Abstract
Bacteria can use branched-chain amino acids (ILV, i.e., isoleucine, leucine, valine) and fatty acids (FAs) as sole carbon and energy sources converting ILV into acetyl-coenzyme A (CoA), propanoyl-CoA, and propionyl-CoA, respectively. In this work, we used the comparative genomic approach to identify candidate transcriptional factors and DNA motifs that control ILV and FA utilization pathways in proteobacteria. The metabolic regulons were characterized based on the identification and comparison of candidate transcription factor binding sites in groups of phylogenetically related genomes. The reconstructed ILV/FA regulatory network demonstrates considerable variability and involves six transcriptional factors from the MerR, TetR, and GntR families binding to 11 distinct DNA motifs. The ILV degradation genes in gamma- and betaproteobacteria are regulated mainly by a novel regulator from the MerR family (e.g., LiuR in Pseudomonas aeruginosa) (40 species); in addition, the TetR-type regulator LiuQ was identified in some betaproteobacteria (eight species). Besides the core set of ILV utilization genes, the LiuR regulon in some lineages is expanded to include genes from other metabolic pathways, such as the glyoxylate shunt and glutamate synthase in Shewanella species. The FA degradation genes are controlled by four regulators including FadR in gammaproteobacteria (34 species), PsrA in gamma- and betaproteobacteria (45 species), FadP in betaproteobacteria (14 species), and LiuR orthologs in alphaproteobacteria (22 species). The remarkable variability of the regulatory systems associated with the FA degradation pathway is discussed from functional and evolutionary points of view.
Proteobacteria comprise one of the largest divisions within prokaryotes and incorporate species possessing a very complex collection of phenotypic and physiological attributes including many phototrophs, heterotrophs, and chemolithotrophs. The proteobacterial group is of great biological significance, as it includes a large number of pathogens and symbionts of animals and plants. Thus, proteobacteria display an amazing versatility in their abilities to use various carbon sources such as carbohydrates, nucleotides, amino acids, and lipids. The degradation of branched-chain amino acids valine, leucine, and isoleucine (ILV) and fatty acids (FAs) is used for ATP and energy production by many proteobacteria.
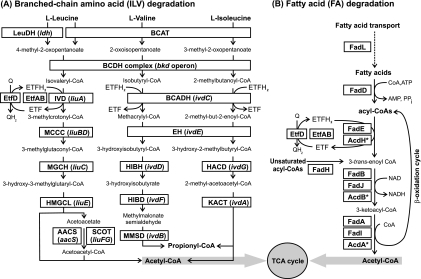
The ILV degradation pathways are outlined in Fig. 1A. The first reaction is transamination to the corresponding α-keto acids using either branched-chain amino acid aminotransferase or leucine dehydrogenase. The second step is oxidative decarboxylation to the corresponding acyl-coenzyme A (CoA) derivative coupled to dehydrogenation, which is carried out by a common branched-chain α-keto acid dehydrogenase (BCDH) (EC 1.2.4.4) complex. The further conversion of branched-chain acyl-CoA derivatives of ILV amino acids, namely, isovaleryl-CoA for leucine, 2-methylbutanoyl-CoA for isoleucine, and isobutyryl-CoA for valine, into acetyl-CoA and propionyl-CoA is mediated by individual ILV catabolic pathways (25).
FIG. 1.
Branched-chain amino acid (A) and FA (B) catabolic pathways. (A) Abbreviations of functional roles of ILV catabolic enzymes were adopted from the respective SEED subsystems (http://seed-viewer.theseed.org/). LeuDH, leucine dehydrogenase (EC 1.4.1.9); BCAT, branched-chain amino acid aminotransferase (EC 2.6.1.42); IVD, isovaleryl-CoA dehydrogenase (EC 1.3.99.10); BCADH, branched-chain acyl-CoA dehydrogenase (EC 1.3.99.12); MCCC, methylcrotonyl-CoA carboxylase (EC 6.4.1.4); EH, enoyl-CoA hydratase (EC 4.2.1.17); MGCH, methylglutaconyl-CoA hydratase (EC 4.2.1.18); HIBH, 3-hydroxyisobutyryl-CoA hydrolase (EC 3.1.2.4); HACD, 3-hydroxyacyl-CoA dehydrogenase (EC 1.1.1.35); HMGCL, hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4); HIBD, 3-hydroxyisobutyrate dehydrogenase (EC 1.1.1.31); KACT, 3-ketoacyl-CoA thiolase (EC 2.3.1.16); MMSD, methylmalonate-semialdehyde dehydrogenase (EC 1.2.1.27). The conversion of acetoacetate to acetoacetyl-CoA is mediated by either of two alternative routes: succinyl-CoA:3-ketoacid-CoA transferase (SCOT) (EC 2.8.3.5) or acetoacetyl-CoA synthetase (AACS) (EC 6.2.1.16). ECF, electron transfer flavoprotein. The assimilation of acetoacetyl-CoA and propionyl-CoA products occurs using central metabolic enzymes that are shared with other pathways (not shown). (B) FAD proteins are abbreviated according to the corresponding gene names in E. coli.
Many enzymes in these downstream ILV degradation pathways belong to large families of paralogs, and thus, most early annotations of the corresponding genes in bacterial genomes were rather nonspecific “general-class” functional assignments. A subsystem-based approach to genome annotations as implemented in the SEED platform (http://theseed.uchicago.edu) was used for the reconstruction of the ILV degradation pathways in bacteria (30). A combination of functional and genome context analysis, as depicted in the SEED Viewer subsystems “leucine degradation,” “isoleucine degradation,” and “valine degradation” (at http://seed-viewer.theseed.org/), provided convincing evidence for the presence of the ILV catabolic pathways in a number of diverse bacteria (30). According to this analysis, the ILV catabolic pathways are present in many lineages of gammaproteobacteria (e.g., in Pseudomonas aeruginosa and Shewanella oneidensis), with a notable exception of Escherichia coli and other enterobacteria.
The liu gene cluster involved in leucine and isovalerate utilization was recently identified and characterized for P. aeruginosa (1, 11, 15). Functional roles of the liuABCDE genes are shown in Fig. 1A. The first gene encodes a hypothetical transcription factor from the MerR family called LiuR (11). Although expression analysis of the liu genes showed their specific induction by leucine (1), a possible role of LiuR in the transcriptional regulation of the liu genes has not yet been investigated. The BCDH-encoding operon bkd in Pseudomonas putida is regulated by an ILV-responsive transcriptional activator, BkdR, from the AsnC family (24).
The FA degradation (FAD) pathway is catalyzed by enzymes encoded by the fad regulon (Fig. 1B) (12). Long-chain FAs are transported across the cell membrane using the outer membrane transporter FadL and the inner membrane-associated CoA ligase FadD. After uptake, FAs can be degraded via the β-oxidation pathway and used as an energy/carbon source via the tricarboxylic acid (TCA) cycle, or alternatively, FAs can be used as precursors for the membrane-phospholipid biosynthesis. The β-oxidative cleavage of acyl-CoAs acts in a cyclic manner and involves their conversion to enoyl-CoAs catalyzed by acyl-CoA dehydrogenase (e.g., FadE), which is followed by hydration, oxidation, and thiolytic cleavage performed by the FA oxidation complex (e.g., FadB-FadA or FadI-FadJ). In addition, FadH is used for the degradation of unsaturated FAs.
The transcriptional control of FA metabolism in E. coli is mediated by the FadR regulatory protein from the GntR family, which recognizes a 17-bp palindromic motif with the consensus sequence AACTGGTCnGACCAGTT, where n denotes any nucleotide (6). FadR senses FA availability in the environment and is released from DNA in the presence of long-chain acyl-CoA (16). FadR acts as a repressor of the FAD operons (fadL, fadD, fadE, fadBA, fadH, and fadIJ) and as an activator of the fabB and fabA genes, involved in unsaturated FA synthesis (5, 36). In addition, E. coli FadR is involved in the regulatory cascade by the activation of the iclR gene, encoding a repressor of the glyoxylate shunt operon aceBAK (14). The comparative genomic analysis of FadR binding sites demonstrated the conservation of the E. coli FadR regulon in enterobacteria and its considerable reduction in other gammaproteobacteria, represented by Haemophilus influenzae and Vibrio cholerae (36). A different transcription factor from the TetR family in Bacillus subtilis (encoded by the ysiA gene) was recently identified as being the master regulator of the fad genes. It recognizes YsiA boxes with the consensus sequence TGAATGAnTAnTCATTCA (26). Apart from E. coli and B. subtilis, the mode of regulation of the FAD genes in other bacterial lineages remains unclear.
In this study, we expanded the use of the comparative genomics approach to regulation (for a recent review, see reference 34) to characterize novel regulons controlling ILV degradation and FAD pathways in the alpha-, beta-, and gammaproteobacteria. The analysis of conserved operons involved in these pathways, initiated in S. oneidensis and related bacteria, led to a tentative identification of two novel regulons characterized by unique DNA motifs. These highly conserved motifs are candidate binding sites of two different subfamilies of transcription factors, which are widely distributed in proteobacteria. Their representatives in P. aeruginosa were previously described as being LiuR and PsrA. In particular, the P. aeruginosa regulator PsrA from the TetR family was originally shown to be involved in the stationary-phase-induced transcriptional regulation of rpoS and some other genes (18, 19, 20). Here, we perform a comparative genomic reconstruction of the respective regulons in proteobacteria and report that their major targets are the ILV degradation and FAD pathways, respectively. Finally, we identified and characterized nonorthologous regulons for ILV degradation and FAD in the Burkholderiales group of betaproteobacteria (named here LiuQ and FadP, respectively). The distribution and partial overlap between three FAD regulons (FadR, PsrA, and FadP) and two ILV degradation regulons (LiuR and LiuQ) in gamma-, and betaproteobacteria and their evolutionary implications are discussed.
MATERIALS AND METHODS
Bacterial genome sequences were downloaded from GenBank (3). The gene identifiers from GenBank are used throughout. A protein similarity search was done using the Smith-Waterman algorithm implemented in the Genome Explorer program (27). Orthologous proteins were initially defined by the best-bidirectional-hits criterion and, if necessary, confirmed by analysis of phylogenetic trees. The phylogenetic trees were constructed by the maximum likelihood method implemented in the PHYLIP package (9) using multiple-sequence alignments of protein sequences produced by ClustalX (37).
A simple iterative procedure implemented in the program SignalX (as described previously in reference 13 and recently reviewed in reference 34) was used for the construction of transcription factor binding motifs in sets of upstream fragments of potentially coregulated genes. For the LiuR regulon, the original training set included the ILV degradation operons in S. oneidensis. Orthologs of these and other candidate members of the predicted S. oneidensis LiuR regulon identified in other gammaproteobacteria (see Table S1 in the supplemental material) were used as a training set for the construction of the “LiuR_gamma” profile. For the PsrA regulon, the training set used for the “PsrA_gamma” profile construction included the FAD operons from S. oneidensis as well as from Vibrio and Pseudomonas species (see Table S5 in the supplemental material). The resulting LiuR and PsrA binding-site profiles were used for the comparative analysis of the respective regulons in gamma-, beta-, and alphaproteobacteria. For the FadR regulon, we started from the training set of known members of this regulon in E. coli and their orthologs in other members of the Enterobacteriales (36). Finally, for each taxonomic group of gammaproteobacteria with the FadR regulon (the Enterobacteriales, Vibrionales, Pasteurellales, and Altermonadales), we used a separate training set of the upstream regions of candidate FadR target operons to construct the FadR binding-site profile (see Table S4 in the supplemental material). For the LiuQ and FadP regulons in betaproteobacteria, the training sets included the ILV degradation and FAD operons, respectively (see Tables S2 and S6 in the supplemental material).
Each genome encoding the studied transcription factor was scanned with the constructed profile using GenomeExplorer software (27), and genes with candidate regulatory sites in the upstream regions were selected. We analyzed only 5′-untranslated gene regions up to 400 nucleotides upstream of the translation start site. z scores of candidate sites were calculated as the sum of the respective positional nucleotide weights. The threshold for the site search was defined as the lowest score observed in the training set (see Tables S1 to S6 in the supplemental material). The consistency check of the predicted members of regulons was used to eliminate false-positive site predictions. This approach is based on the assumption that regulatory events tend to be conserved in closely related species with orthologous regulators (34). The upstream regions of genes that are orthologous to genes containing conserved regulatory sites were examined for candidate sites even if these sites were not detected automatically with a given threshold (weak regulatory sites with scores below the threshold are underlined in Tables S1 to S6 in the supplemental material). Among candidate members of the PsrA and FadP regulons, only genes having candidate sites conserved in at least two other genomes were retained for further analysis. For the PsrA regulon, we also considered several candidate regulon members that did not satisfy this conservation criterion but were functionally related to the FA metabolism. Sequence logos for the derived regulatory motifs were drawn using WebLogo package v.2.6 (7) (http://weblogo.berkeley.edu/).
RESULTS
ILV degradation regulons in proteobacteria. (i) Identification of the LiuR regulon in S. oneidensis.
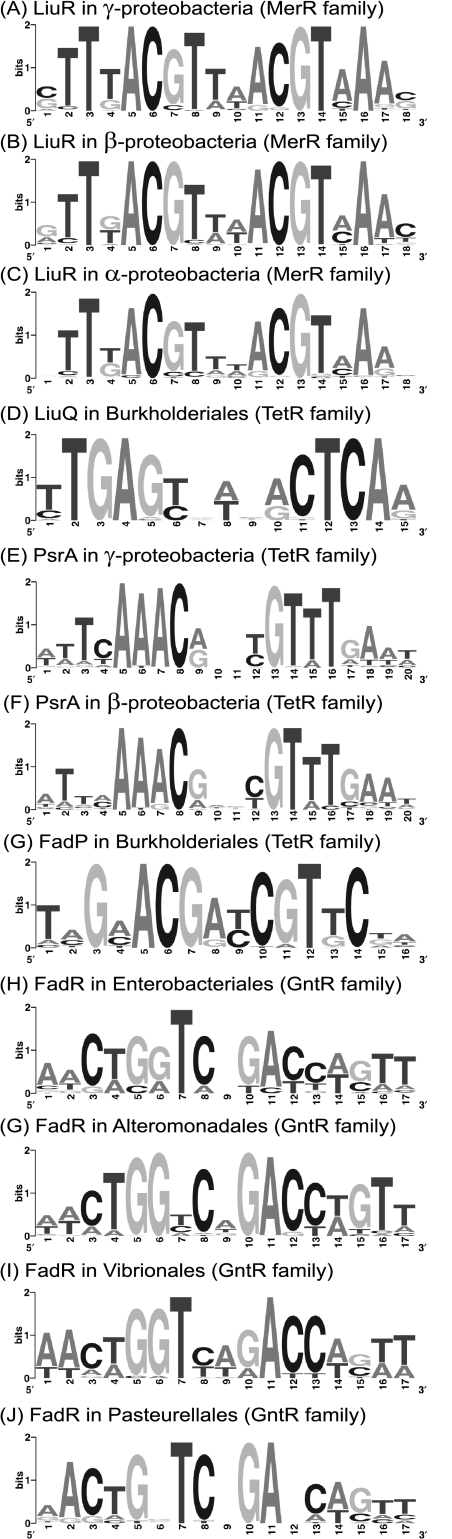
A novel regulon for ILV degradation genes was initially identified by analysis of gene expression data for S. oneidensis. Using microarray data for salt (22) and alkaline (21) stresses, we selected the 15 most upregulated genes constituting three potential operons, namely, SO1898 to SO1891, SO1677 to SO1683, and SO2339 to SO2341. According to the reconstructed metabolic pathways in the SEED database, the above-described three operons belong to the ILV degradation subsystem (see “branched-chain amino acid degradation regulons” subsystem at http://theseed.uchicago.edu/FIG/subsys.cgi). By applying the motif recognition procedure to a training set of upstream regions of these operons n S. oneidensis and orthologous operons in other Shewanella species, we found a common 18-bp DNA motif named the ILV box (see Table S1 in the supplemental material). The consensus sequence for this palindromic motif (ILV box) is sTTTACGTwwACGTAAAs, where “w” and “s” denote “A or T” and “C or G,” respectively (see the motif logo in Fig. 2A). We also identified an additional motif, gTGTAAAnnnnnntTTACAc, of the aromatic amino acid-responsive regulator TyrR, which we studied in detail previously (35). Candidate TyrR binding sites were observed upstream of the three above-mentioned operons and the SO2638 gene. The TyrR regulon was not further analyzed in this study because it does not include the ILV degradation genes in other bacterial species outside of the Shewanella group.
FIG. 2.
Sequence logos for the identified binding sites of LiuR, LiuQ, PsrA, FadR, and FadP regulators in various taxonomic groups of the Proteobacteria.
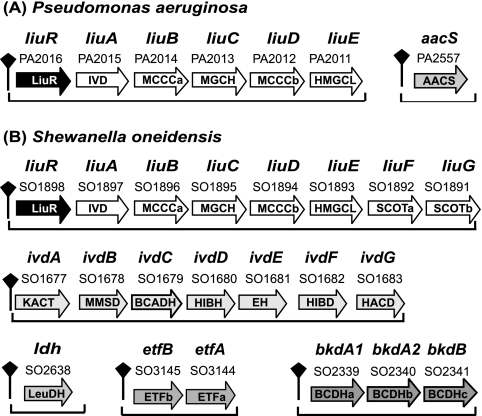
The SO1898 to SO1893 genes are orthologs of the liuRABCDE genes involved in leucine and isovalerate utilization in P. aeruginosa (11) (Fig. 3). The first gene in this cluster encodes a hypothetical transcription factor from the MerR family, named LiuR for a candidate regulator of the liu cluster (11). In the genomes of most gamma- and betaproteobacteria, the scanning with the ILV box recognition profile identified candidate regulatory sites upstream of operons containing orthologs of liuR. We tentatively attributed the ILV box motif to the LiuR transcription factor based on the following comparative genomic evidence: (i) positional clustering on the chromosome of the liuR genes and ILV boxes and (ii) correlation in the phylogenetic pattern of the cooccurrence of liuR and ILV boxes in the genomes of various proteobacteria (see the next section for details).
FIG. 3.
Branched-chain amino acid degradation operons under the control of the LiuR regulons in Pseudomonas aeruginosa (A) and Shewanella oneidensis (B).
The second LiuR-regulated operon in S. oneidensis (SO1677 to SO1683) encodes all enzymes required for the utilization of 2-methylbutanoyl-CoA and isobutyryl-CoA, the products of isoleucine and valine degradation; therefore, it was named the ivd operon (Fig. 1 and 3B). The third candidate member of the LiuR regulon is the BCDH enzyme complex encoded by the bkd operon (SO2339 to SO2341) and involved in the second step of ILV utilization. Scanning of the S. oneidensis genome with the constructed ILV box profile identified six more operons that are likely regulated by LiuR (see Table S1 in the supplemental material). These additional candidate members of LiuR regulon include the leucine dehydrogenase gene ldh, the glyoxylate shunt genes aceBA, the glutamate synthase genes gltBD, the threonine synthesis operon thrABC, the regulator of aromatic amino acid metabolism gene tyrR, and the electron transfer flavoprotein (ETF) operon etfBA. The latter operon is directly connected to the ILV degradation pathway, as isovaleryl-CoA dehydrogenase is known to utilize ETF as an electron acceptor in eukaryotes (10) (Fig. 1A). A search for similar LiuR binding sites in the genomes of 12 other Shewanella species confirmed the conservation of the LiuR regulon in the genus Shewanella, with the only exception being the tyrR gene, which has a LiuR binding site in only eight species (see Table S1 in the supplemental material).
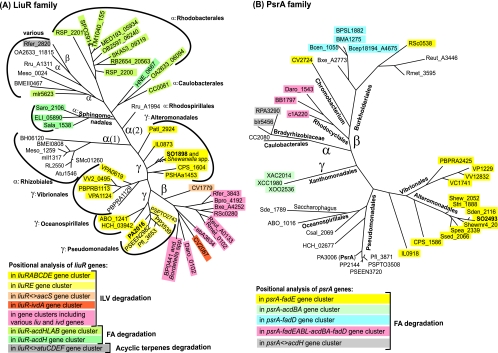
LiuR orthologs show a mosaic distribution in the genomes of gamma-, beta-, and alphaproteobacteria. There are no orthologs in other taxonomic groups (Table 1). The phylogenetic tree of the LiuR family has three main branches corresponding to the three subdivisions of proteobacteria (Fig. 4A); the respective binding motifs are represented in Fig. 2. Most alphaproteobacteria, as well as some beta-, and gammaproteobacteria, have two LiuR paralogs. The analysis of the genome context and the reconstruction of the LiuR regulons are outlined below and summarized in Table 1.
TABLE 1.
Reconstructed ILV degradation regulons in proteobacteriad
| Taxonomic groupa and organism | LiuR/LiuQ regulationb | ILV degradation gene(s)c | Other coregulated gene(s)c |
|---|---|---|---|
| Gammaproteobacteria | |||
| Alteromonadales | |||
| Shewanella spp. (13 genomes) | R | liuABCDEFG, ivdABCDEFG, bkd, ldh, etfBA | aceBA, gltBA, thrABC, tyrR (8) |
| Idiomarina loihiensis | R | liuABCDE, ivdA, bkd, aacS | |
| Colwellia psychrerythraea | R | liuABCDEFG, ivdAEG, bkd, etfBA, etfD | |
| Pseudoalteromonas haloplanktis | R | liuABCDEFG, ivdABCDEFG, bkd, ldh, etfBA, etfD | acdH |
| Pseudoalteromonas atlantica | R | liuABCDEFG, ivdABCDEFG, etfBA, etfD | |
| Vibrionales (3 genomes) | R, R | liuABCDE, ivdABCDEFG, etfBA, etfD | livGHMKF (1) |
| Pseudomonadales (5 genomes) | R | liuABCDE, aacS (4), etfBA, etfD (1) | |
| Oceanospirillales | |||
| Hahella chejuensis | R | liuABCDE, bkd, aacS | |
| Alcanivorax borkumensis | R | liuABCD, aacS | |
| Betaproteobacteria | |||
| Chromobacterium (1 genome) | R, R | liuABCDE, ivdABCDEFG, aacS | |
| Rhodocyclales | |||
| Dechloromonas aromatica | R | liuABCDE, ivdAC | cah, paaI, gloB |
| Azoarcus sp. | R | liuAC, ivdA | cah, paaH, gloB |
| Burkholderiales | |||
| Bordetella spp. (3 genomes) | R | liuABD, ivdACDF | paaI, aceK (2), sdh/glt, mdh |
| Ralstonia solanacearum | R | liuABCDE, ivdAC | cah, paaH, aceK, mcm, gloB |
| Ralstonia eutropha | R, Q | liuABDE-1, ivdAC, liuABCDE-2; aacS | paaI, cah, paaH, aceK, bio, gloB |
| Ralstonia metallidurans | R, Q | liuAC, ivdAC, liuBDE; aacS | cah, paaH, aceK, mcm |
| Burkholderia xenovorans | R, Q | liuABCD, ivdAC, aacS | cah, aceK |
| Burkholderia spp. (4 genomes) | Q | liuABCD | |
| Methylibium petroleiphilum | Q | liuABCD | |
| Polaromonas sp. | R | liuA, ivdAC, etfBA | paaI, cah, paaH, aceK, mcm, acdH |
| Rhodoferax ferrireducens | R, R | liuA, ivdAC, etfBA | cah, paaH, aceK, mcm, acdH |
| Alphaproteobacteria | |||
| Rhodospirillales (1 genome) | R | liuRABCDE, ivdACEF | livGHMKF |
The number of complete genomes with the analyzed regulon(s) is shown in parentheses.
The existence and copy number of candidate regulators of ILV degradation genes, LiuR and/or LiuQ, are shown by “R” and/or “Q,” respectively.
Candidate LiuR-regulated genes are shown in lightface type. Candidate LiuQ-regulated genes are underlined and in boldface. Genes are grouped (form stretches) only when they have the same first three letters in their names. The operon structure of genes is not shown. For candidate regulon members that are not always regulated within a taxonomic group, the number of genomes that have regulated orthologs is given in parentheses.
Functional roles of genes are listed in Table S7 in the supplemental material. The genome-specific locus tags of candidate regulon members are given in Tables S1 and S2 in the supplemental material.
FIG. 4.
Maximum likelihood phylogenetic tree of LiuR (A) and PsrA (B) regulators in various groups of proteobacteria. Background colors denote positional linkages of the regulatory genes.
(ii) Reconstruction of the LiuR regulon in gammaproteobacteria.
Orthologs of liuR (whose DNA motif is given in Fig. 2A) were found in four lineages of gammaproteobacteria: the Alteromonadales, Vibrionales, Pseudomonadales, and Oceanospirillales. They are always located in the leucine degradation liu gene clusters at the first position (see Table S1 in the supplemental material). Furthermore, in the Vibrionales and some members of the Alteromonadales, the liu operons form a supercluster with the isoleucine/valine degradation ivd operons. Genomic identification of LiuR binding sites combined with the comparative regulon consistency check (34) (for details, see Materials and Methods) led to the tentative reconstruction of the LiuR regulons in these genomes (see Table S1 in the supplemental material).
As in Shewanella species, the conserved core of the LiuR regulon in four other Alteromonadales genomes is represented by the liu, ivd, bkd, and etf gene clusters, which are involved in the ILV degradation. However, other predicted members of the Shewanella LiuR regulon (aceBA, thrABC, tyrR, and gltBD) are not controlled by LiuR in these species. The etfBA genes were found in one LiuR-regulated cluster with the ETF-ubiquinone oxidoreductase etfD in Pseudoalteromonas species. Similarly to Shewanella species, candidate LiuR binding sites were found upstream of the ldh gene in Pseudoalteromonas haloplanktis, but this regulatory interaction is not conserved in other members of the Alteromonadales. The liu operon in Idiomarina loihiensis contains an additional gene, IL0879, encoding acetoacetyl-CoA synthase (aacS), which is absent from all Shewanella species. A strong LiuR site was also detected upstream of the acyl-CoA dehydrogenase gene PSHAb0374 (called acdH) in P. haloplanktis, but its orthologs in other members of the Alteromonadales are not regulated by LiuR.
The structure of the LiuR regulon in the Vibrionales is similar to that of the Alteromonadales, although the ldh gene is missing in these genomes, and the bkd operon is not regulated by LiuR. Vibrio parahaemolyticus contains two LiuR-regulated copies of the ivd and liu operons, probably as a result of a recent duplication; a candidate LiuR binding motif was also found upstream of the seven-gene operon consisting of VPA1153 to VPA1147, encoding the ABC transporter for branched-chain amino acid LivGHMKF and two hypothetical enzymes.
The LiuR regulon in the Pseudomonadales is much smaller and includes the liu operon and the acetoacetyl-CoA synthase gene aacS. In addition, a candidate LiuR binding site was found upstream of the etfBA and etfD genes only in Pseudomonas fluorescens. Although the final reaction in the leucine utilization pathway in Pseudomonas aeruginosa and S. oneidensis is represented by two different enzymes, acetoacetyl-CoA synthetase and succinyl-CoA:3-ketoacid-CoA transferase, respectively (Fig. 1A), both these alternative enzymes belong to the respective LiuR regulons (Fig. 3). The P. aeruginosa aacS gene (PA2557) is preceded by a LiuR binding site, whereas succinyl-CoA:3-ketoacid-CoA transferase is encoded by last two genes (liuFG) within the LiuR-regulated liu operon in S. oneidensis.
The LiuR regulon in two Oceanospirillales species, Hahella chejuensis and Alcanivorax borkumensis, includes the liu-aacS operon (see Table S1 in the supplemental material). In addition, a candidate LiuR binding site precedes the bkd gene cluster in H. chejuensis.
(iii) Comparative analysis of the LiuR regulons in betaproteobacteria.
The DNA motif of the betaproteobacterial LiuR is largely similar to that of gammaproteobacteria (Fig. 2B).
Chromobacterium violaceum has two highly similar liuR paralogs likely resulting from a recent duplication (Table 1). The reconstructed LiuR regulon in this genome is most similar to the gammaproteobacterial ones. It includes various ILV degradation genes organized in the liu-aacS operon and aacS2 paralog that are clustered with liuR1 regulatory gene and the ivd operon colocalized with liuR2 (see Table S1 in the supplemental material).
In betaproteobacteria from the orders Rhodocyclales and Burkholderiales, the composition of the reconstructed LiuR regulons is highly variable and differs significantly from the LiuR regulon in gammaproteobacteria (Table 1). Within these taxonomic groups, the liuR gene is always colocalized with operons that include the isovaleryl-CoA dehydrogenase liuA; however, the size and composition of these candidate LiuR-regulated gene clusters vary from just 1 gene, liuA, in Bordetella pertussis to up to 25 genes in Ralstonia eutropha (see Table S1 in the supplemental material). Additional genes within the ILV degradation gene clusters include the carbonic anhydrase gene cah, the isocitrate dehydrogenase phosphatase/kinase gene aceK, the biotin biosynthesis genes bioAFDB, and many hypothetical genes (e.g., paaI and gloB). Other candidate members of the LiuR regulons in betaproteobacteria are the ETF operon etfBA, the methylmalonyl-CoA mutase gene mcm, the malate dehydrogenase gene mdh, the 3-hydroxyacyl-CoA dehydrogenase gene paaH, and the short-chain-specific acyl-CoA dehydrogenase gene acdH (Table 1).
(iv) Identification of the LiuQ regulon in the Burkholderiales.
LiuR orthologs were not identified in most Burkholderia species and Methylibium petroleiphilum. However, the liu operons without liuR genes are present in these genomes, and another transcriptional regulator from the TetR family, named liuQ, was found adjacent to the liu operons. By applying the motif recognition procedure to the training set of upstream regions of these liu operons, we identified a conserved DNA motif with the palindromic consensus sequence TTGAGynnnrCTCAA, where “y” and “r” denote “C or T” and “A or G,” respectively (Fig. 2D). Such sites are present in two copies in the common upstream region of the liuQ and liABCD operons (see Table S2 in the supplemental material). We propose that these palindromes are the binding sites of the LiuQ dimers. The LiuQ and LiuR regulons have overlapping distributions in two Ralstonia species and one Burkholderia species (Table 1). In R. eutropha, LiuR and LiuQ regulate different operons that contain paralogous copies of the liuABDE genes. In Ralstonia metallidurans, the liuAC genes belong to the LiuR-regulated operon, whereas the liuBDE genes are regulated by LiuQ. In Burkholderia xenovorans, the liuABCD operon is under the dual regulation of LiuR and LiuQ (Table 1).
(v) Comparative analysis of the LiuR regulons in alphaproteobacteria.
In Rhodospirillum rubrum, the reconstructed LiuR regulon includes the ILV degradation liu and ivd operons, the liuR gene, and an ortholog of the V. vulnificus operon containing VPA1153 to VPA1147, encoding the branched-chain amino acid ABC transporter LivGHMKF (Table 1).
Besides R. rubrum, the liuR orthologs (and additional paralogs in some genomes) are present in 20 alphaproteobacteria. The consensus of candidate LiuR binding sites in these alphaproteobacteria is very similar to the LiuR consensus in beta-, and gammaproteobacteria (Fig. 2C); however, the composition of the LiuR regulons is completely different, and thus, they were designated LiuRα regulons. Tentative metabolic reconstruction suggests that in four groups of alphaproteobacteria (the Rhizobiales, Caulobacterales, Sphingomonadales, and Rhodobacterales), the LiuRα regulon controls genes from the FAD pathway, such as the FA oxidation complex acdAB and the acyl-CoA dehydrogenase genes acdH and acdL (Table 2 and see Table S3 in the supplemental material). In the Rhizobiales group, additional candidate LiuR binding sites were found upstream of genes involved in FAD (fadD, etfAB, etfD, and hbdA) and the TCA cycle (mdh, sucCDAB, and lpdA).
TABLE 2.
Reconstructed FAD LiuRα regulon in alphaproteobacteriac
| Taxonomic groupa and organism of alphaproteobacteria | FAD gene(s)b | Other coregulated gene(s)b |
|---|---|---|
| Rhizobiales | ||
| Rhizobiaceae (4 genomes) | acdABH, fadD, etfBA, etfD, hbdA, echH | mdh-suc-lpdA, RL0537, RL1312, mll7718 (3) |
| Brucella melitensis | acdABH, fadD, etfBA, etfD, hbdA, echH | mdh-suc-lpdA, RL0537, RL1312, mll7718, BMEI0304 |
| Mesorhizobium loti | acdABH, acdL, echH | mdh-suc-lpdA |
| Mesorhizobium sp. | acdABH, fadD, etfBA, hbdA | mdh-suc-lpdA |
| Bartonella spp. (2 genomes) | - | mdh-suc-lpdA |
| Caulobacterales (1 genome) | acdABH, acdL | |
| Sphingomonadales | ||
| Novosphingobium aromaticivorans | acdH, etfBA, acdQP | Saro_0871-Saro_0867 |
| Erythrobacter litoralis | acdA, acdH, etfBA, acdQP | maoC |
| Sphingopyxis alaskensis | acdABH(multiple copies), acdL, etfBA, acdQP, Sala_0761-Sala_0760 | Sala_0379, Sala_0297, Sala_1639-Sala_1642, Sala_1536 |
| Rhodobacterales (9 genomes) | acdABH, acdL (4), etfBA (1), hbdA (1) |
Numbers of complete genomes with the analyzed regulon(s) are shown in parentheses.
Candidate LiuRα-regulated genes are shown. The operon structure of genes is not shown. For candidate regulon members that are not always regulated within a taxonomic group, the number of genomes that have regulated orthologs is given in parentheses.
Functional roles of genes are listed in Table S8 in the supplemental material. Locus tags are given for groups of orthologous genes that lack a conventional common name. The genome-specific locus tags of candidate regulon members are given in Table S3 in the supplemental material.
Some alphaproteobacteria, such as Brucella, Mesorhizobium, and Rhodobacter spp., have two LiuR paralogs (see the phylogenetic tree in Fig. 4A). We were not able to identify systematic differences between the candidate LiuR binding sites in the genomes harboring these genes. Thus, we were not able to assign the regulated operons to either of the two paralogs. These findings are in line with the observed high level of conservation of the DNA binding (N-terminal) domains of LiuR from alphaproteobacteria and two other groups of proteobacteria and weak similarity of their ligand binding (C-terminal) domains (data not shown). Probably, LiuR paralogs sense different ligands but can recognize similar if not identical sequence motifs.
FAD regulons in proteobacteria. (i) Reconstruction of the FadR regulon in gammaproteobacteria.
The GntR-like transcriptional factor FadR in E. coli is a negative regulator of genes involved in FAD (fadBA, fadD, fadE, and fadIJ) and transport (fadL) and is an activator of two genes involved in unsaturated FA synthesis (fabA and fabB) and also an activator of the iclR gene, encoding the repressor of the glyoxylate shunt operon aceBAK (5, 6, 14, 16, 36). Orthologs of FadR are present in four groups of gammaproteobacteria (the Enterobacteriales, Pasteurellales, Vibrionales, and Alteromonadales). The recognition profile for FadR binding sites in enterobacteria was constructed using the set of upstream regions of known FadR-regulated operons in E. coli and orthologous operons in other enterobacteria. Genomic searches with the constructed profile demonstrated a high level of conservation of the FadR regulon in the Enterobacteriales, whereas the content of the FadR regulon in other groups of gammaproteobacteria differed to some extent (see Table S4 in the supplemental material). A series of taxonomic group-specific FadR recognition profiles was constructed and used for the detailed comparative reconstruction of the FadR regulons in four groups of gammaproteobacteria (Table 3).
TABLE 3.
Reconstructed FAD regulons in gamma- and betaproteobacteriad
| Taxonomic groupa and organism(s) | PsrA/FadR/FadP regulationb | FA degradation gene(s)c | Other coregulated gene(s)c |
|---|---|---|---|
| Gammaproteobacteria | |||
| Enterobacteriales (5 genomes) | R | fadBA, fadH, fadD, fadIJ, fadE, fadL (4) | fabB (4), fabA (3), ybaW (4), iclR (2), aceBA (1) |
| Pasteurellales (7 genomes) | R | fadL | fabA |
| Haemophilus influenzae | R | - | fabA |
| Haemophilus ducreyi | R | fadL | fabA, fabB, fabDG, accA |
| Actinobacillus pleuropneumoniae | R | fadL | fabA, fabB, fabI, accAD |
| Alteromonadales | |||
| Shewanella spp. (13 genomes) | A, R | fadBA, fadH, fadD; [fadIJ], fadE, acdH, etfBA (10); etfD (11); echH; fadL | aceBA, sdh, SO2935 (11), SO0881 (12), SO4716 (11), SO0572, SO0080 (6) |
| Idiomarina loihiensis | A, R | fadBA, fadH, fadIJ, fadE, acdH | SO2935, IL-0656 |
| Colwellia psychrerythraea | A, R | fadBA, fadD, fadIJ, fadE, acdH, etfBA, etfD | SO2935 |
| Pseudoalteromonas haloplanktis | R | fadIJ, fadH | |
| Pseudoalteromonas atlantica | R | fadBA, fadIJ | |
| Saccharophagus degradans | A | fadBA, acdH | |
| Vibrionales | |||
| Vibrio cholerae | A, R | [fadBA], fadH, fadD, fadIJ, fadE1, fadE2 | mdh, plsB |
| Vibrio parahaemolyticus | A, R | fadBA, fadH, fadD, fadIJ, fadE1, fadE2, fadL | mdh, plsB |
| Vibrio vulnificus | A, R | fadBA, fadH, fadD, fadIJ, fadE1, fadE2 | mdh, plsB |
| Photobacterium profundum | A, R | fadBA, fadH, fadD, fadIJ, fadE1, fadE2, fadL1, fadL2 | echH |
| Vibrio fischeri | R | fadBA, fadH, fadE, fadL | plsB |
| Pseudomonadales (5 genomes) | A | fadBA, fadH (2), acdH, etfBA, etfD, scp | rpoS, algQ, PA1831, PP4195 (4) |
| Xanthomonadales (3 genomes) | A | acdAB | fabAB, accBC, aroQ |
| Oceanospirillales | |||
| Hahella chejuensis | A | fadBA, acdH, etfBA, etfD, scp | |
| Chromohalobacter salexigens | A | fadBA, fadH, fadD, fadE, acdH, etfBA, etfD, scp | |
| Alcanivorax borkumensis | A | fadD, acdH, etfBA, etfD | |
| Betaproteobacteria | |||
| Chromobacterium (1 genome) | A | acdAB, fadD, fadE, acdH, etfBA, etfD, fadL | mdh, accBC, fabK, fabF, aroQ |
| Rhodocyclales | |||
| Dechloromonas aromatica | A | fadBA, acdAB, fadD, fadE, acdH, fadL | |
| Azoarcus sp. | A | fadBA, acdAB, fadE, fadL, echH | |
| Burkholderiales | |||
| Bordetella spp. (3 genomes) | A, P | fadBA, acdAB, fadD, fadE, fadL | BP3678 |
| Ralstonia solanacearum | A, P | fadE, fadAB, acdABH, fadD, etfAB, etfD, echIH | acdQP, maoC, bktB, alkK |
| Ralstonia eutropha, R. metallidurans | A, P | fadAB, acdABH, fadD, etfAB, etfD, echIH, acsA | acdQP, maoC, bktB, alkK, fab |
| Burkholderia xenovorans | A, P | fadAB, acdABH, fadD1, etfAB, etfD, echIH, fadA1, fadD2, fadE | acdQP, maoC, fab |
| Burkholderia spp. (4 genomes) | A, P | fadAB, acdABH, fadD1, etfAB, etfD, echIH, fadD2 | acdQP, maoC, fab |
| Methylibium petroleiphilum | P | fadAB, acdABH, etfAB, etfD, echH | |
| Polaromonas sp. | P | fadAB, acdABH, etfAB | |
| Rhodoferax ferrireducens | P | fadAB, acdABH, etfAB, etfD | acdQP |
Numbers of complete genomes with the analyzed regulon(s) are shown in parentheses.
The existence and copy number of candidate regulators of FAD genes, FadR, PsrA, and FadP, are shown by “R,” “A,” and “P,” respectively.
Candidate FadR-regulated genes are underlined. Candidate PsrA-regulated genes are shown in lightface type. Candidate FadP-regulated genes are shown in boldface type. Operon structures of genes are not shown. For candidate regulon members that are not always regulated within a taxonomic group, the number of genomes that have regulated orthologs is given in parentheses. Genes that are regulated by both FadR and PsrA are in square brackets.
Functional roles of genes are listed in Table S8 in the supplemental material. Locus tags are given for groups of orthologous genes that lack a conventional common name. The genome-specific locus tags of candidate regulon members are given in Tables S4 to S6 in the supplemental material.
A highly conserved core of the FadR regulon in the Enterobacteriales is formed by the FAD genes (Table 3). In contrast, FadR binding sites upstream of FA synthesis, transport, and iclR genes are not strictly conserved in the enterobacteria. The FadR-IclR regulatory cascade, where FadR negatively regulates the aceBAK operon by the activation of the IclR repressor, is conserved only in genomes closely related to E. coli, such as Shigella and Salmonella species. Interestingly, the aceBAK operon in Yersinia species is preceded by a candidate FadR binding site, suggesting a rewiring of the regulatory cascade (see Table S4 in the supplemental material).
In the Vibrionales, the conserved core of the FadR regulon is formed by the FAD operons fadBA, fadE, and fadIJ as well as the phospholipid synthesis gene plsB, whereas the FadR-dependent regulation of fadH and fadL is not conserved in two or more species (Table 3). In the Pasteurellales, most FAD genes are absent, and the FadR regulon contains only genes involved in FA transport (fadL) and synthesis (fabA, fabB, fabDG, fabI, and accA). In the Alteromonadales, the FAD operon fadIJ is the only conserved member of the FadR regulon. In Pseudoalteromonas species, there are two additional FadR targets, the fadBA and fadH operons. In most Shewanella species, the FadR regulon also includes the FA transporter fadL and two hypothetical genes encoding probable enoyl-CoA hydratase (SO0572) and acyl-CoA N-acyltransferase (SO4716) genes.
(ii) Identification of the PsrA regulon in proteobacteria.
The FadR regulon was not found in several groups of gammaproteobacteria (e.g., the Pseudomonadales, Xanthomonadales, and Oceanospirillales), whereas in the Alteromonadales (e.g., in Shewanella species), it includes only a small subset of FAD genes. In an attempt to identify a novel FAD regulatory system, we applied the motif detection procedure to the upstream regions of the fad genes from 13 Shewanella species and identified a 20-bp palindromic motif (named the FAD box) (Fig. 2E). Genomic searches with the FAD box profile followed by an intergenomic consistency check allowed us to tentatively reconstruct the novel FAD regulon in Shewanella species. This regulon includes most FAD genes as well as the glyoxylate shunt aceBA operon, the TCA cycle sdh operon, and several hypothetical genes (Table 3). A candidate regulatory gene that encodes a TetR-type transcriptional factor (named PsrA, by the name of its ortholog previously characterized in Pseudomonas aeruginosa) is preceded by a FAD box and forms a putative operon with the fadE genes in the Alteromonadales and Vibrionales (see Table S5 in the supplemental material). Orthologs of the psrA gene preceded by candidate FAD boxes were identified in other groups of the gammaproteobacteria (e.g., the Pseudomonadales) as well as in various groups of the beta- and alphaproteobacteria (Fig. 2F), and in many genomes, they are colocalized with FAD genes (Fig. 4B). The phyletic distribution and genomic colocalization of FAD boxes and psrA genes strongly suggest that PsrA is a regulator that recognizes a FAD box and regulates FAD genes in proteobacteria. An overview of the PsrA regulon reconstructed by the comparative genomics approach in proteobacteria is given below.
The FAD genes fadBA, fadD, fadE, fadIJ, fadH, and acdH are the most conserved members of the PsrA regulon in gammaproteobacteria (Table 3). The ETF operon etfBA and the ETF-ubiquinone oxidoreductase gene etfD were found within the PsrA regulon in the Alteromonadales, the Pseudomonadales, and the Oceanospirillales. The scp gene, encoding a putative sterol carrier protein (COG3255), is an additional member of the PsrA regulon in the Pseudomonadales and Oceanospirillales. A different composition of the PsrA regulon was found in the Xanthomonadales, where psrA forms a regulated operon with the FA oxidation genes acdBA. The additional regulon members are the FA synthesis genes fabBA and accBC (see Table S5 in the supplemental material). The reconstructed PsrA regulons in several lineages are extended by different sets of genes that are either hypothetical genes or genes not directly involved in FA metabolism, e.g., the transcriptional regulator algQ in Pseudomonas species, the TCA cycle genes aceBA and sdhCBA in Shewanella species, and mdh in Vibrio species.
Interestingly, the PsrA regulator was previously described as being the regulator that controls the expression of the alternative sigma factor gene rpoS in P. aeruginosa (18). Experimentally determined PsrA binding sites in the promoter regions of the PsrA-repressed genes psrA, PA0506 (acdH), and PA2952 (etfB) coincide with the predicted PsrA binding sites (20). For the rpoS gene, which is positively regulated by PsrA at the stationary phase, the experimentally determined PsrA binding site is located 411 bp upstream of its translational start point (19) and was thus missed by our procedure.
In the Pseudomonas genomes, we performed an additional search with relaxed thresholds for the FAD box profile and identified candidate PsrA binding sites with scores of between 4.44 and 4.84, upstream of the rpoS genes (see Table S5 in the supplemental material). That candidate PsrA site in P. aeruginosa coincides with the experimentally identified PsrA site (19). Recently, PsrA was shown to bind to the fadBA (PA3014-PA3013) operon promoter region (17). The transcriptome analysis also revealed a PsrA-dependent repression of acdH, etfBA, etf, psrA, PA1830, and other genes (17) for which PsrA binding sites were identified here (see Table S5 in the supplemental material).
Among betaproteobacteria, the psrA gene was found in three lineages, the Rhodocyclales, the Burkholderiales, and the Chromobacterium group (Table 3). Chromobacterium violaceum has the largest PsrA regulon, which includes seven operons involved in FA utilization (psrA-fadE, fadD, fadL, acdBA, acdH, etfBA-acdH2, and etfD), three operons involved in the FA biosynthesis (fabK, fabF, and aroQ-accBC), and the mdh gene from the TCA cycle. In the Rhodocyclales and in Bordetella species, the reconstructed PsrA regulon consists of the FAD gene cluster containing the psrA, fadE, fadAB, fadL, fadD, and acdBA genes. The PsrA regulon in Burkholderia species includes the psrA-fadD gene cluster (in B. xenovorans, this gene cluster includes two additional FAD genes, fadE and fadA) and the FA biosynthesis gene cluster fabH-fabD-fabG-acpP-fabF. In contrast to these betaproteobacteria, the psrA gene and fabH-fabD-fabG-acpP-fabF gene clusters in Ralstonia species are preceded by putative PsrA binding sites with scores just below the threshold.
The only three alphaproteobacterial genomes that encode a PsrA ortholog are Caulobacter crescentus, Bradyrhizobium japonicum, and Rhodopseudomonas palustris. In all three genomes, candidate PsrA binding sites were identified upstream of the acyl-CoA dehydrogenase acdH gene (see Table S5 in the supplemental material). In B. japonicum and R. palustris, PsrA binding sites were also found upstream of the psrA gene itself.
(iii) The FadP regulon of the Burkholderiales.
The above-described analysis left a gap in the regulation of the FA utilization genes in most species from the Burkholderiales lineage. To fill this gap, we performed additional searches for potential regulatory motifs in upstream regions of the FAD genes that are not regulated by PsrA. A conserved 16-bp palindromic motif (Fig. 2G) was identified upstream of the gene cluster containing the acdH, acdBA, and echH genes in three Ralstonia species, in five Burkholderia species, and in Polaromonas sp. strain JS666, Methylibium petroleiphilum, and Rhodoferax ferrireducens. The first gene in this gene cluster (RSc0472 in R. solanacearum) encodes a TetR-like transcriptional regulator (named FadP), which was tentatively proposed to recognize the newly identified FAD regulatory motif in the Burkholderiales. A genomic search for similar candidate FadP binding sites in these species identified 4 to 15 sites per genome (see Table S6 in the supplemental material). Most candidate FadP-regulated genes encode enzymes involved in FAD or metabolism (see Table 3 for details).
In addition to the above-described 11 species, FadP orthologs were found in three Bordetella genomes. However, only two candidate FadP binding sites were found in each of these genomes: a site upstream of the fadP gene, implicating its autoregulation, and a site upstream of the BP3678 gene, encoding an uncharacterized exported protein.
DISCUSSION
We performed a comparative genomic reconstruction of the transcriptional regulatory network for genes involved in ILV degradation and FAD in the gamma- and betaproteobacteria (Fig. 5) and FAD in the alphaproteobacteria. For ILV utilization genes, we report the identification of a novel regulator from the MerR family (LiuR) and its DNA recognition motif (ILV box). The FAD genes in the Enterobacteriales are regulated by the FadR repressor from the GntR family. Here, we report the identification of a novel transcriptional factor from the TetR family (PsrA) and its conserved motif (FAD box) that control FAD genes in other lineages of gammaproteobacteria and in betaproteobacteria. In addition to these major transcription factors, two novel TetR-like regulators were predicted to control the ILV degradation and FAD regulons in the Burkholderiales (FadP and LiuQ, respectively) using other DNA motifs. Finally, we report that the LiuR orthologs in alphaproteobacteria regulate the FAD and TCA cycle genes.
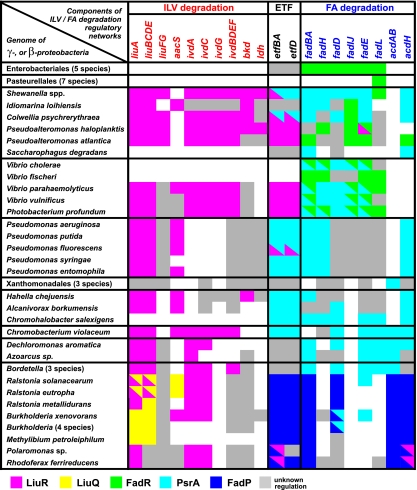
FIG. 5.
Candidate regulatory elements and genes involved in ILV degradation and FAD in gamma- and betaproteobacteria. Only conserved members of the predicted ILV/FA regulons are shown. Genes are arranged by metabolic pathway. Genomes are arranged by taxonomic lineages. When the gene is present in the genome, the background colors denote the presence of the specific recognition motif in its upstream region. If the gene is preceded by two different regulatory motifs, it is shown by a diagonally separated bicolor square. Genes without any candidate regulatory motifs described in this study are indicated by the gray background color. Empty crossings denote the absence of an orthologous gene in the genome.
The gene content of the predicted LiuR regulons in gamma- and betaproteobacteria is considerably variable (Table 1). The identified core of the LiuR regulon includes the liu and ivd genes, which are required for the conversion of CoA ethers of branched-chain carboxylic acids into CoA ethers of linear-chain carboxylic acids for their subsequent utilization through the TCA cycle (Fig. 1A). Although a physiological effector molecule for the LiuR regulator is unknown, we propose that one or several intermediates of the ILV degradation pathway, e.g., the CoA ethers of branched-chain carboxylic acids, might be involved in the modulation of LiuR activity.
Some of the ILV degradation genes, such as aacS, bkd, ldh, etfBA, and etfD, are candidate targets of LiuR regulation in only a fraction of the considered genomes. Thus, the complete leucine degradation pathway is regulated by LiuR only in Shewanella species. Interestingly, the bkd operon in P. aeruginosa is regulated by the ILV-responsive transcriptional activator BkdR (24). The LiuR regulon in the Shewanella lineage includes genes that are involved in glutamate synthesis (gltBA), glyoxylate shunt (aceBA), and threonine biosynthesis (thrABC). This apparent extension of the LiuR regulon could be explained by a metabolic connection of the LiuR-controlled metabolic pathways via acetyl-CoA, a final product of ILV degradation, which could be utilized for the amino acid biosynthesis pathways via the TCA cycle. In betaproteobacteria, the LiuR regulons are extended by a number of genes with unclear functional roles in ILV degradation (e.g., cah, aceK, paaH, gloB, and paaI) as well as the mdh and sdh genes, encoding TCA cycle enzymes, and the bio genes, which are involved in biotin biosynthesis. The latter observation is in line with the role of biotin as a cofactor of methylcrotonyl-CoA carboxylase (LiuBD).
Finally, among alphaproteobacteria, LiuR was found to control the ILV degradation genes only in R. rubrum. In contrast, orthologs of LiuR in other alphaproteobacteria were predicted to control genes involved in FAD and other pathways (Table 2). The changed content of the LiuRα regulon suggests that its physiological effector might be an acyl-CoA intermediate of the FAD pathway.
As seen from previously reported transcriptome analyses (21, 22), most operons regulated by LiuR in S. oneidensis were significantly induced by salt and/or alkaline stresses. The salt stress response and branched-chain amino acid metabolism seem to be linked in Shewanella species. Firstly, leucine was shown to be an important source of branched-chain FA in the cell membrane, as the growth of Shewanella gelidimarina on leucine as a sole carbon source resulted in a twofold increase of the branched-chain FA fraction in the membrane compared with growth on serine or alanine (28). Secondly, the concentration of branched-chain FA in S. gelidimarina was highly regulated by salt stress conditions and resulted in decreases in branched-chain FA content at high salinity (29). Therefore, the LiuR-dependent derepression of ILV degradation genes in Shewanella species reduces the pool of branched-chain acyl-CoA thioesthers (starter units for the biosynthesis of branched-chain FA) and resulted in a decrease in the branched-chain FA proportion in the membrane, thus regulating the membrane fluidity under salt stress conditions.
Unlike most members of the MerR family, LiuR seems to act solely as a repressor. Indeed, promoters activated by MerR-type transcription factors have an extended (19- to 20-bp) spacer between the −35 and −10 promoter boxes, which contains the transcription factor binding site partially overlapping the −35 element (reviewed in reference 4). On the other hand, MerR represses (but does not activate) promoters with standard 17-bp spacers (31, 32). We did not observe candidate promoters with extended spacers for LiuR-regulated operons, whereas in most cases, canonical candidate promoters overlapping the LiuR binding site could be identified (data not shown).
The FAD genes in the Enterobacteriales are regulated by the transcriptional regulatory protein FadR (8). We performed a comparative genomic reconstruction of the FadR regulon in other taxonomic groups of gammaproteobacteria and found that despite the overall conservation of the FadR binding motif (Fig. 2H to J), the regulon composition demonstrated substantial differences (Table 3). We noted that several fad genes in the Vibrionales and many fad genes in Alteromonadales species lack candidate FadR operator sites, suggesting that other factors might be involved in the regulation of these genes.
Using the comparative genomics procedure, we identified the transcriptional factor PsrA as being the master regulator of the FAD genes in five taxonomic groups of gammaproteobacteria (the Alteromonadales, Vibrionales, Xanthomonadales, Pseudomonadales, and Oceanospirillales) and several species of betaproteobacteria (Table 3). The PsrA regulons in different taxonomic groups also demonstrated their significant variability. For example, the FA biosynthesis genes are regulated by PsrA in the Xanthomonadales and several betaproteobacteria, whereas the PsrA regulon in the Pseudomonadales includes a variety of cellular processes. Since FadR and PsrA cooccur in the Alteromonadales and in Vibrionales species, in some genomes, we observed an overlap between two regulons. For instance, the fadH, fadBA, and fadIJ operons in the Vibrionales and the fadIJ operon in Shewanella are coregulated by both FadR and PsrA. In the betaproteobacteria (where only two or three PsrA sites per genome were found), a novel transcriptional factor (FadP) was found to substitute the PsrA function in most of the Burkholderiales. Finally, the FAD genes in alphaproteobacteria were found to be under the candidate regulation of LiuRα. In summary, this work revealed a large diversity in the transcriptional factors controlling FAD pathways in proteobacteria.
An interesting overlap between the LiuR and PsrA regulons was observed in Shewanella species, where candidate binding motifs of both transcription factors were identified in the aceBA and etfBA regulatory regions. The former encodes the acetyl-CoA utilization genes, and thus, this observation could be explained by the fact that acetyl-CoA is a common product of both FAD and ILV degradation pathways. The latter operon (etfBA) encodes electron transfer flavoprotein, which is used as an electron acceptor by LiuR-regulated dehydrogenases of the ILV degradation pathway, as well as PsrA-regulated acyl-CoA dehydrogenase. Similarly, the etfAB and etfD genes were found under the overlapping regulation of LiuR and FadP in Polaromonas and Rhodoferax species.
We also observed a rewiring of regulatory cascades. Indeed, the cascade FadR→iclR plus IclR→aceBAK of E. coli and Salmonella spp. corresponds to a streamlined interaction, FadR→aceBAK in Yersinia spp., whereas the aceBA genes in Shewanella spp. are controlled by the novel regulator PsrA. Three feed-forward loops, LiuR→tyrR, TyrR→(liu, ivd, and bkd), and LiuR→(liu, ivd, and bkd), are present in 8 of 12 Shewanella spp., whereas in the remaining Shewanella spp., the tyrR gene is not regulated by LiuR.
The reconstructed regulatory network suggests that the LiuR and PsrA regulons are the most widespread regulators of the ILV degradation and FAD genes in gamma- and betaproteobacteria (Fig. 5), respectively. In contrast, the LiuQ and FadP regulons have the narrowest phylogenetic distribution, being identified only in the order Burkholderiales. The FadR regulon was identified in four taxonomic orders of gammaproteobacteria, namely, the Enterobacteriales, Pasteurellales, Vibrionales, and Alteromonadales, where it acts either with PsrA (e.g., in Vibrionales) or alone (e.g., in the Enterobacteriales) to control the FAD genes. Based on these observations, we suggest that the most parsimonious evolutionary scenario for the ILV and FA regulons is as follows. LiuQ and PsrA were likely present in the common ancestor of gamma-, and betaproteobacteria, and they have been partially or fully substituted by LiuQ and FadP in the Burkholderiales and by FadR in some groups of gammaproteobacteria.
The results of this comparative genomics study demonstrate significant variability in the design and composition of the regulatory networks for the control of genes from central metabolic pathways. A similar extreme flexibility of transcriptional regulatory networks across various taxonomic groups of bacteria was reported in previous studies (2, 23, 33). The well-characterized FadR regulon that served as a prototype regulon for FAD not only underwent many changes by itself but may not even be the ancestral one. Although the overall picture for the core of the FA and ILV degradation regulons seems to be rather consistent (Fig. 5), many additional members of these regulons whose current functional annotations do not allow us to attribute them to the ILV/FA catabolic pathways were identified (Tables 1 to 3).
The reconstructed regulatory network needs to be integrated with functionally related networks, and few remaining gaps, such as the regulation of ILV degradation in the Xanthomonadales and alphaproteobacteria, need to be filled. Of course, although we are convinced that most computationally identified regulatory interactions reported here are real, each particular prediction requires experimental validation.
Supplementary Material
Acknowledgments
This study was supported by grants from the Howard Hughes Medical Institute (55005610), the Russian Academy of Science (Program Molecular and Cellular Biology), and the Russian Foundation for Basic Research (08-04-01000-a). This work was part of the Virtual Institute for Microbial Stress and Survival (http://VIMSS.lbl.gov) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, Genomics Program: GTL through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U.S. Department of Energy.
The work was initiated during A.E.K.'s visit to the Lawrence Berkeley National Laboratory.
Footnotes
Published ahead of print on 26 September 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aguilar, J. A., A. N. Zavala, C. Diaz-Perez, C. Cervantes, A. L. Diaz-Perez, and J. Campos-Garcia. 2006. The atu and liu clusters are involved in the catabolic pathways for acyclic monoterpenes and leucine in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 722070-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babu, M. M., N. M. Luscombe, L. Aravind, M. Gerstein, and S. A. Teichmann. 2004. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 14283-291. [DOI] [PubMed] [Google Scholar]
- 3.Benson, D. A., I. Karsch-Mizrachi, D. J. Lipman, J. Ostell, and D. L. Wheeler. 2007. GenBank. Nucleic Acids Res. 36D25-D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown, N. L., J. V. Stoyanov, S. P. Kidd, and J. L. Hobman. 2003. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 27145-163. [DOI] [PubMed] [Google Scholar]
- 5.Campbell, J. W., and J. E. Cronan, Jr. 2002. The enigmatic Escherichia coli fadE gene is yafH. J. Bacteriol. 1843759-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cronan, J. E., Jr., and S. Subrahmanyam. 1998. FadR, transcriptional co-ordination of metabolic expediency. Mol. Microbiol. 29937-943. [DOI] [PubMed] [Google Scholar]
- 7.Crooks, G. E., G. Hon, J. M. Chandonia, and S. E. Brenner. 2004. WebLogo: a sequence logo generator. Genome Res. 141188-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DiRusso, C. C., and W. D. Nunn. 1985. Cloning and characterization of a gene (fadR) involved in regulation of fatty acid metabolism in Escherichia coli. J. Bacteriol. 161583-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5164-166. [Google Scholar]
- 10.Finocchiaro, G., M. Ito, and K. Tanaka. 1987. Purification and properties of short chain acyl-CoA, medium chain acyl-CoA, and isovaleryl-CoA dehydrogenases from human liver. J. Biol. Chem. 2627982-7989. [PubMed] [Google Scholar]
- 11.Förster-Fromme, K., B. Höschle, C. Mack, M. Bott, W. Armbruster, and D. Jendrossek. 2006. Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 724819-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita, Y., H. Matsuoka, and K. Hirooka. 2007. Regulation of fatty acid metabolism in bacteria. Mol. Microbiol. 66829-839. [DOI] [PubMed] [Google Scholar]
- 13.Gelfand, M. S., E. V. Koonin, and A. A. Mironov. 2000. Prediction of transcription regulatory sites in Archaea by a comparative genomic approach. Nucleic Acids Res. 28695-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gui, L., A. Sunnarborg, and D. C. LaPorte. 1996. Regulated expression of a repressor protein: FadR activates iclR. J. Bacteriol. 1784704-4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Höschle, B., V. Gnau, and D. Jendrossek. 2005. Methylcrotonyl-CoA and geranyl-CoA carboxylases are involved in leucine/isovalerate utilization (Liu) and acyclic terpene utilization (Atu), and are encoded by liuB/liuD and atuC/atuF, in Pseudomonas aeruginosa. Microbiology 1513649-3656. [DOI] [PubMed] [Google Scholar]
- 16.Iram, S. H., and J. E. Cronan. 2005. Unexpected functional diversity among FadR fatty acid transcriptional regulatory proteins. J. Biol. Chem. 28032148-32156. [DOI] [PubMed] [Google Scholar]
- 17.Kang, Y., D. T. Nguyen, M. S. Son, and T. T. Hoang. 2008. The Pseudomonas aeruginosa PsrA responds to long-chain fatty acid signals to regulate the fadBA5 beta-oxidation operon. Microbiology 1541584-1598. [DOI] [PubMed] [Google Scholar]
- 18.Kojic, M., and V. Venturi. 2001. Regulation of rpoS gene expression in Pseudomonas: involvement of a TetR family regulator. J. Bacteriol. 1833712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kojic, M., C. Aguilar, and V. Venturi. 2002. TetR family member PsrA directly binds the Pseudomonas rpoS and psrA promoters. J. Bacteriol. 1842324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kojic, M., B. Jovcic, A. Vindigni, F. Odreman, and V. Venturi. 2005. Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 246175-181. [DOI] [PubMed] [Google Scholar]
- 21.Leaphart, A. B., D. K. Thompson, K. Huang, E. Alm, X. F. Wan, A. Arkin, S. D. Brown, L. Wu, T. Yan, X. Liu, G. S. Wickham, and J. Zhou. 2006. Transcriptome profiling of Shewanella oneidensis gene expression following exposure to acidic and alkaline pH. J. Bacteriol. 1881633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y., W. Gao, Y. Wang, L. Wu, X. Liu, T. Yan, E. Alm, A. Arkin, D. K. Thompson, M. W. Fields, and J. Zhou. 2005. Transcriptome analysis of Shewanella oneidensis MR-1 in response to elevated salt conditions. J. Bacteriol. 1872501-2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lozada-Chávez, I., S. C. Janga, and J. Collado-Vides. 2006. Bacterial regulatory networks are extremely flexible in evolution. Nucleic Acids Res. 343434-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madhusudhan, K. T., J. Luo, and J. R. Sokatch. 1999. In vitro transcriptional studies of the bkd operon of Pseudomonas putida: l-branched-chain amino acids and d-leucine are the inducers. J. Bacteriol. 1812889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, R. R., V. D. Marshall, J. R. Sokatch, and L. Unger. 1973. Common enzymes of branched-chain amino acid catabolism in Pseudomonas putida. J. Bacteriol. 115198-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuoka, H., K. Hirooka, and Y. Fujita. 2007. Organization and function of the YsiA regulon of Bacillus subtilis involved in fatty acid degradation. J. Biol. Chem. 2825180-5194. [DOI] [PubMed] [Google Scholar]
- 27.Mironov, A. A., N. P. Vinokurova, and M. S. Gelfand. 2000. Software for analysis of complete bacterial genomes. Mol. Biol. 34222-231. [Google Scholar]
- 28.Nichols, D. S., P. D. Nichols, N. J. Russell, N. W. Davies, and T. A. McMeekin. 1997. Polyunsaturated fatty acids in the psychrophilic bacterium Shewanella gelidimarina ACAM 456T: molecular species analysis of major phospholipids and biosynthesis of eicosapentaenoic acid. Biochim. Biophys. Acta 1347164-176. [DOI] [PubMed] [Google Scholar]
- 29.Nichols, D. S., J. Olley, H. Garda, R. R. Brenner, and T. A. McMeekin. 2000. Effect of temperature and salinity stress on growth and lipid composition of Shewanella gelidimarina. Appl. Environ. Microbiol. 662422-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crécy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Rückert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park, S. J., J. Wireman, and A. O. Summers. 1992. Genetic analysis of the Tn21 mer operator-promoter. J. Bacteriol. 1742160-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill, J., and N. L. Brown. 1990. Site-specific insertion and deletion mutants in the mer promoter-operator region of Tn501; the nineteen base-pair spacer is essential for normal induction of the promoter by MerR. Nucleic Acids Res. 185157-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price, M. N., P. S. Dehal, and A. P. Arkin. 2007. Orthologous transcription factors in bacteria have different functions and regulate different genes. PLoS Comput. Biol. 31739-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov, D. A. 2007. Comparative genomic reconstruction of transcriptional regulatory networks in bacteria. Chem. Rev. 1073467-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodionov, D. A., M. S. Gelfand, M. Romine, and A. O. Osterman. 2007. Comparative genomics of transcriptional regulation of metabolic pathways in Shewanella species, abstr. 100. Abstr. Genomics: GTL Awardee Wkshp. V.
- 36.Sadovskaya, N. S., O. N. Laikova, A. A. Mironov, and M. S. Gelfand. 2001. Study of regulation of long-chain fatty acid metabolism using computer analysis of complete bacterial genomes. Mol. Biol. 35862-866. [PubMed] [Google Scholar]
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 254876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.