Abstract
The specific and tightly controlled transport of numerous nutrients and metabolites across cellular membranes is crucial to all forms of life. However, many of the transporter proteins involved have yet to be identified, including the vitamin transporters in various human pathogens, whose growth depends strictly on vitamin uptake. Comparative analysis of the ever-growing collection of microbial genomes coupled with experimental validation enables the discovery of such transporters. Here, we used this approach to discover an abundant class of vitamin transporters in prokaryotes with an unprecedented architecture. These transporters have energy-coupling modules comprised of a conserved transmembrane protein and two nucleotide binding proteins similar to those of ATP binding cassette (ABC) transporters, but unlike ABC transporters, they use small integral membrane proteins to capture specific substrates. We identified 21 families of these substrate capture proteins, each with a different specificity predicted by genome context analyses. Roughly half of the substrate capture proteins (335 cases) have a dedicated energizing module, but in 459 cases distributed among almost 100 gram-positive bacteria, including numerous human pathogens, different and unrelated substrate capture proteins share the same energy-coupling module. The shared use of energy-coupling modules was experimentally confirmed for folate, thiamine, and riboflavin transporters. We propose the name energy-coupling factor transporters for the new class of membrane transporters.
Transport proteins residing in the cytoplasmic membrane allow the selective uptake and efflux of solutes and are essential for cellular growth and metabolism (20). Reflecting the importance of transporters, between 3% and 16% of the genes in prokaryote genomes are predicted to encode transporter proteins (26). These transporters form numerous families that are diverse in structure, energy-coupling mechanisms, and substrate specificities (25). As only a small fraction of predicted transporter proteins have known substrates, the functional prediction and annotation of the specificities of transporter proteins in the rapidly growing number of sequenced genomes represent a substantial challenge (25, 36). For example, the uptake of many cofactors and their precursors is essential for the growth of various pathogenic bacteria whose genomes are sequenced, but the transport proteins involved have not yet been identified. The use of computational comparative genomic techniques including gene colocalization, cooccurrence, and coregulation analyses combined with experimental assays is a powerful approach to identify novel transporters and to uncover their cellular role (for a recent review, see reference 11).
The starting point for the present analysis was our recent discovery of multicomponent transport systems for the vitamin biotin (BioYNM) and the transition metals nickel (NikMNQO) and cobalt (CbiMNQO) (14, 30). These transporters all have substrate-specific components (S components), which are integral membrane proteins, and energy-coupling modules. The S components of the biotin transporter (BioY) and the metal transporters (NikMN and CbiMN) are dissimilar in sequence, but the energy-coupling modules contain similar proteins. These modules consist of an ATPase typical of the ATP binding cassette (ABC) superfamily (A component) and a characteristic transmembrane protein (T component), with the stoichiometry being so far undefined. In many prokaryotes, genes encoding energy-coupling AT modules are unlinked to nikMN, cbiMN, and bioY. This observation prompted us to hypothesize the existence of AT-module-dependent transporters for additional substrates unrelated to transition metal ions and biotin.
In this study, we combined comparative genomics and experimental techniques to identify new transporters with an AT-module-plus-S-component architecture that are specific for various vitamins and related substrates. The majority of these systems are predicted to share a single AT module, a unique design among membrane transporters. The predicted modular design, substrate specificities of representative members, and shared use of the AT module by various substrate capture components were confirmed by biochemical analyses of vitamin transport systems of the Firmicutes.
MATERIALS AND METHODS
Bioinformatics analysis and data sources.
Prokaryotic genome sequences used for the comparative analysis were obtained from GenBank (1). Metabolic reconstruction, genome context analysis, and functional gene annotation were performed using the SEED comparative genomics resource as described previously (11, 24, 27). The results were captured in the “ECF class transporters” subsystem (http://theseed.uchicago.edu/FIG/subsys.cgi). Candidate DNA regulatory motifs were identified using Genome Explorer (9, 23, 27). Candidate RNA regulatory elements such as riboswitches were identified with the RNA-Pattern program (41) using input profiles from the RNA Families (Rfam) database (13). The Protein Families (Pfam) database was used to identify conserved functional domains (8). Transmembrane domains were predicted using the TMPred server (http://www.ch.embnet.org/software/TMPRED_form.html) (18).
Cloning and expression of L. casei transporters.
Lactobacillus casei folT and thiT were amplified using Taq DNA polymerase (Invitrogen, Carlsbad, CA) with L. casei ATCC 334 genomic DNA as the template and ligated into vector pNZ8048. The resulting constructs were used as templates for the amplification of fragments containing the nisinA promoter, folT or thiT, and a terminator using Pfu Ultra DNA polymerase (Invitrogen). Amplicons were ligated into vector pIL252 between the BamHI and XhoI sites. The ecfAA′T operon was amplified using Pfu Ultra DNA polymerase and inserted between the NcoI and SstI sites of vector pNZ8048. The recombinant plasmids were cloned into Lactococcus lactis strain NZ9000. Cells were grown at 30°C in supplemented M17 medium (Difco), and for expression, nisin (0.1% [vol/vol] of a culture supernatant of the nisin A-producing strain NZ9700) was added.
B. subtilis mutants and roseoflavin inhibition assay.
Bacillus subtilis ypaA (ribU), ybaF (ecfT), yuaJ (thiT), and yceI (niaP) disruption strains were obtained from the joint Japanese and European Bacillus subtilis consortium (40). For the roseoflavin inhibition assay, B. subtilis cells were grown overnight at 40°C in chemically defined medium containing glucose (4 g/liter), tryptophan (50 mg/liter), glutamine (2 g/liter), K2HPO4 (10 g/liter), KH2PO4 (6 g/liter), sodium citrate (1 g/liter), MgSO4 (0.2 g/liter), K2SO4 (2 g/liter), FeCl3 (4 mg/liter), and MnSO4 (0.2 mg/liter) in the presence of 0.5 mg/liter erythromycin (pMUTIN2 marker). These cultures were diluted ∼10- to 20-fold to yield the same cell density (optical density at 600 nm [OD600] of 0.05) in fresh medium and grown in duplicate in the absence or presence of roseoflavin (250 μM).
Vitamin uptake assays.
For [3H]5-formyltetrahydrofolate and [3H]thiamine uptake assays, L. lactis cells were washed once with cold phosphate-buffered saline (PBS) and resuspended in PBS at an OD600 of 20. Assays were performed at 30°C with stirring. Cells (500 μl) were preincubated for 5 min with glucose or 2-deoxyglucose (25 mM final concentration). Assays were started by adding 500 μl of PBS containing 2.2 μM [3H]5-formyltetrahydrofolate (specific activity of 0.4 μCi/nmol) or 2.3 μM [3H]thiamine (0.33 μCi/nmol) to the mixture. At intervals, 150-μl aliquots were passed through a Whatman cellulose nitrate membrane filter (0.45 μm). Filters were washed twice with 2 ml of ice-cold PBS, and cell-bound radioactivity was quantitated by liquid scintillation counting. For [3H]riboflavin uptake assays, Bacillus subtilis cells were cultivated in mineral salts medium at 37°C with vigorous shaking. At an OD600 of 0.5, [3H]riboflavin was added (16.6 nM final concentration; 588,000 dpm). Timed aliquots were mixed with 2 ml ice-cold 50 mM potassium phosphate buffer (pH 7.0), and cells were harvested immediately onto membrane filters (see above). The filters were washed twice with 2 ml of ice-cold buffer and dried. Radioactivity was determined by liquid scintillation counting.
Cloning and expression of Leuconostoc mesenteroides transporters.
Leuconostoc mesenteroides strain ATCC 8293 folT, panT, and ribU were amplified and inserted between the NcoI and BglII sites of pARCV (an expression vector containing an ampicillin resistance marker) (29). The resulting plasmids, pLmFolT, pLmPanT, and pLmRibU, encode the respective membrane proteins with a C-terminal FLAG tag. The ecfAA′T operon was likewise amplified and inserted into a variant of pARCV that harbored a streptomycin resistance gene and 10 histidine codons upstream of the insertion site. Plasmid pLmEcf encodes His10-EcfA (with a deca-His tag at the N terminus), EcfA′, and EcfT-FLAG (with a C-terminal FLAG tag). An ampicillin resistance-conferring variant of pLmEcf (pLmEcf-Amp) was constructed by the insertion of the ecfAA′T fragment from pLmEcf into pARCV.
Purification of transport protein complexes.
E. coli BL21 cells containing pLacI-RARE2 (encoding a Lac repressor and the tRNAs for rare codons) were used as the host for the heterologous production of L. mesenteroides proteins. Cells harboring pLmFolT, pLmPanT, pLmRibU, or pLmEcf-Amp individually or pLmEcf in combination with pLmFolT, pLmPanT, or pLmRibU were grown in 2 liters of Luria-Bertani broth supplemented with the appropriate antibiotics and 1 mM isopropyl-β,d-thiogalactopyranoside at 37°C with shaking to an OD578 of ∼2, harvested by centrifugation, washed in 35 mM sodium-potassium phosphate buffer (pH 7.0), resuspended in the same buffer containing a mixture of protease inhibitors (Roche), and disrupted by three passages through a French pressure cell. Membranes were pelleted by ultracentrifugation, resuspended and homogenized in 50 mM Tris-HCl (pH 8.0), and solubilized by agitation for 1 h in the presence of a solution containing 2% (wt/vol) dodecyl-β,d-maltoside, 5% (vol/vol) glycerol, 300 mM NaCl, and protease inhibitors at 4°C. Nonsolubilized material was pelleted by ultracentrifugation. Imidazole was added to the supernatant to a final concentration of 20 mM, and the solution (10 ml) was mixed with 0.5 ml Ni-nitrilotriacetic acid Superflow resin (Qiagen) and incubated for 30 min at 4°C with rotation. After transfer to an empty column, the resin was washed with 50 mM Tris-HCl (pH 7.5) containing 0.05% dodecyl-β,d-maltoside, 5% glycerol, 300 mM NaCl, and 100 mM imidazole. Bound protein was eluted with 4 ml of this buffer containing 500 mM imidazole. The protein solution was concentrated eightfold by Amicon concentrators (30-kDa cutoff), and samples (∼10 μg protein) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were blotted onto nitrocellulose membranes and probed with anti-penta-His (Qiagen) or anti-FLAG (Sigma) antibodies by standard Western blotting protocols. For peptide mass fingerprint analysis, slices were excised from gels and incubated overnight in trypsin solution. Filtered samples were subjected to high-performance liquid chromatography-coupled electrospray ionization time-of-flight mass spectrometry using an Agilent 1100 system. Upon ionization, masses (m/z) were determined in the range of between 20 and 3,500, and peptides were matched using the BIOCONFIRM software package (Agilent).
Chemicals.
[3′,5′,7,9-3H(N)]-(6S)-5-Formyltetrahydrofolic acid diammonium salt (10 Ci/mmol), [3H(G)]thiamine hydrochloride (10 Ci/mmol), and [3H(G)]riboflavin (24 Ci/mmol) were obtained from Moravek Biochemicals (Brea, CA). (6R,6S)-5-Formyltetrahydrofolic acid calcium salt was obtained from Schircks Laboratories (Jona, Switzerland). Roseoflavin was obtained from Toronto Research Chemicals Inc. (North York, Ontario, Canada).
RESULTS
Comparative genomics of AT-module-dependent transport systems.
A comprehensive bioinformatic analysis of 365 prokaryotic genomes using the SEED comparative genomics platform (24) revealed that the abundance and functional diversity of transporters with the novel AT architecture extend far beyond the few cases of metal and biotin transporters noted above. Thus, 432 gene cassettes encoding A and T components (AT modules) were found in 238 genomes, with the A-component genes being very often duplicated (A and A′) (see Tables S1 and S2 in the supplemental material). These AT gene cassettes fall into two groups based on their genomic organizations (Fig. 1). Three hundred thirty-five of them (group I, found in diverse microbes) resemble the previously described nickel, cobalt, and biotin transporters (14, 29) in that they occur next to genes encoding small integral membrane proteins. These membrane proteins are candidate S components, but as fewer than half are related to the S components of the nickel, cobalt, or biotin transporters, it is likely that the majority of them have novel substrate specificities. The remaining 97 AT gene cassettes (group II, found mainly in the Firmicutes, the Thermotogales, and some members of the Archaea) do not have adjacent candidate S-component genes. Almost all these cassettes are accompanied by various candidate S-component genes (459 in total) scattered elsewhere in the genome, with some genomes having as many as 12 distinct S-component genes but only one or two copies of the energy-coupling AT module. We predict that in such cases, multiple S components can use the same AT module to form an active transporter complex. The vast majority of these candidate S components are again unrelated to S components of the nickel, cobalt, or biotin transporters, and hence, most of them probably act on new substrates.
FIG. 1.
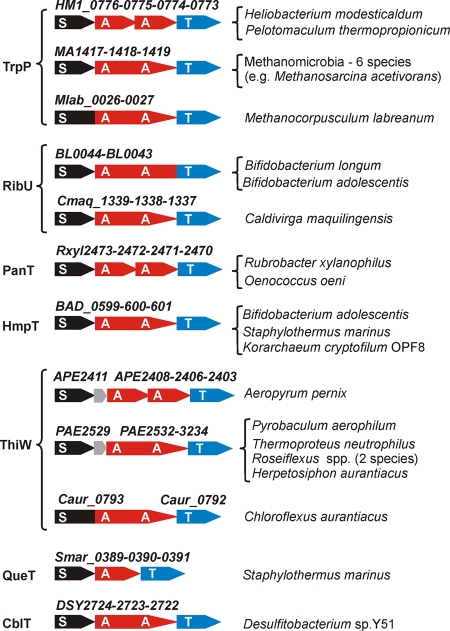
Distribution and comparative genomic analysis of the new class (ECF class) of prokaryotic transporters. (A) Classification and abundance of group I and group II ECF transporters. Group I transporters have a substrate-specific S component and a dedicated AT module encoded by linked genes. Group II transporters have individual S components and shared AT modules that are unlinked to S components. Composite bar colors indicate the contributions of transporters found in different taxa to the total transporter number. Note that the S components BioY, CbrT, HtsT, and QrtT (and, to a lesser extent, RibU, PanT, HmpT, ThiW, QueT, and CblT) occur in both groups. (B) Comparative genomic analysis of the identified transporter families including their domain compositions, names, predicted substrate specificities, and example gene identifications. Substrate-specific integral membrane components (S) are shown by black rectangles, conserved transmembrane components (T) are shown by blue rectangles, and ATPase domains (A) are shown by red circles. Examples of genome context evidence (e.g., gene coregulation or colocalization) supporting the predicted transporter function are shown on the right.
(i) Functional assignments for S components.
The S components were classified into 21 protein families (Table 1). Previously reported experimental data implicated five of these families in transport: the BioY, CbiMN, and NikMN families mentioned above plus one family involved in riboflavin uptake (RibU [YpaA] in Bacillus subtilis and Lactococcus lactis) and another involved in tryptophan uptake (TrpP in B. subtilis) (2, 5, 43, 45). For 15 of the remaining 16 families, we were able to predict substrate specificities by combining genome context analysis and metabolic reconstruction (11, 27). The results show a notable preference for substrates that are vitamins or their precursors, namely, folate, pantothenate, niacin, pyridoxine, lipoate, thiamine, and its precursors, and the precursors of cobalamin, methionine, and queuosine (Fig. 1 and Table 1).
TABLE 1.
Functional roles of substrate capture components of ECF transporters
| Transporter | Pfam accession no.a | Substrateb | Evidence for specificityc | Evidence for AT module dependenced | Reference(s) and/or sourcee |
|---|---|---|---|---|---|
| BioY | PF02632 | Biotin | E, R, C, O | E, C, O | 14, 27, 28, 31 |
| NikMN | PF01891 | Nickel | E, R, C | E, C | 29 |
| CbiMN | PF01891 | Cobalt | E, R, C | E, C | 29, 33 |
| YkoE | Thiamine precursors (HMP?) | R, C | C | 34 | |
| MtsT | PF07155 | Methionine precursors | R, C | C | 32 |
| HtsT | PF09605 | ? | C, O | ||
| QrtT | Queuosine precursor | R, C, O | C, O | ||
| CbrT | Cobalamin precursor | C, O | C, O | ||
| MtaT | Methylthioadenosine | C | C | ||
| RibU | Riboflavin | E, R, O | E, C, O | 2, 5, 10, 21, 42, 43; this study | |
| FolT | Folate | E, C, O | E, O | This study | |
| QueT | PF06177 | Queuosine precursor | R | C, O | |
| PanT | Pantothenate | C, O | E, C, O | This study | |
| HmpT | Thiamine precursor (HMP?) | C, O | C, O | ||
| ThiT | PF09515 | Thiamine | E, R, O | E, O | 7, 34; this study |
| ThiW | Thiazole | R, C, O | C, O | 34 | |
| NiaX | Niacin | R, O | O | 30 | |
| TrpP | Tryptophan | E, R | E, C, O | 37, 41 | |
| PdxT | Pyridoxine | C | O | ||
| CblT | Cobalamin precursor (DMB?) | R, C, O | C, O | 33 | |
| LipT | Lipoate | C | O |
Identifiers of protein families are from the Pfam database (http://pfam.sanger.ac.uk/) (8).
HMP, hydroxymethylpyrimidine; DMB, dimethylbenzimidazole.
Evidences for the function of S components include experimental analysis (E), coregulation of their genes in a regulon (R), colocalization on the chromosome (C), or cooccurrence (O) in the genome with metabolic genes from the same pathway.
Evidences for the AT module dependence of S components. E, experimental; C, colocalization on the chromosome (C), or cooccurrence (O) in the genome with AT module gene cassettes.
References describing genomic predictions and experimental validations are in italics and in bold, respectively.
The transport systems for folate, thiamine, and biotin in Lactobacillus casei were postulated by Henderson et al. in the 1970s to comprise individual substrate-binding membrane proteins plus a common energy-coupling component (named “energy-coupling factor” [ECF]) (15). Below, we provide experimental evidence that these L. casei transporters belong to the class of transporters reported here. In recognition of the pioneering work, we accordingly propose the name “ECF” for the new class and ecfA and ecfT for the genes encoding A and T components of the conserved energy-coupling modules that are shared by multiple S components. A detailed comparative genomic analysis of individual S-component families of the ECF transporters is provided below.
(ii) Nickel and cobalt transporters.
The uptake of the transition metals cobalt and nickel, essential components of various enzymes, is mediated by transporters of several different types (6). Previous comparative genomic analyses identified the homologous CbiMNQO and NikMNQO transport systems as being the most widespread transporters for cobalt and nickel ions, respectively (29). The metal specificities were predicted using a combination of the identification of specific regulatory elements, namely, B12 riboswitches (B12 elements) and binding sites for the nickel-dependent repressor (NikR), and the analysis of colocalization with genes encoding nickel-dependent enzymes or enzymes involved in B12 biosynthesis. The predicted specificities were experimentally confirmed by metal uptake assays upon the recombinant expression of the cbiMNQO and nikMNQO operons (29). In these transport systems, the transmembrane “Q” proteins (T component) and the “O” proteins (A components) energize their cognate metal-specific “MN” modules (S components). Thus, these systems belong to group I of the ECF transporter class. The “M” components constitute a unique family of integral membrane proteins with two separate subfamilies correlating with specificity for cobalt and nickel, respectively.
(iii) Biotin transporter.
A biotin uptake function for the BioY protein family was previously predicted in the comparative genomic reconstruction of the biotin regulon governed by BirA (31). The widespread biotin repressor BirA and two specialized transcriptional factors from different families, BioR in alphaproteobacteria (28) and BioQ in the Actinobacteria (27), control the biotin synthesis genes and/or the biotin transporter gene bioY. In addition to the evidence from multiple regulons, bioY clusters with biotin synthesis genes in some genomes. On the other hand, BirA-regulated bioY genes occur in some organisms without the biotin synthesis pathway. The collective evidence from coregulation, colocalization, and cooccurrence experiments strongly implied that BioY proteins are transporters involved in biotin uptake. Members of the BioY family belong to either the group I transporters that are accompanied by the dedicated AT module BioMN (44 cases, belonging mostly to the Proteobacteria and Actinobacteria) or the group II transporters that depend on the shared EcfAA′T module (81 cases, belonging mostly to the Firmicutes). Some bioY genes (10 cases, mostly in the Archaea but also in Desulfitobacterium sp.) are located adjacent to ecfAA′T gene cassettes. A detailed phylogenetic analysis of the BioY family was performed recently. In a series of biochemical experiments, BioY from Rhodobacter capsulatus was identified as being a biotin capture and transport protein, which is converted into a high-affinity biotin transporter in the presence of its cognate BioMN module (14).
(iv) Riboflavin transporter.
The candidate riboflavin transporter gene ypaA (ribU) was first identified by comparative genomic analysis of the riboflavin biosynthesis regulons governed by RFN (for riboflavin) regulatory elements (10, 21, 42). The RFN-regulated ribU genes were identified both in genomes with the de novo riboflavin synthesis genes and in those of riboflavin auxotrophs that lack the de novo synthesis pathway (e.g., some Streptococcus species). The RFN element serves as the receptor to form a metabolite-dependent riboswitch that directly binds flavin mononucleotide in the absence of proteins (44). The participation of RibU from Bacillus subtilis and Lactococcus lactis in riboflavin uptake was demonstrated by vitamin uptake assays (2, 43). The very high affinity of the RibU protein for riboflavin (Kd [dissociation constant] of 0.6 nM) (5) likely allows the cells to scavenge the vitamin from environments with low concentrations of riboflavin. The group II RibU transporters are widely distributed among the Firmicutes and the Thermotogales (63 cases). Additional RibU transporters attributed to group I were identified in some Archaea and Actinobacteria species, providing genomic evidence for their dependence on an energy-coupling module (Fig. 2). For instance, ribU in Bifidobacterium species forms an operon with a gene encoding a pair of ABC ATPase domains and a transmembrane domain that is homologous to T components of AT modules.
FIG. 2.
Genomic organization of the group I ECF transporters containing S components from the TrpP, RibU, PanT, HmpT, ThiW, QueT, and CblT families. Genes encoding substrate capture S components and A and T components of the dedicated energy-coupling modules are shown by black, red, and blue arrows, respectively.
(v) Tryptophan transporter.
The candidate tryptophan transporter YhaG (TrpP) was previously identified as being a member of the tryptophan regulon governed by the TRAP attenuation protein in B. subtilis (37, 45). On the other hand, trpP orthologs in the Clostridia are regulated by tryptophan-specific T-box RNA structures (41). The involvement of trpP in tryptophan transport was confirmed by the sensitivity of the trpP mutant to growth-inhibiting levels of tryptophan analogs (37). TrpP transporters belong to either the group I transporters that are accompanied by the dedicated energy-coupling module (in methanogenic Archaea and in some of the Firmicutes) (Fig. 2) or the group II transporters that depend on the shared EcfAA′T module (14 cases, in the Firmicutes).
(vi) Transporters of thiamine and thiamine precursors.
Previous comparative genomic analyses of the thiamine regulons governed by THI elements (thiamine-pyrophosphate riboswitches) resulted in the identification of several candidate thiamine-related transporters, including YuaJ/ThiT, YkoEDC, and ThiW (34). Analysis of the distribution of genes involved in thiamine metabolism in the Firmicutes has revealed that thiT is the only thiamine-regulated gene in several pathogens that have no thiamine biosynthesis pathway, suggesting that it is involved in thiamine uptake (34). The YkoEDC system was predicted to be involved in salvaging the thiamine precursor hydroxymethylpyrimidine, based on the positional clustering of the ykoEDC cassette with the thiaminase II gene tenA, and some other thiamine salvage genes (34). Genetic and microarray studies of B. subtilis mutants in the thiT and ykoD genes demonstrated that both mutations result in a derepression of thiamine-regulated genes, confirming the involvement of thiT and ykoEDC in the uptake of thiamine and its precursors (38). The thiamine-related transporter ThiW was predicted to be involved in the uptake of the thiazole precursor of thiamine based on the colocalization of the thiW with thiM genes, with the latter encoding the thiazole salvage enzyme hydroxyethylthiazole kinase (34). In this work, we used the occurrence profile of ThiT transporters, which are present only in the Firmicutes (34 cases), to attribute them to group II ECF transporters. Thiamine binding activity was verified experimentally for the L. casei ThiT protein (7). ThiW transporters belong to either the group I transporters that are accompanied by the dedicated AT module (in some members of the Archaea and Chloroflexi) (Fig. 2) or the group II transporters that depend on the shared EcfAA′T module (21 cases, mostly in members of the Firmicutes but also in Rubrobacter xylanophilus and Korarchaeum cryptofilum). YkoEDC transporters (group I; 44 cases) always have an integral membrane substrate capture component (YkoE), a dedicated AT module composed of duplicated ABC ATPase domains fused in a single protein (YkoD), and a transmembrane T component (YkoC).
(vii) Folate transporter.
The candidate folate transporter FolT was identified using comparative genomic analyses of the folate biosynthesis subsystem and the previously published amino acid composition and molecular mass data (16) for the L. casei folate binding protein (7). The folT genes in several Mycoplasma genomes and in Streptococcus suis cluster on the chromosome with the folC gene, encoding the bifunctional enzyme dihydrofolate synthase/folylpolyglutamate synthase, which can add a polyglutamyl tail to folate molecules. The folT gene is widely distributed in the Firmicutes (55 cases) and is also present in a single Thermotogales species; thus, it occurs only in genomes encoding the shared energy-coupling module. Therefore, FolT was predicted to function as a group II ECF transporter. Folate binding activity was verified experimentally for the L. casei FolT protein (7).
(viii) Pantothenate transporter.
The candidate pantothenate transporter PanT was identified in this study by genome context analysis of the coenzyme A biosynthesis subsystem in the SEED database. Many free-living bacteria are capable of de novo coenzyme A biosynthesis via the precursor pantothenate, whereas numerous pathogens are dependent on exogenous pantothenate. In many cases, the panT gene is colocalized on the chromosome with various pantothenate salvage genes including the phosphopantothenoylcysteine decarboxylase gene coaB, the phosphopantothenoylcysteine synthetase gene coaC, and the pantothenate kinase gene coaX. PanT was found mostly in members of the Firmicutes that require pantothenate for growth (Lactobacillales, Streptococcus, pathogenic members of the Clostridia, and Mycoplasma); thus, it functions presumably as a group II transporter. In contrast, the panT genes in R. xylanophilus and Oenococcus oeni cluster with genes encoding A and T components of the dedicated energy-coupling module (Fig. 2), and consequently, the corresponding PanT transporters were attributed to group I.
(ix) Queuosine precursor transporters.
The candidate queuosine precursor transporters QrtT and QueT were first predicted in this work using the observed coregulation of the qrtT and queT genes by PreQ1 riboswitches that sense the modified nucleobase PreQ1 (7-aminomethyl-7-deazaguanine) and control the expression of queuosine synthesis genes (22, 35). The QrtT transporters belong to either the group I transporters that are accompanied by the dedicated AT module QrtUVW (16 cases, in members of the Enterobacteria, Actinobacteria, and Thermotogales) or the group II transporters that depend on the shared EcfAA′T module (8 cases, in the Firmicutes). The QueT transporters were found to belong mostly to group II (41 cases, in the Firmicutes but also in R. xylanophilus, Petrotoga mobilis, and two Archaea species). However, in a single genome of Staphylothermus marinus, the group I QueT transporter is accompanied by the dedicated energy-coupling module (Fig. 2).
(x) Cobalamin precursor transporter.
The candidate coenzyme B12-regulated transporter CblT was identified in a previous comparative genomic analysis of the B12 regulon in bacteria (33). Orthologs of the cblT gene in members of the Firmicutes are regulated by B12 riboswitches and colocalized with the cobalamin biosynthesis genes (cbi). Analysis of distributions of genes from the B12 synthesis pathway suggested a possible role of CblT in the uptake of dimethylbenzimidazole (33), a compound attached to the cobinamide precursor to form cobalamin. Here, we attributed most of the CblT transporters in the Firmicutes to group II (11 cases). In a single species of the Firmicutes that lacks the shared energy-coupling cassette (Desulfitobacterium sp.), CblT is accompanied by a dedicated energy-coupling module (Fig. 2), thus constituting a group I transporter.
(xi) Cobalamin transporter.
The CbrT transporters belong to either the group I transporters that are accompanied by the dedicated AT module CbrUV (18 cases, in members of the Actinobacteria and some members of the Firmicutes) or the group II transporters that depend on the shared EcfAA′T module (11 cases, in the Firmicutes). The genomic colocalization of cbrT with the cobalamin adenosyltransferase (pduO) and adenosylcobalamin-dependent ribonucleotide reductase (nrdJ) genes in four Lactobacillus species suggests the involvement of CbrT in cobalamin uptake. In addition, the cbrT gene of L. reuteri belongs to a large gene cluster encoding the B12 biosynthesis pathway and the B12-dependent pathway of propanediol utilization. With the exception of L. reuteri, other analyzed Lactobacillus genomes lack all known genes involved in adenosylcobalamin biosynthesis and salvage. Genomic analysis of four Lactobacillus species that possess the nrdJ-cbrT-pduO gene cluster found NrdJ as a single known B12-dependent enzyme, providing an explanation for the observed colocalization of the genes involved in the salvage and utilization of cobalamin.
(xii) Methylthioadenosine transporter.
Methylthioadenosine is a degradation product of S-adenosylmethionine and recycled to form methionine. A group I transporter encoded by the mtaTUV gene cassette was identified in only three bacterial genomes and a single archaeal genome. The colocalization of mtaTUV with the mtnPNKA genes involved in methylthioadenosine salvage implicates MtaTUV in the uptake of external methylthioadenosine.
(xiii) Methionine precursor transporter.
A group I transporter encoded by the mtsTUV gene cluster was identified in members of the Proteobacteria and the Firmicutes and in some members of the Archaea (33 cases) as an optional member of various methionine regulons, suggesting its involvement in the uptake of a methionine precursor. For example, the mtsTUV operon is regulated by the S-adenosylmethionine riboswitch (S box) in Bacillus cereus, by the methionine-specific T-box attenuator in Leuconostoc mesenteroides, and by the methionine regulator MtaR in Streptococcus species (32). In proteobacteria, the mtsTUV cassette was found only in Vibrio species, where it is predicted to be regulated by the methionine repressor MetJ, as it is preceded by candidate MetJ binding sites.
(xiv) Pyridoxine transporter.
The group II transporters of the PdxT family were found only in the Firmicutes (11 cases) and are weakly similar (16 to 18% identity) to BioY, CblT, CbrT, and PanT. The colocalization of pdxT with the pyridoxine kinase gene pdxK and the absence of de novo pyridoxine synthesis in Lactobacillus and Streptococcus suggest a role for PdxT in pyridoxine uptake.
(xv) Thiamine precursor transporter.
Most of the HmpT transporters belong to group II, which depends on the shared energy-coupling module (38 cases, in members of the Firmicutes and the Thermotogales and some members of the Archaea). The group I HmpT transporters accompanied by a dedicated AT module (Fig. 2) are present in two members of the Archaea and a single species of the Actinobacteria. Positional analysis of the hmpT genes suggests their likely involvement in the salvage of thiamine or its precursor hydroxymethylpyrimidine. In Streptococcus and Enterococcus species, hmpT is located in a possible operon with a paralog of the phosphomethylpyrimidine kinase gene thiD, whereas in the Archaea, it is colocalized with the thiamine-monophosphate kinase gene thiL.
(xvi) Niacin transporter.
The candidate niacin transporter NiaX was previously identified in the comparative genomics study of the niacin regulon controlled by NiaR (30). The NiaR-regulated niaX genes are present in Streptococcus and Enterococcus species that lack genes for de novo NAD biosynthesis, suggesting the involvement of NiaX in niacin uptake. Here, we attributed the NiaX transporters to the group II ECF transporters (18 cases) based on their limited distribution only in the Firmicutes and weak similarity (16 to 19% identity) to QrtT and RibU.
(xvii) Lipoate transporter.
The group II transporters of the LipT family were identified only in three Phytoplasma species that possess the shared EcfAA′T module. The LipT proteins are weakly similar (16 to 19% identity) to ThiT, FolT, PdxT, RibU, and HmpT. The colocalization of lipT with the lipoate-protein ligase A gene lplA and the absence of the lipoate synthesis pathway in Phytoplasma suggest a role for LipT in lipoate uptake.
(xviii) Other transporters.
Comparative genomic analysis failed to suggest a specific function for group I transporters encoded by the htsTUV genes (named Hts for hypothetical transport system) that were found mostly in animal-associated members of the Firmicutes (e.g., Streptococcus) and the Actinobacteria. The gram-negative oral spirochete Treponema denticola has nine copies of the htsTUV cassette.
The presence of ecfAA′T genes encoding candidate energy-coupling modules in Methanosarcina species, which lack genes encoding any of the above-described 21 families of S components, points to the existence of additional ECF transporters with unrecognized specificities.
Experimental validation of AT module sharing by vitamin transporters.
To validate our bioinformatic analysis, we tested three of its predictions, namely, that (i) the previously characterized riboflavin transporter RibU depends on the AT module; (ii) the S components FolT and ThiT are specific for folate and thiamine, respectively, and are AT module dependent; and (iii) the multiple S components of group II transporters (Fig. 1) share a common energy-coupling module (EcfAA′T).
(i) RibU interacts with EcfAA′T in B. subtilis.
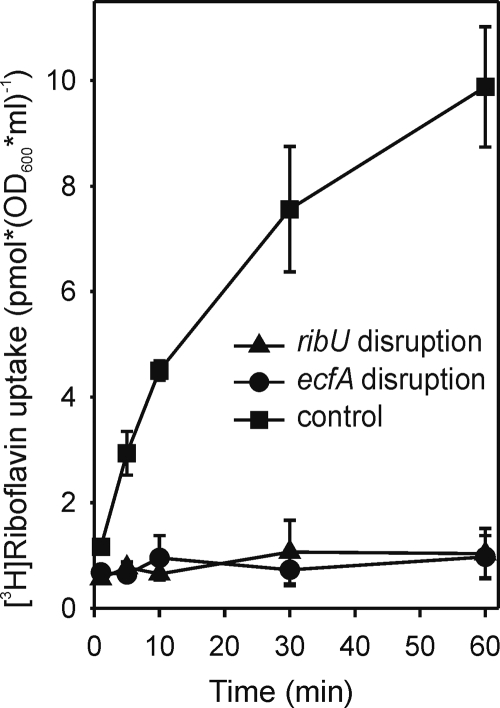
The dependence of riboflavin transport on the S component (RibU) and the energy-coupling module was analyzed in B. subtilis knockout strains. [3H]Riboflavin transport required functional ribU (ypaA) and ecfT (ybaF) genes; the disruption of either gene abolished the uptake of the vitamin (Fig. 3). This result was corroborated by inhibitor studies with the toxic riboflavin analog roseoflavin, which enters cells via the riboflavin transporter (43); disrupting either ribU or ecfT reduced sensitivity to this inhibitor (see Fig. S1 in the supplemental material).
FIG. 3.
Riboflavin uptake in Bacillus subtilis. Shown are data for the effect of disrupting ribU (triangles), ecfT (circles), or yceI (squares) on [3H]riboflavin uptake by B. subtilis. (The yceI gene served as a control; it encodes a protein unrelated to ECF transporters.) Cells were grown without riboflavin to an OD600 value of 0.5, and [3H]riboflavin was added (17 nM final concentration). At the times indicated, cells were harvested by filtration and washed, and their 3H contents were determined. Values are means of duplicates; error bars indicate ranges.
(ii) Substrate specificity and AT module dependency of FolT and ThiT.
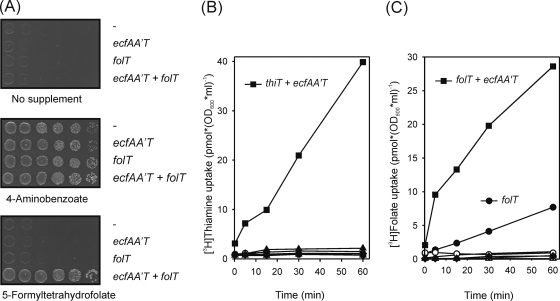
The predicted substrate and the AT module dependence of FolT were tested first using proteins from Leuconostoc mesenteroides. The folT and ecfAA′T genes were expressed in a folate-auxotrophic Escherichia coli strain (pabA abgT), which cannot produce the folate precursor 4-aminobenzoate or take up folates. When analyzed for growth on minimal agar medium (19), only transformants containing both folT and the ecfAA′T cassette were able to utilize 5-formyltetrahydrofolate (Fig. 4A). This establishes that folate is transported and that transport requires the energy-coupling module.
FIG. 4.
Folate and thiamine uptake evidences. (A) Folate uptake by E. coli cells coexpressing L. mesenteroides folT and ecfAA′T. Recombinants containing empty vector or expression plasmids for the production of FolT, EcfAA′T, or FolT plus EcfAA′T were spotted (10 μl) after serial 10-fold dilutions onto nonsupplemented minimal medium and onto minimal medium containing 4-aminobenzoate (3.6 μM) or 5-formyltetrahydrofolate (11 μM). Plates were incubated for 48 h at 37°C. (B and C) Uptake of [3H]thiamine (B) and [3H]5-formyltetrahydrofolate (C) by L. lactis containing empty vectors (triangles) or carrying L. casei thiT or folT (circles), ecfAA′T (diamonds), or thiT or folT coexpressed with ecfAA′T (squares). Cells were energized with glucose (black symbols) or deenergized with 2-deoxyglucose (open symbols).
We then produced the FolT, ThiT, and EcfAA′T proteins from L. casei in various combinations in Lactococcus lactis and assayed [3H]thiamine and [3H]5-formyltetrahydrofolate uptake in energized or deenergized cells (Fig. 4B and C). Deenergized cells acquired neither thiamine nor 5-formyltetrahydrofolate regardless of which proteins they expressed. Among energized cells, only those coexpressing thiT and ecfAA′T acquired significant amounts of thiamine. The situation was similar for 5-formyltetrahydrofolate except that cells expressing folT alone acquired some label; this slow uptake may be due to the functional interaction of FolT with the endogenous L. lactis EcfAA′T module or possibly to the basal activity of the solitary FolT protein. These data confirm activity with the predicted FolT and ThiT substrates and demonstrate the dependence of both thiamine and folate uptake on the same EcfAA′T module, as inferred in the classical work of Henderson et al. (15).
(iii) Physical interaction of an AT module with various S components.
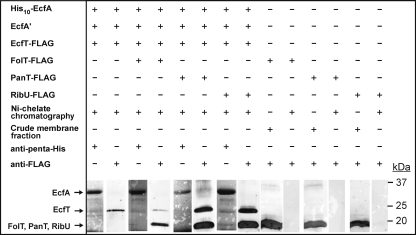
The shared use of the common energy-coupling module was also tested by a series of pullout experiments. His-tagged EcfA, untagged EcfA′, and FLAG-tagged EcfT from L. mesenteroides were coproduced in E. coli with or without FLAG-tagged FolT, PanT, or RibU. Membranes of the recombinant strains were solubilized and subjected to nickel-chelate affinity chromatography, followed by SDS-PAGE and Western blotting. The three EcfAA′T proteins copurified during chromatography (Fig. 5), as indicated by immunodetection (for His-tagged EcfA and FLAG-tagged EcfT) and electrospray ionization time-of-flight peptide mass fingerprint analysis (for EcfA′) (not shown). As expected, FLAG-tagged FolT, PanT, and RibU did not bind to the affinity resin in the absence of the EcfAA′T module (Fig. 5, six right-hand lanes). However, each of these S components copurified with the EcfAA′T complex. The EcfA, EcfA′, and EcfT components thus form a stable tripartite complex and can form quadripartite complexes with each of three different S components.
FIG. 5.
Physical interaction between S and ECFAA′T components from L. mesenteroides. Membranes of recombinant E. coli cells producing the proteins indicated in the upper six lines were solubilized with n-dodecyl-β-d-maltoside and subjected to Ni-chelate affinity chromatography prior to SDS-PAGE and Western blotting or were separated by SDS-PAGE without chromatography. The bottom shows strips of the blots probed with anti-penta-His or anti-FLAG antibodies.
DISCUSSION
The broad distribution, functional versatility, and modular assembly of the energy-coupling module-dependent transport systems are summarized in Fig. 1 and Fig. S2 in the supplemental material. The ECF transporters form a novel class of membrane transporters that can be classified into two groups: group I includes transporters from diverse microbial lineages (170 species) that have a dedicated AT module encoded in the same gene cluster as an S component, and transporters of group II (a total of 459 transporters in 91 species) employ a universal energy-coupling module (EcfAA′T) that is encoded by a separate gene cassette and shared by many different unlinked S components. Group II is ubiquitous in the phyla Firmicutes and Thermotogales and also occurs in some members of the Archaea.
The S components identified in this work could be classified into at least 20 protein families that correspond to different substrate specificities (Table 1). Most of them are integral membrane proteins of comparable sizes (155 to 230 residues) that have six predicted transmembrane domains. The NikM and CbiM proteins, which form a single family in the Pfam database, are larger (210 to 250 residues) and are predicted to have seven transmembrane domains. Apart from the CbiM/NikM family, only five other S-component families (BioY, MtsT, HtsT, QueT, and ThiT) are present in the Pfam database, and all of them are annotated as hypothetical membrane proteins. Sequence comparisons of representative S components from 18 families revealed very little overall pairwise identity between the proteins from different families (see Table S3 in the supplemental material). A detailed phylogenetic comparison will be needed to establish whether different S components are related.
The ECF transporters identified in this study are mechanistically unique. Their substrate specificity is mediated by integral membrane proteins (S components), which form active transporters in the presence of the energy-coupling AT module. The stoichiometry of the transporter components is unknown, but domain fusions in various ECF transport systems give some clues. First, the nik, cbi, and bio gene cassettes encode a single A component (ATPase), but as noted above, dual A components are more common. Second, in some cases, the two A components are fused (Fig. 1B and 2). Third, rare fusions of transporter components include two “SAA” fusions, four “TAA” fusions, and one “ST” fusion in the Archaea, the Chloroflexi, and the Actinobacteria (Fig. 2 and see Table S1 in the supplemental material). On this basis, and because shared EcfAA′T components and specific S components formed quadripartite complexes (Fig. 5), we propose a quadripartite model in which the S component binds and translocates the substrate across the membrane. The translocation process is coupled to ATP hydrolysis mediated by an AT module that contains two ATPase domains and one transmembrane T component.
How do ECF transporters relate to the ABC transporter superfamily? The latter transporters couple ATP hydrolysis to substrate uptake or efflux (3, 4). ABC importers and exporters share a four-component architecture comprised of two transmembrane and two ATP-hydrolyzing domains. Prokaryotic ABC importers have additional extracytoplasmic soluble proteins that mediate substrate binding and delivery to the respective transmembrane components. Fundamental differences between ECF transporters and classical ABC importers include (i) the absence of extracytoplasmic substrate binding proteins and their replacement by integral membrane proteins and (ii) the shared use of energy-coupling AT modules by many highly diverse S components. Such sharing is occasionally seen in classical ABC transporters, but it always involves very similar substrates and substrate binding proteins (17). A less fundamental but nonetheless marked characteristic of ECF transporters is a predilection for vitamins.
Finally, as noted at the outset, numerous human pathogens such as Mycoplasma, Ureaplasma, and Streptococcus strains rely totally upon transporters to obtain vitamins and other essential metabolites due to the absence of the corresponding de novo biosynthetic pathways. Many of these microorganisms use ECF transporters, and indeed, certain ecf genes have been found to be essential for the growth and survival of Streptococcus pneumoniae and Mycoplasma genitalium (12, 39). All components of ECF transporters, especially the unique S and T proteins, are thus potential targets for antibiotic development. In fact, the centrality of the T component to the uptake of multiple compounds makes it a classic Achilles' heel.
Supplementary Material
Acknowledgments
This work was supported by grants from the Deutsche Forschungsgemeinschaft (EI 374/3-1 to T.E. and P.H.), the National Institute of Allergy and Infectious Diseases (R01-AI066244-01A2 to A.L.O. and D.A.R.), the National Institutes of Health (R01-GM071382 to A.D.H.), The Netherlands Organization for Scientific Research (vidi grant to D.J.S. and Toptalent grant to J.T.B.), and the Molecular and Cellular Biology program of the Russian Academy of Sciences and the Howard Hughes Medical Institute (55005610 to M.S.G.).
We thank A. Pohlmann (Berlin, Germany) for peptide mass analysis and E. Dervyn (INRA, France) for B. subtilis disruptants.
Footnotes
Published ahead of print on 17 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2007. GenBank. Nucleic Acids Res. 35D21-D25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgess, C. M., D. J. Slotboom, E. R. Geertsma, R. H. Duurkens, B. Poolman, and D. van Sinderen. 2006. The riboflavin transporter RibU in Lactococcus lactis: molecular characterization of gene expression and the transport mechanism. J. Bacteriol. 1882752-2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73241-268. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, A. L., E. Dassa, C. Orelle, and J. Chen. 2008. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol. Mol. Biol. Rev. 72317-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duurkens, R. H., M. B. Tol, E. R. Geertsma, H. P. Permentier, and D. J. Slotboom. 2007. Flavin binding to the high affinity riboflavin transporter RibU. J. Biol. Chem. 28210380-10386. [DOI] [PubMed] [Google Scholar]
- 6.Eitinger, T., J. Suhr, L. Moore, and J. A. Smith. 2005. Secondary transporters for nickel and cobalt ions: theme and variations. Biometals 18399-405. [DOI] [PubMed] [Google Scholar]
- 7.Eudes, A., G. B. Erkens, D. J. Slotboom, D. A. Rodionov, V. Naponelli, and A. D. Hanson. 2008. Identification of genes encoding the folate- and thiamine-binding membrane proteins of Lactobacillus casei and other firmicutes. J. Bacteriol. 1907591-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelfand, M. S., E. V. Koonin, and A. A. Mironov. 2000. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 28695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gelfand, M. S., A. A. Mironov, J. Jomantas, Y. I. Kozlov, and D. A. Perumov. 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet. 15439-442. [DOI] [PubMed] [Google Scholar]
- 11.Gelfand, M. S., and D. A. Rodionov. 2008. Comparative genomics and functional annotation of bacterial transporters. Physics Life Rev. 522-49. [Google Scholar]
- 12.Glass, J. I., N. Assad-Garcia, N. Alperovich, S. Yooseph, M. R. Lewis, M. Maruf, C. A. Hutchison III, H. O. Smith, and J. C. Venter. 2006. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. USA 103425-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffiths-Jones, S., S. Moxon, M. Marshall, A. Khanna, S. R. Eddy, and A. Bateman. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33D121-D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hebbeln, P., D. A. Rodionov, A. Alfandega, and T. Eitinger. 2007. Biotin uptake in prokaryotes by solute transporters with an optional ATP-binding cassette-containing module. Proc. Natl. Acad. Sci. USA 1042909-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson, G. B., E. M. Zevely, and F. M. Huennekens. 1979. Mechanism of folate transport in Lactobacillus casei: evidence for a component shared with the thiamine and biotin transport systems. J. Bacteriol. 1371308-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson, G. B., E. M. Zevely, and F. M. Huennekens. 1977. Purification and properties of a membrane-associated, folate-binding protein from Lactobacillus casei. J. Biol. Chem. 2523760-3765. [PubMed] [Google Scholar]
- 17.Higgins, C. F., and G. F. Ames. 1981. Two periplasmic transport proteins which interact with a common membrane receptor show extensive homology: complete nucleotide sequences. Proc. Natl. Acad. Sci. USA 786038-6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hofmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler 374166. [Google Scholar]
- 19.Klaus, S. M., E. R. Kunji, G. G. Bozzo, A. Noiriel, R. D. de la Garza, G. J. Basset, S. Ravanel, F. Rebeille, J. F. Gregory III, and A. D. Hanson. 2005. Higher plant plastids and cyanobacteria have folate carriers related to those of trypanosomatids. J. Biol. Chem. 28038457-38463. [DOI] [PubMed] [Google Scholar]
- 20.Konings, W. N. 2006. Microbial transport: adaptations to natural environments. Antonie van Leeuwenhoek 90325-342. [DOI] [PubMed] [Google Scholar]
- 21.Kreneva, R. A., M. S. Gelfand, A. A. Mironov, I. A. Iomantas, Y. I. Kozlov, A. S. Mironov, and D. A. Perumov. 2000. Study of the phenotypic occurrence of ura gene inactivation in Bacillus subtilis. Genetika 361166-1168. [PubMed] [Google Scholar]
- 22.Meyer, M. M., A. Roth, S. M. Chervin, G. A. Garcia, and R. R. Breaker. 2008. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA 14685-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mironov, A. A., N. P. Vinokurova, and M. S. Gel'fand. 2000. Software for analyzing bacterial genomes. Mol. Biol. (Moscow) 34253-262. [PubMed] [Google Scholar]
- 24.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren, Q., and I. T. Paulsen. 2005. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 1e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ren, Q., and I. T. Paulsen. 2007. Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J. Mol. Microbiol. Biotechnol. 12165-179. [DOI] [PubMed] [Google Scholar]
- 27.Rodionov, D. A. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 1073467-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodionov, D. A., and M. S. Gelfand. 2006. Computational identification of BioR, a transcriptional regulator of biotin metabolism in Alphaproteobacteria, and of its binding signal. FEMS Microbiol. Lett. 255102-107. [DOI] [PubMed] [Google Scholar]
- 29.Rodionov, D. A., P. Hebbeln, M. S. Gelfand, and T. Eitinger. 2006. Comparative and functional genomic analysis of prokaryotic nickel and cobalt uptake transporters: evidence for a novel group of ATP-binding cassette transporters. J. Bacteriol. 188317-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodionov, D. A., X. Li, I. A. Rodionova, C. Yang, L. Sorci, E. Dervyn, D. Martynowski, H. Zhang, M. S. Gelfand, and A. L. Osterman. 2008. Transcriptional regulation of NAD metabolism in bacteria: genomic reconstruction of NiaR (YrxA) regulon. Nucleic Acids Res. 362032-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodionov, D. A., A. A. Mironov, and M. S. Gelfand. 2002. Conservation of the biotin regulon and the BirA regulatory signal in Eubacteria and Archaea. Genome Res. 121507-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2004. Comparative genomics of the methionine metabolism in gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 323340-3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2003. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J. Biol. Chem. 27841148-41159. [DOI] [PubMed] [Google Scholar]
- 34.Rodionov, D. A., A. G. Vitreschak, A. A. Mironov, and M. S. Gelfand. 2002. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 27748949-48959. [DOI] [PubMed] [Google Scholar]
- 35.Roth, A., W. C. Winkler, E. E. Regulski, B. W. Lee, J. Lim, I. Jona, J. E. Barrick, A. Ritwik, J. N. Kim, R. Welz, D. Iwata-Reuyl, and R. R. Breaker. 2007. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 14308-317. [DOI] [PubMed] [Google Scholar]
- 36.Saier, M. H., Jr. 2000. A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol. Mol. Biol. Rev. 64354-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sarsero, J. P., E. Merino, and C. Yanofsky. 2000. A Bacillus subtilis gene of previously unknown function, yhaG, is translationally regulated by tryptophan-activated TRAP and appears to be involved in tryptophan transport. J. Bacteriol. 1822329-2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schyns, G., S. Potot, Y. Geng, T. M. Barbosa, A. Henriques, and J. B. Perkins. 2005. Isolation and characterization of new thiamine-deregulated mutants of Bacillus subtilis. J. Bacteriol. 1878127-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thanassi, J. A., S. L. Hartman-Neumann, T. J. Dougherty, B. A. Dougherty, and M. J. Pucci. 2002. Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res. 303152-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 1443097-3104. [DOI] [PubMed] [Google Scholar]
- 41.Vitreschak, A. G., A. A. Mironov, V. A. Lyubetsky, and M. S. Gelfand. 2008. Comparative genomic analysis of T-box regulatory systems in bacteria. RNA 14717-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vitreschak, A. G., D. A. Rodionov, A. A. Mironov, and M. S. Gelfand. 2002. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 303141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vogl, C., S. Grill, O. Schilling, J. Stülke, M. Mack, and J. Stolz. 2007. Characterization of riboflavin (vitamin B2) transport proteins from Bacillus subtilis and Corynebacterium glutamicum. J. Bacteriol. 1897367-7375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Winkler, W. C., S. Cohen-Chalamish, and R. R. Breaker. 2002. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. USA 9915908-15913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yakhnin, H., H. Zhang, A. V. Yakhnin, and P. Babitzke. 2004. The trp RNA-binding attenuation protein of Bacillus subtilis regulates translation of the tryptophan transport gene trpP (yhaG) by blocking ribosome binding. J. Bacteriol. 186278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.