Abstract
Deacetylation of histones is carried out by a corepressor complex in which Sin3A is an essential scaffold protein. Two proteins in this complex, the Sin3A-associated proteins SAP30L and SAP30, have previously been suggested to function as linker molecules between various corepressors. In this report, we demonstrate new functions for human SAP30L and SAP30 by showing that they can associate directly with core histones as well as naked DNA. A zinc-coordinating structure is necessary for DNA binding, one consequence of which is bending of the DNA. We provide evidence that a sequence motif previously shown to be a nuclear localization signal is also a phosphatidylinositol (PI)-binding element and that binding of specific nuclear monophosphoinositides regulates DNA binding and chromatin association of SAP30L. PI binding also decreases the repression activity of SAP30L and affects its translocation from the nucleus to the cytoplasm. Our results suggest that SAP30L and SAP30 play active roles in recruitment of deacetylating enzymes to nucleosomes, and mediate key protein-protein and protein-DNA interactions involved in chromatin remodeling and transcription.
A basic unit of chromatin is the nucleosome, in which 147 bp of DNA is wrapped around a histone octamer core composed of the four histones H2A, H2B, H3, and H4. The N-terminal “tail” domains of these histones project out of the nucleosome core and are the main sites of posttranslational modifications, such as acetylation, methylation, and phosphorylation. These covalent modifications have been proposed to play important roles in regulation of gene expression. According to the “histone code” hypothesis, the modifications function as “marks” which are recognized by various proteins required for the dynamic alterations in chromatin structure that are needed to make a gene accessible to the components of the transcription machinery.
The recruitment of histone deacetylases (HDACs) to chromatin is a common mechanism of transcriptional repression (59). While certain repressors, such as Rb and YY1, are able to recruit HDACs directly (5, 58), others, such as Mad-Max and the nuclear hormone receptor, require association with corepressors (1, 17, 18, 29, 42). Another example of the latter is the Sin3A corepressor complex, in which the Sin3A protein itself does not bind DNA or possess any enzymatic activity. Instead, it is composed of domains that mediate protein-protein interactions and thereby forms a platform for several enzymes (e.g., HDACs and methyltransferases), DNA-binding transcription factors (e.g., Mad family repressors, MeCP2, and Pf1), and other “bridging” proteins (e.g., SDS3) (52). The Sin3A-HDAC corepressor complex contains HDAC1 and HDAC2, the histone-binding proteins RbAp46 and RbAp48, Sin3A-associated protein 18 (SAP18), SAP30, and SDS3 (52).
Mammalian HDAC1 and HDAC2 are almost identical, each containing an N-terminal catalytic domain, which removes acetyl moieties from the ɛ-amino groups of lysine residues, and a C-terminal tail. They are class I HDACs, which share sequence similarity with the Rpd3 (reduced potassium dependency-3) protein in Saccharomyces cerevisiae (59). The corepressor complex components RbAp46 and RbAp48 share 90% sequence identity and belong to the WD repeat family. These proteins have been shown to bind core histones H3 and H4 and are therefore thought to target HDAC-containing complexes to their histone substrates (56). Another member of the complex, SAP18, interacts directly with Sin3A and has multiple functions (60). In addition to regulating transcription, it also participates in mRNA processing (49). SDS3 has been suggested to play a role in stabilizing the Sin3A complex, and a yeast strain lacking SDS3 possesses only residual Sin3A-associated HDAC activity (31). SDS3 also participates in pericentric heterochromatin formation and chromosome segregation (9). Other components, such as SAP25, SAP130, and SAP180, have been reported to associate with the Sin3A-HDAC complex, but their roles in the complex have remained elusive (13, 51).
SAP30 (Sin3A-associated protein 30) was originally identified as a conserved member of the Sin3A corepressor complex (28, 61). It was shown to be required for N-CoR-mediated repression by the antagonist-bound estrogen receptor and the POU domain protein Pit-1 but not by the unliganded retinoic acid receptor or thyroid hormone receptor complexes (28). In Saccharomyces cerevisiae, SAP30 was demonstrated to be important for cell growth and to affect gene expression in a promoter-dependent manner (61). A SAP30-deficient mutant strain exhibited enhanced silencing of the ribosomal DNA, HMR, and telomeric loci, which suggested an antisilencing function for SAP30 (36, 54, 55). Meskauskas et al. observed that mutations in Rpd3p, Sin3p, and Sap30p resulted in a defect in rRNA processing rather than ribosomal DNA transcription (41). Human SAP30, in addition to its associations in the Sin3A-HDAC complex, has been reported to interact with a number of other proteins, such as retinoblastoma-binding protein 1 (RBP1) (30), the CBF1-interacting corepressor (CIR) (20), the YY1 transcription factor (21), and the inhibitor of growth 1b (ING1b) tumor suppressor protein (27, 53). Recently, SAP30-mediated transcriptional repression was shown to play a role in the transmission and propagation of certain viruses (26, 32). The above observations indicate that SAP30 plays a vital role in transcriptional regulation, which can be either negative or positive, depending on local factors, such as chromatin state and the presence of various interacting partners.
Human SAP30L (SAP30-like), thus named because it shares 70% sequence identity with SAP30, is the “newest” member of the Sin3A corepressor complex. It was originally discovered as an expressed transcript in cultured T84 cells induced to differentiate in response to transforming growth factor β (35). Since then, SAP30L has been shown to associate with the Sin3A-HDAC complex and to induce transcriptional repression in a Sin3A- and HDAC-dependent manner (57). Both SAP30 and SAP30L are able to localize to the nucleus or the nucleolus, and we have demonstrated that they can direct Sin3A to the nucleolus (57).
Previous work has led to the view that SAP30 and SAP30L serve mainly a bridging role in various corepressor complexes. In this study, we set out to investigate the functions and the domain structures of these proteins in more detail. We show that both proteins directly bind and bend DNA and interact with core histones 2A/2B. Interestingly, our results suggest that both DNA binding and chromatin association are regulated by nuclear monophosphorylated phosphoinositides (PIs).
MATERIALS AND METHODS
Antibodies, immunoblotting, and immunofluorescence.
The primary antibodies used were those against histone 2B (sc-8650), c-myc (sc-40), Sin3A (sc-767 and sc-5299), HDAC 1 (sc-7872), calregulin (sc-11398), and histone H1 (sc-8030) from Santa Cruz and glutathione S-transferase (GST) (27-4577-01) from GE Healthcare. Immunoblot analyses were done according to standard protocols, and anti-rabbit, anti-mouse, or anti-goat horseradish peroxidase-conjugated secondary antibodies (p0217, p0260, or p0449, respectively) were obtained from Dako. Immunofluorescence was performed as described previously (57), and Alexa fluorophore-conjugated anti-mouse antibody (A11031) was used for detection of immunocomplexes.
Cloning and plasmid constructs.
Full-length SAP30L and deletion mutants were cloned into pcDNA 3.1-myc-his (Invitrogen), pGEX-4T1, and GAL4-DBD (Stratagene) vectors, and some of the constructs have already been described in references 25 and 57. The SAP30L-green fluorescent protein (GFP) and pcDNA3.1-Sin3A1-855 constructs are described in references 35 and 57, respectively. The H2B-GFP construct was a generous gift from P. Peterson (Tartu, Estonia) (46). Point mutations were created using a QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The luciferase reporter vector, under the control of 14D promoters harboring 5× Gal4 sites, was generously provided by D. Ayer (Salt Lake City, UT). The precise coordinates of the constructs will be supplied on request. The integrity of the constructs was confirmed by sequencing.
Cell culture, H2O2 treatment, and transfections.
Human embryonic kidney epithelial cells (HEK293T) were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with penicillin and streptomycin, 5% fetal bovine serum, 1 mM sodium pyruvate, and 50 μg/ml uridine. HeLa cells were cultured in RPMI 1640 (Gibco) supplemented with penicillin and streptomycin, 10% fetal bovine serum, and l-glutamine. Cells were treated for 15 min with 0.5 mM H2O2, washed, and allowed to grow for 4 h before being harvested for analysis. DNA was transfected with FuGENE 6 (for HEK293T cells) and FuGENE HD (for HeLa cells) reagents (Roche) according to the manufacturer's protocol.
GST pulldown experiments.
GST fusion proteins were produced in Escherichia coli (BL-21 strain) and purified with glutathione-Sepharose 4B beads (Amersham Biosciences) according to the manufacturer's instructions. The fusion proteins were eluted from the beads with reduced glutathione, if necessary. The quantity and integrity of the GST fusion proteins were checked using Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel.
GST pulldown experiments with nucleosomes and histones, with the Sin3a 1-855 protein transcribed and translated in vitro, were carried out as described previously (57), except that PI or PI 5-phosphate [PI(5)P] (Echelon, Inc.) was added to the reaction mixtures where indicated.
Electrophoretic mobility shift assays (EMSA) and a novel DNA ladder EMSA (L-EMSA).
A 150-bp DNA probe comprising the mitochondrial tRNA-Leu(UUR) sequence was end labeled with [γ-32P]ATP as described in reference 22. The probe was incubated with 0.5 μg of a GST fusion protein on ice for 30 min in a buffer containing 50 mM Tris-HCl, pH 7.5, 125 mM NaCl, 2.5 mM dithiothreitol, 0.5 mM EDTA, 1 mM MgCl2, and 4% glycerol. The reaction products were analyzed on a 6% nondenaturing polyacrylamide gel, dried, and autoradiographed.
Because of the non-sequence-specific nature of the binding by the SAP30L and SAP30 proteins, a faster and simpler assay, L-EMSA, was designed for the protein-DNA interaction studies. Five micrograms of the fusion protein was incubated with 0.25 μg of a 1-kb DNA ladder (GeneRuler; Fermentas) in phosphate-buffered saline for 10 min at room temperature. The reactions were run on ethidium bromide-containing 1% agarose gel with standard DNA gel loading buffer. Prior to use, this method was validated by comparing the DNA band shifts in L-EMSA to shifts in conventional EMSA. The GST fusion proteins used in Fig. 1A generated identical shifts in both assays (data not shown). Where indicated, PI and PI(5)P were added after the protein-DNA complex formation.
FIG. 1.
The N-terminal domains of SAP30L and SAP30 bind DNA in a sequence-independent manner. (A) A 32P-labeled DNA probe was incubated with GST fusion proteins, and the DNA-protein complexes were analyzed by an EMSA. (B) Median fluorescence intensities of all microarray probe oligonucleotides containing a particular 8-mer sequence, from PBM experiments performed with GST, GST-SAP30L, GST-SAP30, and GST-Cbf1.
PBM experiments and data analysis.
Protein binding microarray (PBM) experiments and analyses were performed as described in reference 3 for four different GST-tagged protein constructs (full-length SAP30, full-length SAP30L, SAP30 residues 1 to 131 [SAP30 1-131], and SAP30L residues 1 to 92 [SAP30L 1-92]) and a GST control (the protein concentration was ∼1uM). An Alexa 488-conjugated anti-GST antibody was applied to the protein-bound microarray to detect bound protein. The feature set of ∼44,000 oligonucleotides present on the custom-designed Agilent microarrays followed the design described in reference 3 but was incorporated on the Agilent 4x44K array platform, allowing four independent PBM experiments to be performed simultaneously on the same microarray. To identify DNA-binding-site motifs, two approaches were used: (i) the approach described in reference 3, based on perturbations of the highest-ranked 8-bp sequence; and (ii) a de novo motif search of the sequences from the 20, 30, and 50 brightest microarray probes, using the program MEME (2).
Mass spectrometry.
Wild-type and mutant SAP30L 1-92 peptides were cleaved from GST by prothrombin, yielding SAP30L 1-94 peptides with two additional amino acids from the GST vector. The samples were desalted on PD-10 columns (Amersham Biosciences, Uppsala, Sweden), and concentrations were estimated from absorbance at 280 nm by using ɛ280 = 2,560 cm−1 M−1. Prior to measurements, the samples were further diluted with the appropriate solvents: CH3CN-H2O-acetic acid (49.5:49.5:1.0, vol/vol, pH 3.2) for denaturing solution conditions and 10 mM ammonium acetate buffer (pH 6.8) for nondenaturing solution conditions. All experiments were performed with a 4.7-T hybrid quadrupole Fourier transform ion cyclotron resonance (Q-FT-ICR) instrument (APEX-Qe; Bruker Daltonics, Billerica, MA) interfaced with an external electrospray ionization (ESI) source (Apollo-II). The samples were infused directly at a flow rate of 1.5 μl min−1, with dry N2 serving as the drying (10 lb/in2, 200°C) and nebulizing gas. ESI-generated ions were externally accumulated in a hexapole ion trap for 0.5 to 1.0 s and transferred to an Infinity ICR cell for Sidekick trapping, conventional “RF-chirp” excitation, and broadband detection. A total of up to 256 coadded (1-megaword) time domain transients were fast Fourier transformed prior to magnitude calculation and external frequency-to-m/z calibration with respect to the ions of an ES tuning mix (Agilent Technologies, Santa Clara, CA). All data were acquired and processed with the use of Bruker XMASS 7.0.8 software.
DNA bending/ligation-mediated circularization assay.
The ligation-mediated circularization assay was essentially performed as described previously (45). See also the legend to Fig. 3.
FIG. 3.
SAP30L bends DNA. (A) Probes of various lengths (170, 150, 130, 110, 90, and 70 bp), labeled internally with 32P and containing EcoRI “sticky ends,” were incubated with GST only (lanes 4, 9, 14, 19, 24, and 29) or with GST-SAP30L (lanes 5, 10, 15, 20, 25, and 30). The reaction mixtures for lanes 2 to 5, 7 to 10, 12 to 15, 17 to 20, 22 to 25, and 27 to 30 were incubated in the presence of T4 DNA ligase at 30°C for 20 min. The reaction mixtures for lanes 3 to 5, 8 to 10, 13 to 15, 18 to 20, 23 to 25, and 28 to 30 were subsequently treated with exonuclease (exo) III to remove any linear ligation products. The reaction products were electrophoresed on a 7% polyacrylamide gel, which was dried and subjected to autoradiography. Mono-, di-, and tricircular DNA ligation products are indicated by the numbered circles. nt, nucleotides or base pairs. (B) Increasing amounts of GST-SAP30L 1-92 were used in the ligation reactions, which were performed as described above.
Protein-lipid blot assays.
PI phosphate (PIP) strips and arrays were purchased from Echelon Biosciences. Protein-lipid blot assays were performed by adding 0.5 μg/ml of GST fusion proteins and were further processed as described in the manufacturer's protocol. Each protein-lipid blot experiment was repeated at least once.
Nucleosome preparations.
Intact nucleosomes and tailless nucleosomes were prepared as described previously (37). The presence of solubilized nucleosomes was confirmed by DNA agarose gel electrophoresis and SDS-PAGE, followed by Coomassie staining (see Fig. S4A in the supplemental material). Calf thymus histones were purchased from Roche. For the GST fusion pulldown experiments, 30 μg of nucleosomes, tailless nucleosomes, or histone proteins was used. For the initial screening experiment (Fig. 4B), 100 μg of histones was used.
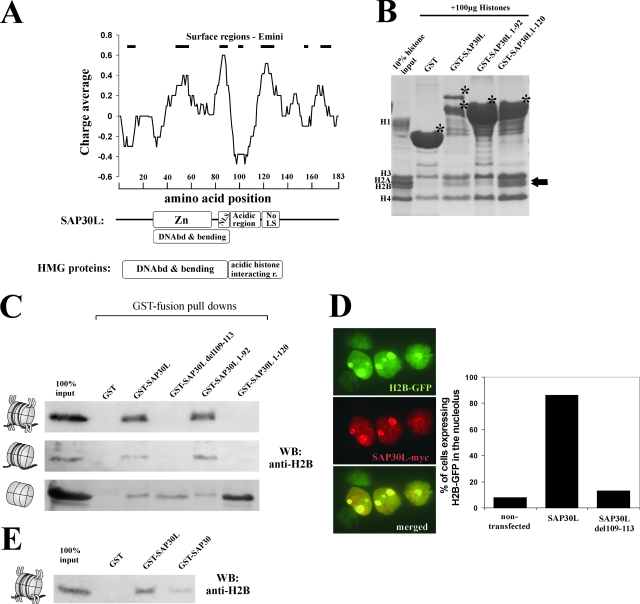
FIG. 4.
SAP30L and SAP30 bind histones and nucleosomes. (A) Schematic representation of the domain architecture of the HMG proteins, SAP30L, and SAP30. The charge average of SAP30L is presented as a sliding window of 10 aa. Surface probability predictions were performed using the approach of Emini (11), and values greater than 2 are shown as black lines. Zn, zinc-binding module; DNAbd & bending, DNA-binding and -bending domain. (B) GST fusion protein pulldowns of calf thymus histones (Roche) were analyzed by SDS-PAGE and stained with Coomassie blue. The asterisks mark the GST fusion proteins, and the arrows indicate interaction with histones 2A/2B. (C) GST fusion protein pulldowns of intact nucleosomes, of nucleosomes from which the tails had been removed with trypsin (see Fig. S4 in the supplemental material), and of calf thymus histones. WB, Western blot. (D) HEK293T cells were cotransfected with an H2B-GFP fusion protein and either wild-type SAP30L or SAP30Ldel109-113 containing a myc-His tag. The cells were stained with the anti-myc antibody, and nuclei positive for both H2B and GFP were scored from 50 cells. The results are illustrated in the histogram. (E) GST fusion pulldowns of intact nucleosomes were analyzed using a Western blot probed with the anti-H2B antibody.
Interphase chromatin spreads, chromatin isolation, and subcellular fractionation.
Chromatin spread preparation for the SAP30L-GFP-transfected HEK293T cells was performed as described previously (38), with the following modifications. Nocodazole was not added, and the collected, phosphate-buffered-saline-washed, hypotonically swollen cells were dropped to a tilted microscope glass. The unfixed, dried drop was counterstained with DAPI (4′,6-diamidino-2-phenylindole) and photographed under a confocal microscope. Chromatin isolation and subcellular fractionation were performed as described previously (39).
Repression analysis.
GAL4DBD-based repression analyses were performed as described previously (57).
RESULTS
SAP30L and SAP30 bind DNA.
It is generally believed that specific repressor proteins are needed to direct the Sin3A-HDAC complex to its target sequence, because Sin3A itself does not bind DNA. Since SAP30L is able to repress transcription in a Sin3A- and HDAC-dependent manner (25, 57), we examined if it could bind DNA directly. An EMSA was carried out using a GST-SAP30L fusion protein incubated in the presence of mitochondrial DNA. As shown in Fig. 1A, a marked shift was detected in the mobility of the DNA after incubation with GST-SAP30L, indicating a direct interaction with DNA. A GST-SAP30 fusion protein also bound DNA, whereas GST alone did not. SAP30L proteins with a C-terminal deletion (GST-SAP30L 1-120) or a mutated nucleolar localization signal (NoLS; 8A mutant) were also able to interact with DNA. Deletion of 25 amino acid residues from the N terminus did not affect DNA binding, whereas a larger deletion of 60 N-terminal residues (GST-SAP30L 61-183) abolished the shift. Similarly, a deletion mutant lacking 40 residues from the N terminus failed to interact with DNA. These results show that both SAP30L and SAP30 are able to bind DNA and that the region between residues 25 to 120 of SAP30L contains the DNA-binding determinants.
DNA binding by SAP30L and SAP30 is not sequence specific.
In order to investigate whether SAP30L and SAP30 can target Sin3A to specific DNA sequences, we took advantage of a recently developed method, the PBM (6a, 41a). GST-tagged proteins were applied to a microarray synthesized with double-stranded DNA oligonucleotides in order to assess the binding preference for all possible contiguous and gapped 8-bp DNA sequence variants. Utilizing the universal microarray design and binding protocol recently described (3) (see Materials and Methods), we performed PBM experiments using four constructs: full-length fusion proteins GST-SAP30L and GST-SAP30 (Fig. 1B) and N-terminally truncated fusion proteins GST-SAP30L (amino acids [aa] 1 to 92) and SAP30 (aa 1 to 131) (see Fig. S1 in the supplemental material). Using two different approaches (see Materials and Methods), we were unable to derive any binding motifs that would demonstrate specific binding behavior. We further examined the median signal intensity distribution of probe sequences containing each 8-bp sequence variant and compared it to the distribution from a PBM experiment in which the sequence-specific transcription factor Cbf1 from S. cerevisiae was used as a positive control (3). The Z-transformed distributions of results from all four experiments with SAP30L were very similar to that for the negative-control experiment (GST only), while the distribution of results from the Cbf1 control experiment showed a narrower distribution and a longer tail at high scores, indicative of specifically bound probes (Fig. 1B). This comparison further suggested a lack of sequence-specific DNA binding of SAP30L and SAP30.
SAP30L and SAP30 contain an N-terminal zinc-coordinating signature.
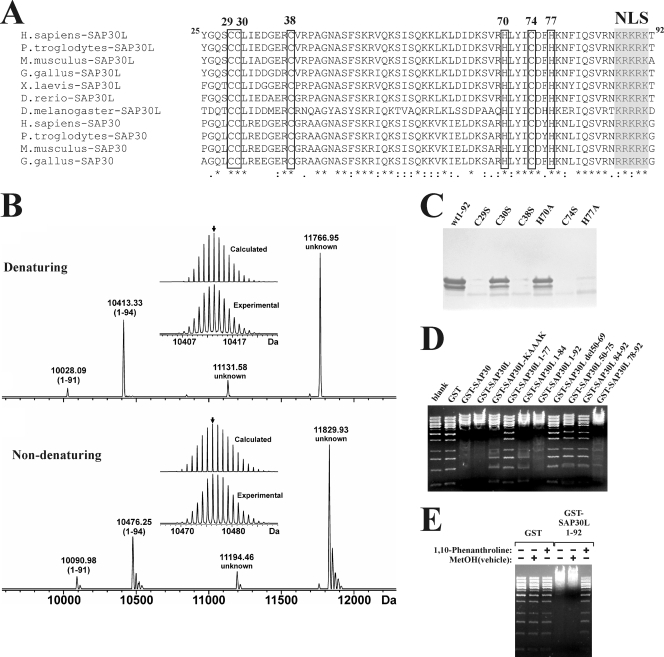
A well-characterized DNA-binding element is the zinc finger, which resembles a finger with a base of four cysteine and/or histidine residues that coordinate a zinc ion through the thiol groups of cysteine residues and/or the imidazole nitrogens of histidine residues (33, 48). In a stretch of 49 residues (aa 29 to 77 in human SAP30L), we identified four cysteine and two histidine residues, which suggested the possibility of a zinc-coordinating motif. These residues are completely conserved in a phylogenetic comparison of SAP30/SAP30L sequences from several species, including the fruit fly and the human (Fig. 2A). To investigate whether SAP30L binds zinc, we determined ESI Q-FT-ICR mass spectra for an N-terminal peptide of SAP30L (aa 1 to 92) and for mutants in which the putative zinc-coordinating cysteine residues were replaced by serines (C29S, C30S, C38S, and C74S) and histidines by alanines (H70A and H77A), one at a time. Figure 2B presents the spectrum measured for SAP30L 1-94 (this peptide contains two additional residues from the GST vector at its N terminus) under denaturing conditions. To aid in the interpretation, the mass spectra were subjected to charge deconvolution (i.e., conversion of m/z to Da). Four major peptide variants, with relative abundance ratios of ∼4:31:7:58, were detected, a result that is consistent with the SDS-PAGE analysis (Fig. 2C). The mass of the second lightest variant (10,413.33 Da) agrees well with the mass calculated for the SAP30L 1-94 construct (Table 1). The species with the smallest mass (10,028.09 Da) is consistent with cleavage of three residues from the C terminus and is presumed to comprise residues 1 to 89 of SAP30L. However, two heavier variants (11,131.58 and 11,766.95 Da) could not be assigned to any peptide sequence, even when all possible additional residues from the expression vector were considered. All peptides appeared as apo peptides, i.e., no zinc binding was detected under denaturing conditions.
FIG. 2.
The N-terminal domains of SAP30L and SAP30 contain an evolutionarily conserved zinc-binding module which is needed for DNA binding. (A) Clustal V alignment of the N-terminal amino acid sequences of SAP30L and SAP30 from various (selected) animals. Conserved cysteine (positions 29, 30, 38, and 74) and histidine (positions 70 and 77) residues in SAP30L are boxed, and the previously identified NLS is shaded in gray. Identical amino acids are marked by asterisks, and conservative substitutions are indicated by punctuation marks (colons and dots). (B) Charge-deconvoluted ESI Q-FT-ICR mass spectra of wild-type SAP30L in denaturing (upper) and nondenaturing (lower) solutions. The most abundant isotopic masses for the detected peptide variants are indicated. The insets show an expanded view of the isotopic distributions for the variant comprising residues 1 to 94 (this construct contains aa 1 to 92 of SAP30L and two amino acids derived from the thrombin cleavage site of the vector). The theoretical isotopic distributions were calculated from the sequence-derived elemental compositions (Table 1). The small arrow indicates the most abundant isotopic peak. (C) The SAP30L C29S, C38S, C74S, and H77A mutant constructs are degraded within 16 h after cleavage from GST, as analyzed by SDS-PAGE and Coomassie blue staining. wt, wild type. (D) L-EMSA with the GST-SAP30/SAP30L fusion proteins. (E) L-EMSA with the GST-SAP30L 1-92 fusion protein in the presence of 50 mM 1, 10-o-phenanthroline, a zinc-chelating agent.
TABLE 1.
Calculated and experimentally determined masses for wild-type SAP30L and the C30S and H70A mutants
| Peptidea | mexp (Da)b | mcalc (Da)c | mexp − mcalc (Da) | Elemental compositiond |
|---|---|---|---|---|
| apo wild type | 10,413.33 | 10,413.28 | 0.05 | C447H726N140O137S5 |
| holo wild type | 10,476.25 | 10,476.20 | 0.05 | C447H724N140O137S5Zn1 |
| apo(C30S) | 10,397.31 | 10,397.35 | −0.04 | C447H726N140O138S4 |
| holo(C30S) | 10,460.24 | 10,460.22 | 0.02 | C447H724N140O138S4Zn1 |
| apo(H70A) | 10,347.26 | 10,347.30 | −0.04 | C444H724N138O137S5 |
| holo(H70A) | 10,410.18 | 10,410.32 | −0.14 | C444H722N138O137S5Zn1 |
The data are presented only for the peptide variants comprising residues 1 to 94.
Most abundant isotopic mass, experimentally determined.
Most abundant isotopic mass, calculated on the basis of the sequence-derived elemental composition.
In the case of zinc binding, a loss of two protons was considered.
To detect zinc binding, the SAP30L 1-94 peptide was analyzed under nondenaturing conditions (Fig. 2B). A 62.94-Da increase in mass was detected for each of the four peptide variants, consistent with the binding of one Zn2+ cation (Table 1). The binding of Zn2+ (average mass = 65 Da) by zinc finger domains is always accompanied by the loss of two protons (deprotonation of two coordinating cysteines), which gives a theoretical 62.92-Da increase in mass, as discussed in detail elsewhere (12). The precision of the mass measurements was approximately 0.01 Da, and such high accuracy provides an unequivocal identification of the incorporated Zn2+ cation in SAP30L. No apo peptides or peptides with higher zinc-binding stoichiometries were detected. Similar results were obtained for the SAP30L C30S and H70A mutants (see Fig. S2A in the supplemental material). The calculated and determined masses for the peptides SAP30L 1-94, C30S, and H70A are listed in Table 1. The SAP30L C29S, C38S, C74S, and H77A mutants had completely degraded into small peptide fragments when expressed in E. coli (Fig. 2C; also see Fig. S2B in the supplemental material) and when transiently transfected into mammalian cells (data not shown). Many of the peptide fragments that could be identified seemed to contain disulfide bridges. In summary, these results indicate that the C2CH motif (C-X8-C-X35-C-X2-H) forms the zinc-binding module in SAP30L (see Fig. 7A) and that disruption of this module destabilizes the protein and leads to its rapid degradation. Similar instability has been reported to occur in the zinc-deficient mutant of the Spt10p zinc finger protein in yeast (40).
FIG. 7.
Domain structure and a proposed mode of action for SAP30L and SAP30. (A) Schematic representation of the N-terminal zinc-coordinating motif of SAP30L. (B) Various domains of SAP30L identified in this and other studies (35, 57). Zn, zinc-coordinating motif; DNAbd, DNA-binding domain; PIPbd, PIP binding domain; protein bd, protein binding domain; acidic region, a central region contributing to histone binding. (C) Proposed model. (Panel 1) When the histones are acetylated, the DNA is loosely packed and therefore accessible to RNA polymerase II. (Panel 2) A sequence-specific transcriptional repressor (TF) recruits the Sin3A complex to its target promoter. SAP30 or SAP30L stabilizes the complex through interactions with DNA and histones 2A/2B. The interaction of SAP30/SAP30L with DNA induces bending of the DNA, as a result of which the nucleosomes are more accessible to HDAC enzymes, and the repressome is fully formed. (Panel 3) Nuclear PIPs (PtdInsPs) interact with the N-terminal domain of SAP30/SAP30L, displacing the DNA, which leads to relocalization of SAP30/SAP30L to the cytoplasm.
The C2CH module is needed for DNA binding.
To further explore the DNA-binding domains of SAP30L, we used a simplified EMSA which utilizes a commercial DNA ladder (L-EMSA). To validate the assay, we incubated the full-length GST-SAP30L and GST-SAP30 fusion proteins with the DNA ladder and observed a marked shift in the mobility of the DNA. The GST-SAP30L 1-92 protein was able to bind DNA and therefore contains all determinants for interaction with DNA. Two polybasic regions (PBRs) were shown to be necessary for DNA binding (Fig. 2D). One of these is in the loop of the zinc-coordinating structure (aa 50 to 69), and the other region (aa 84 to 92) has previously been identified as a nuclear localization signal (NLS) (35). A hydrophobic region (aa 78 to 84) between the zinc-coordinating structure and the NLS is also required for DNA binding, since a construct containing both the zinc-coordinating structure and the hydrophobic region (aa 1 to 84) was able to bind DNA, whereas the zinc-coordinating structure alone (aa 1 to 77) was not. Similarly, a construct which includes both the NLS motif (aa 78 to 92) and the hydrophobic region is able to bind DNA, but the NLS alone (aa 84 to 92) is not. The role of the NLS is further demonstrated by the full-length GST-SAP30L-KAAAK construct, which has markedly reduced affinity for DNA compared to wild-type GST-SAP30L. The importance of the zinc-coordinating structure is best demonstrated in Fig. 1A, where it can be seen that disruption of this structure (constructs GST-SAP30L 61-183 and 40-183) completely abolishes DNA binding.
The mapping experiments described above suggested that the zinc-coordinating structure is necessary for DNA binding. To explore whether DNA binding is dependent on zinc, the GST-SAP30L 1-92 fusion protein was incubated with 1,10-o-phenanthroline, a zinc-chelating agent, and the L-EMSA was performed. As shown in Fig. 2E, 1, 10-o-phenanthroline at a 50 mM concentration abolished the DNA binding, as evidenced by a lack of shift in the mobility of DNA. Neither the GST peptide alone nor the methanol solvent elicited any mobility changes. Addition of different divalent cations showed that only Zn2+ could partially restore the DNA-binding activity of the N-terminal domain of SAP30L (see Fig. S2C in the supplemental material). With DNA fragments less than 1,500 bp in length, we saw reproducible mobility shifts, which evidenced the zinc dependence and specificity of DNA binding by SAP30L. We were unable to test the DNA-binding activity of the specific zinc-coordinating mutants described above, because of extensive degradation of these proteins.
In summary, we have identified a putative zinc-binding C2CH module in SAP30L and SAP30 and shown that DNA binding is dependent on this module. This zinc-dependent structure appears to be important for the stability of the entire protein, and its disruption destabilizes the protein. Our results also show that the loop region in the C2CH module is required for DNA binding. Furthermore, the polybasic motif (NLS) following the zinc-binding module and the intervening hydrophobic pocket are also critical for DNA binding.
SAP30L has DNA-bending activity.
The high-mobility-group (HMG) proteins provide a well-known example of a protein family in which there can exist sequence-independent DNA binding, accompanied by bending of the DNA (19). To test the ability of SAP30L to mediate bending of double-stranded DNA, we examined its effect on the T4 DNA ligase-dependent cyclization of short DNA fragments. This ligation-mediated circularization assay works on the principle that any double-stranded DNA of less than 150 bp in length will not self-circularize in the presence of ligase, because of inherent limitations in the flexibility of DNA. Therefore, circularization is seen only in the presence of a DNA-bending protein. 32P-labeled PCR fragments (see Fig. S3 in the supplemental material) were incubated with the GST-SAP30L fusion protein or the GST-only control as detailed in Materials and Methods. As shown in Fig. 3A, 170-bp and 150-bp DNA fragments were able to self-circularize, whereas shorter fragments were not. In the cases of shorter fragments (110 bp, 90 bp, and 70 bp), addition of GST-SAP30L resulted in the formation of circular monomers, whereas GST alone did not form any circular monomer molecules. The lack of monomer formation in the case of 130-bp fragments suggests that SAP30L introduces such a strong bend to the DNA that cohesive ends are unable to line up. Such a lack of monomer formation in the cases of DNA molecules of certain lengths has also been reported for HMG proteins (45). The GST-SAP30L 1-92 region was able to bend DNA in a dose-dependent manner (Fig. 3B). It is reasonable to infer that SAP30 similarly bends DNA, given its identical zinc-binding module and DNA-binding properties.
SAP30L and SAP30 bind core histones and nucleosomes.
We noticed a similarity in the domain architecture of SAP30L and SAP30 to that of the HMG proteins, both having an N-terminal DNA-binding/bending domain followed by an acidic domain (Fig. 4A). As some reports indicate that the acidic region of HMG proteins mediates interactions with H1 or core histones (4, 7), we set out to examine whether SAP30L may also associate with core histones and nucleosomes. To this end, we used purified histones, isolated nucleosomes, and trypsin-cleaved nucleosomes (see Fig. S4A in the supplemental material). In a pulldown experiment, GST-fused SAP30L was able to associate with purified core histones 2A and 2B (Fig. 4B). GST-SAP30L 1-92, which lacks the central acidic region, interacted with histone 2A/2B slightly less than full-length GST-SAP30L (Fig. 4B and C, lower panels). This comparison is made difficult by the degradation of the full-length GST-SAP30L fusion protein and the possible loss of some of its binding determinants, which may lead to an underestimate of the difference. When the GST-SAP30L 1-120 construct, which contains the central acidic region, was used, histone binding was substantially increased (Fig. 4B and C, lower panels), suggesting that the central acidic region makes a significant contribution to histone binding. The interaction of SAP30L with histones was confirmed for histone 2B by using a specific antibody (Fig. 4C), whereas in the case of histone 2A, the antibody cross-reacted with SAP30L (data not shown), and this hampered the interpretation of the result. GST-SAP30 also interacted with histone 2B (data not shown).
Full-length GST-SAP30L and GST-SAP30L 1-92 were able to interact with DNA-containing nucleosomes. Even though the SAP30L 1-120 construct showed a high affinity for histones without DNA, it did not interact with nucleosomes (Fig. 4C, upper). In the 1-120 construct, the acidic region is likely to be artificially exposed to acidic DNA, and consequently, its interaction with nucleosomes is prevented. Interestingly, the interaction of SAP30L with nucleosomes was not dependent on the protruding N-terminal tails of histones, as demonstrated by a lack of effect after trypsin cleavage of the tails (Fig. 4C, middle). GST-SAP30 was similarly able to interact with nucleosomes (Fig. 4E). Zhang et al. (61) have previously reported that SAP30 is unable to bind nucleosomes and core histones 3 and 4. This is partially in line with our results in that we detected only nonspecific interactions with histones 3 and 4, as the GST moiety alone also bound them (Fig. 4B).
We have previously identified several mRNA isoforms of SAP30L, including an isoform which lacks the entire exon 2 and five residues of exon 3. Specific deletion of these five residues (del109-113) markedly reduced the repression activity of SAP30L, whereas most of the HDAC activity was retained (25). Intriguingly, these residues reside in the central acidic domain and include a single aspartate residue. As shown in Fig. 4C, the del109-113 mutant was still able to interact with purified histone 2B but not with purified nucleosomes containing DNA, a result that may explain the reduced repression capability of this mutant. The lack of nucleosome binding by the del109-113 mutant can be explained only by this mutant's inability to interact with a histone-DNA complex, since it binds naked DNA with the same affinity as does the wild-type protein (see Fig. S4C in the supplemental material).
In confocal microscopy, colocalization of histone 2B and SAP30L was detected, with simultaneous relocalization of histone 2B around the nucleolus in response to overexpression of SAP30L (Fig. 4D). Coexpression with wild-type SAP30L, but not with the SAP30Ldel109-113 mutant, increased the perinucleolar localization of H2B from 10% to over 80% (Fig. 4D). Intriguingly, overexpressed histone 2B was able to direct NoLS-mutated SAP30L (8A) to the nucleolar region and thus overcome the lack of NoLS (data not shown). As a control, endogenous histone 1 showed no changes upon transient overexpression of SAP30L (data not shown).
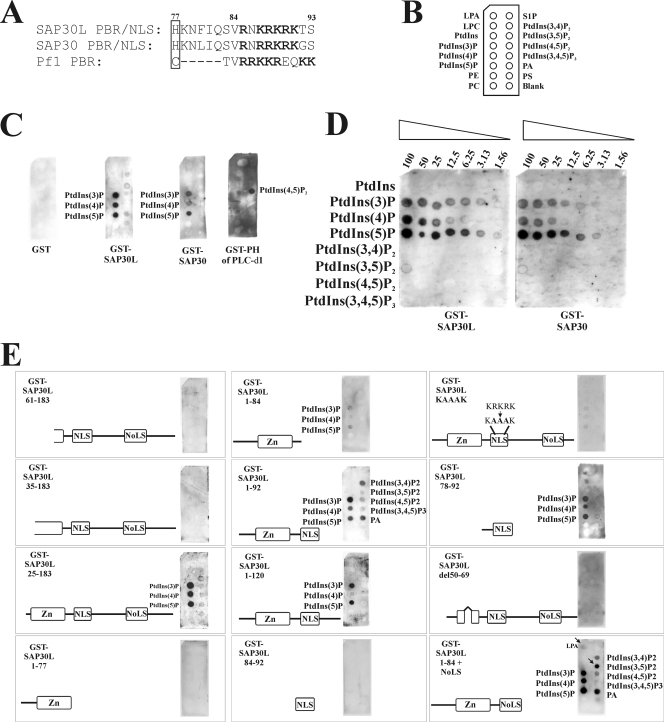
The polybasic NLS binds monophosphoinositides.
Pf1, a recently identified Sin3A-binding protein, has a PI-binding PBR following the first PHD zinc finger (24). A similar organization is found in ING2, in which the PHD zinc finger is also followed by a PBR. We identified a similar modular organization in SAP30L, which also contains a zinc-binding element followed by a PBR (85RNKRKRK91) (Fig. 5A). In SAP30L, the PBR motif has previously been shown to act as an NLS (35). To investigate the PI binding of SAP30L and SAP30, the GST fusion proteins were tested for binding to a variety of immobilized lipids, as depicted in Fig. 5B. Both GST-SAP30L and GST-SAP30 bound the monophosphorylated PIs PI(3)P, PI(4)P, and PI(5)P (Fig. 5C). No lipid binding was detected for GST alone. As a positive control, we used the PH domain of phospholipase C-delta1, which interacted specifically with PI(4,5)P2 in this assay.
FIG. 5.
The zinc-binding structure and the PBR in SAP30L and SAP30 bind monophosphoinositides. (A) Manual alignment of the sequences of the PBRs following the zinc-binding modules in SAP30L, SAP30, and Pf1. The last zinc-coordinating residue is boxed, and the basic residues are indicated with bold letters. (B) Schematic diagram of a lipid blot membrane (PIP strip) containing 20-pmol spots from samples of the following: lysophosphatidic acid (LPA), lysophosphocholine (LPC), PI (PtdIns), PtdIns(3)P, PtdIns(4)P, PtdIns(5)P, phosphatidylethanolamine (PE), phosphatidylcholine (PC), sphingosine 1-phosphate (S1P), PtdIns(3,4)P2, PtdIns(3,5)P2, PtdIns(4,5P)2, PtdIns(3,4,5)P3, phosphatidic acid (PA), phosphatidylserine (PS), and blank. (C, D and E) The indicated GST fusion proteins (0.5 μg/ml) were incubated with PIP strips or with the PIP array as described in Materials and Methods. The lipids which bound most strongly are indicated. Arrows indicate the specificity differences from the 1-92 construct.
To quantify the relative affinities of SAP30L and SAP30 for various PIs, we used their fusion proteins to probe a lipid blot that contained serial dilutions of eight different PIs (Fig. 5D). Full-length GST-SAP30L bound most tightly to PI(5)P, followed by PI(3)P and PI(4)P. The level of PI(5)P binding to GST-SAP30L was fourfold higher than that for PI(3)P and eightfold higher than that for PI(4)P. GST-SAP30 bound to immobilized PIs in an identical manner, though with slightly lower affinities (Fig. 5D).
The determinants for PI binding were analyzed using truncated GST-SAP30L fusion proteins. Strikingly, deletion of 60 residues from the N terminus (SAP30L 61-183) resulted in complete loss of PI binding (Fig. 5E), which was rescued only by inclusion of the entire zinc-binding structure, indicating that the 25 residues in the N terminus are dispensable for this interaction. On the other hand, a construct containing the N terminus and the intact zinc-coordinating structure (aa 1 to 77) was unable to bind PIs. Addition of the hydrophobic region (aa 1 to 84) resulted in weak PI binding, whereas inclusion of the PBR motif following (aa 1 to 92) fully restored the interaction (this construct also exhibited some nonspecific binding, but the specificity was restored in the 1-120 construct). The PBR motif (aa 84 to 92) by itself was not sufficient for PI binding, but a construct which includes both the PBR motif and the preceding hydrophobic region (aa 78 to 92) was sufficient for the interaction. In the case of full-length SAP30L, mutating three basic residues in the PBR motif to alanines markedly reduced its binding activity (KAAAK mutant). It is noteworthy that disruption of the loop in the zinc-coordinating structure by deletion of residues 50 to 69 from otherwise intact SAP30L completely abolished the PI interaction (Fig. 5E). Finally, swapping of basic residues in the PBR motif with the polybasic NoLS region (aa 1 to 84 plus the NoLS) changed the specificity of PI binding (Fig. 5E).
The above-mentioned results give rise to three conclusions. First, SAP30L and SAP30 interact specifically with monophosphorylated PIs. Second, interaction of SAP30L with PIs is mediated by the PBR motif and supported by the preceding hydrophobic region and the zinc-coordinating structure. Third, the specificity of PI binding is partially determined by the composition of the basic sequence of the PBR. It is also noteworthy that the same region in SAP30L interacts with both DNA and PIs, as summarized in Table 2.
TABLE 2.
Summary of mapping studies of DNA and PIP interactions
| Construct | Resulta for:
|
|
|---|---|---|
| DNA interaction | PIP interaction | |
| Wild-type 1-183 | +++ | +++ |
| 61-183 | − | − |
| 40-183 | − | NA |
| 35-183 | NA | − |
| 25-183 | +++ | +++ |
| 1-77 | − | − |
| 1-84 | ++ | ++ |
| 1-92 | +++ | +++ |
| 1-120 | +++ | +++ |
| 84-92 | − | − |
| 78-92 | +++ | +++ |
| del50-69 | − | − |
| 87KAAAK91 | + | + |
NA, not available; −, no interaction; +, weak interaction; ++, moderate interaction; +++, strong interaction.
Monophosphoinositides regulate chromatin association of SAP30L.
GFP-tagged SAP30L associated with chromatin in vivo when hypotonically swollen HEK293 cells were splashed on a microscope slide and counterstained with DAPI (Fig. 6A). It should be noted that SAP30L is not a component of chromatin in the same way as histones, since it is not present in mitotic chromosomes (data not shown). We next explored the domains that regulate the chromatin association of SAP30L. As shown in Fig. 6B, KAAAK and del50-69 mutants of SAP30L associated significantly less than wild-type SAP30L with the chromatin-enriched fraction, as assayed by subcellular fractionation. Also, an intact C-terminal domain was needed, presumably reflecting the importance of protein-protein interactions mediated by the C-terminal region.
FIG. 6.
Chromatin association, subcellular localization, and transcriptional repression activities of SAP30L are regulated by its interactions with DNA and monophosphoinositides. (A) The SAP30L-GFP fusion protein colocalizes with interphase chromatin. (B) HEK293T cells were transfected with the indicated constructs, fractionated into subcellular fractions, and immunoblotted as indicated. Data from three independent experiments are illustrated as histograms, in which the bars represent the ranges of band intensities as measured by a densitometer. wt, wild type. (C) L-EMSA with GST-SAP30L 1-92 in the presence of equivalent molar quantities of PI (PtdIns) or PtdIns(5)P. (D) Cells were transfected with wild-type SAP30L, treated with H2O2, and subjected to subcellular fractionation as described for panel B. (E) Confocal images of cells transfected with SAP30L-GFP and treated with H2O2. (F) (Upper) In vitro-translated, 35S-methionine-labeled Sin3A1-855 was subjected to a pulldown experiment with GST-SAP30L and a GST-only control. PtdIns(5)P was added as indicated, and the results from the experiment were analyzed by SDS-PAGE and autoradiography. (Lower) Pulldown of nucleosomes with GST-SAP30L in the presence of PIs. (G) HEK293T cells were cotransfected with a 5× Gal4-14D luciferase reporter vector, GAL4DBD fusions, and a LacZ vector, as indicated. At 24 h posttransfection, the cells were treated with 500 μM H2O2 for 15 min, washed, and lysed after 4 h of incubation. Luciferase and β-galactosidase activities were measured, and the histograms illustrate the average repressions of the GAL4DBD fusions relative to the level for GAL4 alone. The measurements were done in duplicate in two independent experiments, and the bars represent the ranges of observed values.
As the KAAAK and del50-69 mutants are deficient in both DNA/chromatin and PI binding (Fig. 2D and 5E), we asked if the association of SAP30L with chromatin is regulated by PIs, which could compete for the same binding sites and thus detach SAP30L from chromatin. Two lines of evidence indicate that they do. First, the mobility shift generated by binding of SAP30L to DNA in the L-EMSA was greatly diminished after addition of equivalent molar amounts of monophosphorylated PIs but not other PIs (Fig. 6C). Second, H2O2 treatment, which has previously been shown to increase the amount of intranuclear monophosphorylated PIs (23), led to a significant relocalization of myc-tagged SAP30L as assayed by the chromatin-enriched fraction in HEK293 cells (Fig. 6D). In confocal microscopy of HeLa cells, 9% of nontreated and 41% of H2O2-treated cells expressed cytoplasmic GFP-SAP30L (Fig. 6E). The interaction of SAP30L with nucleosomes and Sin3A remained unchanged after addition of monophosphorylated PIs (Fig. 6F). These results suggest that intranuclear monophosphorylated PIs associate with the PI-binding domain of SAP30L and thereby regulate its association with chromatin.
PI binding decreases the repression activity of SAP30L.
Finally, we tested if association of SAP30L with PIs influenced its repression activity by utilizing a Gal4 fusion system with a luciferase reporter vector as described previously (57). As shown in Fig. 6G, reduced repression activity was observed both in the PBR mutant (SAP30L KAAAK), which lacks DNA binding and mimics PI binding, and after H2O2 treatment, which increases nuclear monophosphorylated PIs (23). Combined, these results suggest that association of SAP30L with chromatin is dependent on intact C-terminal and PI-/DNA-binding domains and that monophosphorylated PIs disrupt this association, leading to decreased transcriptional repression through SAP30L.
DISCUSSION
Although the Sin3A-HDAC corepressor complex has been studied extensively, the roles of the various members of this complex are poorly understood. In this study, we have explored the functions of two members of this complex, the Sin3A-associated proteins SAP30L and SAP30, which share 70% sequence identity. We have discovered three types of interactions that illuminate the functional roles of these proteins. First, both SAP30L and SAP30 interact directly with the core histones 2A/2B. Second, we demonstrate that both proteins have intrinsic DNA-binding activity which is partly mediated through a novel N-terminal zinc-containing structure consisting of a C2CH module and a coordinated zinc ion. Binding to DNA is sequence independent and induces strong bending of the DNA. Third, we have identified a PI-binding site, a basic domain which binds monophosphorylated PIs specifically, adjacent to the zinc-binding element. Intriguingly, we have found that PI binding has a strong influence on the proteins' affinity for DNA in vitro, which leads us to suggest that the DNA binding is actually regulated by PIs. An increase in the concentration of PIs in the nucleus caused by hydrogen peroxide leads to reduced repression activity and cytoplasmic relocalization of SAP30L.
Previously, SAP30 has been assigned the role of a linker protein that mediates interactions of the Sin3-HDAC complex with various transcriptional repressors (e.g., YY1) or corepressors (e.g., N-CoR, CIR, and RBP1). Specifically, the interaction of SAP30 with N-CoR was demonstrated to occur through the N terminus, whereas the C terminus bound mSin3a (28). Our results suggest a second function for the N terminus, which we found to bind DNA. Our results do not necessarily contradict the previous reports, because one can imagine that SAP30 can either bridge different multiprotein complexes or anchor a specific complex to nucleosomes, depending on the circumstances. Functional diversity of this kind is not unprecedented in Sin3A-associated proteins, since Fleischer et al. (13) have observed at least three separate Sin3A-containing complexes. Furthermore, the C terminus also seems to carry multiple functions. Huang et al. (21) identified SAP30 as a binding partner for the transcription factor YY1 and showed that it is able to enhance YY1-mediated repression in a dose-dependent manner. This interaction was mapped to the C-terminal region, i.e., the same region which also binds Sin3A, prompting the authors to suggest that the interactions of SAP30 with YY1 and Sin3A are mutually exclusive. Huang et al. (21) suggested that HDAC activity could be brought to the YY1-SAP30 complex through a direct interaction of SAP30 with HDAC1 (61). If SAP30L and SAP30 were to bind DNA and histones independently of Sin3A, it easy to envision that their N-terminal domains could participate in anchoring the YY1-SAP30-HDAC complex to chromatin to induce repression of transcription.
Sin3A by itself does not bind DNA or repress transcription but instead mediates gene silencing through the enzymes that it associates with (52). Targeting of the Sin3A complex is carried out by DNA sequence-specific repressor proteins. Here, we show that SAP30L and SAP30 are able to bind DNA without any sequence specificity. This binding is dependent on an intact N terminus that contains a C2CH-type zinc module, whose disruption abolishes the DNA binding. Zinc fingers were originally identified as DNA-binding motifs, but they are now known to bind RNA, protein, and lipid substrates as well (6, 14, 16, 24). A zinc finger consists of two antiparallel β strands and an α helix, and the zinc ion is crucial for its stability. Usually, a single zinc finger does not bind DNA with very high affinity and can recognize only two or three base pairs, but when several, up to 60, zinc fingers are strung together, the group binds more tightly and can recognize longer DNA sequences. In the cases of both SAP30L and SAP30, only a single zinc-coordinating element was identified. Moreover, the stability of SAP30L was dependent on the zinc module since mutations in its zinc-coordinating residues led to rapid degradation of the protein. We have previously observed that N-terminally truncated SAP30L is poorly expressed in transient transfections, but this could be overcome by using MG132, a proteasome inhibitor, and now this can be explained by the loss of the stabilizing zinc-dependent module in the N terminus (57). Zinc-binding domains are usually relatively short, i.e., 20 to 30 residues, and the spacing of 35 residues between the C2 and CH coordinating residues in SAP30L is unusually long. There is, however, a precedent for a large zinc-binding module, since THAP domains, which are conserved zinc-dependent modules capable of sequence-specific DNA binding, have a loop of 35 to 53 residues in the middle of the zinc-binding motif (8). The THAP domain, however, contains other conserved elements in addition to the C2CH module, making it distinct from the zinc-binding motif in SAP30L.
The sequence-independent nature of the DNA binding rules out a sequence-specific targeting role for SAP30 and SAP30L and suggests a more general role in anchoring to nucleosomal/linker DNA. In addition, we demonstrated that this DNA binding results in strong bending of the DNA. Classical examples of proteins that bind and bend DNA in a sequence-independent manner are the HMG proteins (45), which interact transiently with DNA. They are thought to antagonize histone H1 binding by competing for the same chromatin sites. Generally, they are thought to open up chromatin, although some HMG proteins may also compact chromatin (43). We find interesting parallels between the HMG proteins and SAP30/SAP30L. Both are small and localized in the nucleus. Their domain structures are also similar, as both contain an N-terminal DNA-binding domain followed by an acidic region which contributes to histone interactions. This could imply functional similarity as well, and it seems likely that SAP30/SAP30L have roles in stabilizing the multiprotein complex on its target, increasing the availability of enzymatic targets to the complex, or promoting the recruitment of interacting proteins, the cumulative effect being increased repression activity.
Perhaps one of the most intriguing features of SAP30L and SAP30 is the presence of a PI-binding site. PIs are known to function in nuclear signaling, and local changes in PI concentrations are sensed by proteins with specific PI-binding domains, such as PH, ENTH, FYVE, and PHOX domains and lysine/arginine-rich patches (34, 44). A number of PI kinases and phosphatases translocate to the nucleus upon activation, and many PI species have been shown to be intranuclear (10, 15). Intervention of chromatin biology by signaling lipids is not unprecedented, since ATP-dependent chromatin-remodeling complexes, such as NURF, ISW2, INO80, and SWI/SNF, are also modulated by specific inositol polyphosphates, the cleavage products generated by PI-specific phospholipase C (50). Additionally, another SWI/SNF-like chromatin remodeling complex, BAF, is targeted to chromatin and the nuclear matrix specifically by a PIP2-dependent mechanism upon lymphocyte activation (62). Pf1, a recently identified nuclear binding partner for the corepressors mSin3A and TLE, has a PBR which binds specific monophosphoinositides (24). The Sin3A-binding tumor suppressor ING2 binds PI(3), PI(4)P, and PI(5)P and shows PI(5)P-dependent association with chromatin and induction of p53-dependent apoptosis (16, 24). In response to cellular stress by UV irradiation or hydrogen peroxide, ING2 associates with chromatin through a PI(5)P-mediated mechanism (23). Initially, the PHD domain of ING2 was reported to be sufficient for PI binding, but later, the PBR motif was demonstrated to be both necessary and sufficient on its own (16, 24). Even though the PBRs of Pf1 and ING2 were deemed critical for the binding activity and specificity, the preceding zinc-binding PHD domain contributed some specificity to the interaction with PIs (24). SAP30 and SAP30L have a number of similarities with the PI-binding proteins Pf1 and ING. First, the domain architecture of SAP30/SAP30L resembles that of Pf1, with a zinc-binding element followed by a basic PI-binding module in both cases. Second, like SAP30/SAP30L, Pf1 and ING2 are part of the Sin3A complex. Third, all three proteins are nuclear and bind monophosphorylated PIs, albeit with different preferences. Fourth, in the cases of SAP30/SAP30L and ING2, the subcellular localization and chromatin association are modified by PI binding. However, in the cases of SAP30/SAP30L (but not in ING2), PI binding competes with DNA binding in vitro so that an increase in the concentration of monophosphorylated PIs causes SAP30L to detach from DNA. Furthermore, an increase in the concentration of nuclear PIs elicited with hydrogen peroxide leads to reduced repression activity and cytoplasmic relocalization of SAP30L. Although we note that these results are preliminary and mostly based on in vitro experiments, they suggest the intriguing possibility that changes in the concentration of nuclear monophosphorylated PIs may regulate transcriptional repression through SAP30/SAP30L in vivo. The site of PI binding was mapped to a region containing a motif previously shown to act as an NLS, and this motif is necessary for the PI-binding activity. However, the adjacent zinc-coordinating module also contributes to this interaction, a result that is in agreement with studies of other proteins (47). The binding interface may reside on one side of the loop region of the zinc module, in a region which contains a stretch of basic residues. The specificity of PI binding is partly determined by the amino acid residue composition of the binding motif, since replacing the NLS motif with another basic motif (NoLS motif) in SAP30L led to changes in PI binding. Differences in binding specificity between different proteins are also evident. SAP30/SAP30L prefer PI(5)P over PI(3)P/PI(4)P, whereas Pf1 prefers PI(3)P, with some binding activity toward PI(3,5)P species (24).
We propose a model in which SAP30L/SAP30 are actively involved in multiple protein-protein and protein-DNA interactions that modulate transcriptional repression. The domain structures of SAP30L/30 and the proposed model are depicted in Fig. 7B and C, respectively. Briefly, we suggest that the DNA-binding activity plays a role in anchoring the Sin3A complex to nucleosomal and/or linker DNA in chromatin and that this binding is further strengthened by the interaction with core histone 2A/2B dimers. One consequence of DNA binding is bending of the DNA, and we envision that this leads to enhanced accessibility of nucleosomes and histone tails to deacetylating enzymes. Moreover, our results provide new evidence for the regulatory role played by nuclear PIs in transcriptional repression and relocalization of nuclear proteins.
Supplementary Material
Acknowledgments
We thank Jorma Kulmala and Ritva Romppanen for technical assistance, Mike Berger for advice and assistance with PBM experiments and data analysis, and Olli Silvennoinen for comments on the manuscript. We are grateful to P. Peterson (Tartu, Estonia) and D. Ayer (Salt Lake City, UT) for H2B-GFP and 14D promoter plasmids, respectively, and to Anne Hyvärinen (Tampere, Finland) for the EMSA probe used in this work.
This work was supported by the Academy of Finland Research Council for Health (funding decision numbers 115260 and 201361) and for Natural Sciences and Engineering (108533), the Foundation for Paediatric Research in Finland, the Competitive Research Funding of the Pirkanmaa Hospital District (EVO), the Nona and Kullervo Väre Foundation, the Päivikki and Sakari Sohlberg Foundation, and grant R01 HG003985 from NIH/NHGRI to M.L.B. T.S. was supported in part by a U.S. National Science Foundation Postdoctoral Research Fellowship in Biological Informatics.
Footnotes
Published ahead of print on 17 November 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alland, L., R. Muhle, H. Hou, Jr., J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 38749-55. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., N. Williams, C. Misleh, and W. W. Li. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34W369-W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, M. F., A. A. Philippakis, A. M. Qureshi, F. S. He, P. W. Estep III, and M. L. Bulyk. 2006. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat. Biotechnol. 241429-1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernues, J., E. Espel, and E. Querol. 1986. Identification of the core-histone-binding domains of HMG1 and HMG2. Biochim. Biophys. Acta 866242-251. [DOI] [PubMed] [Google Scholar]
- 5.Brehm, A., E. A. Miska, D. J. McCance, J. L. Reid, A. J. Bannister, and T. Kouzarides. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391597-601. [DOI] [PubMed] [Google Scholar]
- 6.Brown, R. S. 2005. Zinc finger proteins: getting a grip on RNA. Curr. Opin. Struct. Biol. 1594-98. [DOI] [PubMed] [Google Scholar]
- 6a.Bulyk, M. L., X. H. Huang, Y. Choo, and G. M. Church. 2001. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. USA 987158-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carballo, M., P. Puigdomenech, and J. Palau. 1983. DNA and histone H1 interact with different domains of HMG 1 and 2 proteins. EMBO J. 21759-1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clouaire, T., M. Roussigne, V. Ecochard, C. Mathe, F. Amalric, and J. P. Girard. 2005. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl. Acad. Sci. USA 1026907-6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.David, G., G. M. Turner, Y. Yao, A. Protopopov, and R. A. DePinho. 2003. mSin3-associated protein, mSds3, is essential for pericentric heterochromatin formation and chromosome segregation in mammalian cells. Genes Dev. 172396-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deleris, P., D. Bacqueville, S. Gayral, L. Carrez, J. P. Salles, B. Perret, and M. Breton-Douillon. 2003. SHIP-2 and PTEN are expressed and active in vascular smooth muscle cell nuclei, but only SHIP-2 is associated with nuclear speckles. J. Biol. Chem. 27838884-38891. [DOI] [PubMed] [Google Scholar]
- 11.Emini, E. A., J. V. Hughes, D. S. Perlow, and J. Boger. 1985. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J. Virol. 55836-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabris, D., Y. Hathout, and C. Fenselau. 1999. Investigation of zinc chelation in zinc-finger arrays by electrospray mass spectrometry. Inorg. Chem. 381322-1325. [DOI] [PubMed] [Google Scholar]
- 13.Fleischer, T. C., U. J. Yun, and D. E. Ayer. 2003. Identification and characterization of three new components of the mSin3A corepressor complex. Mol. Cell. Biol. 233456-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamsjaeger, R., C. K. Liew, F. E. Loughlin, M. Crossley, and J. P. Mackay. 2007. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 3263-70. [DOI] [PubMed] [Google Scholar]
- 15.Gozani, O., S. J. Field, C. G. Ferguson, M. Ewalt, C. Mahlke, L. C. Cantley, G. D. Prestwich, and J. Yuan. 2005. Modification of protein sub-nuclear localization by synthetic phosphoinositides: evidence for nuclear phosphoinositide signaling mechanisms. Adv. Enzyme Regul. 45171-185. [DOI] [PubMed] [Google Scholar]
- 16.Gozani, O., P. Karuman, D. R. Jones, D. Ivanov, J. Cha, A. A. Lugovskoy, C. L. Baird, H. Zhu, S. J. Field, S. L. Lessnick, J. Villasenor, B. Mehrotra, J. Chen, V. R. Rao, J. S. Brugge, C. G. Ferguson, B. Payrastre, D. G. Myszka, L. C. Cantley, G. Wagner, N. Divecha, G. D. Prestwich, and J. Yuan. 2003. The PHD finger of the chromatin-associated protein ING2 functions as a nuclear phosphoinositide receptor. Cell 11499-111. [DOI] [PubMed] [Google Scholar]
- 17.Hassig, C. A., T. C. Fleischer, A. N. Billin, S. L. Schreiber, and D. E. Ayer. 1997. Histone deacetylase activity is required for full transcriptional repression by mSin3A. Cell 89341-347. [DOI] [PubMed] [Google Scholar]
- 18.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 38743-48. [DOI] [PubMed] [Google Scholar]
- 19.Hock, R., T. Furusawa, T. Ueda, and M. Bustin. 2007. HMG chromosomal proteins in development and disease. Trends Cell Biol. 1772-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 9623-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang, N. E., C. H. Lin, Y. S. Lin, and W. C. Yu. 2003. Modulation of YY1 activity by SAP30. Biochem. Biophys. Res. Commun. 306267-275. [DOI] [PubMed] [Google Scholar]
- 22.Hyvarinen, A. K., J. L. Pohjoismaki, A. Reyes, S. Wanrooij, T. Yasukawa, P. J. Karhunen, J. N. Spelbrink, I. J. Holt, and H. T. Jacobs. 2007. The mitochondrial transcription termination factor mTERF modulates replication pausing in human mitochondrial DNA. Nucleic Acids Res. 356458-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones, D. R., Y. Bultsma, W. J. Keune, J. R. Halstead, D. Elouarrat, S. Mohammed, A. J. Heck, C. S. D'Santos, and N. Divecha. 2006. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol. Cell 23685-695. [DOI] [PubMed] [Google Scholar]
- 24.Kaadige, M. R., and D. E. Ayer. 2006. The polybasic region that follows the plant homeodomain zinc finger 1 of Pf1 is necessary and sufficient for specific phosphoinositide binding. J. Biol. Chem. 28128831-28836. [DOI] [PubMed] [Google Scholar]
- 25.Korkeamaki, H., K. Viiri, M. K. Kukkonen, M. Maki, and O. Lohi. 2008. Alternative mRNA splicing of SAP30L regulates its transcriptional repression activity. FEBS Lett. 582379-384. [DOI] [PubMed] [Google Scholar]
- 26.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 749637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuzmichev, A., Y. Zhang, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 2002. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33ING1. Mol. Cell. Biol. 22835-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laherty, C. D., A. N. Billin, R. M. Lavinsky, G. S. Yochum, A. C. Bush, J. M. Sun, T. M. Mullen, J. R. Davie, D. W. Rose, C. K. Glass, M. G. Rosenfeld, D. E. Ayer, and R. N. Eisenman. 1998. SAP30, a component of the mSin3 corepressor complex involved in N-CoR-mediated repression by specific transcription factors. Mol. Cell 233-42. [DOI] [PubMed] [Google Scholar]
- 29.Laherty, C. D., W. M. Yang, J. M. Sun, J. R. Davie, E. Seto, and R. N. Eisenman. 1997. Histone deacetylases associated with the mSin3 corepressor mediate mad transcriptional repression. Cell 89349-356. [DOI] [PubMed] [Google Scholar]
- 30.Lai, A., B. K. Kennedy, D. A. Barbie, N. R. Bertos, X. J. Yang, M. C. Theberge, S. C. Tsai, E. Seto, Y. Zhang, A. Kuzmichev, W. S. Lane, D. Reinberg, E. Harlow, and P. E. Branton. 2001. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol. Cell. Biol. 212918-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lechner, T., M. J. Carrozza, Y. Yu, P. A. Grant, A. Eberharter, D. Vannier, G. Brosch, D. J. Stillman, D. Shore, and J. L. Workman. 2000. Sds3 (suppressor of defective silencing 3) is an integral component of the yeast Sin3[middle dot]Rpd3 histone deacetylase complex and is required for histone deacetylase activity. J. Biol. Chem. 27540961-40966. [DOI] [PubMed] [Google Scholar]
- 32.Le May, N., Z. Mansuroglu, P. Leger, T. Josse, G. Blot, A. Billecocq, R. Flick, Y. Jacob, E. Bonnefoy, and M. Bouloy. 2008. A SAP30 complex inhibits IFN-beta expression in Rift Valley fever virus infected cells. PLoS Pathog. 4e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, M. S., G. P. Gippert, K. V. Soman, D. A. Case, and P. E. Wright. 1989. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science 245635-637. [DOI] [PubMed] [Google Scholar]
- 34.Lemmon, M. A. 2003. Phosphoinositide recognition domains. Traffic 4201-213. [DOI] [PubMed] [Google Scholar]
- 35.Lindfors, K., K. M. Viiri, M. Niittynen, T. Y. Heinonen, M. Maki, and H. Kainulainen. 2003. TGF-beta induces the expression of SAP30L, a novel nuclear protein. BMC Genomics 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Loewith, R., J. S. Smith, M. Meijer, T. J. Williams, N. Bachman, J. D. Boeke, and D. Young. 2001. Pho23 is associated with the Rpd3 histone deacetylase and is required for its normal function in regulation of gene expression and silencing in Saccharomyces cerevisiae. J. Biol. Chem. 27624068-24074. [DOI] [PubMed] [Google Scholar]
- 37.Macfarlan, T., S. Kutney, B. Altman, R. Montross, J. Yu, and D. Chakravarti. 2005. Human THAP7 is a chromatin-associated, histone tail-binding protein that represses transcription via recruitment of HDAC3 and nuclear hormone receptor corepressor. J. Biol. Chem. 2807346-7358. [DOI] [PubMed] [Google Scholar]
- 38.McGuinness, B. E., T. Hirota, N. R. Kudo, J. M. Peters, and K. Nasmyth. 2005. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 3e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Méndez, J., and B. Stillman. 2000. Chromatin association of human origin recognition complex, Cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 208602-8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mendiratta, G., P. R. Eriksson, C. H. Shen, and D. J. Clark. 2006. The DNA-binding domain of the yeast Spt10p activator includes a zinc finger that is homologous to foamy virus integrase. J. Biol. Chem. 2817040-7048. [DOI] [PubMed] [Google Scholar]
- 41.Meskauskas, A., J. L. Baxter, E. A. Carr, J. Yasenchak, J. E. Gallagher, S. J. Baserga, and J. D. Dinman. 2003. Delayed rRNA processing results in significant ribosome biogenesis and functional defects. Mol. Cell. Biol. 231602-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41a.Mukherjee, S., M. F. Berger, G. Jona, X. S. Wang, D. Muzzey, M. Snyder, R. A. Young, and M. L. Bulyk. 2004. Rapid analysis of the DNA binding specificities of transcription factors with DNA microarrays. Nat. Genet. 361331-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89373-380. [DOI] [PubMed] [Google Scholar]
- 43.Narita, M., V. Krizhanovsky, S. Nunez, A. Chicas, S. A. Hearn, M. P. Myers, and S. W. Lowe. 2006. A novel role for high-mobility group a proteins in cellular senescence and heterochromatin formation. Cell 126503-514. [DOI] [PubMed] [Google Scholar]
- 44.Overduin, M., M. L. Cheever, and T. G. Kutateladze. 2001. Signaling with phosphoinositides: better than binary. Mol. Interv. 1150-159. [PubMed] [Google Scholar]
- 45.Paull, T. T., M. J. Haykinson, and R. C. Johnson. 1993. The nonspecific DNA-binding and -bending proteins HMG1 and HMG2 promote the assembly of complex nucleoprotein structures. Genes Dev. 71521-1534. [DOI] [PubMed] [Google Scholar]
- 46.Pitkanen, J., A. Rebane, J. Rowell, A. Murumagi, P. Strobel, K. Moll, M. Saare, J. Heikkila, V. Doucas, A. Marx, and P. Peterson. 2005. Cooperative activation of transcription by autoimmune regulator AIRE and CBP. Biochem. Biophys. Res. Commun. 333944-953. [DOI] [PubMed] [Google Scholar]
- 47.Sankaran, V. G., D. E. Klein, M. M. Sachdeva, and M. A. Lemmon. 2001. High-affinity binding of a FYVE domain to phosphatidylinositol 3-phosphate requires intact phospholipid but not FYVE domain oligomerization. Biochemistry 408581-8587. [DOI] [PubMed] [Google Scholar]
- 48.Schwabe, J. W., and A. Klug. 1994. Zinc mining for protein domains. Nat. Struct. Biol. 1345-349. [DOI] [PubMed] [Google Scholar]
- 49.Schwerk, C., J. Prasad, K. Degenhardt, H. Erdjument-Bromage, E. White, P. Tempst, V. J. Kidd, J. L. Manley, J. M. Lahti, and D. Reinberg. 2003. ASAP, a novel protein complex involved in RNA processing and apoptosis. Mol. Cell. Biol. 232981-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen, X., H. Xiao, R. Ranallo, W. H. Wu, and C. Wu. 2003. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science 299112-114. [DOI] [PubMed] [Google Scholar]
- 51.Shiio, Y., D. W. Rose, R. Aur, S. Donohoe, R. Aebersold, and R. N. Eisenman. 2006. Identification and characterization of SAP25, a novel component of the mSin3 corepressor complex. Mol. Cell. Biol. 261386-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silverstein, R. A., and K. Ekwall. 2005. Sin3: a flexible regulator of global gene expression and genome stability. Curr. Genet. 471-17. [DOI] [PubMed] [Google Scholar]
- 53.Skowyra, D., M. Zeremski, N. Neznanov, M. Li, Y. Choi, M. Uesugi, C. A. Hauser, W. Gu, A. V. Gudkov, and J. Qin. 2001. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J. Biol. Chem. 2768734-8739. [DOI] [PubMed] [Google Scholar]
- 54.Smith, J. S., E. Caputo, and J. D. Boeke. 1999. A genetic screen for ribosomal DNA silencing defects identifies multiple DNA replication and chromatin-modulating factors. Mol. Cell. Biol. 193184-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sun, Z. W., and M. Hampsey. 1999. A general requirement for the Sin3-Rpd3 histone deacetylase complex in regulating silencing in Saccharomyces cerevisiae. Genetics 152921-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verreault, A., P. D. Kaufman, R. Kobayashi, and B. Stillman. 1998. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 896-108. [DOI] [PubMed] [Google Scholar]
- 57.Viiri, K. M., H. Korkeamaki, M. K. Kukkonen, L. K. Nieminen, K. Lindfors, P. Peterson, M. Maki, H. Kainulainen, and O. Lohi. 2006. SAP30L interacts with members of the Sin3A corepressor complex and targets Sin3A to the nucleolus. Nucleic Acids Res. 343288-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang, W. M., C. Inouye, Y. Zeng, D. Bearss, and E. Seto. 1996. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc. Natl. Acad. Sci. USA 9312845-12850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang, X. J., and E. Seto. 2008. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell Biol. 9206-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, Y., R. Iratni, H. Erdjument-Bromage, P. Tempst, and D. Reinberg. 1997. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89357-364. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y., Z. W. Sun, R. Iratni, H. Erdjument-Bromage, P. Tempst, M. Hampsey, and D. Reinberg. 1998. SAP30, a novel protein conserved between human and yeast, is a component of a histone deacetylase complex. Mol. Cell 11021-1031. [DOI] [PubMed] [Google Scholar]
- 62.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95625-636. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.