Abstract
Bacteriophage infection has profound effects on bacterial biology. Clustered regular interspaced short palindromic repeats (CRISPRs) and cas (CRISPR-associated) genes are found in most archaea and many bacteria and have been reported to play a role in resistance to bacteriophage infection. We observed that lysogenic infection of Pseudomonas aeruginosa PA14 with bacteriophage DMS3 inhibits biofilm formation and swarming motility, both important bacterial group behaviors. This inhibition requires the CRISPR region in the host. Mutation or deletion of five of the six cas genes and one of the two CRISPRs in this region restored biofilm formation and swarming to DMS3 lysogenized strains. Our observations suggest a role for CRISPR regions in modifying the effects of lysogeny on P. aeruginosa.
Bacteriophages are probably best known for their role as tools used to study bacteria. Phages have also served as important models for the study of mechanisms of transcription, recombination, and transposition (8). Bacteriophages also shape microbial populations both by impacting the size and structure of bacterial communities and through the transfer of genetic material between bacterial strains (6, 39, 52). It is estimated that phages can lyse as many as 20% of all bacterial cells daily; therefore, these infectious particles can have a profound impact on the evolution of microbes (53).
While some bacteriophage infections are primarily lytic, temperate or lysogenic bacteriophages often integrate into the bacterial genome as a prophage causing a chronic infection of the host bacterium (52). In some cases, genes carried by a bacteriophage confer a new function upon a bacterium, typically not directly related to the phage life cycle, through a process known as lysogenic conversion. Lysogenic conversion likely supports phage survival indirectly by increasing the fitness of the host microbe, thus promoting the continued persistence of the phage genome within the host population. Examples of this phenomenon include the phage-mediated introduction of secreted virulence factors such as cholera toxin (30), altered lipopolysaccharide profile (35), and improved adhesion to epithelial cells (50).
Here we report that infection of Pseudomonas aeruginosa PA14 by phage DMS3 results in lysogenized strains unable to form a biofilm or undergo swarming motility—two key group behaviors of this bacterium. Furthermore, we show that the loss of biofilm formation and swarming motility requires clustered regular interspaced short palindromic repeats (CRISPRs) and five of six cas (CRISPR-associated) genes. Our data suggest a complex interaction between microbe and bacteriophage impacts the group behaviors of P. aeruginosa.
MATERIALS AND METHODS
Bacterial and phage culture preparation.
Strains and plasmids used in this study are shown in Table 1. Overnight cultures were streaked from glycerol stocks stored at −80°C onto lysogeny broth (LB) agar (1.5%) and incubated overnight at 37°C to isolated single colonies. These colonies were then used to inoculate planktonic and biofilm assays. Phage stocks were generated by growing lysogenized strains overnight in liquid LB at 37°C. The phage was purified from the lysogenized strains by passage of the culture through a 0.22-μm filter.
TABLE 1.
Strains and plasmids used in this study
| Strain | Relevant phenotype or description | Source or reference |
|---|---|---|
| Strains | ||
| S. cerevisiae strain | ||
| INVSc1 | MATahis3D1 leu2 trp1-289 ura3-52 MAT his3D1 leu2 trp1-289 ura3-52 | Invitrogen |
| E. coli strain | ||
| Top10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (Strr) endA1 nupG | Invitrogen |
| P. aeruginosa strains | ||
| PA14 | WT | 41 |
| SMC3899 | LysNeg2, biofilm-negative DMS3 lysogen | This study |
| SMC3884 | LysNeg6, biofilm-negative DMS3 lysogen | This study |
| SMC3897 | LysNeg8, biofilm-negative DMS3 lysogen | This study |
| SMC3898 | LysNeg10, biofilm-negative DMS3 lysogen | This study |
| SMC3896 | LysPos5, biofilm-positive DMS3 lysogen | This study |
| SMC3885 | LysPos6-3, biofilm-positive DMS3 lysogen | This study |
| SMC3892 | LysNeg6 33310::TnM | This study |
| SMC3886 | PA14_33290::TnM | 23 |
| SMC3887 | PA14_33310::TnM | 23 |
| SMC3888 | PA14_33320::TnM | 23 |
| SMC3889 | PA14_33330::TnM | 23 |
| SMC3890 | PA14_33340::TnM | 23 |
| SMC3891 | PA14_33350::TnM | 23 |
| SMC3893 | ΔCRISPR-1 | This study |
| SMC3895 | ΔCRISPR-2 | This study |
| SMC3894 | ΔPA14_33300 | This study |
| Plasmids | ||
| pCR2.1-TOPO | Invitrogen | |
| pMQ30 | Cloning vector | 44 |
| pMQ70 | Cloning vector | 44 |
| pMQ78 | Cloning vector | 44 |
| pCR2.1-TOPO-DMS3Early | c-repressor construct | This study |
| pDMS3Early | c-repressor expression construct | This study |
| pMQ30-ΔCRISPR-1 | CRISPR-1 deletion construct | This study |
| pMQ30-ΔCRISPR-2 | CRISPR-2 deletion construct | This study |
| pMQ30-ΔPA14_33300 | PA14_33300 deletion construct | This study |
| pMQ30-ΔPA14_33310 | PA14_33310 deletion construct | This study |
| pMQ70-PA14_33310 | PA14_33310 complementation construct | This study |
Generation of phage lysogens and measurements of phage titer.
To generate phage lysogens, 10 μl of filtered lysate containing phage (prepared as described above) was mixed with 10 μl Pseudomonas aeruginosa PA14 cells from an overnight LB-grown culture, and the mixture of PA14 and phage was incubated overnight at 37°C in LB medium. These bacterium-phage mixtures were then struck on plates to obtain individual colonies. Individual colonies were inoculated into liquid LB medium, grown overnight, and then assayed for the presence of phage. Phage production was quantified by spotting 10-μl serial dilutions of the phage lysate on a lawn of wild-type (WT) PA14 grown in LB top agar, and the plates were incubated at 37°C overnight as reported previously (7). Plaques were counted to yield the phage titer expressed as PFU/ml.
Biofilm assays. (i) Ninety-six well dish assay.
Biofilm formation in microtiter dishes was measured as described previously (37). Briefly, microtiter wells were inoculated from overnight LB-grown cultures diluted 1:100. Cells were grown for 24 h at 37°C before they were stained with crystal violet and quantified by dissolution using 30% acetic acid and measured at an absorbance at 550 nm (37).
(ii) CFU of biofilms in microtiter wells.
CFU associated with biofilms attached to the sides of microtiter wells were determined as previously described (18).
(iii) Air-liquid interface assay.
The attachment on plastic was measured using an air-liquid interface assay as reported previously (31).
Planktonic growth assays.
All strains were streaked to single colonies on LB agar plates. A single colony of each strain was picked, inoculated into 3 ml of LB broth, and incubated on a rotating shaker at 37°C overnight. One hundred microliters of each culture was then inoculated into 100 ml of fresh LB broth and incubated at 37°C. At appropriate time points, samples of the culture were obtained and serial dilutions made on LB agar to determine the CFU.
Swarming assays.
Swarming assays were performed as reported previously (19, 49).
Initial phage sequencing.
Phage DNA was isolated from a lysate of DMS3 as reported previously (7). Agencourt Biosciences Corporation performed the construction of the clone library of the phage and sequencing of the clones. Three- or 4-kb-insert-length libraries were constructed in the vector pAGEN-HC by use of the BstXi restriction enzyme. The DNA was cleaved using a Hydroshear device (Genemachines, San Carlos, CA) and ligated to an adapter to facilitate ligation into the BstI site of the vector. DNA templates were then sequenced in a 384-well format with forward and reverse reactions utilizing BigDye version 3.1. The Paracel genome assembler (TIGR) coupled with Agencourt's laboratory information management system was used to assemble the DNA sequence. The average coverage of the genome was approximately 21.5×.
Supplementary phage sequencing.
Preliminary analysis of the phage DMS3 sequence suggested that a portion of the left end of the DMS3 genome was missing from the assembled contig derived from the phage genomic sequence based on the lack of start codon for an otherwise highly conserved gene at the leftmost end of the phage sequence (DMS3-4 in the final annotation). Due to the high degree of similarity between phages D3112 (51) and DMS3 at the left ends of the respective genomes, it was hypothesized that primers could be designed based on the D3112 sequence to amplify their corresponding regions in DMS3 (see Table S1 in the supplemental material). PCR primers to the left end of the known genomic sequence of DMS3 from Agencourt and to the third annotated gene of D3112 yielded an ∼1.4-kb PCR product when DMS3 DNA was used as the template. Determination of the nucleotide sequences of PCR products was performed by first cloning the products into vector pBAD-TOPO, followed by DNA sequencing. The process was repeated with an additional primer set designed to the first and the third annotated genes of D3112.
Finally, arbitrary primed PCR was used to determine the ∼300 bp at the leftmost end of DMS3. Arbitrary primed PCR was performed as reported previously (37) using primers to the known phage genome sequence in conjunction with two arbitrary PCR primers, designated P15 and P16 (see Table S1 in the supplemental material), that were added in equal amounts to the first-round reaction mix. DNA purified from lysogens was used as the template for the first round of PCR. Products of the first-round reaction were used as template in the second round, with the original phage primer and a primer, designated P14, that hybridizes to the 5′ end of primers P15 and P16.
Sequence assembly.
The sequences generated by the PCR, arbitrary PCR, and direct sequencing were assembled with the help of the Lasergene software suite (DNAstar, Inc.). Multiple alignments were generated with GeneInspector version 1.6 (Textco, West Lebanon, NH). The nucleotide sequence of the left end of DMS3 was confirmed by sequencing multiple PCR products (99.88% of bases confirmed by at least two separate PCRs) to eliminate errors during amplification steps.
Genome annotation.
The initial annotation of the DMS3 sequence was performed by searching for similarities between putative translated protein sequences and proteins in the NCBI database by BLASTX (1). The entire genome was also analyzed by GeneMark.hmm for prokaryotes version 2.4 (25; http://opal.biology.gatech.edu/GeneMark/gmhmm2_prok.cgi) with P. aeruginosa as the selected species. The potential open reading frames (ORFs) identified were marked and translated using gene construction kit 2.5 (Textco, West Lebanon, NH). Each putative protein identified was compared to the NCBI database using BLASTP with alignments created using the default settings. Discrepancies between BLASTX and GeneMark.hmm analysis at the beginnings and/or ends of translated regions were resolved by first examining predicted start and stop codons to determine if they matched known start and stop codons, followed by assessing the presence and strength of ribosomal binding sites and determining the lowest expect value compared to those for proteins in the NCBI database. Transcriptional units were predicted with the help of FGENESB (SoftBerry, Mt. Kisco, NY) set to the generic bacterial genome setting and utilizing default settings. Promoters were predicted using the Berkeley Drosophila Genome Project neural network promoter program (http://www.fruitfly.org/seq_tools/promoter.html) set for prokaryotes and including the forward and reverse strands and with BPROM bacterial promoter predictor (SoftBerry, Mt. Kisco, NY; http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb). Neither program was specific for P. aeruginosa. The entire genome was also subjected to BLASTN analysis to search for any regions of similarity at the nucleotide level (1).
Detection of ribosomal binding sites.
Ribosomal binding sites in the area upstream of possible start codons were identified by an algorithm utilizing dynamic programming to align the consensus P. aeruginosa Shine-Dalgarno sequence (TAAGGAGGTGAT) (26) within the area 20 bp upstream of the suspected ORF. The scoring for the dynamic programming was based on the rules developed for changes in RNA duplex free energy by Freier et al. (10). This algorithm, which we named RBSLocator, was written in the Java language and was described and utilized previously (26, 43).
ORF analysis.
Searches for helix-turn-helix motifs were carried out with GYM 2.0 (http://www.cs.fiu.edu/∼giri/bioinf/GYM2/prog.html) (11, 34). Phylogram trees to assess the relatedness of proteins were created for all hypothetical proteins which contained similarity to more than three proteins in the NCBI database as determined by BLASTP expect values under 1e-19. Phylograms were created using ClustalX (version 1.8) (available at http://www.uk.plbio.kvl.dk/bioinfo.htm) utilizing default parameters (48). Neighbor-joining trees were also created with the ClustalX program and bootstrapped with random-number seeds of 111 and 1000 bootstrap replicates. Trees were visualized with Treeview (38). Conserved protein domains were detected as part of the BLASTP, as reported previously (28). A search for potential tRNA genes was carried out with tRNAscan-SE utilizing default settings (http://lowelab.ucsc.edu/tRNAscan-SE/) (24).
G+C analysis of genome.
G+C analysis of the DMS3 genome along with calculations of ORF G+C percent content was performed with Artemis release 7 (http://www.sanger.ac.uk/Software/Artemis/) (42).
Identification of DMS3 insertion in the P. aeruginosa genome.
The PCR products of the arbitrary primed PCRs, using the arbitrary primers and a primer derived from sequence of the leftmost end of the phage genome (P01 reverse 3), were sequenced and aligned to the PA14 genome by use of the BLASTN program from NCBI to indicate a potential site for insertion in the P. aeruginosa chromosome. Once a putative site of DMS3 insertion was established, genomic DNA was purified from the lysogenic strain by use of the Puregene genomic DNA purification kit (Gentra, Minneapolis, MN). Primers were then designed specific to the PA14 region identified and were used to establish whether formation of the PCR product was blocked in the lysogen, indicating the insertion of the phage at that site. Additionally, the appropriate primer was paired with an upstream or downstream primer to DMS3 in order to generate a PCR product containing sequence including the junction point of DMS3 and the flanking P. aeruginosa genome sequence. These products were also sequenced to confirm the presence of DMS3 in a particular insertion site in the PA14 genome and to determine the precise junction point of the phage and bacterial genomes.
Cloning of the DMS3 immunity region.
The region of phage DMS3 containing the predicted DMS3 c-repressor gene was cloned into vector pMQ78, which contains a gfp gene, in order to create a transcriptional fusion (44). The c-repressor ORF and flanking sequence were amplified with primers CRepFor (containing an engineered Acc651 restriction site) and DMS3p03 Reverse and then cloned into vector pCR2.1-TOPO (Invitrogen), yielding plasmid pCR2.1-DMS3Early. Plasmid pCR2.1-DMS3Early was restricted with enzymes Acc651 (New England Biolabs) and EcoRI (Invitrogen), yielding an ∼2 kb fragment which was gel purified and then ligated into pMQ78 to yield plasmid pDMS3Early. Plasmid pDMS3Early was propagated in Escherichia coli JM109 cells and then electroporated into competent P. aeruginosa cells as reported previously (3). The presence of the vector and the transcription of the c-repressor ORF were confirmed by visualization of green fluorescent protein expressed downstream of the c-repressor ORF from the pMQ78 plasmid by epifluorescence microscopy (not shown).
Mariner transposon mutagenesis of LysNeg6.
The biofilm-negative DMS3-lysogenized LysNeg6 in the PA14 background was mutagenized using the mariner transposon (designated TnM) as previously described (22). Briefly, the mariner transposon was conjugated into the LysNeg6 strain. Exconjugants were plated in the presence of 50 μg/ml gentamicin. Individual colonies were picked and grown overnight in 96-well plates under selective conditions. The mutants were then screened for biofilm formation in the microtiter dish assay described above using M63 medium supplemented with 1 mM MgSO4 and 0.4% arginine. Swarming was tested for candidate mutants as described above.
Identification of CRISPR and cas genes.
Identification of the CRISPRs and spacers in PA14 was aided by the use of CRISPERFinder (13) and CRISPRdb (12). Each spacer was also compared against the NCBI database manually.
qRT-PCR.
Quantitative real-time PCR (qRT-PCR) of the cas genes in the WT and in the biofilm-positive and biofilm-negative lysogenized strains was performed as reported previously (20, 21) with the following modifications. The strains of interest were subcultured from a single plate colony into 5 ml LB at 37°C until they reached an optical density at 600 nm of ∼0.4. Cells were normalized to an optical density at 600 nm of 0.4. One milliliter of culture was removed and centrifuged to pellet the cells, RNA was isolated, and cDNA was prepared as previously reported (20, 21). qRT-PCR was performed and the expression levels were quantified and normalized to rplU by use of previously published primers (20, 21). Expression levels are reported in picograms of input cDNA. The primers used in the qRT-PCR assays to measure the expression of the indicated genes were as follows: for PA14_33300, 33300amp.forward and 33300amp.Reverse; for PA14_33310, 33310amp.forward and 33310amp.Reverse; and for PA14_33320, 33320amp.forward and 33320amp.Reverse.
Expression of selected cas genes was also assessed by qRT-PCR in the WT grown planktonically and attached to a surface for 30 min. Fifty-milliliter cultures of the WT were grown in minimal arginine medium until late log phase (∼16 h) and 500-μl cultures transferred to wells of a 24-well polystyrene plate. After a 30-min incubation standing at 37°C, the planktonic cells from six wells were removed and pooled and RNA was prepared using the Qiagen RNAEasy kit, as described by the manufacturer with the recommended concentration of lysozyme. Lysis buffer (350 μl) plus lysozyme was added to the same six wells, the buffer from pairs of cells pooled, and RNA isolated with a Qiagen RNAEasy kit as described by the manufacturer. The RNA of the biofilm cells from all six wells was pooled and represented a single sample. The protocol above was performed three times independently, and the triplicate RNA samples (both from planktonic and biofilm cells) was subjected to qRT-PCR of the indicated genes.
Construction of CRISPR region mutants and complementing plasmids.
Transposon mutations in genes PA14_33290, PA14_33310, PA14_33320, PA14_33330, PA14_33340, and PA14_33350 were obtained from the PA14nr library (23).
Clean deletions of CRISPR-1, CRISPR-2, PA14_33300, and PA14_33310 were also constructed (Table 1). The pMQ30-ΔCRISPR-1, pMQ30-ΔCRISPR-2, pMQS30-Δ33300, and pMQ30-Δ33310 constructs were created utilizing the Saccharomyces cerevisiae recombineering technique described by Shanks et al. (44). Constructs were electroporated into Escherichia coli and analyzed by colony PCR or sequencing. Plasmids pMQ30-ΔCRISPR-1, pMQ30-ΔCRISPR-2, and pMQ30-Δ33300 were propagated in E. coli S17 and conjugated into P. aeruginosa strain PA14, while plasmid pMQ30-Δ33310 was electroporated (3). Exconjugants containing an inserted plasmid were selected on gentamicin. Mutants were isolated as reported previously (20, 21). All mutations were confirmed via PCR amplification and sequencing of the mutated region.
The PA14_33310 complementation construct was cloned utilizing primers PA14_33310 F and PA14_33310 R. The PCR product and vector pMQ70 were restricted with EcoR1 and HindIII at 37°C, gel purified, and then ligated and transformed into competent DH5α cells via electroporation. Colonies were selected based on the ability to grow under ampicillin selection. The PA14_33310 complementation construct or the vector control was purified from DH5α cells by use of a Qiagen miniprep kit and electroporated into the ΔPA14_33310 mutant strain. Transformants were selected on LB agar containing 250 μg/ml carbenicillin.
Nucleotide sequence accession number.
The complete sequence of DMS3 was deposited in GenBank (accession number DQ631426).
RESULTS
Lysogeny by phage DMS3 blocks biofilm formation and swarming by P. aeruginosa PA14.
While working with microbes lysogenized with bacteriophage DMS3 (7) in a P. aeruginosa PA14 background, we observed a loss in the ability of the lysogenized microbe to form biofilms (Fig. 1A, B, C, and D; compare WT with LysNeg6 column). Microscopy studies indicated that the defect in biofilm formation occurred at an early stage in the formation of these communities. At 8 h, there were ∼10-fold fewer bacteria attached for the LysNeg6 strain than for the WT (Fig. 1D). This difference in attachment could be observed as soon as 30 min postinoculation with 50% fewer LysNeg6 cells than WT cells attached at this time point (47 ± 10 versus 109 ± 11, respectively; P < 0.00001). In addition, these lysogenized strains were also inhibited for swarming motility (Fig. 1E). Like biofilm formation, swarming represents a form of bacterial group behavior (15), suggesting that lysogeny by DMS3 interferes with at least two P. aeruginosa group behaviors. The planktonic growth rates of these lysogenized strains were similar to that of uninfected P. aeruginosa PA14 (Fig. 2A). Twitching and swimming, two other forms of motility in P. aeruginosa, were not affected in these lysogenized strains (data not shown).
FIG. 1.

Biofilm and swarming phenotypes of P. aeruginosa PA14 and two DMS3-lysogenized strains. (A) Biofilm formation assessed by crystal violent staining of biofilms grown in microtiter wells for 8 h. (B) Quantification of staining of wells in panel A after solubilization with acetic acid and measurement of optical density at 550 nm (OD550). (C) CFU per well of biofilm bacteria. (D) Phase-contrast micrographs of air-liquid interface of biofilms grown in microtiter wells. Experiments displayed in panels A to D were conducted in a polyvinylchloride microtiter plate assay in M63 minimal medium supplemented with arginine (0.4%) and MgSO4 (1 mM); incubation was at 37°C for 3 h. Bars, 20 μm. (E) Photographs of representative swarming assays after ∼16 h of growth.
FIG. 2.

Quantifying growth and phage production. (A) Planktonic growth of the WT Pseudomonas aeruginosa PA14 (triangles), a biofilm-negative lysogenized strain (LysNeg6; squares), and a biofilm-positive transposon mutant derivative of LysNeg6 (LysNeg6 33310::TnM mutant; diamonds). (B) Phage production in PFU/well of a biofilm-negative lysogenized strain (LysNeg6; squares) and a biofilm-positive transposon mutant derivative of LysNeg6 (LysNeg6 33310::TnM mutant; diamonds). Phage was collected from the same growth assay shown in panel A.
To determine whether altered biofilm formation and swarming were a common consequence of DMS3 infection, we analyzed additional DMS3-lysogenized strains. Eighty-three of 86 lysogenized strains tested were unable to form biofilms or swarm; however, three independently isolated lysogenized strains formed biofilms and swarmed similarly to the WT strain (Fig. 1; LysPos6-3). Biofilm-positive and -negative lysogenized strains show comparable planktonic growth rates, indicating that differences in growth could not account for the change of group behaviors (data not shown). Furthermore, phage harvested from the biofilm-positive lysogenized strains were capable of forming plaques on lawns of WT P. aeruginosa PA14 and produced biofilm- and swarming-inhibited lysogenized strains (data not shown). These results suggest that the biofilm-positive lysogens are not the result of a defective variant of DMS3 but rather represent an altered host response to DMS3 lysogeny. These data indicate that lysogeny alone is insufficient to explain the loss of biofilm formation and swarming. The biofilm-positive lysogenized strains are discussed in more detail below.
Sequencing the genome of DMS3 reveals similarity to Mu-like phages.
To further explore the molecular mechanism by which DMS3 lysogeny inhibits biofilm formation and swarming motility, we determined the sequence and annotated the genome of DMS3 (Fig. 3; also see Table S2 in the supplemental material and http://www.dartmouth.edu/∼gotoole/Phage/index.htm).
FIG. 3.
Map of the DMS3 genome. The arrows represent the predicted phage DMS3 ORFs and the predicted direction of gene transcription, with the ORF number appearing in bold above the arrow (see Table S2 in the supplemental material for more details regarding each ORF). Black arrows represent genes that were at least partially detected by both GeneMark.hmm and BLASTX, while gray arrows represent genes that were detected only by GeneMark.hmm. White areas represent intergenic regions. The organisms and phages included in the similarity profile were selected for having matches based on BLASTP expect values of less than 1e-19. The bacteriophage genomic sequences in the alignment and the abbreviation for each organism can be found in the lower right of the figure. The boxes labeled other (OT) represent similarity with expect values of less than 1e-19 for an organism that is not one of the 13 listed. The shaded boxes under each gene represent different degrees of similarity to detected genes based on the BLASTP expect values (the key is in the lower right of the figure). The “#” symbol represents the organism with the lowest expect value from BLASTP when the corresponding DMS3 ORF was analyzed. The “&” symbol indicates the organism most closely related to the DMS3 ORF by phylogenic analysis. The genes contained in each transcriptional unit are listed in the upper right of the figure.
The complete phage genome of DMS3 is 35,403 bp, which is smaller than the original predicted length of 50 to 65 kb based on pulsed-field gel electrophoresis (7). Fifty-two (52) ORFs were predicted from the genome sequence. It is immediately clear from the analyses of the sequence data that DMS3 has a high degree of similarity to bacteriophage D3112 (Fig. 3; also see Table S2 in the supplemental material), a phage with genomic similarity to Mu-like phages and morphological similarity to λ-like phages (51). Of the 52 putative ORFs identified in DMS3, 42 of the predicted gene products showed similarity by expect values of under 1e-19 to a protein predicted for D3112. There was also a high degree of similarity to proteins from the B3 phage among predicted ORFs of the DMS3 genome.
The majority of the predicted genes are transcribed in the positive orientation, with only ORFs DMS3-1 and −25 transcribed in the reverse direction. This transcriptional organization is common among bacteriophages and is similar to the organization of bacteriophages D3112, B3, MP22, and Mu (4, 17, 33, 51). As was the case with D3112 (51), no tRNA molecules were detected in the genome.
Mapping of the DMS3 lysogens suggests random insertion in the PA14 genome.
Ojha and colleagues have reported that lysogeny by the mycobacterial phage Bxb1 disrupted biofilm formation by insertion of the phage into a specific gene location, a groEL homolog required for mycolic acid synthesis (36). To address the possibility that a similar mechanism was at work in DMS3, we used arbitrary primed PCR to determine the insertion locations of six DMS3 lysogens. Consistent with the sequence similarity of DMS3 to Mu, a phage that inserts randomly into the host genome (32), the phage attachment sites of all DMS3 lysogens mapped to unique insertion locations (Table 2). Therefore, our data argue against a model in which DMS3 attachment to a single genetic locus is responsible for the loss of biofilm formation and swarming motility.
TABLE 2.
Insertion sites of six DMS3 lysogens
| Lysogen | Biofilm phenotype | Insertion site in PA14 genome in intergenic region between: | Corresponding genes in strain PA01 |
|---|---|---|---|
| LysNeg2 | Inhibited | PA14_09410 and PA14_09400 | PA4216 and PA4217 |
| LysNeg6 | Inhibited | PA14_34670 and PA14_34660 | PA2318 and PA2320 |
| LysNeg8 | Inhibited | PA14_13350 and PA14_13340 | PA3908 and PA3909 |
| LysNeg10 | Inhibited | PA14_64450 and PA14_64460 | PA4873 and PA4874 |
| LysPos5 | Intact | PA14_32650 and PA14_32640 | PA2473 and PA2474 |
| LysPos6-3 | Intact | PA14_33380 and PA14_33370 | PA2421 and PA2422 |
Isolation of mutations that restore biofilm formation to a DMS3-lysogenized strain.
To investigate the basis of DMS3-mediated inhibition of biofilm formation and swarming, we transposon mutagenized a biofilm-negative lysogenized strain, LysNeg6, and screened ∼5,000 mutants for the restoration of biofilm formation and swarming. One of these biofilm-restored transposon mutations mapped to the PA14_33310 locus: this mutant is hereafter designated as the LysNeg6 33310::TnM mutant. The planktonic growth of this mutant was similar to that of its parent strain (Fig. 2A).
Levels of phage production were also comparable during the log phase of growth; however, at 24 h we noted PFU of approximately 1 log greater for the parent DMS3 lysogenized strain, LysNeg6, than that for the LysNeg6 33310::TnM mutant (Fig. 2B).
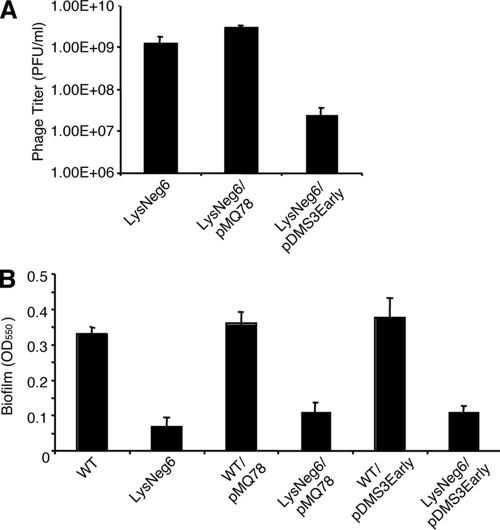
We considered the possibility that the improved biofilm formation noted for the LysNeg6 33310::TnM mutant was the result of a reduced rate of lysis and thus phage production compared with what was seen for LysNeg6. To address this possibility, we reduced the phage production in the parent lysogenized strain, LysNeg6, by expression of the DMS3 c-repressor from a plasmid. The c-repressor protein represses the activation of phage genes associated with the transition of Mu-like bacteriophages to the lytic cycle. Although the expression of the c-repressor resulted in an approximately 2-log reduction in PFU in the transformed strain (Fig. 4A), biofilm formation was not restored (Fig. 4B).
FIG. 4.
Reducing phage production does not rescue the biofilm formation defect of lysogenized strains. (A) Phage titer of LysNeg6 containing the pDMS3Early construct and the empty pMQ78 vector was assayed as described in Materials and Methods. (B) Biofilm formation of WT and LysNeg6 containing the pDMS3Early construct and the pMQ78 vector control was assayed as described in Materials and Methods. OD550, optical density at 550 nm.
We also hypothesized that higher initial levels of free phage in LysNeg6 might account for our observation of reduced biofilm formation compared to that for the LysNeg6 33310::TnM mutant. However, increasing the starting phage concentration to levels exceeding those associated with the parent lysogenized strain did not inhibit biofilm formation by the LysNeg6 33310::TnM mutant (data not shown). We therefore concluded that the differences observed in phage production in late-phase cultures between the parent DMS3-lysogenized strain and the transposon mutant did not explain the restoration of biofilm formation in the mutant.
Analysis of the CRISPR region of P. aeruginosa PA14.
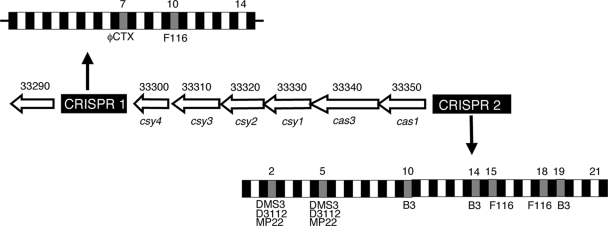
Analysis of PA14_33310 and the surrounding genes revealed six ORFs with sequence similarity to the previously described cas genes (13). Moreover, this region was flanked by two CRISPRs, one in an intergenic region between PA14_33290 and PA14_33300 (designated CRISPR-1) and another CRISPR annotated in the PA14 genome sequence as the PA14_33360 locus (hereafter designated as CRISPR-2) (Fig. 5).
FIG. 5.
CRISPR region of P. aeruginosa PA14. Shown in the center of the figure is the chromosomal organization of the CRISPR genomic region, including the two CRISPRs and the CRISPR-associated (cas) genes. The PA14 gene designation is listed above each gene, and the corresponding homolog from Y. pestis is shown below each gene. The sizes of the ORFs are as follows: for PA14_33300, 504 bp; for PA14_33310, 1,029 bp; for PA14_33320, 984 bp; for PA14_33330, 1,305 bp; for PA14_33340, 3,231 bp; and for PA14_33350, 975 bp. CRISPR-2 is annotated as PA14_33360 in the PA14 genome and is 1,395 bp. The organization of the CRISPRs is also shown. Black boxes indicate repeats, and gray boxes indicate spacers containing identical or partial nucleotide sequence matches from the indicated bacteriophages. The appropriate spacer number is listed above each spacer with known sequence identity to a phage, as is the last spacer number, which indicates the total number of spacers in each CRISPR.
CRISPRs are typically organized as a set of 20- to 30-bp direct repeats alternating with 20 to 30 bp of nonrepeating spacer sequences (45). CRISPR-2 has 22 identical direct repeats and 21 nonrepeated spacer regions. With the exception of a 1-base substitution, the CRISPR-2 direct repeat has the identical sequence of the CRISPR-1 direct repeat, although the two CRISPRs are oriented divergently. The sequence and organization of the P. aeruginosa PA14 CRISPR region are similar to those of the CRISPR region of Yersinia pestis (14, 40).
CRISPR spacers have been previously noted to contain sequences similar to portions of phage genes (45). Notably, our analysis revealed that the entire second 32-bp spacer sequence of CRISPR-2 is identical to a region of the phage gene DMS3-24 (Fig. 3). DMS3-24 is in the predicted fourth transcriptional unit of DMS3 and shows a high degree of similarity to p24 of phage D3112. Although annotated as a hypothetical protein, it is suspected to be an analog of the c gene in bacteriophage Mu (51). The c gene acts as a positive regulator of transcription of four known late gene promoters in Mu that are essential for the cell to enter the lytic cycle (16, 29). In the fifth spacer of CRISPR-2, the first 25 of 32 bp are identical to a region of the phage gene DMS3-13 (Fig. 3). DMS3-13 is in the third predicted transcriptional unit of DMS3. ORFs DMS3-13 to -17 all show relatively weak similarity to their D3112 counterparts by BLASTP expect values and no similarity to any other genes in the protein database. Other spacers with sequence similarity to bacteriophages, such as ΦCTX, F116, and B3, were found in CRISPR-1 and -2 (Fig. 5). However, there were no spacers with sequence similarity to DMS3 identified in CRISPR-1.
Genetic analysis of the CRISPR and cas genes.
The role of the cas genes in DMS3-mediated inhibition of biofilm formation and swarming was first confirmed by analysis of additional transposon mutants of P. aeruginosa PA14 with insertions in this region, which were available from a defined transposon library (23). DMS3-lysogenized strains created from strains carrying mutations in cas genes were isolated and tested for biofilm formation and swarming. Lysogenized strains carrying transposon mutations in PA14_33310, PA14_33320, PA14_33330, and PA14_33340 were intact for biofilm formation and swarming (not shown). However, lysogenized mutants of PA14_33350, a locus with sequence similarity to the cas1 gene, were inhibited for biofilm formation and swarming. Because a transposon mutant of PA14_33300 was not available, we constructed a clean deletion of this gene and found that when lysogenized with DMS3, the corresponding strain was intact for biofilm formation and swarming (not shown).
A clean deletion of PA14_33310 was also constructed, and as predicted, when the corresponding strain was lysogenized with DMS3, biofilm formation and swarming were intact (Fig. 6). The phenotype of the lysogenized PA14_33310 mutant could be complemented by providing the gene on a plasmid (Fig. 6). These data provide strong evidence that the phenotypic changes we observed for the transposon mutants were due to disruptions of the targeted cas genes rather than polar effects on non-cas genes.
FIG. 6.
Biofilm and swarming phenotypes of PA14_33310 mutants and their response to DMS3 lysogeny. (A) Photographs of representative swarming assays after 24 h of growth of WT Pseudomonas aeruginosa PA14, a biofilm-negative lysogenized strain (LysNeg6), a mutant with a clean deletion of PA14_33310 (Δ33310), and the clean deletion of PA14_33310 expressing the PA14_33310 from a plasmid (Δ33310 + pMQ70-33310). DMS3 lysogens of these mutants are noted with the prefix Lys. (B) Biofilm formation assessed by crystal violet staining of biofilms grown in polyvinylchloride microtiter wells. (C) Quantification of staining of microtiter wells after solubilization with acetic acid and measurement of optical density at 550 nm (OD550). Experiments displayed in panels B and C were conducted in a polyvinylchloride microtiter plate assay in M63 minimal medium supplemented with arginine (0.4%) and MgSO4 (1 mM); incubation was at 37°C for 24 h. Similar results were obtained for biofilms assayed at 8 h (not shown). The empty vector pMQ70 had no effect on any of the phenotypes shown (data not shown).
Mutants carrying deletions of CRISPR-1 and CRISPR-2 were also constructed. The CRISPR-1 deletion mutants responded to DMS3 lysogeny in a manner similar to that seen for WT P. aeruginosa PA14, with inhibited biofilm formation and swarming (not shown). In contrast, DMS3-lysogenized cells prepared using the CRISPR-2 deletion mutant displayed intact biofilm formation and swarming (not shown). It is worth noting that CRISPR-2 but not CRISPR-1 contains spacers with sequences derived from DMS3. Taken together, these data indicate that loss or disruption of CRISPR-2, or five of the six cas genes, results in a restoration of biofilm formation and swarming in DMS3-lysogenized cells.
As noted above, we found that planktonic growth of the DMS3-lysogenized strain carrying a transposon in PA14_33310 (this strain is designated the LysNeg6 33310::TnM mutant) was unchanged compared with what was seen for the LysNeg6 parent (Fig. 2A). In contrast, the measured PFU were reduced in the LysNeg6 33310::TnM mutant compared to LysNeg6 (Fig. 2B). To investigate whether or not this pattern was repeated in other lysogenized strains containing mutations in the CRISPR region, we measured CFU/ml and PFU/ml for the DMS3-lysogenized strains containing mutations in CRISPR2 or the cas genes. CFU/ml and PFU/ml were determined both at inoculation and after 24 h of growth in LB. Consistent with our earlier results, there were minimal differences in CFU/ml values among the strains at either time point. However, plaque formation was reduced ∼10-fold for the lysogenized strain carrying mutations in CRISPR2 or the cas genes from what was seen for the LysNeg6 parent, with the exception of the lysogenized strain mutated in the PA14_33350 gene (data not shown). This is noteworthy because a mutation in PA14_33350 also failed to restore biofilm formation and swarming in the presence of DMS3 lysogeny. However, this ∼10-fold reduction in PFU could not explain the restoration of biofilm formation by the LysNeg6 strain, as we have shown that a >100-fold reduction in phage titer upon the expression of the DMS3 c-repressor could not restore biofilm formation to a biofilm-negative lysogenized strain (Fig. 4).
Biofilm-positive lysogenized strains carry deletions of the cas genes.
A role for the cas genes in phage-mediated inhibition of biofilm formation was further supported by the analysis of the two original DMS3-lysogenized strains that were intact for biofilm formation (LysPos5 and LysPos6-3). qRT-PCR analysis revealed that, while highly expressed in nonlysogenized P. aeruginosa PA14 and the biofilm-negative lysogenized strains, the PA14_33300, PA14_33310, and PA14_33320 genes were not expressed in either of the biofilm-positive lysogenized strains (Fig. 7A). Attempts to PCR amplify any part of the CRISPR region of the biofilm-positive lysogenized strains were unsuccessful, indicating a loss or disruption of this region.
FIG. 7.
qRT-PCR analysis of the PA14_33310 gene in DMS3 lysogens. (A) qRT-PCR was performed as described in Materials and Methods, and expression was normalized to the rplU gene. Levels of expression of the cas gene PA14_33310 were similar for WT P. aeruginosa PA14 and two DMS3-lysogenized strains which are inhibited for biofilm formation and swarming, LysNeg8 and LysNeg6. However, no expression was detected for two biofilm- and swarming-positive lysogenized strains, LysPos6-3 and LysPos5. Comparable results were obtained when expression of two other cas genes, PA14_33300 and PA14_33320, was assessed (not shown). (B) qRT-PCR was performed on the WT strain grown planktonically or attached to a surface. Shown is the relative expression for the PA14_33310 gene. The expression of the PA14_33310 gene under planktonic conditions is arbitrarily set at 1. ns, not significant (P > 0.05).
Interestingly, analysis of the P. aeruginosa PAO1 sequence revealed that the entire CRISPR region is absent from this sequenced strain of P. aeruginosa. Consistent with our findings, DMS3-lysogenized strains of P. aeruginosa PA01 are not inhibited for biofilm formation (data not shown). We were unable to generate lysogens to P. aeruginosa PAK because this strain is resistant to DMS3 infection, so we were unable to assess the consequences of lysogeny on biofilm formation and swarming.
Expression of the cas genes in planktonic and biofilm growth conditions.
We assessed the expression of selected cas genes in planktonic and attached bacteria taken from the same culture vessel after 30 min of incubation (see Materials and Methods for details). We found a small, statistically nonsignificant reduction in the expression of PA14_33310 (Fig. 7B) and PA14_33330 (not shown) 30 min after WT P. aeruginosa attached to an abiotic surface. As shown in Fig. 7A, there was also no difference between the cas gene expression of the WT and those of two different biofilm-negative lysogenized strains. Therefore, it appears that transcriptional regulation of the cas genes does not contribute to the differential impact of this region in lysogenized versus nonlysogenized strains.
DISCUSSION
CRISPR regions have been implicated in bacterial resistance to lytic bacteriophage infection (2, 45). Barrangou and colleagues recently described the association of acquired immunity to a lytic bacteriophage infection with the presence of DNA sequences derived from the infecting phage in the spacers of CRISPRs and furthermore demonstrated that mutations in those spacers resulted in a loss of phage resistance (2). They also found that mutation of certain cas genes resulted in the loss of resistance to lytic phage in previously resistant strains, while mutations in different cas genes produced strains that were unable to acquire phage resistance (2). The mechanism by which phage DNA is incorporated into CRISPR spacers, and how these spacers confer resistance to infection by the same phage, have not been established. There is recent evidence that some transcribed CRISPRs are degraded into small RNAs corresponding to each spacer with half of the repeated region on either end (5, 46, 47). Makarova and colleagues have suggested that these small RNAs serve as the basis for an RNA interference system directed at genes expressed by bacteriophages, resulting in resistance to phage infection (27). The existing model for CRISPR-mediated bacteriophage resistance proposes that CRISPR gene products disrupt phage gene transcription or production of these gene products (9, 45).
As discussed above, DMS3-lysogenized strains prepared using a mutation containing a transposon insertion in PA14_33350 (cas1), like WT P. aeruginosa PA14, were unable to form a biofilm or swarm, whereas DMS3-lysogenized strains derived from mutations in all the other cas genes had intact biofilm formation and swarming. The predicted function for PA14_33350 (cas1) is that of an integrase (14). Previous reports have speculated that integrase homologs participate in the process of integration of new phage DNA sequences into CRISPRs (2, 27). One might hypothesize that subsets of cas genes, such as PA14_33350 (cas1), are devoted to the generation of new content in CRISPR spacers, while other cas genes are involved in the response to phage infection. Thus, since P. aeruginosa PA14 already contains CRISPR spacers homologous to DMS3 genes, it is perhaps not unexpected that deletion of PA14_33350 (cas1), a gene presumably involved in the incorporation of new spacers, did not result in alteration of the response to DMS3 lysogeny.
It is important to note that the presence of DMS3 sequences in the second and fifth spacers of CRISPR-2 in unlysogenized Pseudomonas PA14 did not prevent infection by DMS3. Furthermore, we observed that a strain with an intact CRISPR region, LysNeg6, had a high rate of phage production in stationary-phase cultures compared to a strain with an altered CRISPR region, the LysNeg6 33310::TnM mutant, arguing against CRISPR-mediated suppression of the DMS3 lytic cycle in P. aeruginosa PA14. However, the low phage titer produced by the biofilm-positive lysogenized strain carrying a mutation in the cas genes compared to that for the biofilm-negative lysogenized strain could not account for the restored biofilm phenotype of the biofilm-positive lysogenized strain (Fig. 4).
It may be that the current CRISPR spacer content in P. aeruginosa PA14 is ineffective at resisting infection, while incorporation of different spacers would create a resistant isolate. Alternatively, it is possible that CRISPR regions are adapted to different functions in different species and/or play distinct roles in response to infection by lytic versus lysogenic phages. This different role for CRISPRs might not be unexpected given the diversity observed among CRISPR regions. Forty-five different cas genes have been identified; repeat sizes can range from 24 to 47 bp, and the number of repeats can range from 2 to 249 (45). Perhaps the P. aeruginosa PA14 CRISPR region has evolved to alter the effects of lysogeny in addition to, or perhaps instead of, resisting phage infection or lysis. What does seem clear is that CRISPR regions are a genetic locus in which multiple functions related to the bacterial response to phage infection are located and organized.
Why would biofilm formation be altered by P. aeruginosa in response to lysogeny? CRISPR-mediated alteration of biofilm formation and swarming may be a strategy of the bacterium for limiting the effects of bacteriophage dissemination in bacterial communities. In other words, phage-infected bacteria quarantine themselves from biofilms and other group behaviors to reduce the chances of infecting the larger community. Alternatively, inhibition of biofilm formation and swarming may provide an unknown survival advantage to phage DMS3. What is clear is that the changes we observed in biofilm formation and swarming depends both on phage DMS3 infection and on the presence of the P. aeruginosa PA14 CRISPR region. This interaction may represent a novel mechanism of lysogenic conversion. More work is necessary to understand the role of this process in the ecology of P. aeruginosa, as well as to determine the mechanism by which biofilm formation is disrupted during DMS3 lysogeny and the role of the CRISPRs and cas genes in this process.
Supplementary Material
Acknowledgments
These studies were supported by funding from the NIH to G.A.O. (R21-AI068662) and M.E.Z. (5KO8-EY13977), the Harmes Scholarship to M.E.Z., and a grant from the Richter Memorial Fund of Dartmouth College to J.C.W.
We thank Christine Toutain and Alejandro A. Borquez for their assistance on this project and D. Hogan and R. Taylor for their helpful comments on the manuscript. We also thank D. Hogan for her assistance with the biofilm qRT-PCR studies.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou, R., C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero, and P. Horvath. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 3151709-1712. [DOI] [PubMed] [Google Scholar]
- 3.Bloemberg, G., G. O'Toole, B. Lugtenberg, and R. Kolter. 1997. Green fluorescent protein as a marker for Pseudomonas spp. Appl. Environ. Microbiol. 634543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braid, M. D., J. L. Silhavy, C. L. Kitts, R. J. Cano, and M. M. Howe. 2004. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 1866560-6574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brouns, S. J., M. M. Jore, M. Lundgren, E. R. Westra, R. J. Slijkhuis, A. P. Snijders, M. J. Dickman, K. S. Makarova, E. V. Koonin, and J. van der Oost. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Budzik, J. M., W. A. Rosche, A. Rietsch, and G. A. O'Toole. 2004. Isolation and characterization of a generalized transducing phage for Pseudomonas aeruginosa strains PAO1 and PA14. J. Bacteriol. 1863270-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cairns, J. 2000. Phage and the origins of molecular biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Deveau, H., R. Barrangou, J. E. Garneau, J. Labonte, C. Fremaux, P. Boyaval, D. A. Romero, P. Horvath, and S. Moineau. 2008. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J. Bacteriol. 1901390-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freier, S. M., R. Kierzek, M. H. Caruthers, T. Neilson, and D. H. Turner. 1986. Free energy contributions of G.U and other terminal mismatches to helix stability. Biochemistry 253209-3213. [DOI] [PubMed] [Google Scholar]
- 11.Gao, Y., K. Mathee, G. Narashima, and X. Wang. 1999. Motif detection in protein sequences, p.63-72. Proceedings of the 6th SPIRE Conference, Cancun, Mexico.
- 12.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grissa, I., G. Vergnaud, and C. Pourcel. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35W52-W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haft, D. H., J. Selengut, E. F. Mongodin, and K. E. Nelson. 2005. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 1e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harshey, R. M. 2003. Bacterial motility on a surface: many ways to a common goal. Annu. Rev. Microbiol. 57249-273. [DOI] [PubMed] [Google Scholar]
- 16.Hattman, S. 1999. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 84367-388. [DOI] [PubMed] [Google Scholar]
- 17.Heo, Y. J., I. Y. Chung, K. B. Choi, G. W. Lau, and Y. H. Cho. 2007. Genome sequence comparison and superinfection between two related Pseudomonas aeruginosa phages, D3112 and MP22. Microbiology 1532885-2895. [DOI] [PubMed] [Google Scholar]
- 18.Kadouri, D., and G. A. O'Toole. 2005. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 714044-4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohler, T., L. K. Curty, F. Barja, C. van Delden, and J. C. Pechere. 2000. Swarming of Pseudomonas aeruginosa is dependent on cell-to-cell signaling and requires flagella and pili. J. Bacteriol. 1825990-5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuchma, S. L., K. M. Brothers, J. H. Merritt, N. T. Liberati, F. M. Ausubel, and G. A. O'Toole. 2007. BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14. J. Bacteriol. 1898165-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 1871441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55368-380. [DOI] [PubMed] [Google Scholar]
- 23.Liberati, N. T., J. M. Urbach, S. Miyata, D. G. Lee, E. Drenkard, G. Wu, J. Villanueva, T. Wei, and F. M. Ausubel. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 1032833-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lukashin, A. V., and M. Borodovsky. 1998. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 261107-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma, J., A. Campbell, and S. Karlin. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 1845733-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Makarova, K. S., N. V. Grishin, S. A. Shabalina, Y. I. Wolf, and E. V. Koonin. 2006. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchler-Bauer, A., and S. H. Bryant. 2004. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32W327-W331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolin, W., and M. M. Howe. 1986. Localization and DNA sequence analysis of the C gene of bacteriophage Mu, the positive regulator of Mu late transcription. Nucleic Acids Res. 144881-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McLeod, S. M., H. H. Kimsey, B. M. Davis, and M. K. Waldor. 2005. CTXphi and Vibrio cholerae: exploring a newly recognized type of phage-host cell relationship. Mol. Microbiol. 57347-356. [DOI] [PubMed] [Google Scholar]
- 31.Merritt, J. H., D. E. Kadouri, and G. A. O'Toole. 2005. Growing and analyzing static biofilms, p. 1B.1.2. In R. Coico, T. Kowalik, J. Quarles, B. Stevenson, and R. Taylor. (ed.), Current protocols in microbiology. John Wiley & Sons, Hoboken, NJ. [DOI] [PMC free article] [PubMed]
- 32.Mizuuchi, K., and R. Craigie. 1986. Mechanism of bacteriophage Mu transposition. Annu. Rev. Genet. 20385-429. [DOI] [PubMed] [Google Scholar]
- 33.Morgan, G. J., G. F. Hatfull, S. Casjens, and R. W. Hendrix. 2002. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 317337-359. [DOI] [PubMed] [Google Scholar]
- 34.Narasimhan, G., C. Bu, Y. Gao, X. Wang, N. Xu, and K. Mathee. 2002. Mining protein sequences for motifs. J. Comput. Biol. 9707-720. [DOI] [PubMed] [Google Scholar]
- 35.Newton, G. J., C. Daniels, L. L. Burrows, A. M. Kropinski, A. J. Clarke, and J. S. Lam. 2001. Three-component-mediated serotype conversion in Pseudomonas aeruginosa by bacteriophage D3. Mol. Microbiol. 391237-1247. [DOI] [PubMed] [Google Scholar]
- 36.Ojha, A., M. Anand, A. Bhatt, K. Kremer, W. R. Jacobs, Jr., and G. F. Hatfull. 2005. GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell 123861-873. [DOI] [PubMed] [Google Scholar]
- 37.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to the study of biofilms. Methods Enzymol. 31091-109. [DOI] [PubMed] [Google Scholar]
- 38.Page, R. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12357-358. [DOI] [PubMed] [Google Scholar]
- 39.Paul, J. H., M. B. Sullivan, A. M. Segall, and F. Rohwer. 2002. Marine phage genomics. Comp. Biochem. Physiol. B 133463-476. [DOI] [PubMed] [Google Scholar]
- 40.Pourcel, C., G. Salvignol, and G. Vergnaud. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151653-663. [DOI] [PubMed] [Google Scholar]
- 41.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 2681899-1902. [DOI] [PubMed] [Google Scholar]
- 42.Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice, M. A. Rajandream, and B. Barrell. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16944-945. [DOI] [PubMed] [Google Scholar]
- 43.Schurr, T., E. Nadir, and H. Margalit. 1993. Identification and characterization of E. coli ribosomal binding sites by free energy computation. Nucleic Acids Res. 214019-4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shanks, R. M., N. C. Caiazza, S. M. Hinsa, C. M. Toutain, and G. A. O'Toole. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from gram-negative bacteria. Appl. Environ. Microbiol. 725027-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sorek, R., V. Kunin, and P. Hugenholtz. 2008. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat. Rev. Microbiol. 6181-186. [DOI] [PubMed] [Google Scholar]
- 46.Tang, T. H., J. P. Bachellerie, T. Rozhdestvensky, M. L. Bortolin, H. Huber, M. Drungowski, T. Elge, J. Brosius, and A. Huttenhofer. 2002. Identification of 86 candidates for small non-messenger RNAs from the archaeon Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 997536-7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, T. H., N. Polacek, M. Zywicki, H. Huber, K. Brugger, R. Garrett, J. P. Bachellerie, and A. Huttenhofer. 2005. Identification of novel non-coding RNAs as potential antisense regulators in the archaeon Sulfolobus solfataricus. Mol. Microbiol. 55469-481. [DOI] [PubMed] [Google Scholar]
- 48.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toutain, C. M., M. E. Zegans, and G. A. O'Toole. 2005. Evidence for two flagellar stators and their role in the motility of Pseudomonas aeruginosa. J. Bacteriol. 187771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vaca-Pacheco, S., G. L. Paniagua-Contreras, O. Garcia-Gonzalez, and M. de la Garza. 1999. The clinically isolated FIZ15 bacteriophage causes lysogenic conversion in Pseudomonas aeruginosa PAO1. Curr. Microbiol. 38239-243. [DOI] [PubMed] [Google Scholar]
- 51.Wang, P. W., L. Chu, and D. S. Guttman. 2004. Complete sequence and evolutionary genomic analysis of the Pseudomonas aeruginosa transposable bacteriophage D3112. J. Bacteriol. 186400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28127-181. [DOI] [PubMed] [Google Scholar]
- 53.Wommack, K. E., and R. R. Colwell. 2000. Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev. 6469-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.