Abstract
We used microarrays to identify the causes of sporulation deficiencies in Bacillus subtilis after 6,000 generations of evolution. We found that sporulation loss did not result from large-scale deletions; therefore, it must have resulted from smaller indels and/or substitutions. Transcription patterns of one strain versus its ancestor showed that sporulation was not initiated and suggested that sporulation loss may be part of an overall decline in plasticity.
Spore formation by bacteria belonging to the Firmicutes is an ecologically beneficial yet energetically costly developmental process; dormant spores can better withstand harsh environmental insults (e.g., heat, desiccation, radiation, and oxidative and chemical attacks [13, 14, 16, 17]) than vegetative cells, but spore formation is costly in that it requires the coordinated transcription of hundreds of genes throughout several hours (reviewed in references 4 and 22 and references therein). Spore formation is a remarkable example of phenotypic plasticity, where cells are able to tailor their gene expression to specific environments or physiological states. In environments where benefits associated with spores are absent and no longer balance the cost of constructing spores, it would be predicted that sporulation ability should be lost over time (for extensive discussion of the theoretical details, see references 7 and 8). In order to test this prediction, we performed a laboratory evolution experiment in which two sets of five Bacillus subtilis populations were propagated for 6,000 generations in either the presence (populations 628A through 628E) or the absence (populations 624A through 624E) of strong selection for sporulation (7, 8). We observed that indeed, when B. subtilis was evolved without selection for sporulation, the ability to sporulate was either severely reduced (populations 624A and 624C) or completely lost (populations 624B, 624D, and 624E) in the nonsporulating experimental populations (8) (Table 1). It was of interest to know whether sporulation ability was lost because selection favors sporulation mutants in a constant, nutrient-rich environment or whether sporulation had simply become neutral with respect to fitness and mutations in sporulation genes had accumulated over time. These two possibilities were addressed using simulations, which suggested that only one population, 624E, lost sporulation because selection favored its loss (8). Identifying the nature of the selectively advantageous mutation(s) underlying sporulation loss in 624E, and comparing it to the mutation(s) underlying sporulation loss in the other populations, is of interest because such a comparison can facilitate our understanding of why sporulation is adaptive in some natural environments but not in others.
TABLE 1.
Strains used in this study
| Ancestor and evolved strains | Experimental environment | Sporulation frequency |
|---|---|---|
| WN624 (trpC2 spc::amyE) | 0.58 | |
| WN624A | Sporulation repressing | 4.3 × 10−4 |
| WN624B | Sporulation repressing | <2 × 10−6 |
| WN624C | Sporulation repressing | 0.3 |
| WN624D | Sporulation repressing | <2 × 10−6 |
| WN624E | Sporulation repressing | <2 × 10−6 |
| WN628 (trpC2 cat::amyE) | 0.58 | |
| WN628A | Sporulation inducing | 0.6 |
| WN628B | Sporulation inducing | 0.92 |
| WN628C | Sporulation inducing | 1.0 |
| WN628D | Sporulation inducing | 0.83 |
| WN628E | Sporulation inducing | 1.0 |
Two types of mutations were examined: large-scale deletions and small-scale changes. Because sporulation is no longer needed in populations 624A to 624E, genome loss via large-scale deletion might occur, in a manner similar to that observed in bacterial endosymbionts evolving in the relatively constant environment of the insect gut (11, 12). Alternatively, small-scale changes (e.g., small insertions/deletions [indels] or single-nucleotide substitutions) in critical sporulation genes could lead to a blockage of sporulation. A long-standing and extensive literature exists documenting the pleiotropic effects of simple mutations in spo genes (reviewed in references 4, 21, and 22 and references therein). Furthermore, we have previously shown that the rate of spontaneous mutation to rifampin (rifampicin) resistance increased during evolution in populations 624A to -E by as much as 2 orders of magnitude (7), and many of these mutations were determined to be single-nucleotide changes in rpoB (15; H. Maughan and W. L. Nicholson, unpublished data).
To address these two possibilities, we used DNA-DNA microarrays to characterize large-scale changes in genome structure and RNA-DNA (transcription) microarrays to assess small-scale changes leading to alterations in gene expression patterns that have occurred during the loss of sporulation proficiency.
Large-scale deletions.
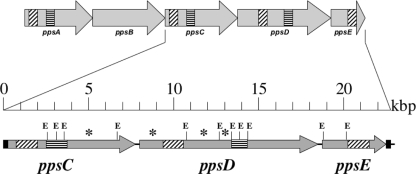
To determine whether large-scale deletions resulted in the loss of sporulation genes, we prepared chromosomal DNA from single colonies isolated from each of populations 624A to 624E, propagated for 6,000 generations under relaxed selective pressure for sporulation, and from each of populations 628A to 628E, propagated for 6,000-generations under strong selective pressure for sporulation. Fluorescently labeled probes generated from the chromosomal DNA of evolved isolates were hybridized to 4,096-open reading frame (ORF) microarray slides prepared from B. subtilis strain 168 (Eurogentec, Belgium), along with differentially labeled probes prepared from DNA of the corresponding ancestral strains, WN624 and WN628, respectively. The intensities of each dye were quantified, and genes were considered to be missing if (i) the intensities differed significantly between the ancestor and the evolved strain (as determined by a false discovery rate-corrected P value from analysis of variance [ANOVA] analyses in R/MAANOVA [http://research.jax.org/faculty/churchill/software/Rmaanova/index.html]) and (ii) the difference between intensities was greater than twofold. No large-scale deletions were detected in isolates from any of the cultures (628A to 628E) that had evolved under strong selection for sporulation (data not shown). However, isolates recovered from two different nonsporulating populations, 624A and 624B, were found by DNA microarray analysis to harbor deletions removing DNA corresponding to the ppsD and ppsC genes, respectively (data not shown). These genes are members of the ppsABCDE operon, which encodes a nonribosomal machine for the synthesis of plipastatin, a lipopeptide antibiotic (24). Because the ppsABCDE operon exhibits repeated domains within the ppsA, ppsC, and ppsD cistrons (23, 24) (Fig. 1), we reasoned that the observed deletions may have resulted from recombination between these homologous domains. To test this notion, the ppsCDE region from ancestral strain WN624 and the evolved 624A strain was amplified by PCR using primers 5′-GCCGCAGCAACCTGAAATACAGGA-3′ and 5′-TGTCGAGGATGATGTCGGCATTC-3′, which amplify essentially the entire ppsC, ppsD, and ppsE coding sequences (Fig. 1). The resulting amplicons were digested with EcoRI and their restriction patterns compared. Analysis of the EcoRI restriction patterns revealed that the evolved 624A strain had suffered a deletion of >9.7 kbp of DNA between the repeat sequences within the ppsC and ppsD cistrons (Fig. 1). This deletion removed the region of DNA corresponding to the Eurogentec ppsD microarray probe, thus explaining the observed DNA microarray result. Although we did not further analyze the deletion removing ppsC from the 624B evolved strain, we assume that ppsC sequences were deleted by a similar recombination event occurring between the repeat sequences located within ppsA and ppsC (Fig. 1).
FIG. 1.
Organization of the ppsABCDE operon in B. subtilis. Shaded arrows represent cistrons. Filled bars mark the locations of PCR primers. Diagonal and horizontal dashes indicate the locations of the probes present on the Eurogentec microarray and the locations of repeat sequences in ppsA, ppsC, and ppsD (5, 6), respectively. The locations of EcoRI restriction sites are marked by the letter E. Asterisks indicate EcoRI restriction fragments missing in the 624A evolved strain but present in the WN624 ancestral strain.
Mutations affecting global transcription patterns.
Because the ppsABCDE operon is not known to affect sporulation, and large-scale deletions were detected only in this operon, the DNA hybridization data suggest that sporulation loss during evolution was not caused by large-scale deletions in sporulation genes. Therefore, small-scale point mutations or indels are likely to have caused sporulation loss in the evolving populations 624A to 624E, and if the regulation of sporulation has been altered, transcriptional profiling may enable us to identify the genes containing such genetic changes. We therefore used transcription microarrays to measure global transcription patterns in the ancestor and evolved strains during the onset of stationary phase, the time at which sporulation is normally initiated under inducing conditions. RNA was isolated as soon as exponential growth started to slow down, which corresponded to optical densities at 600 nm of 0.65 to 0.82 in sporulation-repressing medium and 0.65 to 1.65 in sporulation-inducing medium. The evolved strain chosen for study was a single colony isolate from population 624E obtained after 6,000 generations of evolution under sporulation-repressing conditions, for which simulations suggested that selection had favored the loss of sporulation (8). The 624E isolate exhibited a sporulation frequency of ∼10−6, compared to the ancestral level of ∼58% (8, 9). (Sporulation frequencies were measured by heat shocking the culture at 80°C for 10 min.) This low sporulation frequency for the 624E isolate was consistent with phase-contrast microscopy observations where spores were not seen, and the majority of cells were in long, nonmotile filaments.
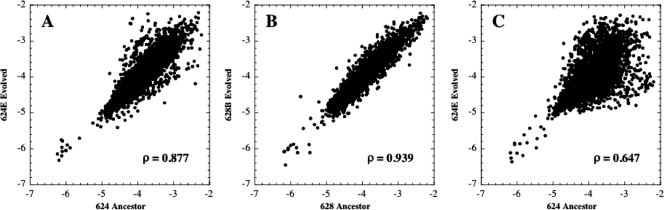
The first set of experiments measured transcription in the ancestral strain WN624 and the 624E evolved strain when both were grown in the glucose-rich, sporulation-repressing experimental environment (Fig. 2A); this would show us how transcription had changed in the evolved strain after evolution for 6,000 generations under sporulation-repressing conditions. Global comparison of the transcription patterns showed a correlation coefficient (ρ) of 0.877 (95% confidence interval [95% CI], 0.869 to 0.884) (Fig. 2A). For purposes of comparison, analysis of a control transcription microarray comparing the two ancestral strains WN624 and WN628, both grown under sporulation-repressing conditions, resulted in a ρ of 0.978 (95% CI, 0.977 to 0.98) (data not shown). In addition, transcription microarrays comparing ancestral strain WN628 with a strain (628B) that had evolved for 6,000 generations under strict selection for sporulation also demonstrated a higher correlation coefficient (ρ = 0.939; 95% CI, 0.935 to 0.942) (Fig. 2B). The data suggested that evolution for 6,000 generations in the absence of selective pressure for sporulation resulted in a higher degree of transcriptome divergence than when selection for sporulation was applied. This transcriptome divergence was particularly dramatic when transcription microarrays compared the ancestor strain WN624 with an evolved strain from population 624E, when both strains were grown under sporulation-inducing conditions (Fig. 2C). This particular condition resulted in the lowest correlation coefficient of all, a ρ of 0.647 (95% CI, 0.629 to 0.665) (Fig. 2C).
FIG. 2.
Scatter plots representing transcription divergence between ancestor strains and the strains that evolved from them. (A) Ancestor WN624 versus an isolate from population 624E, which had evolved for 6,000 generations under sporulation-repressing conditions. RNA was harvested at the onset of stationary phase under sporulation-repressing conditions. (B) Ancestor WN628 versus an isolate from population 628B, which had evolved for 6,000 generations under stringent selection for sporulation. RNA was harvested at the onset of stationary phase under sporulation-inducing conditions. (C) The same strains as in panel A, but RNA was harvested at the onset of stationary phase under sporulation-inducing conditions. Each data point indicates the normalized expression of one gene. The intensities were normalized and log transformed; therefore, each axis has a log scale. Correlation coefficients (ρ) are given. See the text for details.
In addition to global transcriptome patterns, the transcription levels of individual genes were analyzed for ancestor and evolved strains. Transcription differences were considered to be significant if the false discovery rate-corrected P value from ANOVA analyses was less than 0.05 and if transcription levels differed by at least 1.5-fold. We also used quantitative PCR (qPCR) to verify the relative transcription levels of several genes; in all cases, results from the qPCR qualitatively agreed with the microarrays (data not shown). In the transcription microarray experiment in which ancestral strain WN628 was compared to a strain from population 628B, which had evolved for 6,000-generations with stringent selection for sporulation, and RNA was isolated from cultures cultivated in sporulation-inducing medium (Fig. 2B), only 34 out of 3,916 total genes analyzed (0.9% of the total) were significantly differentially expressed (see Table S1 in the supplemental material). Twenty-four and 10 genes were significantly up- or downregulated, respectively, and most of these were genes of unknown function. Interestingly, the gerAB gene was 2.1-fold upregulated in the evolved strain, which might be expected to give a selective advantage to the evolved strain in that it may have enhanced its germination response during daily transfer of spores into fresh medium. In support of this notion, populations that had evolved under strict selection for sporulation indeed exhibited an average spore germination/lag time of 102.5 min, compared to the 182.5 min for the ancestral strain WN628 (7). In the transcription microarray experiment comparing transcription patterns between the ancestor WN624 and the evolved 624E isolate, both grown under sporulation-repressing conditions (Fig. 2A), it was observed that 5% of the total genes spotted onto the microarray (196 out of 3,996 genes analyzed) were transcribed at significantly different levels (see Table S1 in the supplemental material). Under sporulation-repressing conditions, significant upregulation of 107 genes occurred in the 624E evolved strain, most notably genes encoding ribosomal proteins, transport/binding proteins, and metabolic proteins. Upregulation of these functions is presumably related to the continued growth of the 624E strain in stationary phase (data not shown). In the same experiment, significant downregulation was observed for 87 genes, most notably those encoding chemotaxis, motility, and flagellar proteins (see Table S1 in the supplemental material). The decreased transcription of chemotaxis/motility genes has also been observed in Escherichia coli populations that had evolved in the laboratory for 20,000 generations (2) and is presumably associated with the lack of need for chemotaxis or motility in shaking culture, coupled with the large biosynthetic and energetic costs of producing and operating the flagellar apparatus (10); loss of these functions presumably results in a competitively advantageous growth rate.
When we compared the transcription of individual genes in the 624E-derived evolved strain and ancestral strain WN624, both grown under sporulation-inducing conditions (Fig. 2C), we observed that 1,083 genes out of a total of 4,025 genes analyzed (27% of the total) were transcribed at significantly different levels (Fig. 2C; see also Table S1 in the supplemental material). Many of the genes that were upregulated in the evolved strain encoded ribosomal proteins, transport/binding proteins, proteins related to the metabolism of amino acids, and proteins related to cell wall functions, again consistent with the continued growth of the evolved strain in stationary phase. In addition to changes in these functional categories, 34 sporulation-associated genes were differentially transcribed in the ancestor and the evolved strain; 5 of these were upregulated and 29 were downregulated in the evolved strain (Table 2). Notable among the upregulated genes was abrB, a key protein controlling gene expression during the transition from exponential to stationary phase (20). The upregulation of abrB likely explains the concomitant downregulation of numerous genes, such as those involved in competence and the synthesis of extracellular antibiotics and proteases (see Table S1 in the supplemental material). Interestingly, most of the downregulated sporulation-associated genes encode products known to be involved in the initiation of sporulation (4, 5, 18, 22), including one dipeptide permease (dppC), two phosphorelay kinases (kinA and kinE), six rap phosphatases (rapA, -B, -C, -F, -G, and -J), six Rap phosphatase regulators (phrA, -C, -E, -F, -G, and -K), two early sigma factors (sigH and sigF), and three critical stage zero spo genes (spo0A, spo0F, and spo0M) (Table 2). Collectively, these data indicate that the evolved 624E strain is unable to sporulate due to the misregulation of genes involved in the very earliest stages of the vegetative-to-stationary-phase transition state and the initiation of sporulation. In support of this idea, we also noted that many of the flagellar genes that were downregulated when the 624E-derived evolved strain was grown in sporulation-repressing medium were also downregulated when this strain was grown in sporulation-inducing medium (see Table S1 in the supplemental material). Furthermore, several genes whose products are involved in competence for DNA uptake were also downregulated when the 624E-derived evolved strain was grown in sporulation-inducing medium (see Table S1 in the supplemental material). These observations indicate that sporulation is not the only stationary-phase phenotype lost in this strain but that the strain may have lost other transition state functions (e.g., competence, motility) as well.
TABLE 2.
Sporulation-associated genes whose transcription was significantly up- or downregulated in the 624E strain versus the WN624 ancestor grown under sporulation-inducing conditionsa
| Gene | Annotationb | Fold change | P | |||
|---|---|---|---|---|---|---|
| Upregulated genes | ||||||
| abrB | Pleiotropic transcriptional regulator of transition state genes | 7.5 | 0.026 | |||
| spoIID | Required for complete dissolution of asymmetric septum | 5.3 | 0.048 | |||
| spoVC | Probable peptidyl-tRNA hydrolase | 8.7 | 0.016 | |||
| spoVE | Required for spore cortex biosynthesis | 2.8 | 0.038 | |||
| spoVT | Transcriptional regulator of sigma G-dependent genes | 5.3 | 0.016 | |||
| Downregulated genes | ||||||
| dppC | Dipeptide permease (sporulation) | 11.2 | 0.016 | |||
| hpr | Transcriptional repressor of sporulation and extracellular protease genes | 2.0 | 0.048 | |||
| kinA | Two-component sensor kinase, sporulation initiation | 6.2 | 0.042 | |||
| kinE | Two-component sensor kinase, sporulation initiation | 3.0 | 0.014 | |||
| pbpE | Penicillin binding protein 4 (cortex) | 25.6 | 0.010 | |||
| phrA | Phosphatase RapA inhibitor | 52.6 | 0.010 | |||
| phrC | Phosphatase RapC inhibitor | 7.1 | 0.017 | |||
| phrE | Phosphatase RapE inhibitor | 9.6 | 0.041 | |||
| phrF | Phosphatase RapF inhibitor | 12.5 | 0.010 | |||
| phrG | Phosphatase RapG inhibitor | 7.3 | 0.048 | |||
| phrK | Phosphatase RapK inhibitor | 4.6 | 0.048 | |||
| rapA (spoOL) | Response regulator, aspartate phosphatase | 66.7 | 0.015 | |||
| rapB | Response regulator, aspartate phosphatase | 1.7 | 0.048 | |||
| rapC | Response regulator, aspartate phosphatase | 9.7 | 0.010 | |||
| rapF | Response regulator, aspartate phosphatase | 21.3 | 0.010 | |||
| rapG | Response regulator, aspartate phosphatase | 10.4 | 0.010 | |||
| rapJ | Response regulator, aspartate phosphatase | 2.5 | 0.010 | |||
| sigF | Early forespore-specific sigma factor | 26.3 | 0.010 | |||
| sigH | Vegetative and early-stationary-phase sigma factor | 4.8 | 0.010 | |||
| spo0A | Two-component response regulator essential for sporulation initiation | 7.7 | 0.033 | |||
| spoOF | Sporulation initiation phosphotransferase | 14.1 | 0.010 | |||
| spo0M | Sporulation control gene | 6.7 | 0.047 | |||
| spoIIAA | Anti-anti-sigma factor of SpoIIAB | 6.3 | 0.042 | |||
| spoIIAB | Anti-sigma factor serine kinase | 52.6 | 0.017 | |||
| spoIIGA | Protease processing, pro-sigma E to active sigma-E | 4.3 | 0.038 | |||
| spoIIP | Required for dissolution of septal cell wall | 2.8 | 0.017 | |||
| spoVAF | Mutants lead to production of immature spores | 1.7 | 0.048 | |||
| spoVG | Involved in spore cortex synthesis | 50.0 | <0.001 | |||
| spoVS | Involved in forespore engulfmentc | 5.3 | 0.048 | |||
Data are taken from Table S1 in the supplemental material.
Gene annotations are from the BSORF database (http://bacillus.genome.jp/).
See reference 19.
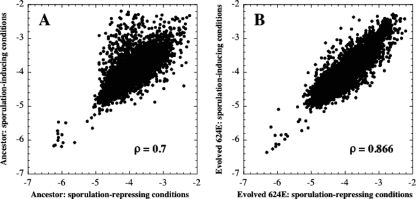
The expression microarray results also suggested that the transcription profile of the 624E-derived evolved strain in sporulation-inducing medium was quite similar to its transcription profile in sporulation-repressing medium. Because we had transcription profiles of the WN624 ancestor and the 624E-derived evolved strain grown under both sporulation-repressing and sporulation-inducing conditions, we were able to analyze how the genes transcribed by each strain differed between the two environments. Comparison of the transcription patterns of the ancestral strain WN624 grown under sporulation-inducing versus sporulation-repressing conditions resulted in a ρ of 0.7 (95% CI, 0.683 to 0.715) (Fig. 3A), indicating a substantial level of transcriptome adjustment in the ancestral strain when it was faced with the two differing environments. In contrast, comparison of the transcription patterns of the evolved strain derived from population 624E, also grown under sporulation-inducing versus sporulation-repressing conditions, resulted in a higher ρ of 0.866 (95% CI, 0.859 to 0.874) (Fig. 3B). This result indicated that fewer genes in the evolved strain were differentially regulated when the strain was faced with two very different environments.
FIG. 3.
Scatter plots of transcription in the sporulation-inducing environment versus the sporulation-repressing environment for ancestral strain WN624 (A) and the population 624E-derived strain (B). Each data point indicates the normalized expression of one gene. The intensities were normalized and log transformed; therefore, each axis has a log scale. Correlation coefficients (ρ) are given. See the text for details.
The similarity of the transcription patterns from the evolved strain when grown in sporulation-repressing versus sporulation-inducing environments shows that the ability of this strain to respond to different environments has declined since its divergence from the ancestor. Phenotypic plasticity, the ability of one organism to express different phenotypes in different environments, is thought to be adaptive (recently reviewed in reference 1) but is also predicted to be lost in constant environments (6). We consider the inability of the 624E-derived evolved strain to tailor its transcription to differing environments to be strong evidence that this strain has lost phenotypic plasticity. Furthermore, the loss of sporulation initiation may be part of a larger decrease in the ability of this strain to respond to different environments.
The 624E population from which the evolved strain was isolated showed a clear fitness advantage over the ancestor when they were competed in sporulation-repressing medium (7). Furthermore, evidence from simulations suggested that the loss of sporulation in this population was selectively advantageous (8). Monitoring the environment and coordinating gene expression to match environmental change is beneficial for cells living in an ever-changing environment, but this monitoring may become costly when cells find themselves in a constant environment (1, 3). Given this evidence, an important question remains: was the loss of phenotypic plasticity also selectively advantageous?
Microarray data accession number.
Complete microarray data and additional methodological details have been deposited in Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE4469.
Supplementary Material
Acknowledgments
We thank Ryan Sprissler of the Genomic Analysis and Technology Core facility at the University of Arizona for nucleic acid labeling and microarray hybridization and scanning; Helen Dunbar for assistance with qPCR; the laboratory of Howard Ochman and Nancy Moran for use of their LightCycler and for discussions concerning this work; and Carlos Machado, Leland S. Pierson III, Michael Nachman, and Nancy Moran for reading an earlier version of this article. We also thank two anonymous reviewers for their suggestions to improve this paper.
This work was funded by two grants from Sigma Xi and an NSF Doctoral Dissertation Improvement Grant (DEB-0412650) to H.M. and by grants from the USDA (FLA-MCS-04602) and NASA (NNX08AO15G) to W.L.N.
Footnotes
Published ahead of print on 24 October 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Callahan, H. S., H. Maughan, and U. K. Steiner. 2008. Phenotypic plasticity, costs of phenotypes, and costs of plasticity: toward an integrative view. Ann. N. Y. Acad. Sci. 113344-66. [DOI] [PubMed] [Google Scholar]
- 2.Cooper, T. F., D. E. Rozen, and R. E. Lenski. 2003. Parallel changes in gene expression after 20,000 generations of evolution in Escherichia coli. Proc. Natl. Acad. Sci. USA 1001072-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DeWitt, T. J., A. Sih, and D. S. Wilson. 1998. Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 1377-81. [DOI] [PubMed] [Google Scholar]
- 4.Eichenberger, P. 2007. Genomics and cellular biology of endospore formation, p. 375-418. In P. Graumann (ed.), Bacillus: cellular and molecular biology. Caister Academic Press, Wymondham, United Kingdom.
- 5.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masel, J., O. D. King, and H. Maughan. 2007. The loss of adaptive plasticity during long periods of environmental stasis. Am. Nat. 16938-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maughan, H., V. Callicotte, A. Hancock, C. W. Birky, Jr., W. L. Nicholson, and J. Masel. 2006. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution 60686-695. [PubMed] [Google Scholar]
- 8.Maughan, H., J. Masel, C. W. Birky, Jr., and W. L. Nicholson. 2007. The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis. Genetics 177937-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maughan, H., and W. L. Nicholson. 2004. Stochastic processes influence stationary-phase decisions in Bacillus subtilis. J. Bacteriol. 1862212-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mitchell, J. G., and K. Kogure. 2006. Bacterial motility: links to the environment and a driving force for microbial physics. FEMS Microbiol. Ecol. 553-16. [DOI] [PubMed] [Google Scholar]
- 11.Moran, N. A. 2003. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 6512-518. [DOI] [PubMed] [Google Scholar]
- 12.Moran, N. A., and A. Mira. 2001. The process of genome shrinkage in the obligate symbiont Buchnera aphidicola. Genome Biol. 2RESEARCH0054. [DOI] [PMC free article] [PubMed]
- 13.Nicholson, W. L. 2002. Roles of Bacillus endospores in the environment. Cell. Mol. Life Sci. 59410-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicholson, W. L. 2004. Ubiquity, longevity, and ecological roles of Bacillus spores, p. 1-15. In E. Ricca, A. O. Henriques, and S. M. Cutting (ed.), Bacterial spore formers: probiotics and emerging applications. Horizon Scientific Press, Norfolk, United Kingdom.
- 15.Nicholson, W. L., and H. Maughan. 2002. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J. Bacteriol. 1844936-4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicholson, W. L., A. C. Schuerger, and P. Setlow. 2005. The solar UV environment and bacterial spore UV resistance: considerations for Earth-to-Mars transport by natural processes and human spaceflight. Mutat. Res. 571249-264. [DOI] [PubMed] [Google Scholar]
- 18.Perego, M., and J. A. Hoch. 2002. Two-component systems, phosphorelays, and regulation of their activities by phosphatases, p. 473-481. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 19.Perez, A. R., A. Abanes-De Mello, and K. Pogliano. 2006. Suppression of engulfment defects in Bacillus subtilis by elevated expression of the motility regulon. J. Bacteriol. 1881159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phillips, Z. E., and M. A. Strauch. 2002. Bacillus subtilis sporulation and stationary phase gene expression. Cell. Mol. Life Sci. 59392-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piggot, P. J., and J. G. Coote. 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40908-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piggot, P. J., and R. Losick. 2002. Sporulation genes and intercompartmental regulation, p. 483-517. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 23.Tosato, V., A. M. Albertini, M. Zotti, S. Sonda, and C. V. Bruschi. 1997. Sequence completion, identification and definition of the fengycin operon in Bacillus subtilis. Microbiology 1433443-3450. [DOI] [PubMed] [Google Scholar]
- 24.Tsuge, K., T. Ano, M. Hirai, Y. Nakamura, and M. Shoda. 1999. The genes degQ, pps, and lpa-8 (sfp) are responsible for conversion of Bacillus subtilis 168 to plipastatin production. Antimicrob. Agents Chemother. 432183-2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.