Abstract
To elucidate the checkpoint mechanism responsible for slowing passage through S phase when fission yeast cells are treated with the DNA-damaging agent methyl methanesulfonate (MMS), we carried out two-dimensional gel analyses of replication intermediates in cells synchronized by cdc10 block (in G1) followed by release into synchronous S phase. The results indicated that under these conditions early-firing centromeric origins were partially delayed but late-firing telomeric origins were not delayed. Replication intermediates persisted in MMS-treated cells, suggesting that replication fork movement was inhibited. These effects were dependent on the Cds1 checkpoint kinase and were abolished in cells overexpressing the Cdc25 phosphatase, suggesting a role for the Cdc2 cyclin-dependent kinase. We conclude that both partial inhibition of the firing of a subset of origins and inhibition of replication fork movement contribute to the slowing of S phase in MMS-treated fission yeast cells.
In response to low levels of the DNA-alkylating agent methyl methanesulfonate (MMS), wild-type yeast cells slow their progression through S phase, while cells lacking the appropriate upstream checkpoint kinase (Mec1 in the budding yeast Saccharomyces cerevisiae; Rad3 in the fission yeast Schizosaccharomyces pombe) or the appropriate downstream checkpoint kinase (Rad53 in budding yeast, Cds1 in fission yeast) fail to do so. Other DNA-damaging agents also cause a checkpoint-dependent slowing of S phase, in vertebrates as well as in yeasts. This slowing of S phase in response to DNA damage is sometimes called the “intra-S-phase” checkpoint (3, 6, 22, 23, 26, 28, 36, 37, 45, 53). Here we shall refer to it as the “S-phase damage” checkpoint.
Prior to this report, the downstream portions of the checkpoint pathway(s) that slow S phase in response to DNA damage in fission yeast were unclear. However, the upstream portions of these pathways in fission yeast and other organisms have been partially elucidated, and downstream mechanisms in other organisms have been partially clarified. In all studied systems, upon detection of DNA damage in S phase, checkpoint proteins initiate a phosphorylation cascade that ultimately leads to slowing of replication. Upstream signaling in these systems involves the activation of one or more of the phosphatidylinositol-3-kinase-like protein kinases (PIK kinases; ATR and/or ATM in humans, Mec1 and/or Tel1 in budding yeast, and Rad3 in fission yeast). The activated PIK kinases then phosphorylate several proteins, including certain Ser/Thr kinases (Chk1 and/or Chk2 in humans, Rad53 in budding yeast, and Cds1 in fission yeast). These kinases, in turn, phosphorylate other substrates that, directly or indirectly, mediate the slowing of S phase (reviewed in reference 3).
In budding yeast, two different mechanisms were shown to slow S phase upon DNA damage by MMS. Of these, one mechanism, inhibition of late-firing origins, depended on the Mec1-Rad53 checkpoint pathway (45, 53), while the other mechanism, inhibition of replication forks, appeared to be a direct consequence of DNA damage rather than a result of checkpoint activation (53). Tercero and Diffley (53) found that, in MMS-treated cells with mutations in the RAD53 gene, unregulated origin firing compensated for checkpoint-independent replication fork slowing, thus permitting a relatively normal overall rate of DNA synthesis. The mechanism by which the Rad53 protein modulates late origin activity is not yet clear, but one possibility is inhibition (by Rad53-catalyzed phosphorylation) of Dbf4, the regulatory subunit of the Cdc7-Dbf4 kinase, which is essential for initiation of replication (7, 8, 14, 55).
In vertebrates, at least three different pathways have been shown to contribute to the slowing of S phase after DNA damage. In some cases checkpoint-mediated phosphorylation of Dbf4 inhibits progression through S phase by downregulating origin firing (7, 14), as may take place in budding yeast. In other cases, checkpoint-mediated phosphorylation leads to inhibition and destruction of the protein phosphatase Cdc25A, which is an activator of Cdk2. Cdk2 is the S-phase-specific cyclin-dependent kinase. Cdk2 activity is crucial for initiation of DNA replication and is modulated by inhibitory phosphorylation at Tyr-15. Cdc25A activates Cdk2 by dephosphorylating Tyr-15. Thus, when Cdc25A is phosphorylated by checkpoint kinases after DNA damage and subsequently destroyed, Cdk2 can no longer promote initiation of DNA replication (9, 27). The third mechanism by which vertebrate cells can slow progression through S phase is inhibition of replication fork movement. In vertebrate cells, slowing of replication forks in response to DNA damage is frequently checkpoint dependent; in contrast, in budding yeast, such slowing appeared to be checkpoint independent. In the tested cases, fork slowing has proved to be dependent on the PIK kinase ATR (homologous to budding yeast Mec1 and fission yeast Rad3) and on the Ser/Thr kinase Chk1 (a functional analogue of budding yeast's Rad53 and fission yeast's Cds1). In each of these cases, the checkpoint response to DNA damage led to inhibition of origin firing as well as to inhibition of replication fork movement (42, 44, 54). The precise mechanism leading to slowing of replication fork movement has not been fully worked out, but the mechanism appears to involve interactions between Chk1 and the proteins Tim and Tipin (54), whose yeast homologues (Swi1 and Swi3 in fission yeast, Tof1 and Csm3 in budding yeast) form a “replication fork protection complex” that is associated with replication forks (19, 33).
Although it is clear that slowing of S phase in response to MMS-induced DNA damage in fission yeast requires both the Rad3 and Cds1 kinases, the pathways operating downstream of Cds1 have been uncertain. We obtained results indicating that Cdc25, which was already known to be a target of Cds1 in hydroxyurea (HU)-treated cells, is also a target of Cds1 in MMS-treated cells, because both overproduction of Cdc25 and conversion of Tyr-15 on Cdc2 (the major cyclin-dependent kinase of fission yeast; also known as Cdk1) to a nonphosphorylatable residue (Cdc2-Y15F; this mutation rendered Cdc2 constitutively active) were sufficient to prevent MMS-induced slowing of S phase (23). We concluded that, in fission yeast, the Rad3→Cds1⊣Cdc25→Cdc2 pathway forms a checkpoint signaling module very similar to the corresponding one of vertebrates. However, Kommajosyula and Rhind were not able to repeat our observations regarding the roles of Cdc25 and Cdc2 (22), so the relevance of Cdc25 and Cdc2 to checkpoint-induced slowing of S phase in fission yeast has remained uncertain until now. In addition, whether S phase in MMS-treated fission yeast cells is slowed by inhibition of origin firing, by reduction in rate of fork movement, or by a combination of these has been equally unclear.
In order to resolve these issues, we initiated the series of experiments reported in this paper. To measure the rate of progression through S phase, we followed S phase by flow cytometry and by two-dimensional (2D) gel electrophoresis in cells released from a G1 block (achieved by incubating cells bearing a cdc10 temperature-sensitive mutation at the restrictive temperature, then releasing to the permissive temperature [21, 23]). We found that, in MMS-treated, checkpoint-competent cells, the firing of early origins near centromeres was partially delayed but that the firing of late origins near telomeres was unaffected. Furthermore, the lifetimes of replication intermediates (RIs) were prolonged, consistent with slowing of replication forks. These effects were completely abrogated both in cells lacking the Cds1 kinase and in cells overproducing the Cdc25 phosphatase, showing that these effects were checkpoint dependent and that the relevant checkpoint pathway probably involved inhibition of Cdc25.
MATERIALS AND METHODS
Strains and growth conditions.
The fission yeast strains used in this study are SZ290 (h+ cdc10-v50 ura4-D18 leu1-32 ade6-M210; a gift from Greg Freyer) (23), SK08 (h− cdc10-v50 cdc25::ura4+ ura4-D18 adh:cdc25+ ade?) (23), and SK12 (h+ cdc10-v50 ura4-D18 cds1::ura4+ leu1-32 ade) (this study). YES medium was used to grow cells, and it consisted of 0.5% yeast extract (Difco); 3.0% glucose; 75 mg/liter each of adenine, histidine, and uracil; and 200 mg/liter of leucine. All supplements were from Sigma. Cells were grown at 25°C unless otherwise mentioned. Where indicated in the figures, MMS (Sigma) was added to a final concentration of 0.015%.
Culture synchronization and sample collection for flow cytometry and 2D gel analyses.
For synchronization in and release from G1 phase, we used the cdc10-v50 block-and-release procedure, as described here. Cells from a 2-liter log-phase culture (0.8 × 107 to 1.0 × 107 cells/ml) were harvested and resuspended in the same volume of fresh medium maintained at 35°C. They were incubated at this temperature with vigorous shaking for different amounts of time depending upon their genotypes. The cdc10-v50 cells required 4.5 h, cdc10-v50 cds1Δ cells needed 4.0 h, and cdc10-v50 cdc25OP cells required 3.5 h at 35°C to efficiently arrest in G1. Arrested cultures were spun down; cell pellets were washed with water and divided into two parts. Half the cells were added to 1.5 liters of fresh medium (maintained at 25°C) containing no MMS. The other half of the cells were added to the same amount of fresh medium and treated with 0.015% MMS. The cultures were incubated at 25°C with vigorous shaking. Every 30 min, 300 μl and 200 ml of culture volume were removed. The 300 μl of culture was fixed by mixture with 900 μl of ethanol in a microcentrifuge tube. The 200-ml sample was mixed with 0.01% (final concentration) sodium azide and 50 mM (final concentration) EDTA and immediately chilled on ice. Samples thus collected were kept on ice in a cold room until the completion of the experiment. Afterwards, the small samples in microcentrifuge tubes were processed for flow cytometry as described previously (21, 23, 28). Cells were harvested from the larger samples, washed with ice-cold water, and pelleted again. Finally, the cell pellets were kept frozen at −80°C until being used for DNA isolation.
DNA isolation and 2D gel electrophoresis.
DNA was isolated and processed for 2D gel electrophoresis as described previously (16) except that benzoylated naphthoylated DEAE-cellulose enrichment was omitted. For these experiments, 5 μg of each DNA sample was digested with a fivefold excess of HindIII (New England Biolabs). The 2D gels were run as described previously (5). The DNA isolation and general experimental procedures involved in 2D gel analysis have also been described at our website: http://hosted2.roswellpark.org/huberman/2D_Gel_Docs_HTML.html.
Hybridization probes, reprobing, autoradiography, and image processing.
Probes for the centromeric K(dg) repeats, ars2-2, and telomere-associated sequences (TAS) were described previously (21). Each 2D gel (representing a single time point from a single strain) was blotted to a GeneScreen Plus (NEN) nylon membrane, then hybridized sequentially with these three probes in the order (i) ars2-2, (ii) centromere, and (iii) telomere. The probe from each earlier hybridization was stripped from the membrane prior to hybridization by a modification of the manufacturer's recommended protocol: the membranes were immersed in stripping buffer and heated to near boiling in a microwave oven, rather than by being boiled for 10 to 30 min. This milder procedure proved sufficient for our purposes, and it reduced loss of target DNA from the membrane. The autoradiograms were captured as 16-bit TIFF images using a Molecular Dynamics Typhoon 8600 phosphorimager (GE Healthcare). The images were subsequently optimized for display of RIs by taking the square roots of all data points using IPLab 3.5 software (originally from Scanalytics; currently updated and sold as iVision 4.0 by BioVision Technologies). Both IPLab 3.5 and Photoshop (Adobe) were used to set upper and lower display limits. Except for normalization to make the signals for 1N spots the same for each 2D gel picture in a single time course, identical operations were performed on all pictures in a single time course.
Quantitation of RIs.
IPLab 3.5 software (see above) was employed for quantitation of RIs. Quantitation was carried out on the raw phosphorimager data files, prior to taking square roots. For each 2D gel, a segment boundary was drawn around the 1N spot(s) and the signals for all pixels inside the boundary were summed. The total was the 1N spot intensity. There was no need to determine background for the 1N spot(s), because background was negligible compared to signal. Then a segment boundary was drawn around the area containing the RIs (both Y and bubble arcs), with care to exclude signals from the 1N spot or from the arc of linear molecules. The segment boundary for RIs was then copied and moved to a nearby portion of the gel that was judged to be signal free and to have about the same amount of background as the RI area. Signals from pixels within each of these two identically shaped segment boundaries were then separately summed, and their difference provided the RI intensity. Finally, the ratios of the RI intensity to the 1N spot intensity were calculated using Microsoft Excel. These ratios (in units of 10−3) are indicated in the upper left or lower right corner of each 2D gel panel in Fig. 2 to 5. The measurements employed for these calculations may be obtained in spreadsheet format from either of the authors.
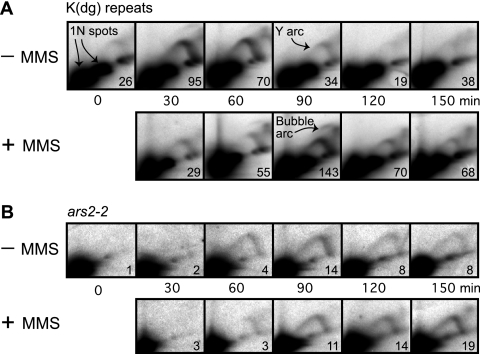
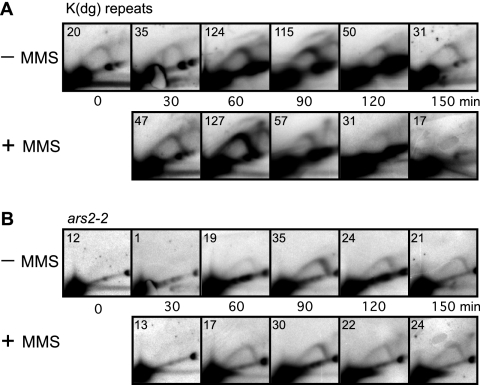
FIG. 2.
2D gel analyses of RIs in the wild-type cdc10-v50 strain at the indicated times after shift to 25°C. In each panel, the ratio of RIs to the 1N spot(s) is shown (in units of 10−3) in the lower right corner of each panel. (A) Replication profile of K(dg) repeats. In the absence of MMS these restriction fragments were passively replicated (indicated by strong Y arcs) or, less frequently, were replicated by the firing of internal origins (indicated by faint bubble arcs). The nonreplicating forms of the restriction fragments formed 1N spots. Replication took place predominantly at 30 and 60 min (−MMS row). MMS treatment delayed maximum replication until 90 min, and RIs persisted at 120 and 150 min. (B) Replication profile of ars2-2. The restriction fragment containing ars2-2 replicated later than the K(dg) repeats in the untreated cells, and MMS treatment further delayed its replication. The abundance of RIs in MMS-treated cells increased at the later time points.
FIG. 5.
2D gel analyses of the terminal HindIII fragments at the telomeres of chromosomes 1 and 2 in untreated and MMS-treated cells. In each panel, the ratio of RIs to the 1N spot(s) is shown (in units of 10−3) in the upper left corner. (A) Wild-type cells prolonged their replication of telomeres upon MMS treatment, and the RIs from these regions persisted at later time points. (B and C) Replication of telomeres in cds1Δ (B) and cdc25OP (C) cells was not confined to 60 to 90 min even in the absence of MMS. Upon MMS treatment, the cds1Δ strain (B) showed a further decay of replication synchrony. These telomeric restriction fragments in the cdc25OP strain (C) replicated asynchronously in the absence as well as the presence of MMS.
RESULTS
Dependence of the MMS-induced S-phase damage checkpoint on Cds1 and on inhibition of Cdc25 in cdc10-synchronized cells.
For experiments to determine the dependence of the MMS-induced S-phase damage checkpoint on Cds1 and on inhibition of Cdc25 in cdc10-synchronized cells, we used three fission yeast strains: the wild type, a cds1Δ strain, and a strain overproducing the Cdc25 protein, which we call the cdc25OP strain (23). All three strains were in the cdc10-v50 background. Use of the cdc10-v50 mutation permitted the cells to be synchronized by arrest in G1 at the restrictive temperature (35°C), followed by release into the cell cycle at the permissive temperature (25°C) (23). The cds1Δ cells are completely defective in the S-phase damage checkpoint, and the cdc25OP cells (which overproduce the Cdc25 protein) are also largely deficient in this checkpoint (23, 26, 28). After synchronization by G1 arrest at 35°C and release at 25°C, we followed the cells' progression into and through S phase by collecting samples for flow cytometry. We also collected larger samples for 2D gel analyses in parallel as described in Materials and Methods. All experiments were performed twice, with essentially identical results.
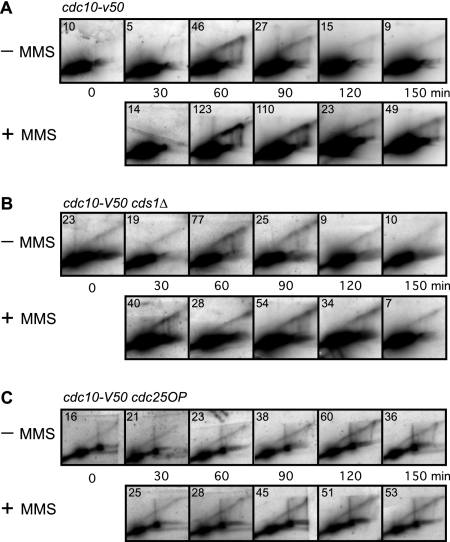
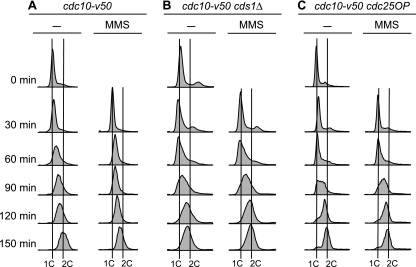
Figure 1 shows the flow cytometry profiles for the three strains. In the absence of MMS, most cdc10-v50 (wild-type) cells completed S phase by 150 min (Fig. 1A, − column). Progression through S phase was slower in MMS-treated wild-type cells (Fig. 1A, MMS column). A significant fraction of these cells were still in S phase at 150 min. In contrast, as shown in Fig. 1B and C, the flow cytometry profiles of untreated and MMS-treated cds1Δ and cdc25OP cells were nearly identical, regardless of MMS treatment. Thus, the MMS-induced slowing of S phase evident in the wild-type cells depended on the checkpoint kinase Cds1 and could be suppressed by overexpression of the Cdc25 phosphatase, as previously observed (23).
FIG. 1.
Assay for the MMS-induced S-phase damage checkpoint. Cells bearing the cdc10-v50 mutation were arrested in G1 by incubation at 35°C. At 0 min, the cells were released into the cell cycle by reducing the temperature to 25°C. Columns labeled MMS show results for cells treated with 0.015% MMS at 0 min. Samples were collected at the indicated times and analyzed by flow cytometry. (A) Wild-type (cdc10-v50) cells retarded their progression through S phase when treated with MMS. (B and C) In contrast to wild-type cells, cds1Δ (B) and cdc25OP (C) cells failed to slow their progression through S phase upon DNA damage. Because fission yeast cells become elongated when cell cycle arrested, which affects their optical properties, the positions of 1C and 2C peaks cannot be determined from log-phase flow cytometry profiles. We determined the positions of the 1C lines from the position of the major peak at 0 min, modified by the position of the major peak at 30 min in the case (cds1Δ cells) (B) where the 30-min peak was left of the 0-min peak. The positions of the 2C lines were determined by the position of the rightmost of the major peaks (without and with MMS) at 150 min. In all panels, the distances separating the 1C lines from the 2C lines for the untreated and MMS-treated samples are identical. That the untreated cds1Δ cells were indeed at the end of S phase at the 150-min time point, despite showing a peak slightly to the left of the 2C line, is evident from the 2D gels in Fig. 3 and 5B, where the RIs from the untreated cells were consistently at background level at 150 min.
Effects of MMS treatment on replication in wild-type cells.
We carried out 2D gel analyses for the samples shown in Fig. 1. We chose to analyze the replication timings of three regions, centromeres, telomeres, and ars2-2, whose replication timings in wild-type cells, in the absence of MMS, have previously been well studied. Centromeres normally replicate in very early S phase, ars2-2 is normally passively replicated in middle or late S phase, and telomeres normally replicate in late S phase (20, 21). In our previous work (20, 21) and here, we analyzed HindIII restriction fragments containing either a repeated segment of centromeric DNA with a potential origin, a repeated segment of telomeric DNA with a potential origin, or a segment of unique DNA containing the potential origin ars2-2. Because we studied only HindIII restriction fragments, we needed to digest the DNA from each strain and time point only with HindIII, and we could analyze all of that DNA in a single 2D gel. Then we were able to probe the single nylon membrane that resulted from blotting each 2D gel sequentially with probes corresponding to ars2-2, centromeres, and telomeres. In this way, each probing served as a control for the other probings of the same membrane. In addition, our use of repeated sequences enabled us to obtain stronger and more easily interpretable RI signals in those cases.
Figure 2 shows the replication patterns in wild-type cells of HindIII restriction fragments containing centromeric K(dg) repeats or ars2-2 (a unique locus on chromosome 2). In Fig. 2 and subsequent figures, in all samples, the main signal appears as a large spot or spots (black area[s] in the lower left corner, called the 1N spot[s]); these spots represent the linear nonreplicating restriction fragments. In the case of the K(dg) repeats shown in Fig. 2A, digestion with HindIII gave rise to 4.3-kb fragments from the centromere of chromosome 2 and 3.3-kb fragments from the centromeres of chromosomes 1 and 3. Thus, there are two 1N spots in each panel of Fig. 2A. RIs generate two arc-shaped signals. A high-rising “bubble arc” signal (example in Fig. 2A) is generated when a restriction fragment contains a functional replication origin in its central third. The family of RIs containing different sizes of internal bubbles collectively produces the bubble arc signal. In contrast, a “Y arc” signal (example in Fig. 2A) is produced by the family of Y-shaped RIs generated by a single replication fork traversing the region (5).
There is a weak Y-arc signal at the 0-min time point in Fig. 2A and in many of the other figures (see Fig. 3A, 4A, and 5B and C). This seems to be due to imperfect synchrony. A few cells appear to enter S phase even at 35°C, and in this small population of cells RIs are generated in early-replicating regions such as the K(dg) repeats.
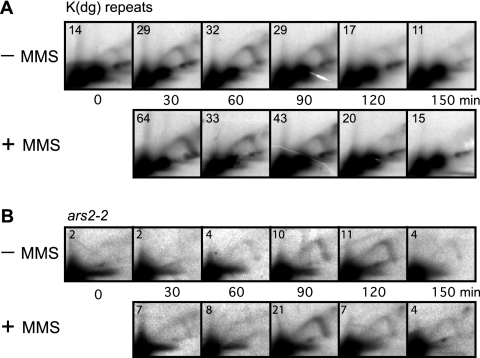
FIG. 3.
2D gel analyses of RIs in the cds1Δ strain. In each panel, the ratio of RIs to the 1N spot(s) is shown (in units of 10−3) in the upper left corner. MMS treatment failed to delay the replication of either the K(dg) repeats (A) or ars2-2 (B), and RIs did not persist at later time points.
FIG. 4.
2D gel analyses of RIs in the cdc25OP strain. In each panel, the ratio of RIs to the 1N spot(s) is shown (in units of 10−3) in the upper left corner. MMS treatment had no significant effect on replication of the K(dg) repeats (A) or ars2-2 (B). The replication profiles appear similar to those for the cds1Δ strain (Fig. 3).
Like most replication origins in fission yeast (10, 13, 15, 32, 35), the origins within the centromeric K(dg) repeats fire with low efficiency, so most of the HindIII restriction fragments from the K(dg) repeats are passively replicated by forks from origins in nearby, flanking fragments. These generate Y arcs. Only a minority of restriction fragments contain active origins and generate bubble arcs (21). When RI signals are averaged over the whole population, the result is a mixed pattern, with a strong Y arc and a weak bubble arc (Fig. 2A). The low ratio of bubble arcs to Y arcs is due to origin inefficiency, not to breakage of bubble arcs during DNA preparation, because broken bubbles do not produce the true Y arc signals observed here (18). Because the passively replicated fragments are always close to fragments with active origins, which fire at about the same time, the relative abundances of Y arc signals and bubble arc signals remain roughly constant during the time course (Fig. 2A). We use the total abundance of signals from all RIs, both bubble arcs and Y arcs, to determine replication timing (21). We quantitated these signals by determining the ratios of signals from RIs to signals from the 1N spot(s), and we show these ratios (in units of 10−3) in the lower right corners of the panels in Fig. 2 (note that the ratios are shown in the upper left corners of the panels in Fig. 3 to 5).
In the absence of MMS, RIs from the early-replicating K(dg) repeats were primarily observed at 30 and 60 min postrelease, consistent with the previously described early replication of K(dg) repeats (20, 21). In the MMS-treated samples, low concentrations of RIs were present at 30 and 60 min, but there was a major accumulation of RIs at 90 min. Thus, upon MMS treatment of these cells, in most cases replication of K(dg) repeats was delayed by at least 30 min. Additionally, RIs were still present at the 150-min time point in MMS-treated cells, whereas they were reduced to background level by 120 min in untreated cells (the stronger signal at 150 min may be due to some cells entering the next cell cycle and beginning to replicate centromeres, which normally replicate very early). Notice that RIs persist in MMS-treated cells for a longer time (60 min past the peak of origin firing at 90 min) than they do in wild-type cells (only 30 min past the peak of origin firing at 60 min). This relative persistence of RIs in the MMS-treated cells is consistent with slowing of replication forks in these cells. These results suggest that retardation of DNA synthesis in the vicinity of centromeres upon MMS treatment in checkpoint-competent cells may be a composite of delay in the firing of a large proportion of origins within K(dg) repeats and reduction in the rate of replication fork movement through these repeats.
In contrast to the K(dg) repeats, which generate HindIII restriction fragments of two different sizes, ars2-2, which is a unique sequence located on the right arm of chromosome 2, generates a single 1N spot (Fig. 2B). The ars2-2 sequence is usually replicated passively in middle to late S phase by replication forks arising from earlier-firing, flanking origins (21). In other words, ars2-2 is an even less efficient origin than the origins in the K(dg) repeats. In Fig. 2B, consistent with previous observations, the ars2-2 RIs from untreated cells appeared at 60 min and peaked at 90 min in middle to late S phase. In contrast, RIs from the MMS-treated cells appeared at 60 min but then persisted with increasing intensities through the time course. The most intense ars2-2 RIs were detected at 150 min. These observations are consistent with the observed slowing of S phase in MMS-treated wild-type cells (Fig. 1A), but they do not distinguish between (i) retardation of the origins responsible for replicating ars2-2, (ii) slowing of the replication forks that travel from those origins to ars2-2, and (iii) a combination of these. Nevertheless, the results obtained with ars2-2 for wild-type cells (Fig. 2B) provide an interesting comparison to results obtained with cds1Δ (Fig. 3B) and cdc25OP (Fig. 4B) cells.
No major effect of MMS treatment on replication in cds1Δ cells.
As shown Fig. 3A, −MMS row, the RIs from K(dg) repeats in the cds1Δ cells were most intense in the 30-, 60-, and 90-min samples, and they lost intensity at later time points, similar to those in untreated checkpoint-competent cells. This replication timing profile remained essentially unaltered upon MMS treatment, consistent with the lack of effect of MMS treatment on the rate of passage through S phase in cds1Δ cells (Fig. 1B). In both the absence and presence of MMS, the ratios of signals from bubble arcs to signals from Y arcs were significantly lower than those for checkpoint-competent cells (Fig. 2A) or for cdc25OP cells (Fig. 4A). This observation suggests that Cds1 may be needed for optimum replication origin efficiency, at least under these synchronization conditions, in both the absence and presence of MMS.
Our comparison of the replication profiles of ars2-2 in cds1Δ cells led to a similar conclusion (Fig. 3B). Neither the time of replication (primarily 90 to 120 min in late S phase) nor the longevities of the RIs were significantly altered by MMS treatment. Thus, Cds1 is required both to delay the firing of replication origins in the K(dg) repeats and to delay the replication of ars2-2 in MMS-treated cells. Cds1 is also required for the persistence of RIs seen in MMS-treated checkpoint-competent cells (compare Fig. 2 and 3), which suggests that Cds1 may be required to slow replication forks after DNA damage.
No major effect of MMS treatment on replication in Cdc25-overexpressing cells.
We recently demonstrated that, under our experimental conditions, the Cdc25-Cdc2 pathway, operating downstream of Cds1, is primarily responsible for MMS-induced slowing of S phase (23). We wanted to know if the same pathway was responsible for inhibiting replication origins and/or replication fork movement upon DNA damage. To test this, we employed a fission yeast strain overproducing Cdc25, which had been shown to have reduced phosphorylation at the Tyr-15 residue of Cdc2 (12), leading to Cdc2 hyperactivity. Figure 1C shows flow cytometry profiles of G1-synchronized, untreated, and MMS-treated cdc25OP cells. The rate of passage through S phase in these cells was not reduced by MMS treatment. Hence, these cells, like the cds1Δ cells, are deficient in the S-phase damage checkpoint.
In contrast to results for wild-type cells (Fig. 2A), but similar to results for cds1Δ cells (Fig. 3A), MMS treatment did not reduce the rate of replication of the K(dg) repeats in cdc25OP cells (Fig. 4A). Indeed, replication of the K(dg) repeats was somewhat faster in MMS-treated cdc25OP cells than in untreated cells. We do not know why MMS treatment accelerated replication of the K(dg) repeats in these cells. It is also interesting that the bubble arc signals in Fig. 4A (cdc25OP cells) are as strong as those in Fig. 2A (wild-type cells), while the bubble arc signals in Fig. 3A (cds1Δ cells) are much weaker. The reasons for this observation are not clear, but the relatively strong bubble arcs generated by the cdc25OP cells (Fig. 4A) could be a result of hyperactive Cdc2, leading to increased origin efficiencies.
In Fig. 4B, the most intense ars2-2 RIs appeared at 90 min (untreated and MMS treated) and the intensities of the RIs at later times in the untreated and MMS-treated cells appeared to be similar. Thus, MMS treatment failed to delay the replication of these regions and failed to prolong the existence of RIs when Cdc25 was overexpressed. The same defects in the S-phase checkpoint response to MMS-induced DNA damage were evident in both the cdc25OP cells (Fig. 4) and the cds1Δ cells (Fig. 3).
Effects of DNA damage on replication of telomeres.
In budding and fission yeasts, telomere regions contain late-firing origins and usually replicate in late S phase (21, 38). Figure 5 shows 2D gel analyses of the four terminal HindIII restriction fragments from the ends of chromosomes 1 and 2. These fragments were detected with a TAS probe that had previously been used in our laboratory for studies of telomere replication timing (21). These fragments contain potential replication origins that sometimes generate detectable bubble arcs, but usually only Y arcs are visible (21). When these fragments are passively replicated, the origins responsible for their replication must be a subset of the numerous potential origins found near fission yeast telomeres (13, 32). As in the case of the K(dg) repeats, the bubble and Y arc signals from telomeres display identical kinetics of appearance and disappearance, so we use the total amount of RIs at each time point as a measure of replication timing (21). In the experiments presented here, only Y arcs were visible (Fig. 5). These Y arcs appear unusual for two reasons. First, the detected HindIII restriction fragments are large (about 8 kb), leading to distorted Y arcs under the standard gel electrophoresis conditions employed here (17). Second, the TAS probe detects HindIII fragments of four different sizes from the four ends of chromosomes 1 and 2, so multiple, usually overlapping, 1N spots and Y arcs are detected.
In the absence of MMS, the telomeric RIs in checkpoint-competent cdc10-v50 cells were most intense at 60 and 90 min, with reduced intensities at later times (Fig. 5A). In these cells, telomeres replicated in middle S phase (note that 60 to 90 min corresponds to middle S phase) (Fig. 1A), in contrast to previously studied cell lines, where telomeres replicated in late S phase (21). Interestingly, the appearance of telomeric RIs was not delayed by MMS treatment, but, in MMS-treated cells, the RIs persisted for longer times: the RI intensities at 90, 120, and 150 min were higher in the MMS-treated cells than in the untreated cells. Thus, the time of telomere replication was not delayed, but it was prolonged by MMS treatment, consistent with reduced rate of fork movement.
In cds1Δ cells (Fig. 5B), in the absence of MMS, the most intense RIs appeared at 60 min but a low-intensity RI signal was visible all through the time course, indicating a reduced synchrony of telomere replication in these cells. Surprisingly, telomere replication synchrony was further reduced in MMS-treated cds1Δ cells, with almost constant RI signals from 30 min to 120 min.
In cdc25OP cells (Fig. 5C), these effects were exacerbated. In these cells, even in the absence of MMS, telomere replication synchrony was significantly reduced and the time of maximum RI signal strength was delayed until 90 to 150 min. MMS treatment did not produce any noticeable change in the RI pattern. In this strain, one of the HindIII fragments detected by the TAS probe was significantly larger than the others, and this larger fragment produced a Y arc that was more angular than the arcs generated by the other fragments (as predicted on the basis of the study by Hyrien and Mechali [17]).
The fact that a Y arc signal was present at the 0-min time point in Fig. 5B and C but not in Fig. 5A is probably a consequence of the loss of replication timing control in the cds1Δ and cdc25OP cells. In these cells, replication of telomeres appeared to begin in early S phase and then continue through the rest of S phase. RIs from early-replicating regions, including telomeres, would be expected to be present at the 0-min time point in the portion of cells that escaped synchrony and entered S phase during the 35°C temperature block. Note that a Y arc signal was also generated by a portion of these cells at the 0-min time point for the early-replicating K(dg) repeats (Fig. 3A and 4A) but not for the later-replicating ars2-2 (Fig. 3B and 4B).
DISCUSSION
Here we have reported the results of measurements of the abundances of RIs from centromeres, telomeres, and ars2-2 (a unique region on chromosome 2) during the course of synchronized S phase in wild-type and checkpoint-mutant fission yeast cells, treated or not with the DNA-damaging agent MMS. These results lead to three surprising conclusions, which are explained in the following three sections.
The checkpoint response to MMS is different from the checkpoint response to HU.
Previous studies indicated that fission yeast checkpoint responses to both MMS (which methylates DNA) and HU (which slows replication forks by inhibiting the synthesis of deoxyribonucleoside triphosphates) are mediated in part by the same pathway, a pathway involving the upstream checkpoint kinase Rad3, the downstream checkpoint kinase Cds1, the phosphatase Cdc25, and the cyclin-dependent kinase Cdc2 (Rad3→Cds1⊣Cdc25→Cdc2; see the introduction) (4, 11, 21, 23, 24, 26, 28, 40). The results presented here show that, although fission yeast checkpoint responses to MMS and HU do, indeed, make use of these proteins, there must be other components of the cellular responses to MMS and HU that distinguish between MMS and HU, because the final results of checkpoint activation appear to be quite different in the two cases.
HU slows replication forks by inhibiting ribonucleotide reductase, which deprives DNA polymerases of their substrates, the deoxyribonucleoside triphosphates. In response to HU treatment, fission yeast cells activate Rad3 and Cds1, and this leads to inhibition of Cdc25. Since Cdc25 is an activator of Cdc2, this results in the inhibition of Cdc2 and prevention of mitosis due to lack of active Cdc2 (40). In addition, HU-treated fission yeast cells delay the firing of replication origins located near ars2-2 and telomeres (both of which normally replicate in middle to late S phase), and this delay is dependent on Rad3 and Cds1 (13, 15, 21, 32). However, it is not known whether this delay is also dependent on inhibition of Cdc25 and Cdc2. In contrast, the firing of early replication origins in the presence of HU is unaffected by the checkpoint proteins Rad3 and Cds1 (13, 15, 21, 32).
When we initiated this 2D gel analysis of origin firing in MMS-treated fission yeast cells, we anticipated that we would obtain results similar to those previously obtained with HU (no effect on early origins, inhibition of late origins). Instead, we found that the firing of a significant portion of the early-firing origins located in the K(dg) repeats of centromeres is delayed, in checkpoint-dependent fashion, by MMS treatment (Fig. 2A, 3A, and 4A). We also found that, in wild-type fission yeast cells, telomeres replicated at their normal middle- to late-S times regardless of the presence of MMS (Fig. 5A). The replication of ars2-2 (which normally replicates in middle to late S phase) was delayed, in checkpoint-dependent fashion, by MMS treatment, but this may have been an effect of checkpoint-dependent retardation of replication fork movement (see below) rather than inhibition of origin firing, since ars2-2 was replicated by forks from flanking origins under the conditions employed here. Thus, at least for two of the three regions that we studied, the effects of MMS treatment in fission yeast cells seem opposite to those of HU treatment: the early centromere origins are unaffected by HU (13, 15, 21, 32) but are partially delayed by MMS, whereas the later-firing telomere origins are delayed by HU (13, 21, 32) but are unaffected by MMS.
Furthermore, our finding that the MMS-induced inhibition of centromeric origins is dependent both on Cds1 and on normal levels of Cdc25 (Fig. 3A and 4A) suggests that this inhibition is mediated by the same Rad3→Cds1⊣Cdc25→Cdc2 pathway that is also activated by HU treatment. In HU-treated cells, this pathway leads to inhibition of mitosis but is not known to affect origin firing. In contrast, in MMS-treated cells, our results suggest that this pathway affects the firing of centromeric origins (and perhaps other early origins), but the pathway is not known to affect entry into mitosis (note that MMS-treated cells do arrest in G2, prior to entry into mitosis, but that the arrest is dependent on Chk1, not Cds1 [47]). We conclude that additional pathways, which differ between MMS and HU, must be activated in MMS- and/or HU-treated cells and that these additional pathways must mediate the different effects of checkpoint activation on origin firing in MMS- and HU-treated cells.
The checkpoint response to MMS leads to inhibition of early-firing origins at centromeres but not late-firing origins at telomeres.
As indicated above, the finding that the checkpoint response to MMS leads to inhibition of early-firing origins at centromeres but not late-firing origins at telomeres was unanticipated, because it is quite different from what had previously been found in HU-treated fission yeast cells (21) or in HU- or MMS-treated budding yeast cells (41, 45). However, inhibition of early origin firing in response to DNA damage by ionizing radiation and by MMS was previously observed in mammalian cells (25, 31), so we should not have been surprised to find it in fission yeast. Furthermore, our laboratory has recently found that, in fission yeast cells bearing mutations in the genes encoding either of the histone deacetylases Clr3 and Clr6, early-firing origins are partially delayed while late-firing origins are unaffected (S. Ramanathan, R. M. Givens, A. Chaudari, G. Jahreis, and J. A. Huberman, unpublished data). Since the effects of these histone deacetylase mutations are similar to the effects of MMS treatment of checkpoint-competent cells, it is possible that MMS-induced inhibition of early origin firing could be mediated in part by modulation of the acetylation levels of histones (or other proteins, since both Clr3 and Clr6 are probably capable of deacetylating some nonhistone proteins).
The checkpoint response to MMS appears to include inhibition of replication fork movement.
The overall rate of passage through S phase is reduced in MMS-treated, checkpoint-competent cells (Fig. 1A). It is uncertain whether partial retardation of early centromeric replication origin firing (Fig. 2A) and, perhaps, retardation of the firing of other origins are sufficient to account for the overall slowing of S phase. An alternative mechanism that could also contribute to the slowing of S phase is reduction in rate of fork movement. Indeed, the observed persistence of RIs in MMS-treated, checkpoint-competent cells (Fig. 2 and 5A) can most easily be explained by reduced fork speed. Another possible alternative explanation for the persistence of RIs would be spreading the firing of origins over a longer time span, as seen for the origins in the centromeric K(dg) repeats in MMS-treated checkpoint-competent cells (Fig. 2A); this should lead to an equivalent increase in RI persistence without requiring a reduction in fork speed. Although staggering the firing of origins may contribute to the observed persistence of RIs, we think that reduced rate of fork movement is a more important contributor for the following reasons. First, the relative abundance of RIs at later time points is higher in MMS-treated wild-type cells than in untreated cells, consistent with the fact that slow-moving replication forks require a greater portion of the cell cycle to replicate a restriction fragment. Thus, RIs will always be more abundant in restriction fragments that are replicated by slow-moving forks. Second, in contrast, staggered origin firing would spread the signal from RIs over more time points, thus diluting the abundance of RIs at any given time point, which is not observed. Third, the time span over which centromeric RIs persist is longer than the delay in origin firing (Fig. 2A). Fourth, RIs persist even in the case of telomeres, for which no initial delay in origin firing is evident (Fig. 5A). Future investigations using microarray or DNA fiber fluorography techniques will be able to provide more precise characterization of the S-phase damage checkpoint effect on fork speed.
The mechanism by which replication forks might be slowed during S-phase damage checkpoint responses is not yet clear. The mechanism might involve uniform reduction in fork speed, or it might involve a combination of normal fork speed in undamaged regions with reduced fork speed, even complete fork stalling, in regions containing still-unrepaired damage. Several proteins in fission yeast both contribute to replication fork stabilization and participate in S-phase checkpoints. In some cases these proteins also bind directly to replication forks. These proteins include Mrc1, Swi1, Swi3, Tel2, and the Dfp1-Hsk1 kinase (1, 30, 33, 43, 46, 47, 50-52). Since these proteins contribute to fork stability, it seems likely that they may also be involved in slowing forks in cells with damaged DNA. Indeed, Tim and Tipin, the mammalian homologues of Swi1 and Swi3, are important for inhibiting replication fork movement after DNA damage by UV light (54). The hypothesis that proteins involved in maintenance of replication fork stability and/or in bypass of DNA lesions might be responsible for checkpoint-dependent damage-induced slowing of replication fork movement in fission yeast was previously proposed by Rhind and Russell (39). So far as we are aware, the persistence of RIs noted in this study (Fig. 2A and 5A) is the first experimental evidence supporting this hypothesis.
The fact that both persistence of RIs (which is evidence for fork slowing) and slowing of passage through S phase are abolished in cells overproducing the Cdc25 protein (Fig. 1C, 4, and 5C) implies that the Cdc25 protein and its target, the Cdc2 cyclin-dependent kinase, are likely to be involved in the mechanism of fork slowing. The activities of any of the above-mentioned proteins (Mrc1, Swi1, Swi3, Tel2, and Dfp1-Hsk1) might be modulated (directly or indirectly) by Cdc2. Another means by which Cdc2 might regulate fork movement is provided by the example of the mammalian cyclin-dependent kinase Cdk2, which phosphorylates several Mcm proteins (29). This phosphorylation may regulate the helicase activity of the Mcm complex and thus may regulate the rate of replication fork movement. Cdk2 also plays a role in chromatin decondensation during S phase (2). If chromatin decondensation is rate limiting for replication fork movement, then regulation of chromatin structure would provide another method by which Cdk2 in mammalian cells or Cdc2 in fission yeast could regulate replication fork rate.
The MMS-induced slowing of replication forks in budding yeast cells was reported to be checkpoint independent (53). However, this may depend on the concentration of MMS employed. We have found that, in fission yeast cells, a higher concentration (0.03%) of MMS frequently leads to checkpoint-independent S-phase slowing (S. Kumar and J. A. Huberman, unpublished observations). In addition, Szyjka et al. (49) recently found that Rad53 (the homologue of Cds1) slowed replication forks in MMS-treated budding yeast cells, presumably in order to stabilize them. Resumption of fork progression required inactivation of Rad53. The authors speculate that the apparent checkpoint independence of the previously observed (53) MMS-induced replication fork slowing might have been the consequence of a fortuitous balance between the fork-slowing effect of increased fork damage in rad53Δ cells and the fork-accelerating effect of loss of Rad53 in those cells.
Although checkpoint-mediated replication fork slowing in budding yeast is still controversial, there is accumulating evidence for its occurrence in mammalian cells. In two independent investigations, Chk1-mediated responses to DNA breaks near replication forks led to significant reductions in fork progression rates (42, 44), and in UV-treated mammalian cells, the Chk1-dependent checkpoint was activated and replication forks were slowed in a process requiring the fork protection proteins Tim and Tipin (54). Thus, combined observations in fission yeast, budding yeast, and mammalian cells suggest that checkpoint-dependent replication fork slowing is probably an evolutionarily conserved phenomenon.
The Cdc25 phosphatase is an important mediator of the S-phase damage checkpoint response to low levels of MMS.
Like the results in this paper (Fig. 1, 4, and 5C), our earlier observations (23) also suggested that the S-phase damage checkpoint response to low levels of MMS involves inhibition of Cdc25 activity, with consequent failure to activate Cdc2. Kommajosyula and Rhind (22) were unable to reproduce our earlier observations and questioned whether Cdc25 and Cdc2 are important for the checkpoint response to MMS treatment during S phase. However, some of their experiments were carried out at 0.03% MMS, which is twice as high as the highest concentration employed by us (0.015%). Furthermore, the effective concentration of the batch of MMS employed by Kommajosyula and Rhind (22) may have been greater than the effective concentration of the batch employed by us, for the following reasons. First, we have observed considerable batch-to-batch variation in effective concentrations of MMS, as judged by the concentration required to produce a given biological effect (which, in our case, is the retardation of progression through S phase in wild-type cells). Second, the Rhind laboratory has reported that, under their conditions, the MRN complex was required for the checkpoint response to MMS treatment (6). Under our conditions, however, deletions of components of the MRN complex had no effect on the checkpoint (28; Kumar and Huberman, unpublished). In mammalian cells and budding yeast, the MRN complex is required for responses to a specific kind of DNA damage, double-strand breaks (reviewed in reference 48), and formation of double-strand breaks is likely to occur primarily at high MMS concentrations, since MMS itself simply methylates DNA. Third, the magnitude of inhibition of progression through S phase was consistently higher in the studies of Kommajosyula and Rhind (22) than in our experiments, even for experiments carried out at the same nominal MMS concentrations. Finally, Kommajosyula and Rhind (22) did not test the effect of deleting the cds1 gene on MMS-induced S-phase slowing; they tested only deletion of the rad3 gene. But Rad3 is needed to activate both the Chk1 kinase and the Cds1 kinase. Thus, it is possible that the checkpoint dependence of MMS-induced S-phase slowing studied by Kommajosyula and Rhind (22) reflected a damage checkpoint pathway mediated by Chk1 rather than an S-phase pathway mediated by Cds1. In our hands, Cds1 but not Chk1 was required for the checkpoint response to low levels of MMS (28), and we have observed that higher levels of MMS lead to Cds1-independent slowing of S phase (Kumar and Huberman, unpublished).
Kommajosyula and Rhind (22) suspected that, in our earlier studies of the importance of Cdc25 for the S-phase checkpoint response to MMS (23), we may have been misled by our use of whole-cell flow cytometry rather than nuclear flow cytometry. However, the 2D gel results presented in this paper (Fig. 4 and 5C) are completely independent of the flow cytometry method. Instead, the abundance of RIs at each time point is a direct measure of the amount of DNA replication taking place within the detected restriction fragment at that time point. These results show clearly that cells overproducing the Cdc25 phosphatase are unable to slow replication when treated with the low level of MMS employed in our studies. In contrast, at the same level of MMS, wild-type cells are able to slow S phase (Fig. 2, 4, and 5A and C).
For these reasons, we suspect that fission yeast cells may employ (at least) two different pathways in their checkpoint responses to MMS damage during S phase. When MMS damage is minimal and few or no double-strand breaks are generated, both Cdc25 and Cdc2 are important downstream components of the checkpoint response (23; this paper). However, when MMS damage is extensive enough to generate double-strand breaks, the MRN complex may mediate a stronger response, which overwhelms the response involving Cdc25 and Cdc2 (6, 22). The involvement of two response pathways to MMS damage in fission yeast is reminiscent of the two pathways responding to UVC damage in mammalian cells: one pathway, involving ATR, Chk1, and Dbf4, responds to low levels of UVC, while a second pathway, involving ATM, ATR, and the MRN complex, responds to higher levels (14, 34).
Loss of telomere replication synchrony in checkpoint mutant cells.
In the absence of MMS, we observed that the temporal replication patterns of the K(dg) repeats and the ars2-2 region in wild-type and checkpoint-mutant cells were similar (Fig. 2 to 4). In all cases, the K(dg) repeats replicated before ars2-2, with their replication times roughly 30 min apart. This conservation of replication timing in untreated checkpoint-competent and checkpoint-mutant cells for the K(dg) repeats and for ars2-2 contrasts with the loss of replication synchrony for the telomere regions in checkpoint-mutant cells (Fig. 5B and C). The contrast is more striking considering that the different results were obtained for the same 2D gel membranes, which were hybridized, stripped, and rehybridized until all three of the probes [ars2-2, K(dg) repeats, and telomeres] produced results (Fig. 2 to 5). Thus, loss of telomere replication synchrony and timing occurred in the very same checkpoint-incompetent cells in which the synchrony and timing of the K(dg) repeats and ars2-2 were preserved.
The reduction in telomere replication synchrony was more severe for the cdc25OP strain than for the cds1Δ strain. Note that cds1Δ cells are thought to be defective in regulation of Cdc2 activity only upon DNA damage. In contrast, the Cdc2 protein is constitutively more active in cdc25OP cells. This difference in Cdc2 regulation may underlie the differences between the replication profiles of telomeric regions in the untreated cds1Δ and cdc25OP cells that we observed. These results suggest that a functional checkpoint mechanism that regulates Cdc2 activity may be required to restrain the replication of telomeres even in the absence of DNA damage, at least under these experimental conditions.
Fission yeast as a model system for understanding the role of cyclin-dependent kinase activity in regulating both replication fork movement and origin firing in response to DNA damage.
Involvement of Cdc25 and Cdk homologues in the S-phase damage checkpoint was first demonstrated in vertebrates. Recent studies of vertebrates indicate that this checkpoint leads to inhibition of both origin firing and replication fork movement. Similarly, our experiments provide evidence for roles for Cdc2 in controlling both origin firing and rates of replication fork movement after DNA damage. In budding yeast, where the effects of DNA damage upon DNA replication have been most extensively studied, the Cdc25 and Cdc2 homologues do not appear to play any role in the DNA damage-induced slowing of S phase. Furthermore, the firing of early origins is not affected in budding yeast (45). Thus, S. pombe could be the model system of choice for asking further questions about the targets of Cdc2 in the replication apparatus that bring about both slowing of replication forks and inhibition of early (and possibly late) replication origins when DNA is damaged.
Acknowledgments
We thank Greg Freyer for the cdc10-v50 wild-type strain, and we thank the referees of both versions of the manuscript for their helpful comments.
This work was supported by Public Health Service grants CA095908 from the National Cancer Institute and GM070566 from the National Institute for General Medical Sciences. This study benefited from Roswell Park Cancer Institute's Flow Cytometry facility, which is supported by Roswell Park Cancer Institute's Cancer Center support grant P30-CA16056 from the National Cancer Institute.
Footnotes
Published ahead of print on 10 November 2008.
REFERENCES
- 1.Alcasabas, A. A., A. J. Osborn, J. Bacchant, F. Hu, P. J. H. Werler, K. Bousset, K. Furuya, J. F. X. Diffley, A. M. Carr, and S. J. Elledge. 2001. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3958-965. [DOI] [PubMed] [Google Scholar]
- 2.Alexandrow, M. G., and J. L. Hamlin. 2005. Chromatin decondensation in S-phase involves recruitment of Cdk2 by Cdc45 and histone H1 phosphorylation. J. Cell Biol. 168875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreassen, P. R., G. P. H. Ho, and A. D. D'Andrea. 2006. DNA damage responses and their many interactions with the replication fork. Carcinogenesis 27883-892. [DOI] [PubMed] [Google Scholar]
- 4.Boddy, M. N., and P. Russell. 2001. DNA replication checkpoint. Curr. Biol. 11R953-R956. [DOI] [PubMed] [Google Scholar]
- 5.Brewer, B. J., and W. L. Fangman. 1987. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell 51463-471. [DOI] [PubMed] [Google Scholar]
- 6.Chahwan, C., T. M. Nakamura, S. Sivakumar, P. Russell, and N. Rhind. 2003. The fission yeast Rad32 (Mre11)-Rad50-Nbs1 complex is required for the S-phase DNA damage checkpoint. Mol. Cell. Biol. 236564-6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costanzo, V., D. Shechter, P. J. Lupardus, K. A. Cimprich, M. Gottesman, and J. Gautier. 2003. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell 11203-213. [DOI] [PubMed] [Google Scholar]
- 8.Duncker, B. P., K. Shimada, M. Tsai-Pflugfelder, P. Pasero, and S. M. Gasser. 2002. An N-terminal domain of Dbf4p mediates interaction with both origin recognition complex (ORC) and Rad53p and can deregulate late origin firing. Proc. Natl. Acad. Sci. USA 9916087-16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410842-847. [DOI] [PubMed] [Google Scholar]
- 10.Feng, W., D. Collingwood, M. E. Boeck, L. A. Fox, G. M. Alvino, W. L. Fangman, M. K. Raghuraman, and B. J. Brewer. 2006. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat. Cell Biol. 8148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furnari, B., A. Blasina, M. N. Boddy, C. H. McGowan, and P. Russell. 1999. Cdc25 inhibited in vivo and in vitro by checkpoint kinases Cds1 and Chk1. Mol. Biol. Cell 10833-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould, K. L., S. Moreno, N. K. Tonks, and P. Nurse. 1990. Complementation of the mitotic activator, p80cdc25, by a human protein-tyrosine phosphatase. Science 2501573-1576. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, M., Y. Katou, T. Itoh, M. Tazumi, Y. Yamada, T. Takahashi, T. Nakagawa, K. Shirahige, and H. Masukata. 2007. Genome-wide localization of pre-RC sites and identification of replication origins in fission yeast. EMBO J. 261327-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heffernan, T. P., K. Unsal-Kacmaz, A. N. Heinloth, D. A. Simpson, R. S. Paules, A. Sancar, M. Cordeiro-Stone, and W. K. Kaufmann. 2007. Cdc7-Dbf4 and the human checkpoint response to UVC. J. Biol. Chem. 2829458-9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heichinger, C., C. J. Penkett, J. Bähler, and P. Nurse. 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J. 255171-5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huberman, J. A., L. D. Spotila, K. A. Nawotka, S. M. El-Assouli, and L. R. Davis. 1987. The in vivo replication origin of the yeast 2μm plasmid. Cell 51473-481. [DOI] [PubMed] [Google Scholar]
- 17.Hyrien, O., and M. Mechali. 1992. Plasmid replication in Xenopus eggs and egg extracts: a 2D gel electrophoretic analysis. Nucleic Acids Res. 201463-1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kalejta, R. F., and J. L. Hamlin. 1996. Composite patterns in neutral/neutral two-dimensional gels demonstrate inefficient replication origin usage. Mol. Cell. Biol. 164915-4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 4241078-1083. [DOI] [PubMed] [Google Scholar]
- 20.Kim, S.-M., D. D. Dubey, and J. A. Huberman. 2003. Early-replicating heterochromatin. Genes Dev. 17330-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim, S. M., and J. A. Huberman. 2001. Regulation of replication timing in fission yeast. EMBO J. 206115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kommajosyula, N., and N. Rhind. 2006. Cdc2 tyrosine phosphorylation is not required for the S-phase DNA damage checkpoint in fission yeast. Cell Cycle 52495-2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumar, S., and J. A. Huberman. 2004. On the slowing of S phase in response to DNA damage in fission yeast. J. Biol. Chem. 27943574-43580. [DOI] [PubMed] [Google Scholar]
- 24.Lambert, S., and A. M. Carr. 2005. Checkpoint responses to replication fork barriers. Biochimie 87591-602. [DOI] [PubMed] [Google Scholar]
- 25.Larner, J. M., H. Lee, and J. L. Hamlin. 1994. Radiation effects on DNA synthesis in a defined chromosomal replicon. Mol. Cell. Biol. 141901-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindsay, H. D., D. J. Griffiths, R. J. Edwards, P. U. Christensen, J. M. Murray, F. Osman, N. Walworth, and A. M. Carr. 1998. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes Dev. 12382-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 2881425-1429. [DOI] [PubMed] [Google Scholar]
- 28.Marchetti, M. A., S. Kumar, E. Hartsuiker, M. Maftahi, A. M. Carr, G. A. Freyer, W. C. Burhans, and J. A. Huberman. 2002. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. Proc. Natl. Acad. Sci. USA 997472-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Masai, H., E. Matsui, Z. You, Y. Ishimi, K. Tamai, and K. Arai. 2000. Human Cdc7-related kinase complex. In vitro phosphorylation of MCM by concerted actions of Cdks and Cdc7 and that of a critical threonine residue of Cdc7 by Cdks J. Biol. Chem. 27529042-29052. [DOI] [PubMed] [Google Scholar]
- 30.Matsumoto, S., K. Ogino, E. Noguchi, P. Russell, and H. Masai. 2005. Hsk1-Dfp1/Him1, the Cdc7-Dbf4 kinase in Schizosaccharomyces pombe, associates with Swi1, a component of the replication fork protection complex. J. Biol. Chem. 28042536-42542. [DOI] [PubMed] [Google Scholar]
- 31.Merrick, C. J., D. Jackson, and J. F. X. Diffley. 2004. Visualization of altered replication dynamics after DNA damage in human cells. J. Biol. Chem. 27920067-20075. [DOI] [PubMed] [Google Scholar]
- 32.Mickle, K. L., S. Ramanathan, A. Rosebrock, A. Oliva, A. Chaudari, C. Yompakdee, D. Scott, J. Leatherwood, and J. A. Huberman. 2007. Checkpoint-independence of most DNA replication origins in fission yeast. BMC Mol. Biol. 8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noguchi, E., C. Noguchi, W. H. McDonald, J. R. Yates III, and P. Russell. 2004. Swi1 and Swi3 are components of a replication fork protection complex in fission yeast. Mol. Cell. Biol. 248342-8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Olson, E., C. J. Nievera, A. Y. L. Lee, L. Chen, and X. Wu. 2007. The Mre11-Rad50-Nbs1 complex acts both upstream and downstream of ataxia telangectasia mutated and Rad3-related protein (ATR) to regulate the S-phase checkpoint following UV treatment. J. Biol. Chem. 28222939-22952. [DOI] [PubMed] [Google Scholar]
- 35.Patel, P. K., B. Arcangioli, S. P. Baker, A. Bensimon, and N. Rhind. 2006. DNA replication origins fire stochastically in fission yeast. Mol. Biol. Cell 17308-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulovich, A. G., and L. H. Hartwell. 1995. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82841-847. [DOI] [PubMed] [Google Scholar]
- 37.Paulovich, A. G., R. U. Margulies, B. M. Garvik, and L. H. Hartwell. 1997. RAD9, RAD17, and RAD24 are required for S phase regulation in Saccharomyces cerevisiae in response to DNA damage. Genetics 14545-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raghuraman, M. K., E. A. Winzeler, D. Collingwood, S. Hunt, L. Wodicka, A. Conway, D. J. Lockhart, R. W. Davis, B. J. Brewer, and W. L. Fangman. 2001. Replication dynamics of the yeast genome. Science 294115-121. [DOI] [PubMed] [Google Scholar]
- 39.Rhind, N., and P. Russell. 2000. Checkpoints: it takes more than time to heal some wounds. Curr. Biol. 10R908-R911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhind, N., and P. Russell. 1998. Tyrosine phosphorylation of Cdc2 is required for the replication checkpoint in Schizosaccharomyces pombe. Mol. Cell. Biol. 183782-3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santocanale, C., and J. F. Diffley. 1998. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature 395615-618. [DOI] [PubMed] [Google Scholar]
- 42.Seiler, J. A., C. Conti, A. Syed, M. I. Aladjem, and Y. Pommier. 2007. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell. Biol. 275806-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shikata, M., F. Ishikawa, and J. Kanoh. 2007. Tel2 Is required for activation of the Mrc1-mediated replication checkpoint. J. Biol. Chem. 2825346-5355. [DOI] [PubMed] [Google Scholar]
- 44.Shimura, T., M. M. Martin, M. J. Torres, C. Gu, J. M. Pluth, M. A. DiBernardi, J. S. McDonald, and M. I. Aladjem. 2007. DNA-PK is involved in repairing a transient surge of DNA breaks induced by deceleration of DNA replication. J. Mol. Biol. 367665-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shirahige, K., Y. Hori, K. Shiraishi, M. Yamashita, K. Takahashi, C. Obuse, T. Tsurimoto, and H. Yoshikawa. 1998. Regulation of DNA-replication origins during cell-cycle progression. Nature 395618-621. [DOI] [PubMed] [Google Scholar]
- 46.Snaith, H. A., G. W. Brown, and S. L. Forsburg. 2000. Schizosaccharomyces pombe Hsk1p is a potential Cds1p target required for genome integrity. Mol. Cell. Biol. 207922-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sommariva, E., T. K. Pellney, N. Karahan, S. Kumar, J. A. Huberman, and J. Z. Dalgaard. 2005. Schizosaccharomyces pombe Swi1, Swi3, and Hsk1 are components of a novel S-phase response pathway to alkylation damage. Mol. Cell. Biol. 252770-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stracker, T. H., J. W. Theunissen, M. Morales, and J. H. Petrini. 2004. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair 3845-854. [DOI] [PubMed] [Google Scholar]
- 49.Szyjka, S. J., J. G. Aparicio, C. J. Viggiani, S. Knott, W. Xu, S. Tavaré, and O. M. Aparicio. 2008. Rad53 regulates replication fork restart after DNA damage in Saccharomyces cerevisiae. Genes Dev. 221906-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takeda, T., K. Ogino, E. Matsui, M. K. Cho, H. Kumagai, T. Miyake, K.-I. Arai, and H. Masai. 1999. A fission yeast gene, him1+/dfp1+, encoding a regulatory subunit for Hsk1 kinase, plays essential roles in S-phase initiation as well as in S-phase checkpoint control and recovery from DNA damage. Mol. Cell. Biol. 195535-5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeda, T., K. Ogino, K. Tatebayashi, H. Ikeda, K.-I. Arai, and H. Masai. 2001. Regulation of initiation of S phase, replication checkpoint signaling, and maintenance of mitotic chromosome structures during S phase by Hsk1 kinase in the fission yeast. Mol. Biol. Cell 121257-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka, K., and P. Russell. 2001. Mrc1 channels the DNA replication arrest signal to checkpoint kinase Cds1. Nat. Cell Biol. 3966-972. [DOI] [PubMed] [Google Scholar]
- 53.Tercero, J. A., and J. F. Diffley. 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412553-557. [DOI] [PubMed] [Google Scholar]
- 54.Unsal-Kacmaz, K., P. D. Chastain, P. P. Qu, P. Minoo, M. Cordeiro-Stone, A. Sancar, and W. K. Kaufmann. 2007. The human Tim/Tipin complex coordinates an intra-S checkpoint response to UV that slows replication fork displacement. Mol. Cell. Biol. 273131-3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weinreich, M., and B. Stillman. 1999. Cdc7p-Dbf4p kinase binds to chromatin during S phase and is regulated by both the APC and the RAD53 checkpoint pathway. EMBO J. 185334-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]