Abstract
Infections by the bacterium Aeromonas salmonicida subsp. achromogenes cause significant disease in a number of fish species. In this study, we showed that AsaP1, a toxic 19-kDa metallopeptidase produced by A. salmonicida subsp. achromogenes, belongs to the group of extracellular peptidases (Aeromonas type) (MEROPS ID M35.003) of the deuterolysin family of zinc-dependent aspzincin endopeptidases. The structural gene of AsaP1 was sequenced and found to be highly conserved among gram-negative bacteria. An isogenic ΔasaP1 A. salmonicida subsp. achromogenes strain was constructed, and its ability to infect fish was compared with that of the wild-type (wt) strain. The ΔasaP1 strain was found to infect Arctic charr, Atlantic salmon, and Atlantic cod, but its virulence was decreased relative to that of the wt strain. The 50% lethal dose of the AsaP1 mutant was 10-fold higher in charr and 5-fold higher in salmon than that of the wt strain. The pathology induced by the AsaP1-deficient strain was also different from that of the wt strain. Furthermore, the mutant established significant bacterial colonization in all observed organs without any signs of a host response in the infected tissue. AsaP1 is therefore the first member of the M35 family that has been shown to be a bacterial virulence factor.
Proteolytic enzymes are essential factors in the life cycles of microorganisms, and peptidases produced by pathogenic bacteria may also be toxic to the host. Many toxic peptidases are metallopeptidases that have a zinc ion in the catalytic site (19). The gram-negative bacterium Aeromonas salmonicida is a well-known pathogen of various fish species worldwide. Five subspecies within A. salmonicida have been described: A. salmonicida subsp. salmonicida, A. salmonicida subsp. achromogenes, A. salmonicida subsp. masoucida, A. salmonicida subsp. smithia, and A. salmonicida subsp. pectinolytica (15, 24). Furthermore, strains that do not fulfill the criteria for these subspecies are frequently isolated. A. salmonicida subsp. salmonicida, the causative agent of classical furunculosis of salmonid fish, is often referred to as typical A. salmonicida, whereas the other strains have been referred to as atypical. A. salmonicida subsp. salmonicida has been described as a homogenous species, indicating that it is a host-adapted pathogen. One group of atypical A. salmonicida strains, A. salmonicida subsp. achromogenes, has recently been found by many researchers to be a homogeneous group of strains that cause atypical furunculosis, a systemic disease, in many species of fish (8).
Several secreted peptidases have been identified in A. salmonicida strains, yet only two have been associated with toxic activity. The 64-kDa serine peptidase AspA (P1), which has a 50% lethal dose (LD50) of 2.4 μg protein/g fish in Atlantic salmon (Salmo salar L.), is considered to be a major toxic factor of typical and various atypical strains (5) but has not been detected in the extracellular products (ECP) of A. salmonicida subsp. achromogenes strains (10).
A. salmonicida subsp. achromogenes protease 1 (AsaP1) has been described as a monomeric polypeptide of approximately 20 kDa and a major toxic factor of strains belonging to A. salmonicida subsp. achromogenes, but it has not been detected in the ECP of typical A. salmonicida strains (9, 10). When purified AsaP1 is injected into salmon or mice at low concentrations, it leads to death within 24 h. The LD50 of AsaP1 in salmon is as low as 0.03 μg protein/g fish when injected intraperitoneally (i.p.) but is six times higher in mice (9, 12). When a sublethal dose of purified AsaP1 is injected intramuscularly into salmon, all pathological changes seen in salmon infected by the AsaP1-producing bacterium are observed (9).
The AsaP1 toxin is caseinolytic and weakly gelatinolytic. It is not hemolytic against horse, sheep, or fish erythrocytes, nor is it cytotoxic against RTG-2 cells, bluegill fry (BF-2) cells, or EPC cells (9, 14). Furthermore, AsaP1 does not cleave the C3 complement component of either cod or halibut (17). In vitro studies have shown that AsaP1 is a powerful mitogen of Atlantic salmon leukocytes (13) and induces mouse peritoneal monocytes to produce the acute-phase cytokines tumor necrosis factor alpha, interleukin-6, and interleukin-1β (12). AsaP1 is immunogenic in salmonid fish, and anti-AsaP1 antibodies are known to provide Atlantic salmon protection against infection by A. salmonicida subsp. achromogenes, indicating its importance as a protective antigen (11).
In this study we isolated, cloned, characterized, and inactivated the gene encoding the AsaP1 peptidase. An AsaP1-negative mutant was constructed, and its virulence in three species of fish was studied.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
All bacterial strains and plasmids used in cloning, growth, and challenge assays are described in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Properties | Source or reference |
|---|---|---|
| Strains | ||
| A. salmonicida subsp. achromogenes | ||
| Keldur265-87 | Fish-pathogenic isolate, ampicillin and cephalothin resistant | 9 |
| Keldur265-87-2 | ΔasaP1::Km, derived from 265-87 | This study |
| E. coli | ||
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Clontech |
| DH5α | endA1 hsdR17(rK− mK+) supE44 thi-1 recA1 gyrA96 relA1 Δ(lacZYA-argF)U169 (φ80lacZΔM15) | Invitrogen |
| BM25.8 | supE44 thi Δ(lac-proAB) [F′ traD36 proAB+ lacIqZΔM15] λimm434 (Kanr)P1 (Camr) hsdR (rK12-mK12−), cre-lox-mediated excision of pTriplEx from λpTriplEx | Clontech |
| S17.1 | thi pro hsdR hsdM+recA [RP4 2-Tc::Mu-Km::Tn7(Tpr Smr)Tra+], mobilizing donor for conjugation | 26 |
| Plasmids | ||
| pTriplEx | Triple-reading-frame translation preceding the EcoRI site; cloning and sequencing | Clontech |
| pGEM-T and pGEM-T-Easy | Cloning of modified blunt-ended DNA fragments | Promega |
| pUC19 | Apr, cloning vector | Fermentas |
| pSSVI186-IN | Kmr determinant from Tn903, located on a 1.3-kb PstI fragment | 28 |
| pSUP202sac | Mobilizable suicide vector, SucS (Mob+sacRB+ Tetr) | 27 |
A. salmonicida subsp. achromogenes strain Keldur265-87 was isolated and identified, in our laboratory, from an Atlantic salmon suffering from atypical furunculosis (9). A. salmonicida strains were routinely cultured at 15°C on blood agar (BA). Liquid cultures of A. salmonicida were grown in brain heart infusion (BHI) broth with agitation (200 rpm), and the growth was measured as log10 CFU/ml. For genetic modifications, the bacteria were grown on Luria-Bertani (LB) agar plates (2). For isolation of ECP, the strains were cultivated on cellophane-covered BHI agar plates and ECP were then washed off the cellophane with minimal amounts of phosphate-buffered saline and isolated by centrifugation (10). Escherichia coli strains were routinely grown on LB agar or broth at 37°C, except for strain BM25.8, which was grown at 31°C. When required for the selection and maintenance of recombinant plasmids, antibiotics were added to the culture media at the following final concentrations: ampicillin (Sigma), 100 μg/ml; tetracycline, 25 μg/ml; cephalothin, 200 μg/ml; and kanamycin, 40 μg/ml.
Growth curves of A. salmonicida strains were obtained by monitoring CFU/ml in BHI broth following incubation at 15°C. In order to monitor the proteolytic activity during growth, 1-ml aliquots were sampled at different time intervals and the supernatants were isolated by centrifugation (10,000 × g for 15 min at 4°C).
Amino acid sequencing of AsaP1.
Isolation of the AsaP1 peptidase from the ECP of strain Keldur265-87 was performed by anion-exchange chromatography followed by gel filtration using MonoQ HR 5/5 and Superose 12 HR 10/30 columns (Amersham Biosciences) and GE Healthcare's (formerly Amersham Biosciences) ÄKTA fast protein liquid chromatographic system, as described previously (9). Purified enzyme was transferred from a sodium dodecyl sulfate (SDS)-polyacrylamide gel (1 μg well−1) to a polyvinylidene fluoride membrane by Western blotting. N-terminal and internal amino acid sequences were obtained from the purified protein by automated Edman degradation at the Oxford Centre for Molecular Sciences protein sequencing facility.
DNA preparation and PCR amplification.
Genomic bacterial DNA was obtained by the phenol-chloroform extraction method. Plasmid DNA and DNA fragments were purified from agarose gels using the QIAprep spin miniprep kit (Qiagen).
PCRs were used to amplify the asaP1 gene for cloning, to confirm positive colonies on agar plates after transformation, and for DNA sequencing. Details of primer sequences are listed in Table 2. When DNA fragments were produced by PCR for subsequent cloning and expression, the Expand-Long-Template PCR kit (Roche, Molecular Biochemicals) containing a polymerase with proofreading capacity was used.
TABLE 2.
Oligonucleotide primers and their application in this study
| Primer | Sequence (5′→ 3′) | Application |
|---|---|---|
| oligo 11 | TGCACCAACACCCAGAAGA | Amplification of asaP1 ORF and sequencing |
| oligo 12 | GGATAGACATAGGCAAAGTAA | |
| AsaP1TOPO2 | CCATGGTGAAAGTGACTCCAATAG | |
| AsaP1TOPO3 | TCAGTTTTCGCTCGGGGTATTC | |
| Up-190-asaP1 | TCAGCTCACCCTGCACTATG | Sequencing of sequences flanking asaP1 ORF |
| Down-1954-asaP1 | CTCGAGGTGTTGAGCTGGTT | |
| Down1814asaP1 | GGAGGCTCCATGATGACTC | |
| Down1814cont | CACTTGCTGCCAGGATAATCC | |
| asaP1_pGEM_f | CGGACTCCACAACAGCTCGG | Cloning and sequencing |
| asaP1_pGEM_r | GCCCTGCATGAGATGGGTGC | |
| asaP1_EcoRI_f | CCGAATTCGGTGGTGGAAAATTTCTGC | |
| asaP1_HindIII_r | CCAAGCTTGGTGGTGAGCAAGAGTGTC | |
| asaP1_pGEM_f | CGGACTCACAACAGCTCGG | |
| T7 | GTAATACGACTCACTATAGGGC | Sequencing |
| 5′ Clontech | TCCGAGATCTGGACGAGC |
Construction of an A. salmonicida subsp. achromogenes gene library and isolation of the asaP1 gene.
The sequencing of 40 N-terminal amino acids of purified AsaP1 revealed 85% similarity to a corresponding sequence in the EprA1 protease of Aeromonas hydrophila (Table 3). Primers binding to the N-terminal and C-terminal ends of the erpA1 gene were then designed (oligo 31EcoRI and oligo 24EcoRI [Table 2]) and found to amplify a 1,032-bp sequence in the genome of strain Keldur265-87. This sequence was cloned into EcoRI-digested pUC18 vector and used as a probe in Southern blotting and screening of recombinant phage.
TABLE 3.
Bacterial peptidases with highest amino acid sequence similarity to the mature 172-aa AsaP1 peptidase of A. salmonicida subsp. achromogenesa
| Homologous protein | Organism | Strain | GenBank accession no. | %
|
E value | ||
|---|---|---|---|---|---|---|---|
| Gaps | aa identity | aa positive | |||||
| Extracellular protease | Aeromonas hydrophila | ATCC 7966 | ABK37697 | 0 | 92 | 97 | 1e−87 |
| Extracellular protease, EprA1 | Aeromonas hydrophila | AH-1 | AAB88221 | 0 | 91 | 95 | 4e−86 |
| Epr protein | Aeromonas hydrophila | J-1 | AAY21066 | 0 | 88 | 96 | 6e−63 |
| Metalloprotease | Aeromonas punctata | NRb | BAE72122 | 0 | 91 | 97 | 2e−69 |
| Peptidyl-Lys metalloendopeptidase | Shewanella loihica | PV-4 | ABO23113 | 0 | 60 | 81 | 4e−57 |
| Peptidyl-Lys metalloendopeptidase | Shewanella woodyi | ATCC 51908 | EAV37755 | 0 | 60 | 81 | 3e−57 |
| Peptidyl-Lys metalloendopeptidase | Shewanella denitrificans | OS217 | ABE55895 | 1 | 59 | 80 | 2e−56 |
| Peptidyl-Lys metalloendopeptidase | Shewanella amazonensis | SB2B | ABL99412 | 2 | 59 | 80 | 9e−53 |
| Extracellular protease | Pseudoalteromonas tunicata | D2 | EAR28299 | 2 | 62 | 79 | 5e−49 |
| Probable protease | Chromobacterium violaceum | ATCC 12472 | AAQ61167 | 1 | 56 | 73 | 8e−49 |
According to BLAST analyses (1).
NR, not reported.
A λ phage gene library from strain Keldur265-87 was constructed. Chromosomal DNA was digested with EcoRI, and 3- to 10-kb fragments were purified from agarose gel and ligated to EcoRI-digested λ _TriplEx vector (Clontech). The ligated DNA was packed into λ prophage with the Gigapack III Gold packaging extract (Stratagene) and used to transduce E. coli XL1-Blue (Table 1). Phage plaques were transferred to nylon membranes (HybondN+; Amersham), and recombinant plaques were screened with a digoxigenin (DIG)-labeled probe constructed with the DNA labeling and detection kit from Roche. The in vivo excision of plasmids from selected phagemid plaques was performed according to the instructions with the λ TriplEx kit (Clontech).
Southern blot analysis.
One to two micrograms of DNA was digested with various restriction enzymes, separated on 0.7% agarose gels, and blotted onto Hybond N+ membranes. DIG-labeled probes were created with the DNA labeling and detection kit (Roche), and hybridization was performed overnight at 42°C. The DIG-labeled probe was detected with anti-DIG (Fab fragment conjugated to alkaline phosphatase), and the bound antibody conjugate was then visualized with nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate as specified by the manufacturer.
DNA sequencing and sequence analysis.
All sequences were determined on both DNA strands. Sequence alignment and editing were performed using the software Sequencher (Gene Codes Corp.). Analyses of sequence similarity and conserved protein domains were performed using the BLAST (1) network server of GenBank, National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). Multiple alignments of protein sequences were generated by CLUSTALW (http://align.genome.jp/) and the FASTA and SSEARCH programs (http://www.ebi.ac.uk/Tools/fasta33/index.html).
Prediction of promoter sequences was performed with the software Prediction of Bacterial Promoters (BPROM) (Softberry) and Prokaryotic Promoter Prediction (PPP) (http://bioinformatics.biol.rug.nl/websoftware/ppp). The Expasy ProtParam program (7) (http://expacy.org) was used to calculate the molecular mass and theoretical pI of AsaP1. Signal sequence prediction was performed using the SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP/).
Construction of a ΔasaP1 A. salmonicida subsp. achromogenes mutant.
The asaP1 gene of A. salmonicida subsp. achromogenes was inactivated by marker replacement mutagenesis. A 3.2-nucleotide fragment containing the asaP1 gene and its flanking regions was amplified from phagemid pTriplEx-asaP1 with primers pGEMf and pGEMr (Table 2) and cloned into the pGEM-T-Easy vector. A 313-bp Eco47III/BamHI fragment was then removed from the asaP1 gene and replaced with the kanamycin cassette from plasmid pSSVI186-IN, located on a ∼1.2-kb StuI/BamHI fragment. The inactivated asaP1 gene and flanking sequences were subsequently cloned into the mobilizable suicide vector pSUP202sac digested with PstI. The resulting plasmid was transformed into E. coli strain S17.1 and conjugated into A. salmonicida subsp. achromogenes strain Keldur265-87 by filter mating (26). Selection of double-crossover AsaP1-negative mutants was performed by cultivation on BHI agar supplemented with cephalothin, kanamycin, and sucrose (15%, wt/vol) at 15°C for 7 days. Cephalothin was used to select against the E. coli donor strain, as strain Keldur265-87 is cephalothin resistant. Double-crossover ΔasaP1 kanamycin-resistant mutants (Keldur265-87-2) were confirmed by PCR and cultivation on BHI agar supplemented with kanamycin.
Detection of the AsaP1 peptidase.
A spectrophotometric azo-casein assay and casein SDS-polyacrylamide gel electrophoresis (SDS-PAGE) were used for detection of caseinolytic activity as previously described (10). Western blotting using a polyclonal mouse anti-AsaP1 antibody was used to confirm the presence of the AsaP1 peptidase.
Experimental fish and challenge experiments.
Arctic charr (Salvelinus alpinus) (30 ± 8 g [mean ± standard deviation]), Atlantic salmon (25 ± 5 g), and Atlantic cod (Gadus morhua) (33 ± 11 g) were used in challenge experiments. The fish were free of infections according to standard routine diagnostic procedures. Fish were kept in 200-liter tanks supplied with continuously running fresh aerated water at 9 ± 1°C, except for bath immersion challenges, where fish were kept in closed systems containing 30 liters of aerated liquid that was changed daily. Charr and salmon were kept in brackish water (0.35% salinity) and cod in seawater (3.5% salinity). The fish were fed a commercial fish diet at 1% body weight per day. The oxygen concentration, temperature, and mortality in each tank were monitored daily throughout the experiments.
The fish were acclimatized to laboratory conditions for 2 weeks before experimental treatments. Prior to treatment, the fish were anesthetized with tricaine methanesulfonate (50 mg/liter) and marked along fin margins with Visible Implant Fluorescent Elastomer dye (Northwest Marine Technology).
A. salmonicida subsp. achromogenes strain Keldur265-87 and its isogenic AsaP1− mutant Keldur265-87-2 were passaged three times in the respective fish species before their use in challenge experiments. Challenges were performed by i.p. injection or bath immersion. Injection was performed with 0.1 ml of 10−1 to 10−5 10-fold dilutions of the bacterial suspensions. Control fish received phosphate-buffered saline only. Six fish were in each injection group. Bath immersion was performed by bathing 30 fish per tank for 1 h in 30 liters of an aerated bacterial suspension and control fish in aerated diluted culture medium. The fish were then kept in 30-liter tanks and observed for 2 weeks. Mortalities were recorded daily. The head kidneys from all dead and surviving fish were sampled, inoculated onto blood agar, and incubated for 7 days at 15°C. The identity of A. salmonicida subsp. achromogenes isolated from dead fish was established by PCR and cultivation on BHI agar with or without kanamycin.
Fish experiments were approved and performed according to the Icelandic Animal Research Authority (approval no. YDL03080041/023BE).
Pathological examinations.
The gross pathological changes of dead and moribund fish were described. Moribund salmon from the bath challenge were sampled for microscopic examination. Samples of skin and underlying muscle, brain, kidney, liver, spleen, pancreas, gut, heart, and gills were fixed in 10% buffered formalin, embedded in paraffin wax, cut into 4-μm sections, and stained with Giemsa stain before microscopic examination and photography.
Calculations, statistical analysis, and image processing.
StatView software for Windows (version 5.0.1), Microsoft Excel 2003, and Fig.P 2006 were used for statistical analysis and table and image processing. A chi-square test (Fisher's exact) was used to analyze the significance of differences in mortality between groups. Adobe PhotoshopCS (version 8.0) was used for processing of photographs.
The LD50 was calculated according to the method of Reed and Muench (25). The mean number of days to death (MDD) were calculated using the formula MDD = [Σ(number of mortalities × number of days postchallenge)]/total number of mortalities. A two-sample t test was used to compare the MDDs of different groups. The threshold level for significance was 0.05.
Nucleotide sequence accession number.
The nucleotide sequence of the asaP1 gene and its flanking regions has been deposited in the GenBank nucleotide sequence database under accession no. AF550405.
RESULTS
Molecular cloning of the asaP1 gene.
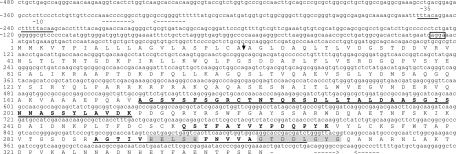
Forty amino acids (aa) were identified at the amino-terminal end of the active AsaP1 enzyme, as were two internal sequences consisting of 14 and 20 aa, respectively (Fig. 1). Sequence analysis indicated 91% amino acid identity between AsaP1 and the EprA1 protease of A. hydrophila (Table 3). Primers based on the eprA1 sequence were therefore designed and were able to amplify a 1,032-bp sequence from strain Keldur265-87. Screening of the Keldur phage library using a DIG-labeled probe binding to this 1,032-bp sequence revealed an open reading frame (ORF) of 1,029 bp, which translates into a 343-aa propeptide with a calculated molecular mass of 37 kDa (Fig. 1). Predicted promoter sequences, −35 TTTACA and −10 TTTTACAAG, were detected upstream of the AsaP1 ORF (bases −230 to −251). A Shine-Dalgarno ribosomal binding site was found immediately upstream of the start codon (bases −4 to −7). Palindrome sequences with characteristics of a Rho-independent transcription terminator were located upstream of the ORF (AACCCCGGCNNNNNGCCGGGGTT −315 to −337) and downstream of the stop codon (AGGGGCCNNNNGGCCCCT +1042 to +1059).
FIG. 1.
Nucleotide sequence of the asaP1 gene and flanking regions and amino acid sequence of the AsaP1 prepropeptide. Predicted −35 (−246 to −251) and −10 (−230 to −239) promoter sequences are underlined. In frame is a potential Shine-Dalgarno sequence (−4 to −7). - indicates the TGA termination codon. Amino acids 1 to 21 (vertical arrow) form a predicted signal sequence. Double-underlined boldface aa 172 to 211 correspond to the amino-terminal sequence obtained by Edman degradation. The active AsaP1 enzyme consists of aa 172 to 343. Internal amino acid sequences deduced from the purified peptidase are underlined and in boldface. Two highly conserved motifs of deuterolysin metallopeptidases (M35) that form the zinc binding site are highlighted in gray. Convergent dashed arrows show palindrome sequences characteristic of a Rho-independent transcription terminator upstream of the ORF (−315 to −337) and downstream of the stop codon (1042 to 1059).
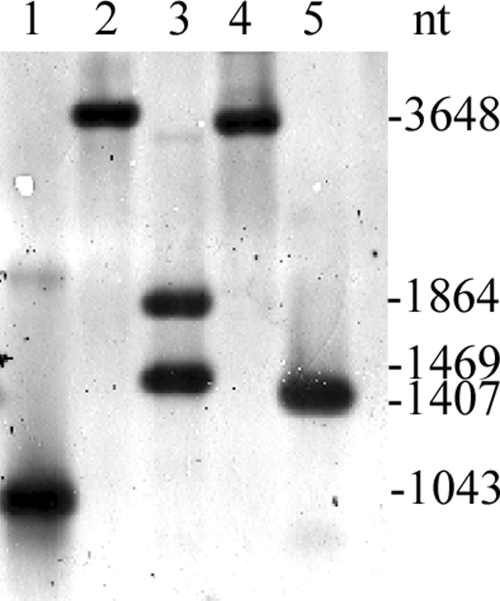
Southern blot analysis of genomic DNA digested with various restriction enzymes and probed with a 1,032-bp fragment containing the AsaP1 gene revealed the presence of only one copy of the gene in A. salmonicida subsp. achromogenes strain Keldur265-87 (Fig. 2).
FIG. 2.
Southern blot analysis of the asaP1 gene. Chromosomal DNA from Aeromonas salmonicida subsp. achromogenes strain Keldur265-87 was digested with the indicated restriction enzymes and probed with a 1,043-bp fragment of the asaP1 gene. Lane 1, 1,043-bp asaP1 fragment used as a probe; lane 2, XmaI; lane 3, SfiI (the probe contains a SfiI restriction site); lane 4, BstXI; lane 5, BstBI.
The first 21 aa of the translated AsaP1 peptide were determined to encode a Sec-dependent signal sequence (22). A long propeptide of 150 aa was located between the signal peptide and the mature peptidase sequence. Amino acid sequencing of the mature AsaP1 protein indicated that the N terminus corresponds to aa 172 in the translated ORF (Fig. 1). Mature AsaP1 is therefore composed of aa 172 to 343 and has a calculated molecular mass of 19 kDa. The aa sequence was found to contain two highly conserved motifs, HExxH and GTxDxxYG, of aspzincin metalloendopeptidases in the deuterolysin family (MEROPS ID M35) whereby His293, His297, and Asp306 coordinate to the catalytic zinc ligands and Tyr309 acts as a proton donor during catalysis (16).
AsaP1 is a highly conserved protein.
BLAST analysis of the amino acid sequence of the mature AsaP1 peptidase with all nonredundant GenBank CDS translations revealed 78 significant similarities with proteins from both bacteria and fungi. The bacterial peptidases found to have the highest similarity with AsaP1 peptidase are shown in Table 3.
Characterization of a ΔasaP1 A. salmonicida subsp. achromogenes mutant.
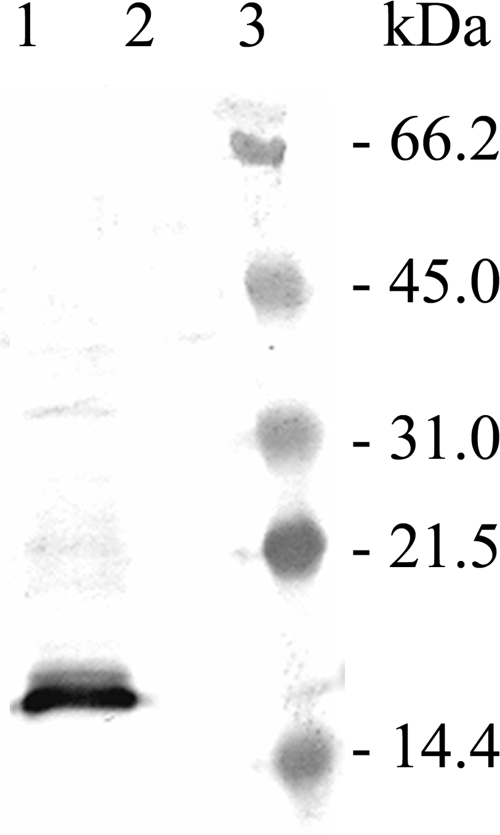
Anti-AsaP1 antibodies detected the toxin in ECP from the wild-type (wt) strain but not the ΔasaP1 strain (Fig. 3). Furthermore, caseinolytic activity could not be detected in the ECP or cells of the ΔasaP1 mutant in casein zymograms or in the azocasein assay, confirming that AsaP1 is not secreted by the ΔasaP1 strain.
FIG. 3.
Immunostaining of AsaP1 with anti-AsaP1 antibodies. Lane 1, ECP of the wt strain Keldur265-87; lane 2, ECP of the isogenic ΔasaP1 mutant Keldur265-87-2; lane 3, prestained protein molecular size marker.
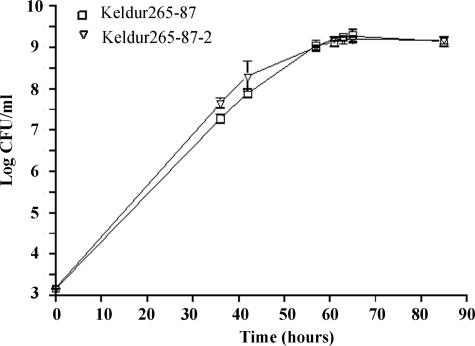
Comparison of the growth kinetics of strain Keldur265-87 and its isogenic AsaP1-deficient mutant Keldur265-87-2 did not reveal any significant differences (P < 0.001) (Fig. 4).
FIG. 4.
Logarithmic growth of Aeromonas salmonicida subsp. achromogenes strain Keldur265-87 and its isogenic ΔasaP1 mutant Keldur265-87-2 (▿) cultured in BHI broth at 15°C and assayed by viable counts. All values are the means and standard deviations of five determinations.
Infection of Arctic charr, Atlantic salmon, and Atlantic cod.
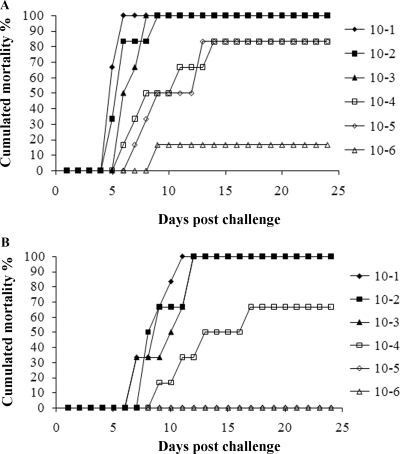
Injection challenges were performed with charr and salmon. Mortality rates in all groups of challenged fish were dose dependent. The calculated LD50 for the wt strain (Keldur265-87) in charr was 1.6 × 103 CFU/fish, and that in salmon was 1.4 × 104 CFU/fish (Fig. 5). In contrast, the LD50 of the ΔasaP1 strain (Keldur265-87-2) was 10-fold higher in charr (1.7 × 104 CFU/fish) and approximately 5 times higher in salmon (6.5 × 104 CFU/fish) than that of the wt strain (Fig. 5). Calculated MDDs were always higher when the fish were challenged with the ΔasaP1 strain than when they were challenged with the wt strain (P < 0.05). The MDDs in challenges with log5 doses of the respective bacteria were 7 and 10 days in charr and 8 and 10 days in salmon.
FIG. 5.
Challenge of Arctic charr by i.p. injection of Aeromonas salmonicida subsp. achromogenes. (A) Strain Keldur265-87 (wt strain diluted from 5 × 109 CFU/ml); (B) strain Keldur265-87-2 (ΔasaP1 mutant strain diluted from 3 × 109 CFU/ml). The calculated LD50s were 1.6 × 103 CFU fish−1 for strain Keldur265-87 and 1.7 × 104 CFU fish−1 for strain Keldur265-87-2.
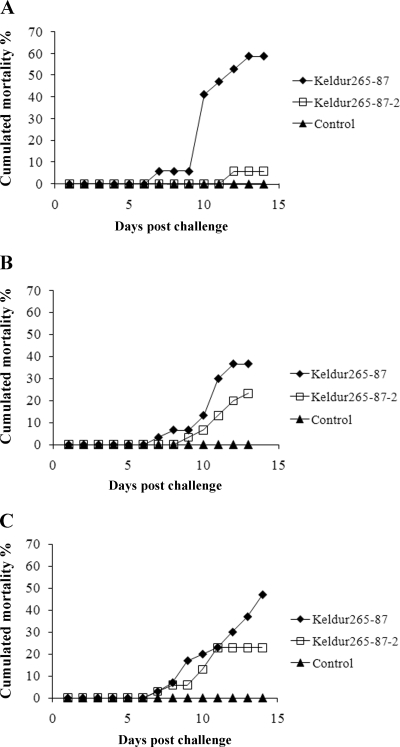
Bath immersion was performed on charr, salmon, and cod. Fewer fish of all three species died following challenge with the ΔasaP1 mutant than following challenge with the wt strain (Fig. 6). The difference was significant between the two groups of charr (P < 0.0001) but not between the cod (P = 0.0620) or salmon (P = 0.3985) groups. No mortalities were observed in the control groups.
FIG. 6.
Cumulative mortality following bath immersion with the A. salmonicida subsp. achromogenes wt strain (Keldur265-87) and the ΔasaP1 mutant strain (Keldur265-87-2). (A) Arctic charr (30 g) immersed in a bacterial dose of 5 × 108 CFU/ml. (B) Atlantic salmon (25 g) immersed in a bacterial dose of 4 × 106 CFU/ml. (C) Atlantic cod (33 g) immersed in a bacterial dose of 9 × 106 CFU/ml. For all panels, n = 30.
Clinical signs and pathological changes in bath-infected fish.
Pathological changes were detected in all fish that died following challenge, and both the wt and the ΔasaP1 mutant strains were isolated from the kidneys of infected fish. In general, the clinical signs in all three fish species included darkening in color, fin rot, diffuse skin hemorrhage, hemorrhage at fin bases, and loss of appetite. In more severe cases, lethargy, empty intestines, and ascites in the body cavity, often with generalized hemorrhages, were observed.
Skin ulcers or lesions and degenerative changes with liquefactive necrosis in underlying muscle and gills pallor were commonly detected in all three species of fish infected with the wt strain but not in any fish infected with the ΔasaP1 mutant. Pronounced scale loss and skin erosion without detectable hemorrhage were detected only in fish infected with the ΔasaP1 mutant. Autopsies of charr, salmon, and cod infected with the wt strain revealed general anemia, which was not seen in any fish infected with the ΔasaP1 mutant. Detection of blood-stained ascites was more prominent in fish infected with the ΔasaP1 mutant.
Microscopic examination was performed on salmon. Two moribund fish infected with the wt strain and four infected with the ΔasaP1 mutant were sampled for histological examination. Bacterial colonies were detected in all examined organs of fish infected with either strain, except the muscle. Infections by both strains were found to induce hyperemia and hemorrhages in major organs, including liver, kidney, heart, brain, and muscle. Sloughing off of the gill's secondary lamellae was also frequently observed.
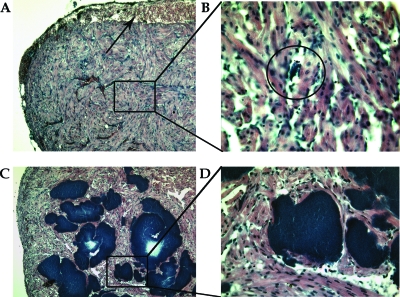
The major difference detected between fish infected with the wt bacterium and fish infected with the ΔasaP1 mutant was that in salmon infected with the wt strain, tissue degeneration and infiltration of mononuclear cells were detected around small bacterial colonies in the various tissues observed. In salmon infected with the ΔasaP1 mutant, severe bacterial colonization was seen in all organs examined without any signs of a host response in the infected tissue. Figure 7 demonstrates the pathological changes seen in heart muscle of salmon infected with the wt and the mutant strains.
FIG. 7.
(A) Subepicardial hemorrhages (arrow) in the heart muscle of salmon infected with the wt A. salmonicida subsp. achromogenes strain (Keldur265-87) (magnification, ×10). Black box, bacterial colony. (B) Enlarged view of a bacterial colony (inside the circle) from panel A (magnification, ×40). (C) Massive accumulation of bacterial colonies in the heart muscle of salmon infected with the ΔasaP1 mutant A. salmonicida subsp. achromogenes strain (Keldur265-87-2) (magnification, ×10). (D) Enlarged view of bacterial colonies from panel C (magnification, ×40).
DISCUSSION
In this study, a toxic 19-kDa metallopeptidase of A. salmonicida, AsaP1, was characterized as a metalloendopeptidase belonging to the group of extracellular peptidases (Aromonas type) (MEROPS ID M35.003) of the deuterolysin family of zinc-dependent endopeptidases. It was also shown that the virulence of a ΔasaP1 mutant was different from that of its parental wt strain. AsaP1 is therefore the first member of the M35 family that has been shown to be a bacterial virulence factor.
The asaP1 gene was found to contain 1,032 nucleotides, encoding a 343-aa prepropeptide. Amino acid sequencing of the N terminus of the mature protein showed that it is composed of 172 aa (aa 172 to 343) with a deduced molecular mass of 19 kDa. The size of the mature protein is in agreement with results from gel filtration chromatography and SDS-PAGE analysis (9) describing a protease of approximately 20 kDa and composed of a single polypeptide. The molecular mass of the mature AsaP1 is also in accordance with what has been described for the AP19 metallopeptidase of Aeromonas caviae (21) and the peptidyl-Lys metalloendopeptidases of Grifola frondosa and Pleurotus ostreatus, respectively (23). The AsaP1 sequence was found to be 91% identical to the sequence of A. hydrophila extracellular protease (Table 3), and Chang et al. (3) reported that the A. hydrophila extracellular protease is detected as a 29-kDa protein in casein SDS-PAGE. This is in accordance with our previous reports of the migration distance of purified native AsaP1 in casein SDS-PAGE (10). The asaP1 gene may form its own transcriptional unit, as promoter sequences as well as a Shine-Dalgarno ribosomal binding site were found upstream of the asaP1 start codon. Furthermore, palindrome sequences with characteristics of a Rho-independent transcription terminator were located upstream of the ORF and downstream of the stop codon.
The AsaP1 mutation was found to significantly affect the virulence of the bacterium. The mutant established a systemic infection in charr, salmon, and cod; however, the LD50 of the mutant was higher than that of the wt strain, and fewer mortalities were observed at 14 days after bath infection in fish infected with the isogenic mutant strain than in fish infected with the wt strain. The difference in survival of fish following bath challenge with the two strains was found to be statistically significant in charr but not in salmon for cod. The incubation time of the ΔasaP1 mutant in all three fish species studied was also longer than that of the wt strain, yet the growth rates of the two strains in vitro were found to be identical. This confirms that loss of the AsaP1 toxin delays onset of the disease.
In this study, five separate challenge experiments were performed, i.e., two injection challenges of charr and salmon and three bath challenges of charr, salmon, and cod. Charr were found to be more susceptible than salmon to the injection challenge. The LD50 of the wt strain in salmon in this study was about fivefold higher than has been previously reported (11), but this is the first report of an LD50 of A. salmonicida subsp. achromogenes in charr. Comparison of the susceptibilities of the different fish groups is, however, problematic when based on separate challenge experiments, as seasonal variability in fish immunity is well reported (4, 18, 20) and in this study the challenges were performed at different times of the year.
The pathology induced by the mutant strain was different from that induced by the wt strain. Skin ulcers, a characteristic of A. salmonicida subsp. achromogenes infection, were not detected on fish infected with the mutant. Furthermore, general anemia in various organs, which is a symptom of infection by this bacterium (8), was not seen in any fish infected with the ΔasaP1 mutant. This shows the involvement of AsaP1 in disease development, as has previously been suggested (9, 14).
Microscopic examination was performed on salmon and revealed that a loss of the AsaP1 toxin resulted in much more severe accumulation and dispersion of the bacterium in salmon than were seen in fish infected with the wt strain. A massive accumulation of the ΔasaP1 mutant strain was seen in skin and underlying muscle, brain, kidney, liver, spleen, pancreas, gut, heart, and gills, yet no visible signs of tissue degradation or host response were detected in tissue surrounding these bacterial colonies.
The mortality rates and incubation times for fish infected with the mutant were lower than those for fish infected with the wt strain even though the accumulation of the mutant in the tissue was so much greater. This and the lack of tissue degradation around the large colonies of the mutant suggest that the mortality of fish infected with the mutant was due to general tissue failure rather than secreted bacterial or host factors.
The results obtained in this study show that the AsaP1 peptidase plays an important role in the virulence of A. salmonicida subsp. achromogenes and that it has a major role in the fish innate immune response, as the host was not found to recognize the ΔasaP1 mutant. This observation makes further studies of the interaction of AsaP1 with host immunity particularly interesting.
Acknowledgments
This project was funded by the Icelandic Centre for Research (RANNIS) and the University of Iceland Research Found.
Special thanks are due to Slavko H. Bambir and Margret Jonsdottir for carrying out the histological work.
Footnotes
Published ahead of print on 24 October 2008.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Chang, T. M., C. C. Liu, and M. C. Chang. 1997. Cloning and sequence analysis of the gene (eprA1) encoding an extracellular protease from Aeromonas hydrophila. Gene 199225-229. [DOI] [PubMed] [Google Scholar]
- 4.Coteur, G., N. Corriere, and P. Dubois. 2004. Environmental factors influencing the immune responses of the common European starfish (Asterias rubens). Fish Shellfish Immunol. 1651-63. [DOI] [PubMed] [Google Scholar]
- 5.Ellis, A. E. 1997. The extracellular toxins of Aeromonas salmonicida subsp. salmonicida, p. 248-268. In E.-M. Bernoth, A. E. Ellis, P. J. Midtlyng, G. Olivier, and P. Smith (ed.), Furunculosis: multidisciplinary fish disease research. Academic Press, London, United Kingdom.
- 6.Reference deleted.
- 7.Gasteiger, E., C. Hoogland, A. Gattiker, S. Duvaud, M. R. Wilkins, R. D. Appel, and A. Bairoch. 2005. Protein identification and analysis tools on the ExPASy server, p. 571-607 In J. M. Walker (ed.), The proteomics protocols handbook. Humana Press Inc., Totowa, NJ.
- 8.Gudmundsdóttir, B. K., and B. Björnsdóttir. 2007. Vaccination against atypical furunculosis and winter ulcer disease of fish. Vaccine 255512-5523. [DOI] [PubMed] [Google Scholar]
- 9.Gudmundsdóttir, B. K., T. S. Hastings, and A. E. Ellis. 1990. Isolation of a new toxic protease from a strain of Aeromonas salmonicida subsp. achromogenes. Dis. Aquat. Org. 9199-208. [Google Scholar]
- 10.Gudmundsdóttir, B. K., Í. Hvanndal, B. Björnsdóttir, and U. Wagner. 2003. Analysis of exotoxins produced by atypical isolates of Aeromonas salmonicida, by enzymatic and serological methods. J. Fish Dis. 2615-29. [DOI] [PubMed] [Google Scholar]
- 11.Gudmundsdóttir, B. K., and B. Magnadóttir. 1997. Protection of Atlantic salmon (Salmo salar L.) against an experimental infection of Aeromonas salmonicida ssp. achromogenes. Fish Shellfish Immunol. 755-69. [Google Scholar]
- 12.Gudmundsdóttir, S., and B. K. Gudmundsdóttir. 2001. Induction of inflammatory cytokines by extracellular products and LPS of the fish pathogen Aeromonas salmonicida ssp. achromogenes in mice and mouse cell cultures. Vet. Immunol. Immunopathol. 8171-83. [DOI] [PubMed] [Google Scholar]
- 13.Gudmundsdóttir, S., B. Magnadóttir, and B. K. Gudmundsdóttir. 1995. Effects of antigens from Aeromonas salmonicida ssp. achromogenes on leukocytes from primed and unprimed Atlantic salmon (Salmo salar L.). Fish Shellfish Immunol. 5493-504. [Google Scholar]
- 14.Gunnlaugsdóttir, B., and B. K. Gudmundsdóttir. 1997. Pathogenicity of atypical Aeromonas salmonicida in Atlantic salmon compared with protease production. J. Appl. Microbiol. 83542-551. [DOI] [PubMed] [Google Scholar]
- 15.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Facultatively anaerobic Gram-negative rods, p. 175-289. In W. R. Hensyl (ed.), Bergey′s manual of determinative bacteriology, 9th ed. Williams & Wilkins, Baltimore, MD.
- 16.Hori, T., T. Kumasaka, M. Yamamoto, N. Nonaka, N. Tanaka, Y. Hashimoto, U. Ueki, and K. Takio. 2001. Structure of a new ‘aspzincin’ metalloendopeptidase from Grifola frondosa: implications for the catalytic mechanism and substrate specificity based on several different crystal forms. Acta Crystallogr. D 57361-368. [DOI] [PubMed] [Google Scholar]
- 17.Lange, S., A. W. Dodds, and B. Magnadóttir. 2004. Isolation and characterization of complement component C3 from Atlantic cod (Gadus morhua L.) and Atlantic halibut (Hippoglossus hippoglossus L.). Fish Shellfish Immunol. 16227-239. [DOI] [PubMed] [Google Scholar]
- 18.Magnadottir, B. 2006. Innate immunity of fish (overview). Fish Shellfish Immunol. 20137-151. [DOI] [PubMed] [Google Scholar]
- 19.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 291-98. [DOI] [PubMed] [Google Scholar]
- 20.Mjaaland, S., T. Markussen, H. Sindre, S. Kjoglum, B. H. Dannevig, S. Larsen, and U. Grimholt. 2005. Susceptibility and immune responses following experimental infection of MHC compatible Atlantic salmon (Salmo salar L.) with different infectious salmon anaemia virus isolates. Arch. Virol. 1502195-2216. [DOI] [PubMed] [Google Scholar]
- 21.Nakasone, N., C. Toma, T. Y. Song, and M. Iwanaga. 2004. Purification and characterization of a novel metalloprotease isolated from Aeromonas caviae. FEMS Microbiol. Lett. 237127-132. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen, H., J. Engelbrecht, S. Brunak, and G. Von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 101-6. [DOI] [PubMed] [Google Scholar]
- 23.Nonaka, T., N. Dohmae, T. Hashimoto, and K. Takio. 1997. Amino acid sequences of metalloendopeptidases specific for acyl-lysine bonds from Grifola frondosa and Pleurotus ostreatus fruiting bodies. J. Biol. Chem. 27230032-30039. [DOI] [PubMed] [Google Scholar]
- 24.Pavan, M. E., S. L. Abbott, J. Zorzopulos, and J. M. Janda. 2000. Aeromonas salmonicida subsp. pectinolytica subsp. nov., a new pectinase-positive subspecies isolated from a heavily polluted river. Int. J. Syst. Evol. Microbiol. 501119-1124. [DOI] [PubMed] [Google Scholar]
- 25.Reed, L. J., and H. Muench. 1937. A simple method of estimating fifty per cent endpoints. Am. J. Hyg. 27493-497. [Google Scholar]
- 26.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology 1784-791. [Google Scholar]
- 27.Vipond, R., I. R. Bricknell, E. Durant, T. J. Bowden, A. E. Ellis, M. Smith, and S. MacIntyre. 1998. Defined deletion mutants demonstrate that the major secreted toxins are not essential for the virulence of Aeromonas salmonicida. Infect. Immun. 661990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viret, J. F. 1993. Meganuclease I-SceI as a tool for the easy subcloning of large DNA fragments devoid of selection marker. BioTechniques 14325-326. [PubMed] [Google Scholar]