Abstract
The yeast Atg1 serine/threonine protein kinase and its mammalian homologs ULK1 and ULK2 play critical roles during the activation of autophagy. Previous studies have demonstrated that the conserved C-terminal domain (CTD) of ULK1 controls the regulatory function and localization of the protein. Here, we explored the role of kinase activity and intramolecular interactions to further understand ULK function. We demonstrate that the dominant-negative activity of kinase-dead mutants requires a 7-residue motif within the CTD. Our data lead to a model in which the functions of ULK1 and ULK2 are controlled by autophosphorylation and conformational changes involving exposure of the CTD. Additional mapping indicates that the CTD contains other distinct regions that direct membrane association and interaction with the putative human homologue of Atg13, which we have here characterized. Atg13 is required for autophagy and Atg9 trafficking during autophagy. However, Atg13 does not bind the 7-residue dominant-negative motif in the CTD of ULK proteins nor is the inhibitory activity of the CTDs rescued by Atg13 ectopic expression, suggesting that in mammalian cells, the CTD may interact with additional autophagy proteins.
During macroautophagy in mammalian cells, a membrane cisterna wraps around cytoplasmic material to form a nascent autophagosome, which then fuses with late endosomal structures to initiate the degradation of autophagosomal contents. The targets of macroautophagy (herein referred to as autophagy) can include long-lived proteins, organelles, ubiquitinated cellular substrates, and aberrant protein aggregates (7, 13, 15, 24, 26). Autophagy has been implicated in a number of medical contexts, such as cancer, neurodegeneration, and immunity (as recently reviewed in references 16, 19, and 21), raising interest in understanding its basic regulatory mechanisms.
The serine-threonine protein kinase Atg1 was originally identified as a critical autophagy regulator in genetic screens performed with the yeast Saccharomyces cerevisiae (17, 29, 31, 34). Autophagy in yeast is induced by nitrogen starvation or rapamycin treatment, and studies with yeast have shown that Atg1 functions at a regulatory step downstream of the nutrient-sensing signaling kinase TOR (target of rapamycin). Atg1 forms part of a complex that includes additional autophagy (Atg) proteins, such as Atg13, Atg17, Atg29, Atg31, Atg11, Atg20, and Atg24 (9-12, 22). While the proteins Atg17, Atg29, and Atg31 have autophagy-specific functions, Atg11, Atg20, and Atg24 function in the yeast autophagy-related CVT (cytoplasm-to-vacuole targeting) pathway, raising the possibility that a number of different Atg1-containing subcomplexes may exist in vivo to regulate distinct pathways. Atg1 also regulates the recycling of Atg9 between the preautophagosomal structure (PAS) and a peripheral pool via a mechanism involving Atg2, Atg18, and the phosphatidylinositol-3 kinase complex I (25). In addition, the localization of yeast Atg1 to the PAS can be regulated by cyclic AMP-dependent protein kinase-mediated phosphorylation in a starvation-dependent manner (2).
Although the role of Atg1 kinase activity has been controversial (1, 8), recent data have demonstrated that Atg1 acts in distinct kinase-dependent and kinase-independent roles (5, 11). Kinase-inactivated Atg1 was capable of directing the assembly of the PAS containing Atg8, Atg17, and Atg29, consistent with the proposal that Atg1 plays a structural role. However, the kinase activity of Atg1 was required to drive protein or membrane dynamics through disassembly or dissociation of Atg proteins from the PAS, a step which is required to produce fully formed autophagosomes.
Full kinase activity of Atg1 in yeast requires its binding partners Atg13 and Atg17 (10). Interestingly, the kinase activity of Atg1 is stimulated when autophagy is induced in yeast, and this correlates with increased binding of Atg1 to Atg13 and Atg17 upon autophagy induction (10, 11). Furthermore, the efficiency of Atg1-Atg13 binding is inversely correlated with levels of phosphorylation on Atg13 (10), leading to the model in which Atg13 is dephosphorylated upon autophagy induction, thereby promoting its ability to bind to and act as a cofactor for Atg1. The autophagy-dependent kinases and phosphatases controlling yeast Atg13 phosphorylation remain to be determined. However, Atg13 dephosphorylation has been shown to be TOR dependent (10).
The single Atg1 homologues of Dictyostelium discoideum, Caenorhabditis elegans, and Drosophila melanogaster have been confirmed to be key autophagy regulators (20, 23, 27). In contrast, mammals have at least two Atg1 homologues (6), Unc-51-like kinase 1 (ULK1) and ULK2, that share strong homology with the C. elegans Atg1 homologue Unc (uncoordinated)-51 (32). Whether ULK1 and ULK2 play similar roles for autophagy induction remains unclear.
We previously found that, in HEK293A cells, small interfering RNA (siRNA)-mediated knockdown of ULK1 was sufficient to reduce starvation-induced autophagy and inhibit the starvation-dependent redistribution of mammalian Atg9 (mAtg9) to a dispersed, peripherally localized pool (3, 37). In this cell system, knockdown of ULK2 had no effect on the induction of autophagy or mAtg9 traffic, suggesting a preferential role of ULK1 in autophagy. ULK1 and -2 have overlapping widespread mRNA expression patterns (35, 36). However, only ULK1 mRNA was upregulated in maturing reticulocyte cultures to promote autophagic clearance of mitochondria, indicating that some specificity exists in vivo (15). These authors went on to show that mice lacking ULK1 displayed abnormal erythrocyte maturation but were viable and without a developmental phenotype, in contrast with other models of mice deficient in autophagy genes (13, 14). These data suggest that ULK2 can support autophagic function in the absence of ULK1, implying a more specialized role for ULK1 in vivo (15). Although the precise roles of ULK1 and ULK2 require further clarification, our data on ULK1 and recent other data show that both of these proteins can localize to mammalian PASs (called isolation membranes, or phagophores) in a starvation-dependent manner (3, 6).
All Atg1 homologs share similar domain structures in which the kinase catalytic domain comprises the N-terminal one-third of the protein and the remaining two-thirds contain regulatory sequences. Comparison of the mouse ULK1 and ULK2 sequences with that of C. elegans Unc-51 has allowed the definition of a C-terminal domain (CTD) (222 residues long in mouse ULK1) that shows relatively high levels of conservation (32), suggestive of an important biological function. We previously described how the deletion of a three-residue PDZ binding motif at the CTD C terminus transforms ULK1 into a potent dominant inhibitor of autophagy (3). In D. discoideum, defects in development and autophagy found in Atg1 null cells could not be rescued with a mutant D. discoideum Atg1 containing a deletion of the last 40 amino acids of the CTD (30). A recent study has identified a novel ULK1-binding protein called FIP200 (focal adhesion kinase family-interacting protein of 200 kDa), which was required for starvation-induced autophagy, proper ULK1 phosphorylation, and ULK1 stability (6). FIP200 binding to ULK1 required the CTD, further supporting a role of the CTD in autophagy regulation.
In this study, we aimed to gain insight into the function of ULK1 and ULK2 for the regulation of autophagy by studying the role of their kinase activities and the regulation of these activities by the CTD. We demonstrated that complete ablation of ULK1 kinase activity results in decreased protein autophosphorylation and a potent dominant-negative effect on autophagy. Our further analysis has led to a working model in which autophosphorylation is critical for promoting a closed molecular conformation that regulates interactions of the CTD. In further support of this model, expression of the CTD from either ULK1 or ULK2 was sufficient to inhibit autophagy, and additional analysis has identified a 7-residue motif critical for this effect. We determined that this motif was distinct from other signals within the CTD that direct membrane binding of ULK1 and ULK2 and incorporation into large protein complexes.
To explore other mechanisms involved in the function of the CTD, we identified a putative human Atg13 orthologue that we confirmed is essential for autophagy by using siRNA depletion. In addition, we found that loss of human Atg13 affected the trafficking of mAtg9, as was previously observed after the loss of ULK1 (37). Although mAtg13 bound the CTDs of both ULK1 and ULK2, this interaction utilized sequences that were distinct from the dominant-negative 7-residue motif, and overexpression of Atg13 did not rescue the dominant-negative activity of the overexpressed CTDs. Thus, our data have identified a 7-residue motif in the ULK1 and ULK2 CTDs that engages a novel dominant-negative mechanism which is independent of Atg13 and membrane-associated components.
MATERIALS AND METHODS
Cell culture.
HEK293A cells and their derivative cell line stably expressing enhanced green fluorescent protein (EGFP)-rat LC3 (293/GFP-LC3) have been described previously (3). Cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, which also served as the full nutrient medium in the analytical experiments. Starvation medium consisted of Earle's balanced saline solution (EBSS), containing 0.5 mM leupeptin where indicated.
DNA constructs.
Expression plasmids for N-terminal Myc epitope-tagged wild-type and kinase-dead (K46R) murine ULK1 (ULK1-K46R) have been described previously (3). ULK2 containing an N-terminal Myc tag was constructed by PCR from a full-length clone containing murine ULK2 (IMAGE:5709559) as the template. The CTDs of ULK1 and -2 were amplified from full-length cDNAs using PCR and cloned with an N-terminal Myc tag into a modified pcDNA3.1 or pRK5 backbone, respectively. Substitutions in the kinase domains of ULK1 and -2 were generated using a PCR site-directed mutagenesis kit (Stratagene), and additional deletion constructs were generated by isolating internal regions using PCR. We obtained a full-length cDNA containing the human putative Atg13 homolog KIAA0652 (pF1KSDA0652; Kazusa DNA Research Institute, Kisarazu, Japan). A C-terminal FLAG epitope tag was added onto the Atg13 open reading frame using PCR and cloned into a modified pcDNA3.1 vector. For biochemical analyses, cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocols and analyzed 24 h later.
siRNAs and knockdown determination.
A pool of four siRNA duplexes (On-Target plus SMARTpool) targeting the putative human Atg13 (KIAA0652, Entrez nucleotide accession no. NM_014741) was obtained (Dharmacon). Two duplexes within this pool specific for KIAA0652 that were confirmed in multiple assays correspond to Duplex1 (5′-AGA CCA UCU UUG UCC GAA A-3′) (sense sequence 989 to 1007) and Duplex2 (5′-GAA GAA UGU CCG CGA GUU U-3′) (sense sequence 1398 to 1416). The siRNAs targeting human ULK1 have been described previously (3). Experimental and control siRNAs were transfected into HEK293 and 293/GFP-LC3 cells by use of a wet/reverse-knockdown protocol in which trypsinized cells in suspension are plated directly into an siRNA-Lipofectamine 2000 (Invitrogen) mixture. Fresh medium is replenished 24 h before the day of the experiment.
Confirmation of siRNA knockdown by reverse transcription-PCR was carried out by assaying message RNA levels in cell samples following 72 h of knockdown, as described previously (3), using Sybr green real-time PCR. QuantiTect (Qiagen Biotech) primer sets were obtained for detection of the putative Atg13 (KIAA0652) transcript and the beta-actin control.
Immunoprecipitations and in vitro kinase assays.
Singly or doubly transfected cells were lysed in ice-cold TNTE (20 mM Tris, pH 7.5, 150 mM NaCl, 0.3% [vol/vol] Triton X-100 [TX-100], 5 mM EDTA) containing Complete EDTA-free protease inhibitor cocktail (Roche) and PhosStop phosphatase inhibitor cocktail (Roche). Lysates cleared by centrifugation were incubated with protein G-Sepharose beads coupled to anti-Myc monoclonal 9E10 or anti-FLAG monoclonal M2 (Sigma-Aldrich) antibody for 1 h and then washed three times with TNTE. For coimmunoprecipitation studies, proteins were eluted using 3× sodium dodecyl sulfate (SDS) sample buffer (with heating to 65°C for 5 min) and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
For in vitro kinase assays, following the three TNTE washes, immunoprecipitates were further washed once with kinase reaction buffer (KRB) (20 mM HEPES, pH 7.5, 20 mM MgCl2, 25 mM beta-glycerophosphate, 2 mM dithiothreitol, 100 μM sodium orthovanadate) and then incubated at a final volume of 20 μl in KRB containing 20 μM ATP, 5 μg myelin basic protein (MBP), and 5 μCi [γ-32P]ATP at 30°C for 15 min. For mixed-bead kinase reactions, immunoprecipitates of ULK1 and -2 and Atg13-FLAG were combined after the TNTE wash step, further washed in KRB, and then incubated in a 20-μl final volume of KRB containing 20 μM ATP at 30°C for 30 min. Reactions were stopped with the addition of 3× SDS sample buffer during heating to 65°C for 5 min. [32P]-labeled reaction products were resolved on NuPAGE 4 to 12% bis-Tris gels in morpholinepropanesulfonic acid (MOPS)-SDS running buffer (Invitrogen). Mixed-bead reaction products were resolved by Laemmli SDS-PAGE. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes and then analyzed using a phosphorimager scanner and anti-Myc immunoblotting.
Immunoblotting.
To detect GFP-LC3 lipidation, 293/GFP-LC3 cells were lysed following various treatments in 1× SDS sample buffer. Lysates were then heated to 65°C for 5 min and passed through a 27-gauge needle five times to reduce viscosity before analysis by 10% Laemmli SDS-PAGE. To detect lipidation of endogenous LC3, HEK293A cells were lysed following treatments in ice-cold TNTE containing Complete EDTA-free protease inhibitor cocktail (Roche). Lysates cleared by centrifugation were mixed with 3× SDS sample buffer, heated to 100°C for 5 min, and then analyzed on either 12% Laemmli SDS-PAGE gels or 4 to 12% bis-Tris NuPAGE gels (morpholineethanesulfonic acid [MES]-SDS running conditions) (Invitrogen). Proteins were transferred to PVDF membranes. GFP-LC3 and endogenous LC3 were detected with 5F10 anti-LC3 monoclonal antibody (Nanotools, Teningen, Germany). Additional immunoblotting antibodies included anti-ULK1 polyclonal H-240 (Santa Cruz), anti-Myc 9E10 monoclonal, β-tubulin polyclonal (6046; Abcam, Cambridge, United Kingdom), anti-FLAG M2 monoclonal (Sigma-Aldrich), and antiactin monoclonal AC-40 (Sigma-Aldrich). Rabbit polyclonal antibodies toward human Atg13 (KIAA0652) were generated using the peptide sequence LAVHEKNVREFDAFVETLQ. Following incubation with primary antibodies, signals were detected and quantified using secondary antibodies coupled to infrared chromophores and a two-channel scanning method (Licor Odyssey), as described previously (3). Statistical analyses for various pairwise comparisons were performed using Student's two-tailed t test on sample sets with equal variances.
Limited proteolysis.
Cells were lysed in ice-cold TNT (50 mM Tris, pH 8.0, 150 mM NaCl, 0.5% TX-100) (without protease inhibitors) and then incubated with protein G-Sepharose beads coupled to 9E10 anti-Myc monoclonal antibody for 1 h. Immunoprecipitates were washed three times with TNT and then incubated in a 20-μl final reaction volume of TNT containing the concentrations of sequencing-grade chymotrypsin (Roche) indicated in Fig. 3 on ice for 15 min. Reactions were stopped (addition of 3× SDS sample buffer and heating to 100°C) and analyzed by SDS-PAGE and anti-Myc immunoblotting.
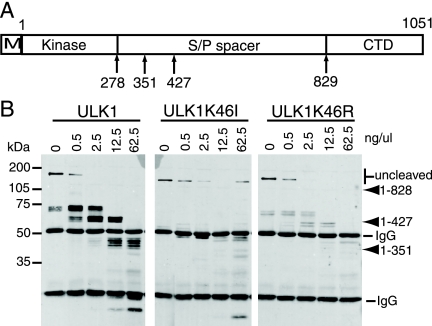
FIG. 3.
Differential sensitivities of ULK1 kinase mutants to limited proteolysis. (A) Schematic showing the domain structure of ULK1, including the N-terminal Myc tag (M), the kinase domain, the serine- and proline-rich spacer domain (S/P spacer), and the conserved CTD. Positions of stop codons used by Chan et al. (3) to generate various deletion mutants are indicated at the bottom. (B) ULK1 constructs transfected into 293A cells were immunoprecipitated and then incubated with the indicated concentrations of chymotrypsin for 15 min on ice. Reaction products were resolved on SDS-PAGE gels and detected by anti-Myc immunoblotting. Molecular mass markers are indicated on the left. The relative mobilities of the uncleaved proteins, previously described ULK1 truncation mutants, and the heavy and light immunoglobulin G (IgG) chains are shown on the right.
Long-lived protein degradation.
Autophagy-dependent degradation of [14C]valine-labeled cellular proteins in transfected 293/GFP-LC3 cells was measured as previously described (3).
Membrane association.
Following transfection and treatments as indicated, 293/GFP-LC3 cells were scraped into ice-cold homogenization buffer (HB) (20 mM HEPES, pH 7.2, 1 mM EGTA, 5 mM MgCl2, 150 mM KCl) containing Complete EDTA-free protease and PhosStop phosphatase inhibitors (Roche). Cells were disrupted by being passed 20 times through a 27-gauge needle and then cleared using a low-speed (1,000 × g) centrifugation. These cleared lysates were then centrifuged at 100,000 × g for 1 h (4°C) to obtain a crude membrane pellet and the resulting supernatant fraction. Where indicated, membrane pellets were further washed/extracted in HB containing either high salt concentrations or 1% TX-100 and then recentrifuged at 100,000 × g. Membrane pellets were finally resuspended in HB containing 1% TX-100. Membrane and supernatant fractions were mixed with 3× SDS sample buffer, heated to 65°C for 5 min, and resolved by SDS-PAGE for immunoblotting.
Native gel electrophoresis.
Following transfection and treatments as indicated, HEK293A cells were lysed in ice-cold 1× native PAGE sample buffer (Invitrogen) containing 1% TX-100 and Complete EDTA-free protease and PhosStop phosphatase inhibitors. Cell lysates were precleared by centrifugation and then mixed with Coomassie G-250 dye additive (0.25% final) (Invitrogen) before being run on 3 to 12% Novex NativePAGE bis-Tris gels (Invitrogen) according to the manufacturer's protocols. Resolved proteins were transferred to PVDF membranes and detected with an anti-Myc monoclonal antibody, horseradish peroxidase-coupled secondary antibodies, and chemiluminescence.
Microscopic analysis of cell structures.
For analysis of GFP-LC3-labeled autophagosomes using a Cellomics ArrayScan VTI high-content screening system (Thermo Scientific), 293/GFP-LC3 cells were seeded into 96-well tissue culture plates coated with poly-d-lysine-containing Lipofectamine-siRNA mixtures. Seventy-two hours after transfection, cells were starved, fixed, and analyzed using an automated calibrated quantitation algorithm (spot total intensity per object) (Thermo Scientific).
For confocal analyses, 293/GFP-LC3 cells or parental HEK293 cells were either plated onto glass coverslips and transfected with expression constructs or plated onto glass coverslips in an siRNA-Lipofectamine mixture. Following various treatments, cells were fixed and then further stained using 9E10 monoclonal Myc antibody, M2 monoclonal FLAG antibody, or a hamster anti-Atg9 monoclonal antibody generated by immunizing animals with a previously described C-terminal peptide (37). Samples were further analyzed using a confocal laser scanner fitted onto a Zeiss Axioplan 2 microscope.
RESULTS
Kinase function of ULK1.
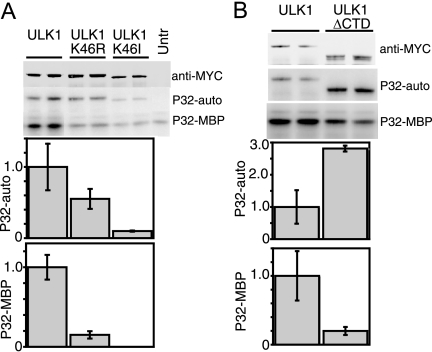
It was recently shown that in NIH 3T3 cells kinase-inactivated ULK1-K46N had dominant-negative properties on starvation-induced autophagy (6). Our previous data using a HEK293 cell system showed that the kinase-inactivated ULK1-K46R mutant behaved similarly to wild-type ULK1 in modulating autophagy (3). Our results also contrasted with the reported dominant-negative properties of ULK1-K46R in neurite outgrowth assays (32). Since ULK1-K46R displayed normal electrophoretic mobility (3), we hypothesized that ULK1-K46R had remaining kinase activity and we generated an additional kinase-inactivated mutant, ULK1-K46I. We examined the effects of these mutations by assaying the efficiency of immunoprecipitated ULK1 proteins to undergo autophosphorylation and to phosphorylate the generic substrate MBP. The K46R substitution inactivated these in vitro kinase activities only partially, while the K46I mutation produced stronger inactivation (Fig. 1A).
FIG. 1.
In vitro kinase activities of ULK1 mutants. (A and B) HEK293A cells were transfected with Myc-tagged ULK1 constructs, lysed, and used for immunoprecipitation reactions. Immunoprecipitates were incubated in an in vitro kinase reaction mixture containing [32P]ATP and the generic substrate MBP. Reaction products were resolved on SDS-PAGE gels (top). Anti-Myc immunoblotting was performed as a control to quantify the amount of ULK1 protein precipitated. Phosphorimager analysis detected levels of ULK1 autophosphorylation (P32-auto) and MBP phosphorylation (P32-MBP). Specific activities (in relative units) are expressed as the amount of phosphorylation normalized to the amount of Myc-tagged ULK1 per reaction (corrected for background activities detected in a parallel control reaction performed with untransfected cells [Untr] [not shown in panel B]). Each bar represents results from three independent samples ± standard deviations, and each experiment was reproduced a minimum of two times.
Kinase function in ULK1ΔCTD.
We previously demonstrated that deletion of the ULK1 CTD generates a potent dominant-negative molecule, ULK1ΔCTD (3). We questioned whether the CTD modulated kinase activity, possibly via an intramolecular interaction. We observed that deletion of the CTD in ULK1 increased autophosphorylation levels (approximately 2.8-fold), while phosphorylation of MBP was reduced approximately 80% following deletion of the CTD (Fig. 1B). From these data, we speculated that loss of the CTD produced a kinase with an altered conformation in which the kinase domain has increased accessibility to the spacer domain for autophosphorylation and a decreased ability to phosphorylate substrates.
Kinase function in full-length ULK1.
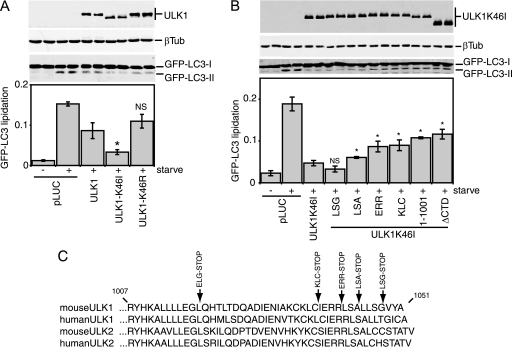
Since our in vitro data indicated that the K46R and K46I substitutions had various severities, we retested these mutants for their effect on starvation-induced autophagy in 293/GFP-LC3 cells (3). In this cell system, we can assess autophagy activation by measuring the conversion of its unmodified (GFP-LC3-I) form to its modified, lipidated species (GFP-LC3-II). Strikingly, we observed that ULK1-K46I exhibited strong dominant-negative properties compared to both the wild-type control and ULK1-K46R (Fig. 2A). In addition, ULK1-K46I displayed faster electrophoretic mobility, consistent with decreased autophosphorylation in vivo, than wild-type ULK1 and ULK1-K46R. These data demonstrate that strong inactivation of catalytic activity is required to transform ULK1 into a dominant-negative protein.
FIG. 2.
Inhibition of GFP-LC3 lipidation by ULK1-K46I. (A and B) 293/GFP-LC3 cells were transfected with a control plasmid expressing luciferase (pLUC) or the indicated Myc-tagged ULK1 constructs. Twenty-four hours after transfection, cells were either left alone or starved for 2 h in EBSS containing leupeptin and lysed for SDS-PAGE analysis of GFP-LC3 lipidation (conversion of GFP-LC3-I to GFP-LC3-II) by use of anti-LC3 antibodies. The membrane was also probed with anti-Myc and -β-tubulin (βTub) antibodies. GFP-LC3 lipidation is quantified as follows: GFP-LC3-II/(GFP-LC3-I + GFP-LC3-II). Each bar represents results from three independent samples ± standard deviations. (A) *, P < 0.05; NS, P = 0.20 (in pairwise comparisons with wild-type ULK1). (B) *, P < 0.05; NS, P = 0.18 (in pairwise comparisons with full-length ULK1-K46I). (C) Alignments show C-terminal amino acid residues from CTDs of ULK1 and ULK2. Positions of stop codons used to generate various C-terminal truncations are indicated at the top. ULK1(1-1001) is missing its C-terminal 50 residues; ΔCTD is a deletion of the entire CTD and corresponds to amino acids 1 to 828 of ULK1. The EGL-STOP mutant is further studied in Fig. 6F.
To understand further this dominant inhibitory effect, we performed a deletion analysis of ULK1-K46I. We determined that ULK1-K46I lost approximately 50% of its potency to inhibit GFP-LC3-II upon deletion of its CTD (Fig. 2B). An equivalent loss of potency was seen with ULK1-K46I lacking the C-terminal 50 amino acids [ULK1-K46I(1-1001)]. Finer mapping determined that dominant-negative potency was similarly lost after the deletion of 10 residues from the ULK1 C terminus (ERR-STOP mutant). However, deletion of the PDZ binding motif from ULK1-K46I (LSG-STOP) had no effect on dominant-negative function. Thus, the ability of kinase-inactive, nonautophosphorylated ULK1 to inhibit autophagy depended upon a small region just inside its C terminus. In conjunction with our in vitro kinase data showing altered autophosphorylation efficiency following CTD deletion, we proposed a working model in which the CTD is normally folded back to keep the protein in a more closed conformation. Full kinase inactivation of ULK1 leads to a loss of autophosphorylation and a conformational change that causes the CTD to be abnormally exposed, revealing cryptic binding sites within the CTD that then recruit and titrate out other essential autophagy proteins.
Limited proteolysis reveals a conformational change in ULK1-K46I.
To probe for possible conformational changes, we performed limited proteolysis on immunoprecipitated ULK1 compared to ULK1-K46I and ULK1-K46R. Treatment of immunoprecipitated wild-type Myc-ULK1 with low concentrations of chymotrypsin resulted in the loss of the full-length protein and generated distinct protected fragments ranging between 60 and 70 kDa in apparent molecular mass (Fig. 3B). Using the relative mobilities of previously described ULK1 deletion constructs, such as ULK1ΔCTD [ULK1(1-828)], ULK1(1-427), and ULK1(1-351), for comparison (Fig. 3A), we observed that the larger predominant digestion products are approximately 420 to 500 residues in length. Digestion of wild-type Myc-ULK1 with higher concentrations of enzyme generated smaller protected digestion products of approximately 350 to 380 residues. Digestion of Myc-ULK1-K46I also resulted in the loss of the full-length protein. However, distinct protected fragments were not detected efficiently, which is consistent with the idea that internal regions of the kinase-inactivated mutant were more accessible to protease. Digestion of Myc-ULK1-K46R yielded an intermediate phenotype, suggesting that the closed molecular conformation of our working model is altered only mildly when the kinase is partially inactivated.
Kinase inactivation in full-length ULK2.
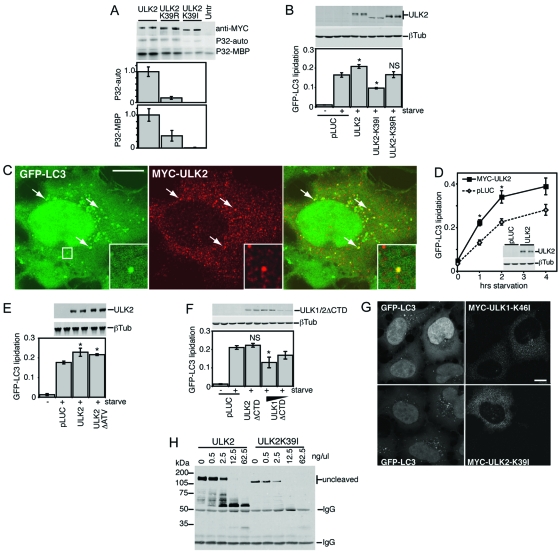
Hara et al. (6) recently reported that kinase-inactivated ULK2 also exhibited dominant-negative properties on autophagy. Given our observed differential effects of kinase inactivation in the context of ULK1, we performed a similar analysis using ULK2. We confirmed that the mutation of K39 in ULK2 to R or I resulted in the partial or full inactivation, respectively, of in vitro autophosphorylation and MBP phosphorylation activities (Fig. 4A). We next tested the effects of wild-type ULK2 and these kinase mutants on starvation-induced autophagy in 293/GFP-LC3 cells (Fig. 4B). Surprisingly, overexpression of wild-type ULK2 enhanced the levels of GFP-LC3-II following amino acid starvation. This enhancing effect was not seen with the ULK2-K39R mutant (which contains partially inactivated kinase activity). In agreement with our ULK1 results, ULK2-K39I produced strong inhibitory effects on starvation-induced GFP-LC3 lipidation. Strong and weak effects on autophosphorylation could also be observed as differences in electrophoretic mobilities for overexpressed ULK2-K39I and -K39R, respectively (Fig. 4B).
FIG. 4.
Autophagy regulatory roles of wild-type and kinase-dead ULK2. (A) In vitro autophosphorylation and MBP kinase activities for ULK2 constructs were measured as described in the legend for Fig. 1. (B, D, E, and F) Modulation of GFP-LC3 lipidation by ULK constructs was detected as described in the legend for Fig. 2. (B) *, P < 0.05; NS, P = 0.94 (in pairwise comparisons with pLUC-transfected, starved cells). (D) *, P < 0.03 in pairwise comparisons with pLUC-transfected cells at the same time point. (E) *, P < 0.06 in pairwise comparisons with pLUC-transfected, starved cells. (F) *, P < 0.02; NS, P = 0.42 (in pairwise comparisons with pLUC-transfected, starved cells). (C) In 293/GFP-LC3 cells starved in EBSS-leupeptin for 2 h, Myc-tagged ULK2 could be observed on multiple cytoplasmic structures, a portion of which colocalized with GFP-LC3, as indicated by arrows and the boxed inset. Bar = 10 μm. (G) Myc-tagged ULK1-K46I or ULK2-K39I inhibited the formation of cytoplasmic GFP-LC3-labeled autophagosomes in starved 293/GFP-LC3 cells. Bar = 10 μm. (H) Limited proteolysis of wild-type ULK2 and ULK2-K39I, assayed as described in the legend for Fig. 3. Untr, untransfected; P32-auto, ULK1 autophosphorylation; P32-MBP, MBP phosphorylation; βTub, β-tubulin; IgG, immunoglobulin G.
Analysis of ULK2 reveals both differences and similarities to ULK1.
Consistently with recent observations showing the localization of ULK2 on Atg16L-positive-forming autophagosomes (also termed isolation membranes, or phagophores) (6) and our results with ULK1 (3), we could detect a proportion of ULK2 on GFP-LC3-labeled autophagosomes in starved 293/GFP-LC3 cells (Fig. 4C). Overexpressed ULK2 did not promote GFP-LC3 lipidation under full nutrient conditions (zero time point) but appeared to enhance the response following amino acid starvation (Fig. 4D). Using deletion analysis, we observed that ULK2 lacking its C-terminal three residues (ATV) similarly enhanced starvation-induced GFP-LC3 lipidation (Fig. 4E). Thus, while deletion of the C-terminal PDZ motif modulated ULK1 function (3), residues at the extreme C terminus of ULK2 were not required to enhance GFP-LC3 lipidation. A ULK2ΔCTD mutant had no effect on GFP-LC3 lipidation, in contrast with ULK1ΔCTD (Fig. 4F), which has dominant inhibitory properties (3). These data on wild-type and ΔCTD deletion mutants indicate that overexpression of ULK1 and ULK2 may have distinct effects on autophagy. In contrast, kinase-inactivated forms of ULK1 and ULK2 are both strong dominant-negative inhibitors of autophagy, as evidenced by their ability to block starvation-induced GFP-LC3 puncta (Fig. 4G). Finally, similarly to ULK1, limited proteolysis of immunoprecipitated wild-type Myc-ULK2 generated protected fragments of roughly 60 to 75 kDa in apparent molecular mass (Fig. 4H). Immunoprecipitated Myc-ULK2-K39I could also be digested by chymotrypsin (note the loss of the full-length protein), but smaller protected fragments did not accumulate, consistent with our model that kinase-inactivated ULK2, like ULK1, has an altered protease-sensitive conformation.
Dominant-negative activity of the ULK CTDs depends on a short conserved sequence within the C terminus.
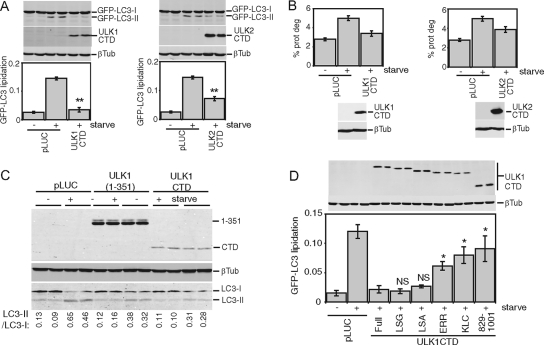
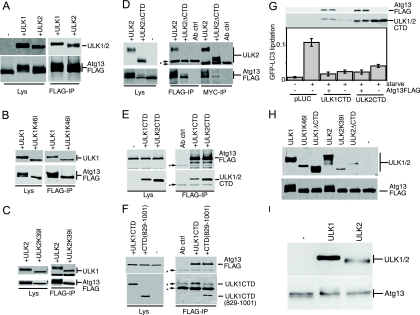
In our model, as a result of a decreased kinase activity, an abnormally exposed CTD engages in uncontrolled interactions with essential autophagy regulatory components, thereby halting the pathway. Supporting this model, overexpression of the CTD alone from either ULK1 (amino acids 829 to 1051) or ULK2 (amino acids 811 to 1037) resulted in a strong dominant-negative effect on starvation-induced GFP-LC3 lipidation (Fig. 5A). The CTD of ULK1 or ULK2 also inhibited starvation-induced protein degradation (Fig. 5B). Overexpression of the ULK1 CTD inhibited lipidation of endogenous LC3 to similar extents, as previously described for N-terminal ULK1 dominant-negative fragments (3), such as ULK1(1-351) (Fig. 5C).
FIG. 5.
CTDs of ULK1 and ULK2 inhibit autophagy. (A) Control (pLUC) and expression constructs for Myc-tagged ULK1 and ULK2 CTDs were transfected into 293/GFP-LC3 cells. Inhibition of starvation-induced GFP-LC3 lipidation was measured as described in the legend for Fig. 2. **, P < 0.006 in pairwise comparisons with pLUC-transfected, starved cells. (B) 293/GFP-LC3 cells transfected as described for panel A were labeled overnight with [14C]valine. Cells were then either left untreated or starved in EBSS for 2 h, and autophagic degradation of long-lived proteins (prot deg) was analyzed. Cell samples transfected in parallel were used as a control to detect overexpressed constructs. Each bar represents the average from three independent samples ± the standard deviation. (C) HEK293A cells were transfected for 24 h with control pLUC plasmid, a Myc-tagged ULK1(1-351) deletion mutant as an internal control, or Myc-tagged ULK1 CTD. Cells were then left untreated or starved in EBSS-leupeptin for 2 h and then lysed for SDS-PAGE analysis of LC3 lipidation. The ratio of LC3-II/LC3-I measured for each sample is indicated at the bottom. (D) Myc-tagged ULK1 CTD constructs with C-terminal truncations were transfected into 293/GFP-LC3 cells, and GFP-LC3 lipidation was measured as described in the legend for Fig. 2. *, P < 0.05; NS, P > 0.4 (in pairwise comparisons with full-length-CTD-transfected, starved cells). A schematic of the truncations is shown in Fig. 2. βTub, β-tubulin.
To investigate this dominant-negative mechanism, we performed a deletion analysis of the ULK1 CTD. Similarly to observations from a deletion analysis of full-length ULK1-K46I, removal of the PDZ binding motif (LSG-STOP) and the next 4 amino acid residues (LSA-STOP) had no effect on the dominant-negative potency of the CTD (Fig. 5D). However, removal of the C-terminal 10 residues (ERR-STOP) began to rescue the dominant-negative activity, and further truncation (KLC-STOP) produced a stronger reversion of the dominant-negative activity. This region contains a 7-residue motif (IERRLSA) that is entirely conserved between the CTDs of ULK1 and ULK2 from human and mouse (Fig. 2C). Using the CTD of ULK2 (which is likely more ancestral than ULK1 [E. Y. W. Chan and S. A. Tooze, unpublished observations]) as a reference, we determined that the IERRLSA motif is perfectly conserved in a predicted avian ULK2 orthologue but displays clearly less conservation in the predicted ULK2 orthologues of fish and is unrecognizable in the Atg1 orthologues of D. melanogaster and C. elegans. Thus, our deletion analysis demonstrated that the ability of the ULK1/2 CTDs to act as dominant-negative molecules is highly dependent upon a well-conserved 7-residue motif at the C terminus.
Membrane binding signal within the CTD.
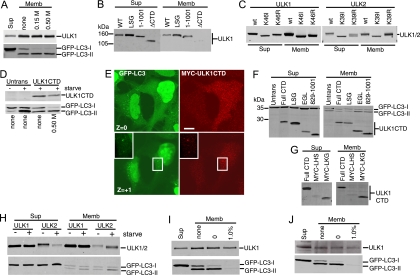
We next aimed to elucidate the dominant-negative mechanism of the ULK CTDs, which by deletion analysis appeared to require the same amino acid residues as kinase-inactivated ULK1 and -2. Our previous data showed that localization of ULK1 to GFP-LC3-labeled structures was dependent on the CTD (3), so we investigated the relationship between dominant-negative activity and membrane association by using biochemical assays. We first confirmed that approximately 30% of the total overexpressed Myc-tagged ULK1 was associated with the crude membrane pellet in starved 293/GFP-LC3 cells (Fig. 6A), which was consistent with previous fractionation data from brain extracts (33). For control purposes, we observed that lipidated GFP-LC3-II was associated exclusively with crude membranes (relative to the soluble supernatant). Some GFP-LC3-I was also detected on the membranes, but this association could be disrupted by an additional salt wash of the pellet. Importantly, ULK1 and GFP-LC3-II were stably membrane bound even after a 0.5 M salt wash. We next confirmed that the association of ULK1 to crude membrane fractions was lost upon deletion of the CTD (Fig. 6B). However, deletion of the PDZ binding motif (LSG-STOP) or deletion of the last 50 amino acids [ULK1(1-1001)] of ULK1 did not disrupt membrane binding. Thus, ULK1 could still associate with membranes even after removal of C-terminal residues that are required for dominant-negative activity, suggesting that these functions were independent. Supporting the idea that altered membrane binding was not involved in the dominant-negative mechanism, kinase-inactivated forms of ULK1 and ULK2 were detected in the membrane fraction with an efficiency similar to that of wild-type proteins (Fig. 6C).
FIG. 6.
Membrane targeting signal within the CTDs of ULK1 and ULK2. (A) 293/GFP-LC3 cells were transfected with Myc-ULK1 and starved in EBSS-leupeptin for 2 h. Cell homogenates were centrifuged at 100,000 × g to isolate membrane (Memb) and supernatant (Sup) fractions. Aliquots of the supernatant (representing 2% of the cell sample) and membrane (representing 10% of the cell sample) fractions were analyzed by SDS-PAGE. The top half of the blot was probed with anti-Myc and the bottom with anti-LC3. Where indicated, the membrane fraction was washed with HB containing 0.15 M or 0.5 M KCl. (B and C) Myc-tagged ULK1 and -2 constructs were transfected into cells and analyzed for membrane association as described above. (D) After transfection with Myc-tagged ULK1 CTD, cells were left untreated or starved in EBSS-leupeptin for 2 h. Where indicated, the membrane pellet was washed in HB containing 0.5 M KCl and analyzed as described above. (E) 293/GFP-LC3 cells were transfected with Myc-ULK1 CTD for 24 h and then starved in EBSS-leupeptin for 2 h before fixation and immunostaining with anti-Myc monoclonal antibody. In a Z section close to the substratum (Z = 0), ULK1 CTD strongly inhibited GFP-LC3 punctum formation. In the Z + 1 section, Myc-ULK1 CTD could be detected colocalizing with GFP-LC3-positive structures (inset). Bar = 10 μm. (F) Membrane associations of ULK1 CTD (Full CTD) and various C-terminal deletion constructs in starved 293/GFP-LC3 cells. See the schematic in Fig. 2 for the positions of inserted stop codons. The expression of ULK1 CTD was detected with anti-Myc, and GFP-LC3 was detected with anti-LC3. (G) Membrane associations of ULK1 CTD and two N-terminal deletion constructs, Myc-LHS (845-1051) and Myc-LKG (864-1051), in starved 293/GFP-LC3 cells. (H) 293/GFP-LC3 cells were transfected with full-length Myc-ULK1 or -ULK2 and then left untreated or starved in EBSS-leupeptin for 2 h before isolation of supernatant and membrane fractions. Expressed proteins were detected as described above. (I) Membrane association of transfected Myc-ULK1 in starved 293/GFP-LC3 cells. Membrane pellets were analyzed before washes (none) or following extraction in HB (0) or HB supplemented with 1.0% TX-100. (J) Membrane association of endogenous ULK1 was analyzed in untransfected, starved 293/GFP-LC3 cells following the fractionation and wash procedures described for panel I. WT, wild type; Untrans, untransfected.
Since deletion of the entire CTD largely abolished membrane association, we questioned whether this region alone could bind membranes. Indeed, the CTD of ULK1 could be detected in the membrane pellets derived from extracts of starved 293/GFP-LC3 cells, and this association was salt resistant (Fig. 6D). As expected, an overexpressed ULK1 CTD reduced the amount of membrane-associated GFP-LC3-II. Expression of the ULK1 CTD clearly inhibited the formation of starvation-induced GFP-LC3-positive autophagosomes (Fig. 6E). Besides existing as a diffuse cytosolic pool, the ULK1 CTD could be observed on discreet puncta that colocalized with the remaining GFP-LC3 puncta, consistent with the idea that the CTD contains autophagosomal targeting signals. Membrane association was also confirmed biochemically for the ULK2 CTD (data not shown).
In agreement with deletion analysis of full-length ULK1, the last 50 residues of the ULK1 CTD C terminus were not required for membrane binding (Fig. 6F), suggesting that membrane targeting signals were distinct from the region involved in dominant-negative interactions. To map further the minimal membrane targeting region, we analyzed the hydropathy profile of the ULK1 CTD to identify regions amenable for deletion. Up to 35 amino acid residues (containing both a hydrophobic and a hydrophilic stretch) (MYC-LKG) could be removed from the N terminus of the ULK1 CTD without disruption of membrane binding (Fig. 6G). Thus, deletion analysis of the CTD N and C termini has further defined the membrane association region to amino acids 864 to 1001.
Although membrane binding did not appear to be critically involved in dominant-negative activity, we explored whether membrane association of ULK proteins was regulated upon induction of autophagy. Overexpressed ULK1 binding to membranes was unaltered following amino acid starvation (Fig. 6H). In contrast, overexpressed ULK2 exhibited both faster electrophoretic mobility and increased membrane association following amino acid starvation. These data suggest that, upon autophagy induction, ULK2 becomes partially dephosphorylated and more strongly associated with autophagosomal membranes, providing more evidence that ULK1 and ULK2 may have distinct roles in autophagy.
Further investigation unexpectedly revealed that the association of ULK1 to membrane fractions was resistant to extraction with 1% TX-100 (in contrast to lipidated GFP-LC3-II, which was no longer membrane bound) (Fig. 6I). This tight association of ULK1 to detergent-resistant membranes was confirmed for endogenous ULK1 (Fig. 6J). Note that membrane binding of endogenous ULK1 was not altered following amino acid starvation (data not shown).
ULK1 and -2 form distinct protein complexes.
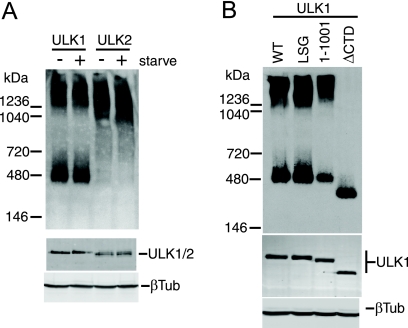
Since altered membrane binding did not appear to explain the dominant-negative effects of the ULK1/2 CTD or kinase-dead proteins, we investigated whether these mutant ULK proteins displayed altered protein complex formation. Using native polyacrylamide gels, we detected overexpressed ULK1 in two major species: a smaller population with an apparent molecular mass of 400 to 500 kDa, and a larger complex migrating with an apparent molecular mass of >1,200 kDa (Fig. 7A). By comparison, overexpressed ULK2 in native gels was detected primarily as one protein species migrating at >1,000 kDa. Although these protein complexes were not altered following amino acid starvation, these results suggest that ULK1 and ULK2 enter into distinct molecular complexes. Interestingly, and in contrast with wild-type ULK1, kinase-inactivated ULK1-K46I and ULK1-K46R existed primarily as a larger 1,200-kDa species (data not shown), raising the possibility that altered protein complexes were related to our dominant-negative mechanism. However, ULK1 lacking its C-terminal 50 residues still entered into two molecular complexes, similarly to the wild type, indicating that the C-terminal dominant-negative IERRLSA motif was not involved in these molecular interactions (Fig. 7B). Further deletion of the entire CTD prevented ULK1 from entering into the high-molecular-mass complex, indicating a requirement for sequences within the CTD between amino acids 829 and 1001.
FIG. 7.
Molecular complexes of ULK1 and ULK2 detected using native PAGE. HEK293A cells were transfected with ULK1 and ULK2 and starved for 2 h in EBSS where indicated (A) or ULK1 constructs (B) before lysis in native gel sample buffer. Complexes were resolved on native PAGE gels and then detected by immunoblotting with anti-Myc antibody (top). Positions of native-gel molecular mass markers are indicated on the left. As a control, aliquots of lysates were resolved by conventional SDS-PAGE to detect overexpressed proteins (anti-Myc) and total protein (β-tubulin [βTub]) (bottom). WT, wild type.
Putative human homologue of Atg13.
Our deletion analyses indicated a requirement of amino acid residues within the CTD C terminus for dominant-negative activity. Recent findings have shown that C-terminal residues of yeast Atg1 direct binding to Atg13 (5). Thus, we questioned whether an Atg13 orthologue was involved in the dominant inhibitory mechanism of the ULK1/2 CTDs. Using both Schizosaccharomyces pombe and Saccharomyces cerevisiae Atg13 protein sequences, we searched the genomes of higher organisms using repetitive rounds of PSI-BLAST for an mAtg13 orthologue. In the first round of iteration, we identified a putative orthologue in D. melanogaster (CG7331) in addition to orthologues in mice (Entrez protein database accession no. NP_663503.1) and humans (Entrez protein database accession no. NP_055556.2/KIAA0652), which have both been annotated as the gene products of harbinger transposase derived 1 (HARBI1). Harbi1 (Entrez protein database accession no. AAH02378) had previously been identified as the putative human Atg13 orthologue (18). The D. melanogaster CG7331 protein was found in a systematic characterization of protein interactions to bind D. melanogaster Atg1 (28), supporting its role as a putative Atg13 orthologue. Bioinformatic analysis of current genome information revealed that the putative vertebrate Atg13 orthologues are tightly conserved (94% amino acid identity for mouse versus human), while the putative orthologues in D. melanogaster and C. elegans are clearly divergent.
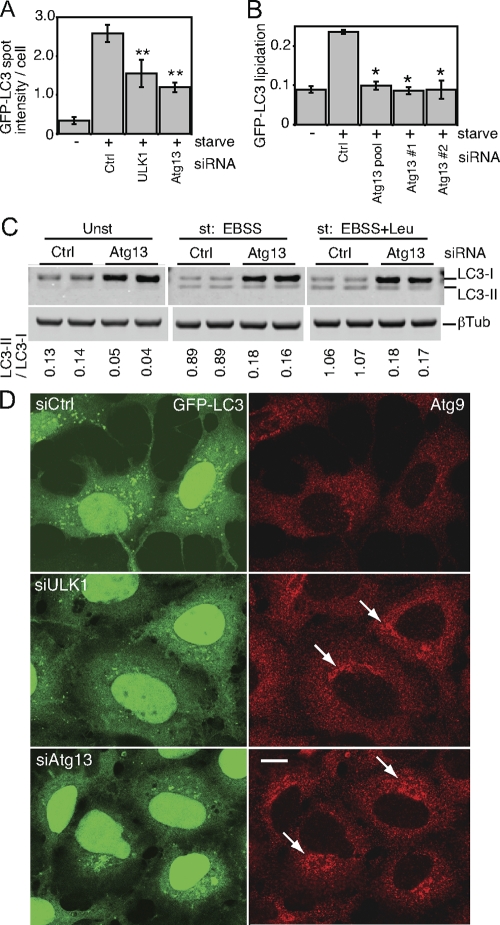
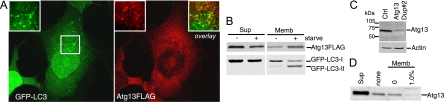
To confirm the role of this putative Atg13 orthologue in autophagy, we obtained an siRNA pool targeting the human transcript NM_014741. In 293/GFP-LC3 cells, knockdown of Atg13 inhibited the formation of starvation-induced GFP-LC3 punctum formation to an extent similar to that of ULK1 knockdown (Fig. 8A). We identified two individual siRNA duplexes within this pool that could be confirmed to knock down Atg13 mRNA (data not shown) and inhibit starvation-induced lipidation of GFP-LC3 (Fig. 8B). We further confirmed that knockdown of Atg13 in parental HEK293 cells inhibited lipidation of endogenous LC3 during unstarved and starvation conditions, both without and with lysosomal protease inhibition by leupeptin (Fig. 8C).
FIG. 8.
A putative human Atg13 homologue is required for autophagy. (A) 293/GFP-LC3 cells were transfected with control siRNA (Ctrl) or siRNA targeting ULK1 or a SMARTpool targeting KIAA0652 (Atg13) for 72 h. Cells were then left unstarved or starved in EBSS-leupeptin for 2 h before fixation and morphological analysis. The quantification represents the total GFP-LC3 spot intensity per cell (arbitrary units), and each bar represents the average of 60 image fields (from six independent cell samples) ± the standard deviation. **, P < 0.0007 in pairwise comparisons with Ctrl knockdown starved cells. (B) 293/GFP-LC3 cells were transfected for 72 h with control siRNA, the Atg13 SMARTpool, or individual duplexes (#1 or #2) specific for Atg13. Cells were left unstarved or starved as described for panel A and then lysed for immunoblot analysis of GFP-LC3 lipidation, as described in the legend for Fig. 2. *, P < 0.02 in pairwise comparisons with siRNA Ctrl starved cells. (C) HEK293A cells were transfected with control siRNA or Atg13 duplex 2 for 72 h. Cells were then left unstarved (Unst) or starved (st) for 2 h in EBSS (with or without leupeptin [Leu]) before lysis for immunoblot analysis of endogenous LC3 lipidation. The quantified ratio of LC3-II/LC3-I is shown below each sample. βTub, β-tubulin. (D) 293/GFP-LC3 cells were transfected for 72 h with control siRNA (siCtrl) or siRNAs targeting ULK1 (siULK1) or Atg13 (duplex 2) (siAtg13), starved in EBSS-leupeptin for 2 h, and then fixed and immunostained to detect endogenous mAtg9 localization (red). After starvation in siCtrl-treated cells, mAtg9 becomes redistributed predominantly to a diffuse cytoplasmic pool. After knockdown of Atg13, mAtg9 redistribution is blocked and mAtg9-positive vesicles remain in juxtanuclear clusters (arrows) to an extent similar to that after knockdown of ULK1. Inhibition of starvation-induced GFP-LC3 autophagosome formation after knockdown of ULK1 and Atg13 is shown as a control. Bar = 10 μm.
As an independent means to confirm the function of Atg13 in autophagy, we investigated its role in mAtg9 trafficking under starvation conditions (Fig. 8D). mAtg9 is a multispanning membrane protein required for autophagy, and after amino acid starvation or rapamycin treatment, mAtg9 redistributes from a juxtanuclear clustered pool to a dispersed peripheral cytosolic pool (37). We have previously shown that siRNA knockdown of ULK1 inhibits the starvation-induced redistribution of mAtg9 (37). Using this assay, we find that knockdown of human Atg13 similarly prevented the starvation-induced dispersal of mAtg9, which supports the proposed identification of KIAA0652 as human Atg13 and demonstrates the function of Atg13 in a ULK1-dependent autophagic pathway.
We expressed the putative human Atg13 orthologue with a C-terminal FLAG tag in 293/GFP-LC3 cells (Fig. 9A). Ectopically expressed Atg13-FLAG existed primarily as a diffuse cytoplasmic pool in both unstarved (data not shown) and amino acid-starved conditions. In some cells, Atg13-FLAG could be observed on small cytoplasmic puncta that were often juxtaposed, but not substantially colocalized, with GFP-LC3-labeled structures. Consistently with this immunofluorescence analysis, approximately 5% of Atg13-FLAG (migrating at a 70-kDa apparent molecular mass by Laemmli SDS-PAGE) was membrane associated (Fig. 9B) in unstarved conditions. Amino acid starvation enhanced the membrane association of Atg13-FLAG. We generated polyclonal antisera toward Atg13 and confirmed the loss of the endogenous protein following knockdown (Fig. 9C). Endogenous Atg13 bound to membranes, although this association was sensitive to 1% TX-100 extraction (Fig. 9D), which contrasts with the TX-100-resistant membrane association of ULK1.
FIG. 9.
Human Atg13 is membrane associated. (A) 293/GFP-LC3 cells expressing Atg13-FLAG were starved for 2 h in EBSS-leupeptin, fixed, and then immunostained using anti-FLAG antibody. Atg13-FLAG can be found both in a diffuse cytosolic pool and localized to punctate structures that do not colocalize with GFP-LC3. Bar = 10 μm. (B) Membrane association of Atg13-FLAG was analyzed as described in the legend for Fig. 6. (C) HEK293A cells were transfected with control or Atg13 duplex 2 (Dup#2) for 72 h. Cell lysates were then immunoblotted with a polyclonal antibody for endogenous Atg13 or actin as a loading control. (D) Membrane fractions analyzed as described for Fig. 6J from untransfected 293/GFP-LC3 cells were probed for endogenous Atg13. Sup, supernatant; Memb, membrane fraction; Ctrl, control.
Overexpression of Atg13 is not sufficient to rescue inhibition of autophagy caused by ULK dominant-negative proteins.
Data from yeast indicate that Atg13 binds Atg1 in a phosphorylation-dependent manner (10). We found that, in cotransfected HEK293 cells, Atg13-FLAG could readily coimmunoprecipitate both Myc-tagged ULK1 and ULK2 (Fig. 10A). With control immunoblots of the lysates, we consistently observed that Atg13-FLAG singly overexpressed migrated as a sharp band close to 70 kDa. Strikingly, coexpression with either ULK1 or ULK2 led to the reduced mobility of Atg13, suggesting that ULK1 and ULK2 were regulating the phosphorylation of human Atg13. In yeast, both Atg13 phosphorylation and Atg1-Atg13 binding could be regulated by rapamycin treatment (10). In contrast, amino acid starvation in our overexpression cell system did not robustly affect either the electrophoretic mobility of Atg13 or its ability to coimmunoprecipitate with ULK1 or ULK2 (data not shown). In contrast to wild-type ULK proteins, coexpressed ULK1-K46I or ULK2-K39I did not promote a hypershift of Atg13-FLAG. However, Atg13-FLAG still coimmunoprecipitated with both ULK1-K46I and ULK2-K39I, suggesting that binding was not regulated by Atg13 phosphorylation (Fig. 10B and C).
FIG. 10.
Binding of Atg13 to the CTD is required for phosphorylation by ULK1 and ULK2. (A, B, and C) HEK293A cells were transfected with Atg13-FLAG alone (−) or in combination (+) with Myc-ULK1, Myc-ULK2, Myc-ULK1-K46I, or Myc-ULK2-K39I, as indicated. An aliquot of cell lysate was analyzed by SDS-PAGE and immunoblotting with Myc (top) and FLAG (bottom) antibodies. Lysates of doubly transfected cells were incubated with anti-FLAG beads to coimmunoprecipitate Atg13 and ULK proteins. (D) Myc-ULK2 and Myc-ULK2ΔCTD were coexpressed with FLAG-Atg13 in HEK293A cells. As a control, Atg13-FLAG was expressed alone (−). Cell lysates were immunoprecipitated with anti-Myc or anti-FLAG antibody and then immunoblotted as described above. (E) Coimmunoprecipitation of the Myc-tagged CTDs of ULK1 and ULK2 with Atg13-FLAG. Lysates and immunoprecipitates were analyzed as described above. (F) Coimmunoprecipitation of Myc-ULK1CTD(829-1001) with Atg13-FLAG. Lysates and immunoprecipitates were analyzed as described above. The asterisks in panels D, E, and F indicate nonspecific bands arising from the antibody used for immunoprecipitation, as demonstrated by the antibody-plus-beads-alone control (Ab ctrl). (G) 293/GFP-LC3 cells were cotransfected with Myc-ULK1 CTD or Myc-ULK2CTD with or without Atg13-FLAG for 24 h and then either left unstarved or starved in EBSS-leupeptin for 2 h before lysis for analysis of GFP-LC3 lipidation, as described in the legend for Fig. 2. Protein expression controls (top) using anti-FLAG and anti-Myc antibodies are shown. (H) Immunoprecipitated Atg13-FLAG protein was used as a substrate for immunoprecipitated Myc-ULK proteins in a mixed-bead in vitro kinase reaction. Phosphorylation of Atg13-FLAG is detected as a decrease in mobility. A control reaction without the addition of ULK proteins (−) shows no mobility shift. (I) HEK293A cells were either left untransfected (−) or transfected with Myc-ULK1 or Myc-ULK2 before cell lysates were immunoblotted to detect hypershifting on endogenous Atg13. Lys, lysates; IP, immunoprecipitate.
Since the last 20 residues of yeast Atg1 are critical for binding to yeast Atg13 (5), we predicted that the ULK CTD would be involved in the binding of human Atg13, and we tested this idea using coimmunoprecipitation experiments. While coexpression with ULK1ΔCTD resulted in the loss of the Atg13-FLAG protein, this effect was less pronounced when the ULK2ΔCTD mutant was used. As shown in Fig. 10D, when ULK2ΔCTD and Atg13 were coexpressed, Atg13 did not hypershift or coimmunoprecipitate with ULK2ΔCTD. Importantly, the CTDs of both ULK1 and ULK2 readily coimmunoprecipitated with Atg13-FLAG (Fig. 10E). Surprisingly, the ULK1 CTD lacking 50 residues from its C terminus [ULK1CTD(829-1001)] still coimmunoprecipitated with Atg13-FLAG with an efficiency similar to that of the full-length CTD (Fig. 10F). We similarly observed that the C-terminal 50 residues of ULK2 were dispensable for Atg13 binding (data not shown). Thus, the C-terminal portions of the ULK1 and -2 CTDs (including the dominant-negative IERRLSA motif) could be removed without affecting Atg13 binding.
Earlier, we hypothesized that the dominant-negative mechanism initiated by the CTDs and kinase-dead versions of ULK1 and -2 might involve the titration of an essential Atg13-dependent function, perhaps through Atg13 recruitment. However, our mapping data suggested that the region of the CTD binding Atg13 was separable from the residues that participated in dominant-negative interactions. In agreement with this conclusion, coexpression with Atg13-FLAG did not produce any rescuing effects toward the dominant-negative activities of the ULK1 and -2 CTDs on GFP-LC3 lipidation (Fig. 10G). Atg13-FLAG coexpression similarly could not rescue the inhibition of autophagy caused by ULK1-K46I or ULK2-K39I (data not shown). Thus, we conclude that the dominant inhibitory mechanism involves an essential factor independent of Atg13.
Phosphorylation of Atg13 by ULK proteins.
Last, because the kinase that phosphorylates Atg13 in yeast remains unclear, we investigated whether ULK1 and -2 could directly phosphorylate Atg13. We asked whether immunoprecipitated ULK proteins could phosphorylate immunoprecipitated Atg13-FLAG in a mixed-bead in vitro kinase reaction. Immunoprecipitated Atg13-FLAG incubated alone under reaction conditions did not undergo a mobility hypershift in SDS-PAGE, demonstrating that there was no associated kinase activity in the immunoprecipitate (Fig. 10H). However, incubation of Atg13 together with immunoprecipitated ULK1 or ULK2 did result in a decreased mobility of Atg13. This mobility change was not observed when Atg13 was incubated with kinase-dead ULK1 and -2 or ULK proteins lacking their CTDs. These data support the conclusion that Atg13 binds ULK1 and -2 via their CTDs, promoting phosphorylation of Atg13 via the catalytic kinase domain. Immunoblotting of lysates from cells under normal medium conditions revealed that endogenous Atg13 migrates as a doublet, suggestive of distinct species with different levels of phosphorylation (Fig. 10I). Expression of ULK1 or ULK2 promoted substantially more Atg13 with reduced mobility, further supporting the phosphorylation of Atg13 by ULK1 and -2 in vivo.
DISCUSSION
ULK proteins adopt a closed conformation to prevent the CTD from forming abnormal interactions.
Our previous data showed unexpectedly that the ULK1-K46R kinase mutant (32) did not show a strong dominant inhibitory effect in our autophagy cell system (3). Here, we confirmed that while the K46R substitution significantly impaired catalytic function in vitro, the charge-disruptive K46I substitution fully ablated activity and produced a mutant protein that displayed less autophosphorylation and inhibited starvation-induced autophagy. These data agree with a recent report that showed a ULK1-K46N mutant to inhibit autophagy in NIH 3T3 fibroblasts (6). Our data also showed that the CTD was required for kinase-inactive ULK1 to inhibit autophagy. Similarly, in D. discoideum, deletion of CTD sequences rescued the dominant-negative effects of the Atg1-K36A kinase mutant on autophagy-dependent survival (30). Deletion of the CTD also stimulated in vitro autophosphorylation but decreased the ability of the kinase to phosphorylate exogenous substrates.
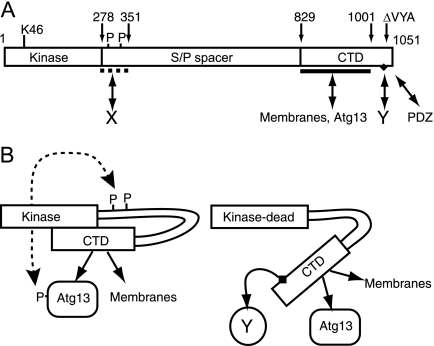
These findings and our other results led us to propose a working model in which the CTD is normally folded back onto the N-terminal region to keep the protein in a closed molecular conformation (Fig. 11). It is still unclear whether the interaction of the CTD with N-terminal regions is controlled by the kinase domain (perhaps via a protein-protein interaction) or via an interaction at the autophosphorylation sites. The K46I mutation triggers a robust conformational change leading to exposure of the CTD, potentially revealing a binding site in the CTD that interacts with essential autophagy regulatory factors.
FIG. 11.
Conformational changes of ULK proteins modulated by autophosphorylation status. (A) Domain structure of ULK1 showing the N-terminal kinase domain and the critical lysine 46 residue (K46) targeted in our mutant constructs. Our data are consistent with the existence of multiple, but so far unidentified, autophosphorylation (P) sites within residues 278 to 351 of the serine- and proline-rich spacer domain (S/P spacer). Our previous findings indicate a region spanning residues 278 to 351 that is critical for inhibition of autophagy, presumably via interaction with an unknown protein (X). Our previous data also indicated that the last three residues of ULK1 (VYA) are critical for regulatory function, possibly by binding a PDZ domain-containing protein. Our findings here indicate multiple functions directed by sequences within the conserved CTD. A region between residues 829 and 1001 contains signals for binding to membranes and interaction with human Atg13. Additional sequences between residues 1038 and 1044 (Fig. 2) contain a conserved motif (IERRLSA, black diamond) that is required for dominant inhibitory activity by interacting with another unknown factor (Y). (B) Proposed model showing phosphorylation-dependent conformational changes. In normal resting cells, ULK proteins are autophosphorylated, and these modifications help maintain a closed conformation that (i) brings Atg13 in closer proximity to the kinase domain for phosphorylation and (ii) keeps the dominant-negative motif hidden or inaccessible. Full ablation of kinase activity results in a low degree of autophosphorylation and a more open molecular conformation that exposes the dominant inhibitory CTD motif, allowing it to bind the unknown factor (Y) that is critical for autophagy.
Our deletion analysis has identified a 7-residue motif (IERRLSA) within the last 14 C-terminal amino acids of the CTD that was required for dominant-negative function, which we propose constitutes a binding site for a critical autophagy regulatory factor (“Y” in Fig. 11). Interestingly, the IERRLSA motif is well conserved in ULK1/2 proteins from mice, humans, and other vertebrates but not in Atg1 homologs of S. cerevisiae, D. melanogaster, and C. elegans. Based on this sequence information, we speculate that the factor binding to the IERRLSA motif might be specific to vertebrates.
Could conformational changes represent a physiological mechanism for the regulation of ULK proteins? The in vitro limited proteolysis assay that we employed could not detect significant changes after amino acid starvation (Chan and Tooze, unpublished), although this could represent a limitation of our system. However, our findings have established that an almost complete ablation of the kinase activity is required to produce readily detectable effects in protease cleavage and autophagy assays. It was recently demonstrated that the kinase activity of endogenous ULK1 in mouse embryonic fibroblasts was modestly activated following amino acid starvation (6), which suggests that subtle conformational alterations could possibly represent a normal means of ULK1 regulation.
Signals within the ULK1 CTD direct membrane binding and protein complex formation.
We and others have seen that ULK1 localizes to GFP-LC3- or Atg16L1-positive autophagosomal structures (3, 6) and that this localization depends upon the CTD. In order to define the downstream effectors of the CTD (i.e., to identify factor “Y”), we used biochemical fractionation and imaging techniques to characterize the ULK CTD. The CTDs of both ULK1 and ULK2 contained signals that could direct membrane binding, and further deletion mapping identified N- and C-terminal sequences that were dispensable for membrane association. Importantly, CTD deletion mutants lacking the IERRLSA dominant-negative motif still retained normal membrane binding efficiencies, which suggests that the critical inhibitory interactions initiated by the CTD are not membrane associated (Fig. 11B). Our data on overexpressed and endogenous ULK1 also suggest that association to membranes was resistant to detergent extraction, unlike results for other autophagy proteins, such as the lipidated form of LC3, and Atg13. Our preliminary data using floatation in detergent-containing gradients suggest that ULK1 cofractionates with markers of lipid rafts (A. Longatti and S. A. Tooze, unpublished observations), although the molecular basis of these observations requires further investigation.
In S. cerevisiae, Atg1 enters into a number of protein complexes (9-12, 22). Using native gel electrophoresis, we observed ULK1 in two distinct populations suggestive of different subcomplexes. Inactivation of kinase function shifted the ULK1-K46I protein toward the higher-molecular-weight complex, with a corresponding loss of the low-molecular-weight species (Chan and Tooze, unpublished). Importantly, while entry of ULK1 into the higher-molecular-weight population required the entire CTD, up to 50 C-terminal residues, including the IERRLSA dominant-negative motif, could be removed from the CTD C terminus without affecting entry into either protein complex. Thus, the dominant-negative motif proposed to bind the critical factor “Y” is separate from other CTD sequences that direct formation of higher-molecular-weight ULK1 protein complexes.
Our data further suggest the existence of two types of ULK1 complexes: a small one containing proteins that bind to ULK1 in a CTD-dependent manner, and a large complex containing proteins that can still bind kinase-inactive ULK1. It has been demonstrated that in S. cerevisiae Atg1 plays two roles in bulk nonselective autophagy: one being structural, facilitating recruitment of Atg proteins to the PAS, and the second being responsible for recycling proteins from the PAS during formation (4, 5). The former role requires the CTD, while the latter requires the kinase domain. While our data do not provide any support for a dual role of the mAtg1 orthologues, further analysis of the small and large complexes may shed light on whether ULK1 or ULK2 behaves similarly to yeast Atg1. Further investigation is also required to understand the differences between ULK1 and ULK2 complexes revealed by native gel analysis.
Human Atg13 binds to the CTD independently of the IERRLSA motif.
Since binding of yeast Atg13 depends upon the C-terminal 20 residues of yeast Atg1 (5), we asked whether the ULK1 and ULK2 CTDs could bind the human homologue of Atg13 and whether this binding occurred via the IERRLSA motif. We identified and characterized a putative human Atg13 orthologue and showed that it was required for autophagy by using LC3 lipidation and the mAtg9 subcellular redistribution assays. Consistent with a role in autophagy, tagged versions of Atg13 became more enriched on membranes following the activation of autophagy. Alteration of mAtg9 trafficking caused by the loss of Atg13 could be due to either decreased Atg13 function or an indirect modulation of ULK1 function or stability. In yeast, Atg1 kinase activity is not required for Atg9 trafficking, so it is unlikely that the loss of ULK1 kinase activity underlies the aberrant mAtg9 trafficking. Rather, as we previously showed, siRNA-mediated depletion of ULK1 inhibits mAtg9 trafficking (37), and our preliminary data suggest that siRNA depletion of Atg13 causes a decrease in ULK1 levels (N. C. McKnight and S. A. Tooze, unpublished observations). Further work is required to clarify the role of human Atg13 in mAtg9 trafficking.
Despite their sequence differences, ULK1 and ULK2 both bound human Atg13 via their CTDs. The IERRLSA motif that we identified here, while not well conserved in yeast, includes an R residue (position 1041 in ULK1) which corresponds to the conserved R885 in yeast Atg1 that constitutes part of an Atg1-Atg13 interaction motif (5). Furthermore, within the C-terminal 20 amino acids of yeast Atg1 is a conserved Y residue at position 878, and substitution of both R885 and Y878 with alanine (Y878A/R885A) in Atg1 results in the loss of recruitment of Atg17 to the PAS (5). Interestingly, this Y residue is conserved in mouse and human ULK2 but not ULK1. However, our data using CTD deletions of ULK1 and ULK2 show that the IERRLSA motif (and also the last 20 amino acids) is not required for human Atg13 binding. We speculate either that another binding site has evolved in the ULKs for human Atg13 or possibly that the Y878A/R885A mutations, which were made in the context of the whole yeast Atg1 protein, altered the conformation or accessibility of the true Atg13 binding site.
In support of our model and binding data, coexpression with Atg13 did not rescue the inhibition of autophagy caused by ULK CTDs (or kinase-dead mutants [data not shown]), suggesting that Atg13 is not the protein being titrated out by the 7-residue CTD motif. A possible candidate for this binding protein (and perhaps factor “Y”) is FIP200, which recently has been shown to bind ULK1 and ULK2 in a CTD-dependent manner and to regulate ULK1 kinase function and protein stability (6). Interestingly, Hara et al. suggest that FIP200 may be the human homologue of yeast Atg17 and that, although there is no sequence conservation between Atg17 and FIP200, both proteins have coiled-coil domains, suggesting a conservation of function as a scaffold protein (6).
Phosphorylation of Atg13 by ULK1 and ULK2.
In the course of our studies, we observed that Atg13 displayed a decreased electrophoretic mobility when coexpressed with ULK1 or ULK2, suggestive of a modification by phosphorylation. Consistent with direct phosphorylation of Atg13 by ULK1 and ULK2, Atg13 did not show an altered migration when coexpressed with ULK proteins lacking Atg13 binding sites (ΔCTD) or kinase-inactivated ULK proteins. Furthermore, both ULK1 and ULK2 could promote Atg13 phosphorylation in a kinase-dependent fashion following an in vitro kinase reaction. These data suggest that Atg13 binds to ULK1 and ULK2 via the CTDs and is phosphorylated by the kinase domain (Fig. 11). In contrast with findings for yeast, binding of Atg13 to ULK proteins in our HEK293 cell model was not modulated by starvation of cells or by the overall phosphorylation status of Atg13. The definition of Atg13 phosphorylation sites and their regulatory function requires further investigation.
Differences between ULK1 and ULK2.
Several lines of evidence have shown that ULK1 and ULK2 behave similarly. Kinase mutants of both ULK1 and ULK2 have dominant-negative effects, and our findings suggest that this is via exposure of a conserved motif within their CTDs. In addition, both ULK1 and ULK2 localize to autophagosomal structures and both proteins bind Atg13 and FIP200.
However, multiple aspects of our findings also highlight that ULK1 and ULK2 may have independent functions or modes of regulation. Deletion of CTD sequences in overexpressed ULK1 and ULK2 appears to promote different downstream effects. ULK1 and ULK2 can enter into different molecular complexes, and the binding of ULK2 to membranes was increased following amino acid starvation, in contrast to results with ULK1. Finally, we previously observed that knockdown of ULK1 but not ULK2 in HEK293 cells inhibited starvation-induced autophagy and starvation-induced mAtg9 redistribution. We also previously observed that overexpression of ULK1 produced a mild dominant inhibitory effect on autophagy, while here we show that overexpression of ULK2 in the same cell system enhanced GFP-LC3 lipidation following starvation. We have not yet been able to confirm the ability of overexpressed ULK2 to modulate other measures of autophagic flux by use of assays such as degradation of long-lived proteins or p62/SQSTM-1 stability (Chan, Longatti, and Tooze, unpublished), possibly due to the limited capabilities of downstream pathways. While additional experiments are required to fully understand the functional differences between ULK1 and ULK2, our biochemical data presented here are in general agreement with recent knockout mouse data that suggest ULK1 and ULK2 could be performing distinct roles in specialized and general autophagy. In addition, to further understand the mammalian equivalent of the yeast Atg1 signaling complex, it will be important to dissect the binding site within the ULK CTD for FIP200 and Atg13 as well as to examine the relationship between FIP200 and Atg13 for regulating the kinase activity and function of ULK proteins.
Acknowledgments
We thank Mike Howell (LRI High Throughput Screening Unit) for advice and assistance in the Cellomics array scan experiments and Mike Mitchell (CRUK Bioinformtics Unit) for help with Atg13 bioinformatics. We thank the Secretory Pathways Laboratory for advice and encouragement and Cancer Research UK for support.
Footnotes
Published ahead of print on 20 October 2008.
REFERENCES
- 1.Abeliovich, H., C. Zhang, W. A. Dunn, Jr., K. M. Shokat, and D. J. Klionsky. 2003. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol. Biol. Cell 14477-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Budovskaya, Y. V., J. S. Stephan, S. J. Deminoff, and P. K. Herman. 2005. An evolutionary proteomics approach identifies substrates of the cAMP-dependent protein kinase. Proc. Natl. Acad. Sci. USA 10213933-13938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan, E. Y., S. Kir, and S. A. Tooze. 2007. siRNA screening of the kinome identifies ULK1 as a multidomain modulator of autophagy. J. Biol. Chem. 28225464-25474. [DOI] [PubMed] [Google Scholar]
- 4.Cheong, H., and D. J. Klionsky. 2008. Dual role of Atg1 in regulation of autophagy-specific PAS assembly in Saccharomyces cerevisiae. Autophagy 4724-726. [DOI] [PubMed] [Google Scholar]
- 5.Cheong, H., U. Nair, J. Geng, and D. J. Klionsky. 2008. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 19668-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hara, T., A. Takamura, C. Kishi, S. Iemura, T. Natsume, J. L. Guan, and N. Mizushima. 2008. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181497-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwata, A., B. E. Riley, J. A. Johnston, and R. R. Kopito. 2005. HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 28040282-40292. [DOI] [PubMed] [Google Scholar]
- 8.Kabeya, Y., Y. Kamada, M. Baba, H. Takikawa, M. Sasaki, and Y. Ohsumi. 2005. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 162544-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kabeya, Y., T. Kawamata, K. Suzuki, and Y. Ohsumi. 2007. Cis1/Atg31 is required for autophagosome formation in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 356405-410. [DOI] [PubMed] [Google Scholar]
- 10.Kamada, Y., T. Funakoshi, T. Shintani, K. Nagano, M. Ohsumi, and Y. Ohsumi. 2000. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 1501507-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawamata, T., Y. Kamada, Y. Kabeya, T. Sekito, and Y. Ohsumi. 2008. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 192039-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, J., Y. Kamada, P. E. Stromhaug, J. Guan, A. Hefner-Gravink, M. Baba, S. V. Scott, Y. Ohsumi, W. A. Dunn, Jr., and D. J. Klionsky. 2001. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153381-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Komatsu, M., S. Waguri, T. Ueno, J. Iwata, S. Murata, I. Tanida, J. Ezaki, N. Mizushima, Y. Ohsumi, Y. Uchiyama, E. Kominami, K. Tanaka, and T. Chiba. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuma, A., M. Hatano, M. Matsui, A. Yamamoto, H. Nakaya, T. Yoshimori, Y. Ohsumi, T. Tokuhisa, and N. Mizushima. 2004. The role of autophagy during the early neonatal starvation period. Nature 4321032-1036. [DOI] [PubMed] [Google Scholar]
- 15.Kundu, M., T. Lindsten, C. Y. Yang, J. Wu, F. Zhao, J. Zhang, M. A. Selak, P. A. Ney, and C. B. Thompson. 2008. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed]
- 16.Levine, B., and G. Kroemer. 2008. Autophagy in the pathogenesis of disease. Cell 13227-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuura, A., M. Tsukada, Y. Wada, and Y. Ohsumi. 1997. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192245-250. [DOI] [PubMed] [Google Scholar]
- 18.Meijer, W. H., I. J. van der Klei, M. Veenhuis, and J. A. Kiel. 2007. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy 3106-116. [DOI] [PubMed] [Google Scholar]
- 19.Melendez, A., and T. P. Neufeld. 2008. The cell biology of autophagy in metazoans: a developing story. Development 1352347-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melendez, A., Z. Talloczy, M. Seaman, E. L. Eskelinen, D. H. Hall, and B. Levine. 2003. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 3011387-1391. [DOI] [PubMed] [Google Scholar]
- 21.Mizushima, N., B. Levine, A. M. Cuervo, and D. J. Klionsky. 2008. Autophagy fights disease through cellular self-digestion. Nature 4511069-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nice, D. C., T. K. Sato, P. E. Stromhaug, S. D. Emr, and D. J. Klionsky. 2002. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the pre-autophagosomal structure is required for selective autophagy. J. Biol. Chem. 27730198-30207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otto, G. P., M. Y. Wu, N. Kazgan, O. R. Anderson, and R. H. Kessin. 2004. Dictyostelium macroautophagy mutants vary in the severity of their developmental defects. J. Biol. Chem. 27915621-15629. [DOI] [PubMed] [Google Scholar]
- 24.Ravikumar, B., C. Vacher, Z. Berger, J. E. Davies, S. Luo, L. G. Oroz, F. Scaravilli, D. F. Easton, R. Duden, C. J. O'Kane, and D. C. Rubinsztein. 2004. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat. Genet. 36585-595. [DOI] [PubMed] [Google Scholar]
- 25.Reggiori, F., K. A. Tucker, P. E. Stromhaug, and D. J. Klionsky. 2004. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev. Cell 679-90. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez-Enriquez, S., L. He, and J. J. Lemasters. 2004. Role of mitochondrial permeability transition pores in mitochondrial autophagy. Int. J. Biochem. Cell Biol. 362463-2472. [DOI] [PubMed] [Google Scholar]
- 27.Scott, R. C., G. Juhasz, and T. P. Neufeld. 2007. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 171-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stanyon, C. A., G. Liu, B. A. Mangiola, N. Patel, L. Giot, B. Kuang, H. Zhang, J. Zhong, and R. L. Finley, Jr. 2004. A Drosophila protein-interaction map centered on cell-cycle regulators. Genome Biol. 5R96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Straub, M., M. Bredschneider, and M. Thumm. 1997. AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J. Bacteriol. 1793875-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tekinay, T., M. Y. Wu, G. P. Otto, O. R. Anderson, and R. H. Kessin. 2006. Function of the Dictyostelium discoideum Atg1 kinase during autophagy and development. Eukaryot. Cell 51797-1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thumm, M., R. Egner, B. Koch, M. Schlumpberger, M. Straub, M. Veenhuis, and D. H. Wolf. 1994. Isolation of autophagocytosis mutants of Saccharomyces cerevisiae. FEBS Lett. 349275-280. [DOI] [PubMed] [Google Scholar]
- 32.Tomoda, T., R. S. Bhatt, H. Kuroyanagi, T. Shirasawa, and M. E. Hatten. 1999. A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24833-846. [DOI] [PubMed] [Google Scholar]
- 33.Tomoda, T., J. H. Kim, C. Zhan, and M. E. Hatten. 2004. Role of Unc51.1 and its binding partners in CNS axon outgrowth. Genes Dev. 18541-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukada, M., and Y. Ohsumi. 1993. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 333169-174. [DOI] [PubMed] [Google Scholar]
- 35.Yan, J., H. Kuroyanagi, A. Kuroiwa, Y. Matsuda, H. Tokumitsu, T. Tomoda, T. Shirasawa, and M. Muramatsu. 1998. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem. Biophys. Res. Commun. 246222-227. [DOI] [PubMed] [Google Scholar]
- 36.Yan, J., H. Kuroyanagi, T. Tomemori, N. Okazaki, K. Asato, Y. Matsuda, Y. Suzuki, Y. Ohshima, S. Mitani, Y. Masuho, T. Shirasawa, and M. Muramatsu. 1999. Mouse ULK2, a novel member of the UNC-51-like protein kinases: unique features of functional domains. Oncogene 185850-5859. [DOI] [PubMed] [Google Scholar]
- 37.Young, A. R., E. Y. Chan, X. W. Hu, R. Kochl, S. G. Crawshaw, S. High, D. W. Hailey, J. Lippincott-Schwartz, and S. A. Tooze. 2006. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 1193888-3900. [DOI] [PubMed] [Google Scholar]