Abstract
Sam68 (Src-associated in mitosis, 68 kDa) is a KH domain RNA binding protein implicated in a variety of cellular processes, including alternative pre-mRNA splicing, but its functions are not well understood. Using RNA interference knockdown of Sam68 expression and splicing-sensitive microarrays, we identified a set of alternative exons whose splicing depends on Sam68. Detailed analysis of one newly identified target exon in epsilon sarcoglycan (Sgce) showed that both RNA elements distributed across the adjacent introns and the RNA binding activity of Sam68 are necessary to repress the Sgce exon. Sam68 protein is upregulated upon neuronal differentiation of P19 cells, and many Sam68 RNA targets change in expression and splicing during this process. When Sam68 is knocked down by short hairpin RNAs, many Sam68-dependent splicing changes do not occur and P19 cells fail to differentiate. We also found that the differentiation of primary neuronal progenitor cells from embryonic mouse neocortex is suppressed by Sam68 depletion and promoted by Sam68 overexpression. Thus, Sam68 controls neurogenesis through its effects on a specific set of RNA targets.
Alternative splicing allows multiple functionally distinct mRNAs and proteins to be generated from a single gene, greatly enhancing the coding potential of the genome. Splicing patterns are controlled by RNA binding proteins that recognize specific splicing enhancer and silencer elements in the pre-mRNA to alter spliceosome assembly, and variation in the expression of these proteins leads to tissue-specific exon use (5, 38). Splicing within a single cell can also be dynamically controlled by extracellular stimuli, although how this information is transmitted to splicing regulatory proteins is not yet known (55).
Sam68 is a nuclear RNA binding protein implicated in various aspects of mRNA metabolism, including splicing, nuclear export, somatodendritic transport, and translation. Sam68 belongs to the family of GSG (GRP33, Sam68, GLD1) or STAR (signal transduction and activation of RNA) domain proteins. This domain includes a central KH (hnRNP K homology) RNA binding domain flanked by conserved N and C termini (36, 54, 59). The GSG domain in Sam68 binds to RNA motifs that are rich in A or U, such as UAAA or UUUA, and also mediates homodimerization (9, 31). Sam68 contains a variety of other protein domains that allow its interaction and modification by multiple signaling pathways. These many regulatory interactions and posttranslational modifications affect the RNA binding activity and localization of Sam68 and make it an appealing molecule for transducing information from signaling systems to pathways of mRNA metabolism (13, 33, 46, 51, 60).
Sam68 is localized primarily in the nucleus as observed by immunofluorescence, consistent with its role in alternative splicing. Sam68 helps regulate the splicing of CD44 variable exon v5 in response to phosphorylation by extracellular signal-regulated kinase in T lymphoma cells (39). Sam68 binds to exonic regulatory elements of v5 and cooperates with the splicing coactivator SRm160 (10, 39). In other studies, the Brm1 subunit of the SWI/SNF chromatin remodeling complex was shown to induce CD44 v5 inclusion upon mitogen-activated protein kinase activation. This activation is required for Sam68 association with v5 (3). Sam68 also regulates the alternative splicing of the apoptotic regulator Bcl-x, where it cooperates with hnRNPA1 to induce a switch from the antiapoptotic (Bcl-xL) to the proapoptotic (Bcl-xS) isoform (45). Sam68 was also shown to cross-link to an intronic regulatory sequence of the β-tropomyosin pre-mRNA, although its effect on splicing is not yet clear (22). The small number of confirmed Sam68 pre-mRNA targets has limited our understanding of both its mechanisms of action and its cellular role.
Here, we identify a novel set of target RNAs whose splicing is regulated by Sam68. We show that Sam68 promotes neuronal differentiation, a process during which many of these new target mRNAs undergo Sam68-dependent changes in splicing regulation.
MATERIALS AND METHODS
Cell culture.
HEK 293T cells and N2A cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. P19 cells were cultured and differentiated as described previously (63). Embryonic day 11.5 mouse central nervous system tissues were dissected and mechanically dissociated with Pasteur pipettes (17). Dissociated cells were plated on coverslips coated with polyornithine (10 μg/ml) and fibronectin (10 μg/ml) with serum-free Dulbecco's modified Eagle's medium-F-12 medium supplemented with B27, penicillin-streptomycin (50 U/ml and 50 μg/ml, respectively; Invitrogen, La Jolla, CA). Cultures were fed with 10 ng/ml basic fibroblast growth factor (bFGF) at the time of plating(18). The cells were passaged by gentle dissociation with Accutase (Innovative Technology). Neuronal progenitor cells (NPCs) were passaged at least three times before lentiviral transduction.
RT-PCR assay.
Cytoplasmic RNA was isolated using NP-40 lysis, followed by proteinase K digestion and phenol-chloroform extraction. Reverse transcription-PCR (RT-PCR) was performed as described previously (7). The primers used for RT-PCR are listed in Table S3 of the supplemental material.
Plasmid constructs.
The cDNA for the mouse Sam68 was cloned from BAC clone 3490667 (Image Clones) into pcdna 3.1(-) as an EcoRI-BamHI fragment using the primers listed in Table S3 of the supplemental material. The G178E and I184N mutations were made by PCR. The wild-type Sgce mini-gene construct (Sgce WT) contains the complete genomic sequence of the SGCE gene from exon 7 to exon 9 (Sgce_2, seg 15 to seg 19). The DNA fragment was generated from the bacmid clone RP23-281I15 (IMAGE Consortium) by PCR. Following restriction digestion, the PCR product was cloned in the EcoRV-Not1 site of the mammalian expression vector pCDNA3.1. Deletion mutants were generated by PCR and verified by sequencing.
Short hairpins were designed to target the 3′ untranslated regions of Sam68. The hairpin constructs were generated by annealing two synthetic oligonucleotides and cloning into the Bbs1 site of psiRNA-W (H1.4). The oligonucleotides are listed in Table S3 of the supplemental material. The H1-RNA cassette was excised from the psiRNA-W (H1.4) construct and cloned into the Nhe1 site of a lentiviral vector (LVCMV GFP) (41). The cDNAs encoding the wild-type mouse Sam68 was subcloned into the EcoRI site of the lentiviral vector FUCGW (61).
Microarrays.
Oligonucleotides and microarray design have been described previously (8, 11, 56). The cDNA labeling, microarray hybridization, and scanning were performed as described previously (8). Scanned image data were analyzed using GenePix Pro6 software (Axon) as described previously (56). Briefly, each spot was defined within a grid layout over the image. Flagging criteria were applied to exclude bad spots, as detailed elsewhere (56). For each channel, the median values of the qualified spots were determined after subtraction of local background intensities. The result files generated from the GenePix Pro 6 were then analyzed in R using Bioconductor as described previously (8).
Word frequency analysis.
To derive the control set of exons, a set of 10,000 constitutive exons defined earlier (28) were mapped to the mouse genome using BLAST. Exons with canonical 5′ and 3′ splice sites (GT-AG) and their upstream and downstream sequences were retrieved. Word frequency analysis (Wilcoxon rank-sum test) was applied to the experimental and control exon sets to assess the UAAA and UUUA word enrichment in the Sam68-regulated exons and their flanks (48).
Western blot analysis.
Cell lysates were prepared by resuspending pelleted cells in Phospho-Safe extraction reagent (Novagen) containing Benzonase (Sigma). The lysates were diluted with Laemmli buffer and resolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels. Gels were run under standard electrophoresis conditions and were transferred to a nitrocellulose membrane (Protran) using the Novex gel transfer system (Invitrogen). Primary antibody dilutions were as follows: anti-Sam68 (1:1,000; AD1 peptide polyclonal; Upstate), anti-Flag (1:5,000; Sigma), anti-TuJ1 (1:500; Covance), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH; 1:400,000; Fitzgerald). Immunoblots were developed using enhanced chemiluminescence. Quantitative immunoblot analysis was performed with Cy5- or Cy3-labeled secondary antibodies (GE Healthcare). The blots were scanned on a Typhoon PhosphorImager and quantified using ImageQuant 5.1 software (Molecular Dynamics).
Flag-tagged protein purification.
The Flag-Sam68 wild-type and mutant expression plasmids (G178E and I184N) were transfected into 293T cells. The cells were harvested after 48 h for total protein isolation. The cells were washed in phosphate-buffered saline (PBS)-EDTA solution and lysed in 5 volumes of lysis buffer (20 mM HEPES-KOH, pH 7.5, 500 mM KCl, 2 mM EDTA, Complete EDTA-free protease inhibitor, 0.5 mM dithiothreitol [DTT], 0.6% NP-40). The lysates were centrifuged for 10 min at 12,000 × g at 4°C and the pellets were discarded. The supernatants were incubated with preequilibrated anti-Flag M2 agarose beads (Sigma) for 3 to 4 h at 4°C with shaking. The volume of beads was determined according to the manufacturer's instructions. After incubation the resin was centrifuged at 10,600 × g for 1 min and the supernatants were removed. The beads were washed three times with wash buffer (20 mM HEPES-KOH, pH 7.5, 300 mM KCl, 0.05% NP-40) and one time with a similar buffer with 150 mM KCl. The Flag-tagged proteins were eluted under native conditions with a buffer containing Flag peptide (150 ng/μl, final concentration). Elution was carried out at 4°C for 1 h. Following incubations the resin was centrifuged for 5 s at 10,600 × g, and the supernatants were collected and dialyzed in buffer DG (20 mM HEPES-KOH, pH 7.9, 20% glycerol, 80 mM potassium glutamate, 0.2 mM EDTA, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM DTT).
Electrophoretic mobility shift assay.
A constant concentration of radiolabeled RNA (50 pM) was equilibrated with various concentrations of Flag-Sam68 (50 nM to 1.5 μM) in binding buffer (20 mM HEPES at pH 7.6, 20% glycerol, 80 mM potassium glutamate, 100 ng/μl tRNA, 0.02% NP-40, 6 mM MgCl2, 1 mM DTT). All reaction components except the RNA probe were mixed and incubated for 5 min at 30°C; the RNA probe (50,000 cpm, ∼100 fmol) was then added and incubated for another 25 min at 30°C. After incubation an equal volume of RNA loading dye (glycerol containing 0.25% [wt/vol] bromophenol blue, 0.25% [wt/vol] xylene cyanol) was added to each reaction mixture. Each reaction mixture was loaded onto a prerun native polyacrylamide gel (6% [wt/vol] 29:1 acrylamide-bis-acrylamide, 0.5× Tris-borate-EDTA). Gels were resolved at 100 V for 4 h before they were dried and exposed to a PhosphorImager screen and visualized as described above.
Immunofluorescence.
The cells were plated on poly-l-lysine-coated coverslips, fixed with 4% paraformaldehyde-PBS for 20 min, permeabilized with 0.5% Triton-PBS for 10 min, and blocked with 1% goat serum in PBS for 1 h. The cells were then incubated with primary antibodies at 4°C overnight. The coverslips were rinsed with PBS followed by incubation with Alexa-conjugated secondary antibodies for 1 h at room temperature. Following incubation the coverslips were rinsed with PBS and mounted (Vectashield). Primary antibodies were used at the following concentrations: anti-Sam68 (1:2,000) and anti-Tuj1 (1:500). Secondary antibodies Alexa 350 anti-mouse immunoglobulin G and Alexa 568 anti-rabbit immunoglobulin G (Molecular Probes) were used at a 1:1,000 dilution. Images in Fig. 4A, 5A, 6A, and 7A, below, were taken with an inverted microscope (Eclipse 2000-S; Nikon) using a Plan Fluor 20×/0.50 objective. Images were acquired at room temperature using an RT-KE Slider (diagnostic) camera and Spot advanced software. Images were acquired as TIFF files, and Canvas 9 software was used for composing the panel. For measuring the β-tubulin III index in the shSam68 versus control cells, ∼1,000 green fluorescent protein (GFP)-positive cells were scored from three experiments. In the case of the Sam68 overexpression experiment, ∼300 cells were counted. Statistical analysis was performed with the paired two-sample Student t test.
FIG. 4.
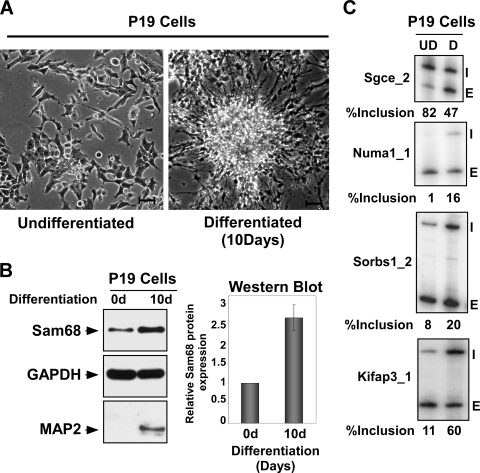
Sam68 is upregulated upon neuronal differentiation. (A) Bright-field image of P19 cells before and after differentiation. Bar, 10 μm. (B) Protein lysates from undifferentiated and 10-day differentiated P19 cells immunoblotted for Sam68 and GAPDH as loading control. The bar graph on the right shows the induction of Sam68 protein upon differentiation. (C) Alternative splicing of Sam68 target exons as assayed by RT-PCR in undifferentiated and differentiated P19 cells. The gene names are on the left and the exon included (I) and exon skipped (E) forms are indicated on the right. The percent inclusion [I/(I + E) × 100] is shown below.
FIG. 5.
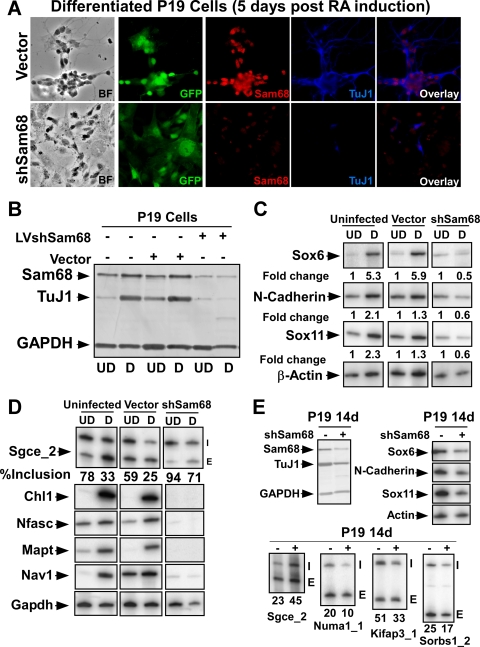
Sam68 regulates alternative splicing events during neurogenesis. (A) Immunofluorescence of Sam68 (red) and Tuj1 (blue) in P19 cells 9 days after RA treatment. Prior to differentiation, cells were infected with lentiviruses expressing shRNA to knock down Sam68 (bottom panels) or an empty vector control (top panels). Enhanced GFP (EGFP) marks the infected cells. The bright-field image, EGFP fluorescence, and staining for Sam68 (red) and TuJ1 (blue) are shown along with the Sam68/TuJ1 overlay. (B) Immunoblot analysis of lysates from undifferentiated (UD) and differentiated (D) cells. Uninfected, control virus-infected, and shSam68 lentivirus-infected cells were probed for Sam68, TuJ1, and GAPDH. (C) RT-PCR analysis of neuronal marker mRNAs from uninfected, control vector-infected, and shSam68-infected P19 cells. Expression of Sox6, Sox11, and N-cadherin was examined and the change relative to β-actin is shown below each panel. (D) RT-PCR analysis of Sam68-dependent mRNAs in uninfected, control vector-infected, and shSam68-infected P19 cells. Splicing of the Sgce exon 8 is shown in the top panel with the percent exon inclusion. The lower panel shows expression of mRNA previously found to change upon Sam68 knockdown in N2A cells. (E) P19 cells were infected with Sam68 shRNA lentivirus after differentiation (day 9). (Top left panel) Immunoblot for Sam68, TuJ1 (neuronal marker), and GAPDH in infected and uninfected cells. (Top right panel) RT-PCR for neuronal marker expression in uninfected and shSAM68-treated cells. (Bottom panel) RT-PCR analysis of target exons regulated by Sam68 in control and shSam68-treated cells.
FIG. 6.
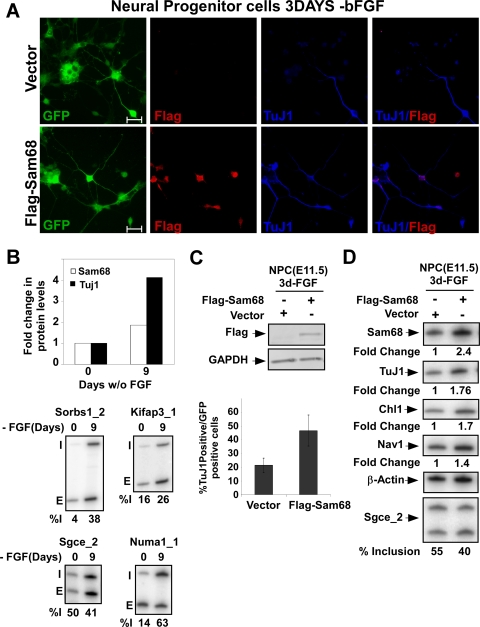
Overexpression of Sam68 enhances neuronal differentiation of NPCs. (A) Immunostaining of Flag tag (red) and Tuj1 (blue) in NPCs infected with lentiviruses expressing Flag-Sam68 (bottom panels) or the empty vector control (top panels). Enhanced GFP (EGFP) marks the infected cells. These cells were induced to differentiate by bFGF withdrawal 2 days after transduction, and the morphology of cells was examined after 3 days. EGFP fluorescence and individual staining for Flag-Sam68 (red) and TuJ1 (blue) are shown along with the overlay of the two stains. Bar, 20 μm. (B) The bar graph at the top shows the induction of Sam68 and TuJ1 protein upon differentiation as measured by immunoblotting after 0 and 9 days. RT-PCR analysis of Sam68-dependent exons before and after neuronal differentiation of NPCs is shown at the bottom. (C, top panel) Immunoblot analysis of lysates prepared from control vector-infected and Flag-Sam68 lentivirus-infected NPCs, probed for Flag-tag and GAPDH. (C, bottom panel) Percentage of infected cells expressing the neuronal marker TuJ1 (β-tubulin III) for Flag-Sam68 virus and control virus infections. (D) RT-PCR analysis of Sam68 and TuJ1 expression in NPCs in Flag-Sam68- or control virus-infected cells. Similar analysis of Sam68 target RNAs (Chl1 and Nav1) and the splicing of Sgce_2 is shown in the lower panels; indicated below is the percent inclusion, [I/(I + E) × 100].
FIG. 7.
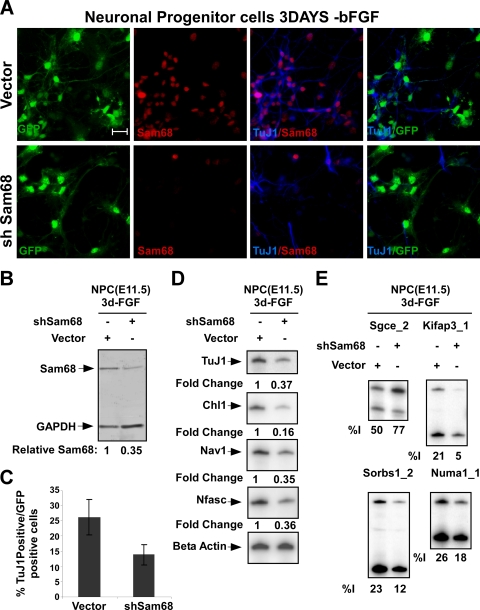
Sam68 knockdown in NPCs impairs neuronal differentiation. (A) Immunostaining of NPCs 5 days after lentiviral transduction and 3 days after FGF withdrawal. Expression of Sam68 (red) and Tuj1 (blue) in NPCs infected with lentivirus expressing Sam68-targeted shRNA (bottom panels) or empty vector (top panels). Enhanced GFP marking the infected cells and staining for Sam68 (red) and TuJ1 (blue) are shown along with the Sam68/TuJ1 overlay. Bar, 20 μm. (B) Immunoblot analysis of lysates from control vector- and shSam68 lentivirus-infected NPCs, probed for Sam68 and GAPDH as a loading control. (C) Graph of the percentage of infected (GFP-positive) cells that express the neuronal marker β-tubulin III (TuJ1) for shSam68 and control virus infections. (D) RT-PCR analysis of Sam68 target RNA expression (Chl1, Nav1, and Nfasc) and TuJ1 mRNA in control vector- and shSam68-infected NPCs. (E) RT-PCR analysis of Sam68 target exons in control vector- and shSam68-infected NPCs.
Transfections.
N2A cells were plated at a density of 2.5 × 105 cells/ml. DNA/Lipofectamine 2000 (Invitrogen) complexes were assembled according to the manufacturer's protocol using 20 μg of DNA and 50 μl of Lipofectamine 2000. The complexes were added to plated cells. Cells were harvested after 3 days for isolation of protein and RNA, and knockdown was tested by immunoblotting. For transient coexpression assays 293T cells were plated at a density of 1 ×105 to 2 ×105 cells in 1 ml of growth medium 1 day before transfection. The transfection was performed in a 12-well format. The transfection was carried out with Lipofectamine 2000 as per the manufacturer's instructions. Each transfection mixture contained 1.5 μg of DNA. In each transfection reaction mixture, 750 ng of reporter plasmid, 500 ng of expression plasmid, and 250 ng of pEGFPN1 plasmid was used. Cells were harvested 22 to 24 h after transfection for isolation of total protein and cytoplasmic RNA.
Lentivirus preparation and infection.
Recombinant lentiviruses were produced as described previously (32, 47). Infections were performed with lentivirus (titer, 2 × 107) in the presence of 4 μg/ml Polybrene (Sigma). All procedures were performed under the University's safety regulations for lentivirus usage.
RESULTS
Microarray analysis identifies many novel splicing targets for Sam68.
Exons whose splicing depends on Sam68 were identified using splicing-sensitive microarrays (6, 8, 56). Mouse N2A neuroblastoma cells were transfected with a plasmid expressing a Sam68 complementary shRNA (shSam68) or with an empty vector control plasmid. The shSAM68-treated cells exhibited a substantial loss of Sam68 expression (Fig. 1A). RNA isolated from these and control cells was used to probe microarrays and analyzed as described previously (8, 56). Significant splicing changes between the knockdown and the control were identified by comparing the log2 ratios of the exon inclusion probe set and the exon-skipping probe set using two-sample t tests. Exons were chosen whose t test indicated that the likelihood was less than 0.05 that the inclusion signals and the skipping signals were from the same distribution. The signals from these exons were further assessed for their likely false discovery rate (FDR) using BH-FDR analysis and then ranked by FDR value (see Materials and Methods) (8). Of the 61 exons with an FDR value below 0.05, 35 exons showing the largest change were tested by RT-PCR. Of these 35, 24 exons were confirmed to show splicing changes by RT-PCR and were designated true positives (68.5%), and 14 exons (31.5%) showed no change by RT-PCR and were designated as false positives (Table 1). To calculate the false-negative rate, 19 exons with FDR values greater than 0.05 were tested by RT-PCR. Six exons from this set showed changes by RT-PCR and were designated as false negatives (Table 1; see also Table S1 in the supplemental material). We also identified a set of transcripts whose overall expression was affected by Sam68 depletion, by performing one-sample t tests on the average log2 ratios for the constitutive probes of each gene (see Table S2 in the supplemental material).
FIG. 1.
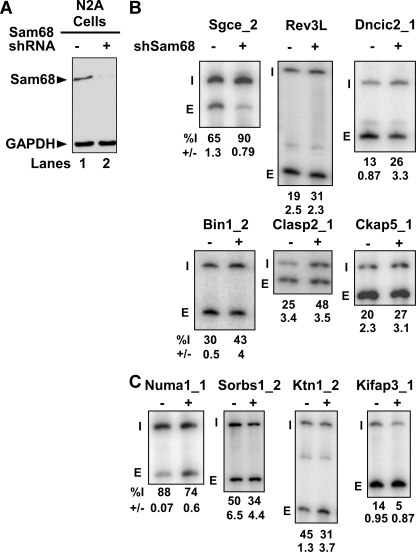
Microarray analysis identifies new splicing targets for Sam68. (A) RNA interference against Sam68 was induced in N2A cells by using an shRNA expression vector, and empty vector was transfected as negative control. Western blot analysis was performed with Sam68 antibody and GAPDH as a loading control. (B) RT-PCR analysis of some exons repressed by Sam68. (C) RT-PCR analysis of some exons enhanced by Sam68. RT-PCR was performed with end-labeled primers in the exons flanking constitutive exons. The exon ID is indicated on the top of each panel with the percent exon inclusion (Inc) and standard deviation (n = 3) shown at the bottom of each panel. The included (I) and the skipped (E) forms are indicated on the left of each panel.
TABLE 1.
Summary of RT-PCR-validated target exons regulated by Sam68a
| ID | Difference | P value | Adjusted P value |
|---|---|---|---|
| Sorbs1_3 | −0.972287436 | 0.007256965 | 0.068 |
| Ktn1_2 | −0.450238873 | 5.46E-05 | 0.005 |
| Jarid1c_2 | −0.416139755 | 0.002175416 | 0.037 |
| Ppt1_1 | −0.391045892 | 0.000485751 | 0.015 |
| Spag9_2 | −0.382333422 | 0.000102919 | 0.006 |
| Snx14_1 | −0.350253415 | 0.107386198 | 0.334 |
| Dlgh4_3 | −0.30688661 | 5.29E-05 | 0.005 |
| Epb4.1l3_3 | −0.299083176 | 0.000480547 | 0.015 |
| Tcerg1_2 | −0.298358484 | 0.002747823 | 0.041 |
| chr4.2054_1 | −0.296165141 | 0.004477784 | 0.054 |
| Kifap3_1 | −0.288516237 | 0.05757102 | 0.231 |
| Opa1_2 | −0.263647361 | 0.001580715 | 0.031 |
| chr12.218_1 | −0.199200787 | 0.001917359 | 0.034 |
| Numa1_1 | −0.117730254 | 2.23E-05 | 0.003 |
| Pum1_5 | 0.169227377 | 0.003411662 | 0.048 |
| Clta_2 | 0.183418881 | 0.001318903 | 0.027 |
| Pdia4_1 | 0.20031599 | 0.003712491 | 0.049 |
| Bin1_2 | 0.216851792 | 0.002155608 | 0.037 |
| Hnrpa1_4 | 0.295854539 | 0.005673684 | 0.061 |
| Igsf4a_1 | 0.31040488 | 0.001205238 | 0.026 |
| Pbx1_1 | 0.332196193 | 0.000106328 | 0.006 |
| Tpp2_2 | 0.343501805 | 0.002704292 | 0.041 |
| Ckap5_1 | 0.347213687 | 0.000331816 | 0.014 |
| Clasp2_1 | 0.433469485 | 0.001114589 | 0.025 |
| Tpm2_1 | 0.450872519 | 0.000849197 | 0.022 |
| Sgce_2 | 0.496503118 | 0.000539215 | 0.016 |
| Asph_2 | 0.52525427 | 5.39E-08 | 0 |
| Rev3l_2 | 0.632094424 | 0.000275253 | 0.013 |
| Dncic2_3 | 0.74684407 | 0.000245762 | 0.012 |
| Abcc9_1 | 1.044675879 | 0.04605215 | 0.206 |
The ID is the exon identifier. P values were obtained with a t test (P values of less than 0.05 were considered significant). Difference is the difference between the mean log ratio of the inclusion and the skipping probes. The adjusted P value was obtained by BH-FDR analysis. Exons were considered to show a splicing change if the adjusted P value was less than 0.05.
The RT-PCR validation of some of these exons is shown in Fig. 1. Interestingly, some exons displayed increased inclusion upon knockdown of Sam68 (Sgce_2, Rev3l_1, Bin1_2, Clasp2_1, Ckap5_1, and Dncic2_1), indicating that they are normally repressed by the protein (Fig. 1B). Other exons (Numa1_1, Ktn1_2, Kifap3_1, and Sorbs1_2) showed increased skipping upon Sam68 knockdown, implying positive regulation by the protein (Fig. 1C). Of the 30 Sam68-regulated exons in this new set, Sam68 functions as an enhancer for 16 and a repressor for 14 exons. To assess potential off-target effects from the original shRNA, exons were also tested after Sam68 knockdown with a different shRNA (see Fig. S1 in the supplemental material). Although one shRNA had stronger effects on Sam68 expression and splicing, all the tested exons responded similarly to both Sam68 shRNAs, indicating that the number of off-target effects is relatively low.
If Sam68 directly regulates splicing of these exons, the pre-mRNAs containing them should have binding sites for Sam68. Selex experiments with recombinant Sam68 showed that the protein binds to a variety of A- and U-rich sequences, often including the motifs UAAA and UUUA (31). The frequency of these two sequence motifs within the validated set of Sam68-regulated exons was compared to a control set of 8,995 constitutive exons. The UAAA motif was found to be enriched in the 500 nucleotides downstream of the Sam68 exons relative to the control exon set (Wilcoxon rank sum test, P = 0.00017996). The UUUA motif was enriched both in the 200 nucleotides upstream (P = 0.000913635) and in the 200 nucleotides downstream (P = 0.011298) of the Sam68 target exons.
We also used the Improbizer algorithm to search more generally for motifs that were enriched in the SAM68-regulated exons and their flanking sequences (1, 2). The searched sequences included 500 nucleotides upstream and downstream of the exon, excluding 15 nucleotides of the 3′ splice site and 6 nucleotides of the 5′ splice site. These were compared to a control set of exons in transcripts whose expression was detected on the microarray but whose splicing did not change upon Sam68 knockdown. Two of the top five high-scoring motifs from this analysis were additional AU-rich sequences (UUUUAA and AUUUUU) that were enriched in the introns upstream of the Sam68-regulated exons compared to the control set. The similarity of these elements to the known Sam68 binding sequences and their presence adjacent to nearly all the exons altered by Sam68 knockdown indicate that most exons in the Sam68 target set are likely to be directly regulated by the protein. However, it is possible that some exons are affected indirectly through Sam68 effects on other factors.
Sam68 regulates splicing of sarcoglycan epsilon exon 8 by binding RNA.
Demonstration of regulation by direct RNA binding by Sam68 requires detailed analysis of each exon. To examine Sam68 regulation of one exon in more detail, we chose epsilon-sarcoglycan (Sgce) exon 8, which exhibits a particularly strong increase in inclusion upon Sam68 knockdown (Fig. 1B). Epsilon-sarcoglycan is an atypical component of the sarcoglycan complex involved in anchoring the dystrophin protein complex in the muscle cell membrane (44). Mutations in human Sgce cause the movement disorder myoclonus dystonia, which often presents with a variety of psychiatric symptoms likely derived from a neuronal function of the protein in addition to its role in muscle (65). There are several different splice variants of Sgce, including brain-specific transcripts that skip exon 8 (43). Since knockdown of Sam68 resulted in increased exon 8 inclusion (Fig. 1B), we tested whether overexpression of Sam68 would increase skipping. Indeed, transfection of N2A cells with a Sam68 expression plasmid repressed the inclusion of exon 8 in the endogenous Sgce mRNA by 13% (Fig. 2A). Thus, both increasing and decreasing Sam68 expression alters Sgce exon 8 inclusion.
FIG. 2.
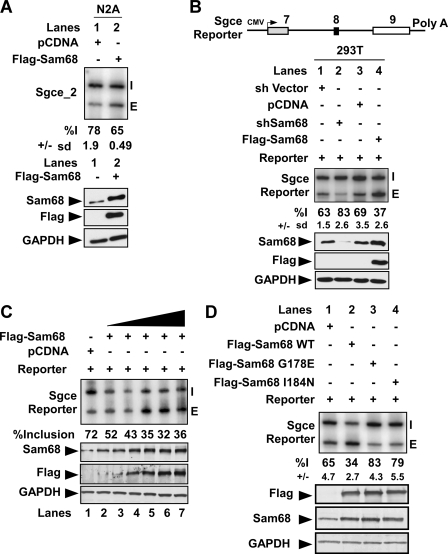
Sam68 regulates splicing of the Sgce mini gene reporter. (A) Flag-tagged Sam68 was overexpressed in the N2A cell line. RNA and protein were extracted from the cell lysates. Splicing was assayed by RT-PCR analysis using end-labeled primers against the flanking constitutive exons. The quantification of percent inclusion and standard error calculated from three separate experiments is indicated below. (Bottom panel) Western blots were probed with antibodies to detect Sam68, Flag tag, and GAPDH as a loading control. (B) Schematic diagram of the Sgce mini gene. Exons are shown as boxes and introns as lines. RT-PCR analysis of 293T cells transiently transfected with the Sgce reporter along with control plasmid (lane 1), shSam68 plasmid (lane 2), pcDNA (lane 3), or Flag-Sam68 (lane 4) results are shown. The percent inclusion with the standard deviation from three different experiments is shown below. (Bottom panel) Western blots were probed with antibodies to detect Sam68, Flag, and GAPDH as a loading control. (C) A 0.75-μg amount of the Sgce mini gene and different amounts of Flag-Sam68 (0.1 μg to 2 μg) were transfected into 293T cells. Cells were harvested after 20 h, and splicing was assayed by RT-PCR. (Bottom panel) Quantitative Western blot probed with antibodies to Sam68, Flag peptide, and GAPDH, used as a loading control. (D) RT-PCR analysis of Sgce mRNA from 293T cells transiently transfected with either wild-type Sam68 or Sam68 mutant (G178E or I184N) constructs, as indicated at the top. Cells were harvested after 24 h for isolation of RNA and protein. The percent inclusion of the Sgce reporter and the standard deviation from three different experiments is shown below. (Bottom panel) Immunoblot analysis of the expressed proteins.
To analyze sequence requirements for Sam68 regulation of Sgce exon 8, we constructed a minigene reporter construct containing the entire alternatively spliced exon 8 region of the Sgce gene with its flanking exons (Fig. 2B). Upon expression of the minigene in HEK293T cells, we observed partial inclusion of exon 8 (63 to 70%), similar to the endogenous Sgce RNA (data not shown). Cotransfection of short hairpin (shSam68), or protein overexpression constructs (Flag-Sam68), recapitulated the splicing changes observed for the endogenous Sgce RNA in N2A cells (Fig. 2B). Expression of Flag-Sam68 enhanced exon 8 skipping in a dose-dependent manner (Fig. 2C), demonstrating that even modest changes in intracellular Sam68 concentration were sufficient to alter Sgce splicing.
To examine whether Sam68 regulates Sgce exon 8 splicing by binding to RNA, Sam68 mutants defective in RNA binding were tested. Two point mutations in the Sam68 KH domain, G178E and I184N, were previously shown to abolish its binding to a UAAA motif and are analogous to loss-of-function mutations in Caenorhabditis elegans Gld1 and human Fmr1 (15, 27, 31). Flag-tagged versions of G178E- or I184N-Sam68 were coexpressed with the Sgce minigene in HEK293T cells. Unlike the wild-type protein, neither the G178E nor the I184N mutants induced repression of the reporter exon, indicating that Sam68 RNA binding activity is needed for alternative splicing regulation (Fig. 2D). Immunostaining with anti-Flag antibodies confirmed that both G178E and I184N exhibit diffuse nucleoplasmic staining similar to that of wild-type Sam68 (data not shown). Interestingly, expressing either of the RNA binding mutants induced greater exon inclusion than the empty vector control (Fig. 2B, lanes 1 and 2; compare with D, lanes 3 and 4). This is similar to what is observed after Sam68 knockdown and indicates that the mutant proteins likely possess dominant negative Sam68 activity. This suggests that in the absence of RNA binding, other domains of the mutant proteins remain functional and may compete with the endogenous wild-type protein for interactions with itself or with essential cofactors. Thus, in addition to RNA binding, Sam68-dependent repression of Sgce splicing likely requires these additional interactions.
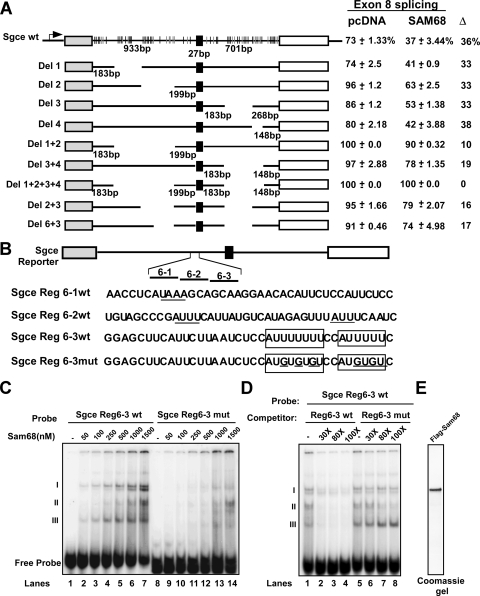
To investigate the RNA elements involved in the regulation of Sgce exon 8 splicing, mutations in the Sgce reporter were constructed (Fig. 3A). As seen with many functionally important alternative exons, the intronic sequences surrounding Sgce exon 8 are highly conserved across multiple mammalian species. These regions contain many putative Sam68 binding motifs. We deleted multiple UAAA/UUUA motifs present in the introns flanking the Sgce exon 8, many of which are conserved between the human and mouse Sgce genes. Individual deletions 1, 2, 3, and 4 show variable levels of basal exon inclusion, but all exhibit similar responses to cotransfected Sam68 (Fig. 3A; see also Fig. S2 in the supplemental material). Thus, none of the deleted sequences alone is essential for Sam68 repression. Combinations of multiple deletions all enhanced overall inclusion and generally reduced the ability of Sam68 to repress exon inclusion. For example, combining deletions 1 and 2 yielded 100% inclusion without Sam68 but only 90% inclusion with Sam68 expression (Fig. 3; see also Fig. S2 in the supplemental material). These and additional results indicate that there is no single Sam68-responsive element that controls Sgce exon 8. Instead, these sequences are distributed across the intronic regions flanking the exon, as might be expected from the distribution of the UAAA/UUUA elements.
FIG. 3.
Sam68 regulates Sgce alternative splicing by binding to intronic RNA repressor elements. (A) Schematic of splicing reporter constructs with exons shown as boxes and introns as lines. The UAAA (black) and the UUUA (gray) in the flanking introns are represented as vertical lines, with deletions indicated by blanks. Intron and exon lengths are indicated. Each splicing reporter was expressed in 293T cells in the absence or presence of Sam68. Cells were harvested 24 h after transfection for RNA and protein isolation. The difference in percent inclusion [inclusion/(inclusion + exclusion) × 100] between vector control (pCDNA) and Sam68 coexpression is indicated to the right. (B) Sequences of probes for regions 6-1, 6-2, and 6-3 and the Region 6-3 mutant that were used for the electrophoretic mobility shift assay. The element identified by Improbizer as enriched adjacent to Sam68 target exons is boxed; mutated residues are underlined. (C) Electrophoretic mobility shift assay results using Flag-tagged Sam68 and 32P-labeled RNA probes corresponding to region 6-3, and the region 6-3 mutant (indicated at the top). (D) In lanes 1 to 8, 500 nM of Flag-Sam68 was used. Unlabeled cold RNA was used as competitor (30-fold or 80-fold excess over the labeled probe). (E) Coomassie-stained gel of the purified Flag-Sam68.
To further map elements that were responding to Sam68, we combined upstream deletion region 6 with downstream deletion 3. This reduced the response to Sam68 by about half compared to the deletion 3 construct alone (16% versus 33%) (see Fig. S2 in the supplemental material). Thus, at least one Sam68-responsive element is likely in region 6, which contains several A- and U-rich sequence elements.
To demonstrate whether Sam68 binds to region 6, Flag-tagged Sam68 was purified from HEK 293T cells and tested for RNA binding (Fig. 3C and E). Three different RNA probes (40 nucleotides), together spanning the entire region 6, were incubated with Sam68, and the resulting RNP complexes were resolved in gel shift experiments (Sgce region 6-1, region 6-2, and region 6-3) (Fig. 3B and C). Only weak complexes were observed in gel shift assays with the region 6-1 and 6-2 probes (see Fig. S3 in the supplemental material). However, region 6-3 formed several Sam68-dependent RNP complexes (I, II, and III). This probe did not show complex formation with mutant Flag-Sam68 G178E (data not shown). Region 6-3 has several A- and U-rich motifs, including two elements (AUUUUUUU and AUUUUU) that match one of the motifs identified by Improbizer as enriched in Sam68 target exons (Fig. 3B). Introducing several U-to-G mutations into these elements abolished Sam68 complexes I and III and reduced complex II. Complex II may result from binding to other AU elements in the probe. The mutant probe also lost the ability to compete with the wild-type probe for Sam68 binding in complexes I and III (Fig. 3D, lanes 5 to 8). Thus, Sam68 directly binds to AUUUU elements in Sgce region 6 and presumably to additional sites distributed along the Sgce transcript, as would be expected for a direct target of Sam68 regulation.
Sam68-dependent splicing during neurogenesis.
Many exons change in splicing during neurogenesis due to changes in the expression of RNA binding proteins, such as PTB, nPTB, and others (8, 29, 37). The Sam68 gene was shown to be induced during adult subventricular zone and olfactory bulb neurogenesis in mice (30). Sam68 is primarily nuclear in neurons but has also been implicated in dendritic mRNA localization (21). We find that Sgce exon 8 is repressed by Sam68, and in vivo this exon repression is specifically seen in the brain (43). We thus wanted to test if neuron-specific changes in Sgce and other transcripts were in part due to changes in Sam68.
We examined the expression of Sam68 in P19 embryonal carcinoma cells induced to differentiate in the presence of retinoic acid (RA). Five days after induction, most cells had small, round cell bodies with extensive processes and expressed a variety of neuronal markers, including MAP2, TuJ1, Sox6, Sox11, and N-cadherin (Fig. 4B and 5A, B, C, and E). Immunoblot analysis showed that Sam68 protein levels increase 2.5- to 3-fold upon P19 differentiation (Fig. 4B). This increase was accompanied by changes in the splicing of multiple exons identified as Sam68 dependent in N2A cells (Fig. 4C). Other exons affected by Sam68 overexpression or knockdown in N2A cells did not change during neurogenesis in P19 cells and are presumably affected by other proteins whose activities differ in the two cell lines.
To test whether Sam68 is required for P19 cell differentiation and/or for changes in the splicing of particular exons during this process, we infected P19 cells with a recombinant lentivirus carrying a Sam68-targeted shRNA (Fig. 5A). This viral infection reduced Sam68 levels by 60 to 80% relative to a control virus (Fig. 5B). P19 cells stably expressing the Sam68 hairpin failed to differentiate as indicated by the absence of neuronal processes and the lack of TuJl expression after RA treatment (Fig. 5A, bottom panel, and B). Similar results were obtained with a second Sam68-targeted shRNA, indicating that they were not due to off-target effects of the shRNA (data not shown). In contrast, P19 cell lines generated with the empty vector control virus showed extended neurites and differentiated normally into TuJ1-positive neurons (Fig. 5A, top panel). Immunoblot analysis of TuJ1 confirmed that it is not expressed after Sam68 knockdown (Fig. 5B). Other neuronal cell markers, including Sox6, Sox11, and N-cadherin, also failed to be activated in the Sam68 shRNA-expressing cells after RA treatment (Fig. 5C). Similarly, mRNAs seen as Sam68 dependent on the microarray are also upregulated in P19 cells by RA-induced neuronal differentiation. These also failed to be induced after Sam68 depletion (Fig. 5D). Depletion of Sam68 from P19 cells also affected the splicing patterns of Sam68 target exons. As seen for Sgce, exon 8 was not properly repressed in the Sam68-depleted P19 cells after RA treatment (Fig. 5D). Thus, Sam68 is required for proper neuronal differentiation of P19 cells.
Since the Sam68-depleted cells do not differentiate after RA induction, the loss of splicing regulation observed for particular transcripts could result indirectly from the failure to proceed down this developmental pathway rather than the specific loss of Sam68 (Fig. 5A, B, and C). We thus examined whether Sam68 expression also contributes to maintaining splicing patterns after differentiation. We infected P19 cells with the shSam68 lentivirus after they had differentiated and assayed them for splicing changes. Sam68 protein was reduced by 50% in these cells, but they continued to show neuronal morphology 5 days after infection and continued to express neuronal markers, including TuJ1, Sox6, Sox11, and N-cadherin (Fig. 5E). RT-PCR analysis of several Sam68-dependent exons showed that Sam68 knockdown reversed the splicing change observed during differentiation (Fig. 5E). Thus, Sam68 is required for maintaining these patterns of splicing even after differentiation.
To further evaluate the role of Sam68 in the context of neurogenesis, we used NPCs derived from mouse cortex at embryonic day 11.5. These multipotent neural progenitors can self-renew when cultured with bFGF2 but differentiate into neurons upon bFGF withdrawal (Fig. 6A and B) (8, 17, 18, 20, 24, 26, 50). After explant and culture in vitro for 7 days, bFGF was withdrawn, and differentiation into postmitotic neurons was monitored by cell morphology and Tuj1 protein expression (Fig. 6A and B). Similar to the observations in P19 cells, Sam68 expression increased about twofold after neuronal induction (Fig. 6B). This differentiation also induced changes in the splicing of several Sam68-dependent exons (Fig. 6B, bottom panel).
To test whether Sam68 overexpression could influence neuronal differentiation in this system, NPCs were cultured for 7 days in vitro and then transduced with a lentivirus-expressing Flag-tagged Sam68 (Fig. 6A and C). Two days after transduction, neuronal differentiation was initiated by bFGF withdrawal and the cells were cultured for an additional 3 days. Overexpression of Sam68 led to a twofold increase in TuJ1 mRNA expression (Fig. 6D, right panel). The number of infected cells exhibiting TuJ1 staining also doubled in the Sam68-overexpressing cultures compared to the vector control (Fig. 6C, bottom panel). Overexpression of Sam68 also stimulated expression of mRNAs (Chl1 and Nav1) seen to be Sam68 responsive in the microarray experiment and again stimulated the neuronal splicing pattern of Sgce exon 8 (Fig. 6B and D).
To examine the effect of Sam68 knockdown on NPC differentiation, cells in 7-day-old cultures were transduced with lentiviruses expressing shRNAs that target Sam68 or an empty vector control virus (Fig. 7A). Two days after transduction, neuronal differentiation was initiated with bFGF withdrawal and the cells were cultured for an additional 3 days. Sam68 was depleted by 65% in the shRNA-expressing cells (Fig. 7B). This Sam68 depletion significantly decreased the number of TuJ1-positive neurons in the culture (∼50%) (Fig. 7A and C). The knockdown of Sam68 also reduced expression of TuJ1 mRNA as well as Sam68-dependent mRNAs compared to β-actin mRNA (Fig. 7D). Sam68 knockdown again resulted in changes in the splicing of Sam68 target exons (Fig. 7E). Thus, as seen for P19 cells, Sam68 is required for proper neuronal differentiation of NPCs in vitro.
DISCUSSION
Novel mRNA splicing targets of Sam68.
We have identified a new set of target exons whose splicing is dependent on Sam68. This exon set will be very useful both for characterizing the mechanisms of Sam68 function and for understanding its biological roles. We also find that changes in Sam68 expression during neuronal differentiation affect a large number of specific splicing events. Sam68 controls splicing of some of these targets through direct binding to RNA elements, as seen with Sgce and CD44 (39). Sam68 can also potentially modulate splicing through its interactions with other factors, such as PTB (unpublished data) or hnRNP A1 (45). Either of these activities could be modulated in response to particular cellular stimuli.
Our analysis indicates that Sam68 regulates Sgce splicing by binding to numerous intronic regulatory elements distributed across the highly conserved region of the target exon. Splicing regulatory elements are frequently clustered at positions adjacent to a target exon, and this multimerization can be needed for proper activity. However, the binding elements in Sgce are unusual in their large number and broad distribution in the sequence surrounding the exon. Thus, Sam68 may use unusual mechanisms to affect spliceosome assembly at this and possibly other exons. This does not preclude it from more typical actions through individual binding sites and in cooperation with other factors, such as that seen with CD44. The brain-specific repression of Sgce exon 8 in response to Sam68 will be interesting to investigate with regard to its role in the sarcoglycan complex and myoclonus dystonia.
The functions of its target transcripts give important clues to the cellular processes in which Sam68 acts. One interesting example is Sorbs1 (Ponsin/SH3P12/CAP), which belongs to a family of adapter proteins containing a sorbin homology domain and three SH3 domains. Sorbs1 is widely expressed and interacts with the insulin receptor, Sos, flotillin, and focal adhesion kinase and functions in insulin signaling as well as in ephrin reverse signaling in neuronal cells (4, 14, 52). Sorbs1 produces a number of splice variants. The exon inclusion stimulated by Sam68 creates an isoform containing a nuclear localization signal and part of a coiled-coil domain. How this splicing affects Sorbs1 activity is not clear, but it will presumably alter its intracellular localization. It is interesting that Sam68 itself is both phosphorylated by the insulin receptor and upregulated by insulin stimulation, in addition to targeting this component of the insulin signaling complex.
Another Sam68 target is kinesin superfamily-associated protein 3 (Kifap3/Kap3), which forms a heterotrimeric complex with kinesin intermediate filament proteins 3A and 3B (Kif3A and Kif3B). This complex plays a role in anterograde transport of membrane-bound organelles in neurons and other cells (25, 62). The two different carboxy termini of Kifap3 arising from Sam68 regulation may allow recognition of different cargoes (57). Interestingly, this C-terminal domain of Kifap3 is phosphorylated by BRK, which also targets Sam68 (34). Thus, similar to Sorbs1 and the insulin receptor, Kifap3 is both a target of Sam68 splicing activity and a target along with Sam68 of a common upstream signaling pathway (BRK).
A third Sam68 splicing target is Numa1 (nuclear/mitotic apparatus protein, also referred to as centrophilin), which is a long coiled-coil protein with functions in the cell cycle. During interphase Numa1 plays a structural role in the nucleoskeleton, and during mitosis it is associated with spindle microtubules (12, 58, 64). Numa1 is also a component of somatodendritic microtubule arrays in postmitotic neurons, where it appears as small somatodendritic particles that increase in number during dendritic maturation (19). Numa1 has multiple spliced isoforms, and it is not yet clear how these isoforms may differ in activity.
Another interesting transcript whose expression decreases upon Sam68 knockdown is Chl1 (close homolog of L1). Chl1 is a transmembrane cell adhesion receptor shown to promote neurite outgrowth and branching (23, 42). Mice deficient in Chl1 display alterations in motor coordination (49), a defect recently associated with ablation of Sam68 in mice (35).
Sam68 as a modulator of neuronal differentiation.
We found that the loss of Sam68 in P19 cells eliminates their ability to form neurons. Similarly, Sam68 knockdown in neuronal progenitor cells reduces the number of TuJ1-positive neurons that differentiate in the culture. However, Sam68 null mice do not show a neurogenesis defect but instead show defects in bone mesenchymal cell differentiation (53). Only about one-third of Sam68−/− mice survive to adulthood, with the survivors showing defects in motor coordination (35, 53). Sam68 is widely expressed and plays roles in multiple cell types. It is possible that the neurogenesis defect we observed in vitro results from the loss of a general function that is not specific to neurons, possibly through Sam68 effects on the cell cycle (16). Alternatively, it may be that in vivo other Sam68 family members, such as Slm-1 and Slm-2, can substitute for Sam68 during neurogenesis. Interestingly, recently Sam68 overexpression was shown to cause a significant reduction in cell proliferation in neural stem cell cultures (40).
We found that Sam68 protein upregulation upon P19 or NPC differentiation affects the splicing and/or expression of a variety of transcripts. Genes carrying Sam68 target exons are involved in a variety of cellular processes important in neurogenesis, including cytoskeletal organization (Numa1, Clasp2, and Sgce), organellar biogenesis and transport (Bin1, Ktn1, Kifap3, and Opa1), and synaptogenesis (Cadm1, Dlgh4, and Sorbs1). Thus, although it may not drive the differentiation, Sam68 is another RNA binding protein involved in defining the differentiated state of the neuron through its effects on splicing and other processes of mRNA metabolism. It will be interesting to examine the splicing of the Sam68 targets identified here, such as Sgce, in Sam68 null mice. Besides its effects on splicing, the loss of differentiation seen with Sam68 knockdown could result from the loss of the cytoplasmic functions of the protein. Although primarily nuclear, Sam68 can be found in the soma and dendrites of neurons in the hippocampus and cortex, where it cosediments with polysomes and binds to specific mRNAs (21). In the future it will be important to understand how this protein can coordinate splicing of certain pre-mRNAs in the nucleus and then follow these RNAs to the cytoplasm to interact with a variety of signaling pathways.
Supplementary Material
Acknowledgments
We thank members of the Black lab for helpful discussions and extend special thanks to Ji Ann Lee and Peter Stoilov for advice on microarray experiments. We thank John Paul Donohue for help with the data analysis and Owen Witte and Inder Verma's labs for the gift of lentiviral vectors.
This work was supported by NIH grants RO1 GM49662 to D.L.B. and R24 GM070857 to D.L.B. and M.A. D.L.B. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 20 October 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ao, W., J. Gaudet, W. J. Kent, S. Muttumu, and S. E. Mango. 2004. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 3051743-1746. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 228-36. [PubMed] [Google Scholar]
- 3.Batsche, E., M. Yaniv, and C. Muchardt. 2006. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 1322-29. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, C. A., V. Ribon, M. Kanzaki, D. C. Thurmond, S. Mora, S. Shigematsu, P. E. Bickel, J. E. Pessin, and A. R. Saltiel. 2000. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature 407202-207. [DOI] [PubMed] [Google Scholar]
- 5.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 6.Blencowe, B. J. 2006. Alternative splicing: new insights from global analyses. Cell 12637-47. [DOI] [PubMed] [Google Scholar]
- 7.Boutz, P. L., G. Chawla, P. Stoilov, and D. L. Black. 2007. MicroRNAs regulate the expression of the alternative splicing factor nPTB during muscle development. Genes Dev. 2171-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutz, P. L., P. Stoilov, Q. Li, C. H. Lin, G. Chawla, K. Ostrow, L. Shiue, M. Ares, Jr., and D. L. Black. 2007. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes Dev. 211636-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, T., F. M. Boisvert, D. P. Bazett-Jones, and S. Richard. 1999. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol. Biol. Cell 103015-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, C., and P. A. Sharp. 2006. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol. Cell. Biol. 26362-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, T. A., C. W. Sugnet, and M. Ares, Jr. 2002. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science 296907-910. [DOI] [PubMed] [Google Scholar]
- 12.Compton, D. A., and D. W. Cleveland. 1994. NuMA, a nuclear protein involved in mitosis and nuclear reformation. Curr. Opin. Cell Biol. 6343-346. [DOI] [PubMed] [Google Scholar]
- 13.Cote, J., F. M. Boisvert, M. C. Boulanger, M. T. Bedford, and S. Richard. 2003. Sam68 RNA binding protein is an in vivo substrate for protein arginine N-methyltransferase 1. Mol. Biol. Cell 14274-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cowan, C. A., and M. Henkemeyer. 2002. Ephrins in reverse, park and drive. Trends Cell Biol. 12339-346. [DOI] [PubMed] [Google Scholar]
- 15.De Boulle, K., A. J. Verkerk, E. Reyniers, L. Vits, J. Hendrickx, B. Van Roy, F. Van den Bos, E. de Graaff, B. A. Oostra, and P. J. Willems. 1993. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 331-35. [DOI] [PubMed] [Google Scholar]
- 16.Di Fruscio, M., T. Chen, and S. Richard. 1999. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc. Natl. Acad. Sci. USA 962710-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, G., K. Martinowich, M. H. Chin, F. He, S. D. Fouse, L. Hutnick, D. Hattori, W. Ge, Y. Shen, H. Wu, J. ten Hoeve, K. Shuai, and Y. E. Sun. 2005. DNA methylation controls the timing of astrogliogenesis through regulation of JAK-STAT signaling. Development 1323345-3356. [DOI] [PubMed] [Google Scholar]
- 18.Feng, J., H. Chang, E. Li, and G. Fan. 2005. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J. Neurosci Res. 79734-746. [DOI] [PubMed] [Google Scholar]
- 19.Ferhat, L., C. Cook, R. Kuriyama, and P. W. Baas. 1998. The nuclear/mitotic apparatus protein NuMA is a component of the somatodendritic microtubule arrays of the neuron. J. Neurocytol. 27887-899. [DOI] [PubMed] [Google Scholar]
- 20.Ge, W., K. Martinowich, X. Wu, F. He, A. Miyamoto, G. Fan, G. Weinmaster, and Y. E. Sun. 2002. Notch signaling promotes astrogliogenesis via direct CSL-mediated glial gene activation. J. Neurosci Res. 69848-860. [DOI] [PubMed] [Google Scholar]
- 21.Grange, J., V. Boyer, R. Fabian-Fine, N. B. Fredj, R. Sadoul, and Y. Goldberg. 2004. Somatodendritic localization and mRNA association of the splicing regulatory protein Sam68 in the hippocampus and cortex. J. Neurosci. Res. 75654-666. [DOI] [PubMed] [Google Scholar]
- 22.Grossman, J. S., M. I. Meyer, Y. C. Wang, G. J. Mulligan, R. Kobayashi, and D. M. Helfman. 1998. The use of antibodies to the polypyrimidine tract binding protein (PTB) to analyze the protein components that assemble on alternatively spliced pre-mRNAs that use distant branch points. RNA 4613-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillenbrand, R., M. Molthagen, D. Montag, and M. Schachner. 1999. The close homologue of the neural adhesion molecule L1 (CHL1): patterns of expression and promotion of neurite outgrowth by heterophilic interactions. Eur. J. Neurosci. 11813-826. [DOI] [PubMed] [Google Scholar]
- 24.Hirabayashi, Y., Y. Itoh, H. Tabata, K. Nakajima, T. Akiyama, N. Masuyama, and Y. Gotoh. 2004. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 1312791-2801. [DOI] [PubMed] [Google Scholar]
- 25.Hirokawa, N. 2000. Stirring up development with the heterotrimeric kinesin KIF3. Traffic 129-34. [DOI] [PubMed] [Google Scholar]
- 26.Israsena, N., M. Hu, W. Fu, L. Kan, and J. A. Kessler. 2004. The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev. Biol. 268220-231. [DOI] [PubMed] [Google Scholar]
- 27.Jones, A. R., and T. Schedl. 1995. Mutations in gld-1, a female germ cell-specific tumor suppressor gene in Caenorhabditis elegans, affect a conserved domain also found in Src-associated protein Sam68. Genes Dev. 91491-1504. [DOI] [PubMed] [Google Scholar]
- 28.Lee, J. A., Y. Xing, D. Nguyen, J. Xie, C. J. Lee, and D. L. Black. 2007. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 5e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Q., J. A. Lee, and D. L. Black. 2007. Neuronal regulation of alternative pre-mRNA splicing. Nat. Rev. Neurosci. 8819-831. [DOI] [PubMed] [Google Scholar]
- 30.Lim, D. A., M. Suarez-Farinas, F. Naef, C. R. Hacker, B. Menn, H. Takebayashi, M. Magnasco, N. Patil, and A. Alvarez-Buylla. 2006. In vivo transcriptional profile analysis reveals RNA splicing and chromatin remodeling as prominent processes for adult neurogenesis. Mol. Cell. Neurosci. 31131-148. [DOI] [PubMed] [Google Scholar]
- 31.Lin, Q., S. J. Taylor, and D. Shalloway. 1997. Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J. Biol. Chem. 27227274-27280. [DOI] [PubMed] [Google Scholar]
- 32.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295868-872. [DOI] [PubMed] [Google Scholar]
- 33.Lukong, K. E., D. Larocque, A. L. Tyner, and S. Richard. 2005. Tyrosine phosphorylation of sam68 by breast tumor kinase regulates intranuclear localization and cell cycle progression. J. Biol. Chem. 28038639-38647. [DOI] [PubMed] [Google Scholar]
- 34.Lukong, K. E., and S. Richard. 2008. Breast tumor kinase BRK requires kinesin-2 subunit KAP3A in modulation of cell migration. Cell Signal. 20432-442. [DOI] [PubMed] [Google Scholar]
- 35.Lukong, K. E., and S. Richard. 2008. Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav. Brain Res. 189357-363. [DOI] [PubMed] [Google Scholar]
- 36.Lukong, K. E., and S. Richard. 2003. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta 165373-86. [DOI] [PubMed] [Google Scholar]
- 37.Makeyev, E. V., J. Zhang, M. A. Carrasco, and T. Maniatis. 2007. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27435-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 6386-398. [DOI] [PubMed] [Google Scholar]
- 39.Matter, N., P. Herrlich, and H. Konig. 2002. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420691-695. [DOI] [PubMed] [Google Scholar]
- 40.Moritz, S., S. Lehmann, A. Faissner, and A. von Holst. 2008. An induction gene trap screen in neural stem cells reveals an instructive function of the niche and identifies the splicing regulator Sam68 as a tenascin-C regulated target gene. Stem Cells 262321-2331. [DOI] [PubMed] [Google Scholar]
- 41.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272263-267. [DOI] [PubMed] [Google Scholar]
- 42.Naus, S., M. Richter, D. Wildeboer, M. Moss, M. Schachner, and J. W. Bartsch. 2004. Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J. Biol. Chem. 27916083-16090. [DOI] [PubMed] [Google Scholar]
- 43.Nishiyama, A., T. Endo, S. Takeda, and M. Imamura. 2004. Identification and characterization of epsilon-sarcoglycans in the central nervous system. Brain Res. Mol. Brain Res. 1251-12. [DOI] [PubMed] [Google Scholar]
- 44.Ozawa, E., Y. Mizuno, Y. Hagiwara, T. Sasaoka, and M. Yoshida. 2005. Molecular and cell biology of the sarcoglycan complex. Muscle Nerve 32563-576. [DOI] [PubMed] [Google Scholar]
- 45.Paronetto, M. P., T. Achsel, A. Massiello, C. E. Chalfant, and C. Sette. 2007. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 176929-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paronetto, M. P., J. P. Venables, D. J. Elliott, R. Geremia, P. Rossi, and C. Sette. 2003. Tr-kit promotes the formation of a multimolecular complex composed by Fyn, PLCgamma1 and Sam68. Oncogene 228707-8715. [DOI] [PubMed] [Google Scholar]
- 47.Pfeifer, A., M. Ikawa, Y. Dayn, and I. M. Verma. 2002. Transgenesis by lentiviral vectors: lack of gene silencing in mammalian embryonic stem cells and preimplantation embryos. Proc. Natl. Acad. Sci. USA 992140-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pozzoli, U., L. Riva, G. Menozzi, R. Cagliani, G. P. Comi, N. Bresolin, R. Giorda, and M. Sironi. 2004. Over-representation of exonic splicing enhancers in human intronless genes suggests multiple functions in mRNA processing. Biochem. Biophys. Res. Commun. 322470-476. [DOI] [PubMed] [Google Scholar]
- 49.Pratte, M., G. Rougon, M. Schachner, and M. Jamon. 2003. Mice deficient for the close homologue of the neural adhesion cell L1 (CHL1) display alterations in emotional reactivity and motor coordination. Behav. Brain Res. 14731-39. [DOI] [PubMed] [Google Scholar]
- 50.Qian, X., A. A. Davis, S. K. Goderie, and S. Temple. 1997. FGF2 concentration regulates the generation of neurons and glia from multipotent cortical stem cells. Neuron 1881-93. [DOI] [PubMed] [Google Scholar]
- 51.Rho, J., S. Choi, C. R. Jung, and D. S. Im. 2007. Arginine methylation of Sam68 and SLM proteins negatively regulates their poly(U) RNA binding activity. Arch. Biochem. Biophys. 46649-57. [DOI] [PubMed] [Google Scholar]
- 52.Ribon, V., J. A. Printen, N. G. Hoffman, B. K. Kay, and A. R. Saltiel. 1998. A novel, multifuntional c-Cbl binding protein in insulin receptor signaling in 3T3-L1 adipocytes. Mol. Cell. Biol. 18872-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Richard, S., N. Torabi, G. V. Franco, G. A. Tremblay, T. Chen, G. Vogel, M. Morel, P. Cleroux, A. Forget-Richard, S. Komarova, M. L. Tremblay, W. Li, A. Li, Y. J. Gao, and J. E. Henderson. 2005. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 1e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saccomanno, L., C. Loushin, E. Jan, E. Punkay, K. Artzt, and E. B. Goodwin. 1999. The STAR protein QKI-6 is a translational repressor. Proc. Natl. Acad. Sci. USA 9612605-12610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin, C., and J. L. Manley. 2004. Cell signalling and the control of pre-mRNA splicing. Nat. Rev. Mol. Cell Biol. 5727-738. [DOI] [PubMed] [Google Scholar]
- 56.Srinivasan, K., L. Shiue, J. D. Hayes, R. Centers, S. Fitzwater, R. Loewen, L. R. Edmondson, J. Bryant, M. Smith, C. Rommelfanger, V. Welch, T. A. Clark, C. W. Sugnet, K. J. Howe, Y. Mandel-Gutfreund, and M. Ares, Jr. 2005. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods 37345-359. [DOI] [PubMed] [Google Scholar]
- 57.Takeda, S., H. Yamazaki, D. H. Seog, Y. Kanai, S. Terada, and N. Hirokawa. 2000. Kinesin superfamily protein 3 (KIF3) motor transports fodrin-associating vesicles important for neurite building. J. Cell Biol. 1481255-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang, T. K., C. J. Tang, Y. L. Chen, and C. W. Wu. 1993. Nuclear proteins of the bovine esophageal epithelium. II. The NuMA gene gives rise to multiple mRNAs and gene products reactive with monoclonal antibody W1. J. Cell Sci. 104249-260. [DOI] [PubMed] [Google Scholar]
- 59.Vernet, C., and K. Artzt. 1997. STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet. 13479-484. [DOI] [PubMed] [Google Scholar]
- 60.Wang, L. L., S. Richard, and A. S. Shaw. 1995. P62 association with RNA is regulated by tyrosine phosphorylation. J. Biol. Chem. 2702010-2013. [DOI] [PubMed] [Google Scholar]
- 61.Xin, L., M. A. Teitell, D. A. Lawson, A. Kwon, I. K. Mellinghoff, and O. N. Witte. 2006. Progression of prostate cancer by synergy of AKT with genotropic and nongenotropic actions of the androgen receptor. Proc. Natl. Acad. Sci. USA 1037789-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamazaki, H., T. Nakata, Y. Okada, and N. Hirokawa. 1996. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc. Natl. Acad. Sci. USA 938443-8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yao, M., G. Bain, and D. I. Gottlieb. 1995. Neuronal differentiation of P19 embryonal carcinoma cells in defined media. J. Neurosci. Res. 41792-804. [DOI] [PubMed] [Google Scholar]
- 64.Zeng, C., D. He, S. M. Berget, and B. R. Brinkley. 1994. Nuclear-mitotic apparatus protein: a structural protein interface between the nucleoskeleton and RNA splicing. Proc. Natl. Acad. Sci. USA 911505-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zimprich, A., M. Grabowski, F. Asmus, M. Naumann, D. Berg, M. Bertram, K. Scheidtmann, P. Kern, J. Winkelmann, B. Muller-Myhsok, L. Riedel, M. Bauer, T. Muller, M. Castro, T. Meitinger, T. M. Strom, and T. Gasser. 2001. Mutations in the gene encoding epsilon-sarcoglycan cause myoclonus-dystonia syndrome. Nat. Genet. 2966-69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.