp, P., L. R. Garcia, and I. J. Molineux. 2005. Changes in bacteriophage T7 virion structure at the initiation of infection. Virology 340307-317. [DOI] [PubMed] [Google Scholar]
- 19.Kemp, P., M. Gupta, and I. J. Molineux. 2004. Bacteriophage T7 DNA ejection into cells is initiated by an enzyme-like mechanism. Mol. Microbiol. 531251-1265. [DOI] [PubMed] [Google Scholar]
- 20.Labedan, B., and E. B. Goldberg. 1979. Requirement for membrane potential in injection of phage T4 DNA. Proc. Natl. Acad. Sci. USA 764669-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Labedan, B., K. B. Heller, A. A. Jasaitis, T. H. Wilson, and E. B. Goldberg. 1980. A membrane potential threshold for phage T4 DNA injection. Biochem. Biophys. Res. Commun. 93625-630. [DOI] [PubMed] [Google Scholar]
- 22.Letellier, L., P. Boulanger, L. Plancon, P. Jacquot, and M. Santamaria. 2004. Main features on tailed phage, host recognition and DNA uptake. Front. Biosci. 91228-1339. [DOI] [PubMed] [Google Scholar]
- 23.Maloy, S. R., V. J. Stewart, and R. K. Taylor. 1996. Genetic analysis of pathogenic bacteria, 1st ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Moak, M., and I. J. Molineux. 2000. Role of the gp16 lytic transglycosylase motif in bacteriophage T7 virions at the initiation of infection. Mol. Microbiol. 37345-355. [DOI] [PubMed] [Google Scholar]
- 25.Molineux, I. J. 2001. No syringes please, ejection of phage T7 DNA from the virion is enzyme driven. Mol. Microbiol. 401-8. [DOI] [PubMed] [Google Scholar]
- 26.Muro-Pastor, A. M., P. Ostrovsky, and S. Maloy. 1997. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J. Bacteriol. 1792788-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poteete, A. R., and J. King. 1977. Functions of two new genes in Salmonella phage P22 assembly. Virology 76725-739. [DOI] [PubMed] [Google Scholar]
- 28.Pryde, J. G. 1986. Triton X-114—a detergent that has come in from the cold. Trends Biochem. Sci. 11160-163. [Google Scholar]
- 29.Purohit, P. K., M. M. Inamdar, P. D. Grayson, T. M. Squires, J. Kondev, and R. Phillips. 2005. Forces during bacteriophage DNA packaging and ejection. Biophys. J. 88851-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed., vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Simon, L. D., and T. F. Anderson. 1967. The infection of Escherichia coli by T2 and T4 bacteriophages as seen in the electron microscope. I. Attachment and penetration. Virology 32279-297. [DOI] [PubMed] [Google Scholar]
- 33.Struthers-Schlinke, J. S., W. P. Robins, P. Kemp, and I. J. Molineux. 2000. The internal head protein gp16 controls DNA ejection from the bacteriophage T7 virion. J. Mol. Biol. 30135-45. [DOI] [PubMed] [Google Scholar]
- 34.Susskind, M. M., D. Botstein, and A. Wright. 1974. Superinfection exclusion by P22 prophage in lysogens of Salmonella Typhimurium. 3. Failure of superinfecting phage DNA to enter sieA+ lysogens. Virology 62350-366. [DOI] [PubMed] [Google Scholar]
- 35.Szoka, F., Jr., and D. Papahadjopoulos. 1978. Procedure for preparation of liposomes with large internal aqueous space and high capture by reverse-phase evaporation. Proc. Natl. Acad. Sci. USA 754194-4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, D., and P. Prevelige. 1991. A pilot protein participates in the initiation of P22 procapsid assembly. Virology 182673-681. [DOI] [PubMed] [Google Scholar]
- 37.Umlauf, B., and B. Dreiseikelmann. 1992. Cloning, sequencing, and overexpression of gene 16 of Salmonella bacteriophage P22. Virology 188495-501. [DOI] [PubMed] [Google Scholar]
- 38.Zhu, M., J. Li, and A. L. Fink. 2003. The association of alpha-synuclein with membranes affects bilayer structure, stability, and fibril formation. J. Biol. Chem. 27840186-40197. [DOI] [PubMed] [Google Scholar]