Abstract
The majority of spontaneous chromosome breakage occurs during the process of DNA replication. Homologous recombination is the primary mechanism of repair of such damage, which probably accounts for the fact that it is essential for genome integrity and viability in mammalian cells. The Mre11 complex plays diverse roles in the maintenance of genomic integrity, influencing homologous recombination, checkpoint activation, and telomere maintenance. The complex is essential for cellular viability, but given its myriad influences on genomic integrity, the mechanistic basis for the nonviability of Mre11 complex-deficient cells has not been defined. In this study we generated mice carrying a conditional allele of Rad50 and examined the effects of Rad50 deficiency in proliferative and nonproliferative settings. Depletion of Rad50 in cultured cells caused extensive DNA damage and death within 3 to 5 days of Rad50 deletion. This was not associated with gross telomere dysfunction, suggesting that the telomeric functions of the Mre11 complex are not required for viability. Rad50 was also dispensable for the viability of quiescent liver and postmitotic Purkinje cells of the cerebellum. These findings support the idea that the essential functions of the Mre11 complex are associated with DNA replication and further suggest that homologous recombination is not essential in nondividing cells.
The Mre11 complex regulates both DNA damage checkpoint function and repair. Its checkpoint functions appear to be primarily related to its role as a DNA double-strand break (DSB) sensor which binds DNA damage and activates ATM (ataxia-telangiectasia [AT] mutated). The ATM kinase transduces the damage signal via phosphorylating mediators of the damage response (30, 42), which promotes cell cycle arrest, DNA repair, and apoptosis. Mre11 complex functions are compromised in the human chromosome instability syndromes Nijmegen breakage syndrome and AT-like disorder, which are caused by hypomorphic mutations in Nbs1 and Mre11. Cells derived from patients and from mouse models of these diseases exhibit spontaneous DNA damage, ionizing radiation (IR) sensitivity, and checkpoint defects (25, 27, 48, 52, 57).
The complex's primary role in DNA repair is in recombinational DSB repair, and this role likely underlies its essential nature. In Saccharomyces cerevisiae, the complex governs homologous recombination (HR) and nonhomologous end joining (NHEJ) (19), whereas in vertebrate systems it primarily functions in HR (51, 61, 62). In fact, studies of Nbs1-deficient cells suggest that the Mre11 complex may inhibit NHEJ in mammals (62). Data from several species also implicate the Mre11 nuclease in the metabolism of topoisomerase adducts (40, 43, 49). This highly conserved function could also explain why the Mre11 complex is essential.
The Mre11 complex's function at telomeres may also be required for viability. Telomeres protect the ends of linear chromosomes from being recognized as DSBs and thereby activating the DNA damage response (DDR) (9). In S. cerevisiae the Mre11 complex influences telomere length maintenance (5, 28), whereas in mammals the complex interacts with the telomere binding protein Trf2 and localizes to telomeres (63). Loss of Trf2 results in telomere uncapping, causing activation of the DDR, telomere fusions, and senescence (7). Given the association of Mre11 with Trf2, it is conceivable that acute Mre11 complex deficiency in the mouse would phenocopy Trf2 loss and similarly lead to cell death as a result of telomere uncapping.
Conclusions regarding the essential nature of HR in general (33, 47, 53) and the Mre11 complex specifically (10, 17, 45, 59, 62) have been derived from the analysis of proliferating cells in vitro or in vivo. The coincidence of DNA replication and the formation of spontaneous DSBs prompted us to test whether the Mre11 complex and, by extension, HR would be essential in quiescent or postmitotic tissues in which the frequency of spontaneous DSBs is significantly reduced. To examine this issue, we generated mice containing a conditional Rad50 allele in which the Rad50 gene could be inactivated in quiescent and postmitotic cells.
Our results indicate that Rad50 is not required for homeostasis or viability of quiescent hepatocytes of the adult liver; nor does it appear to be required for maintenance of postmitotic Purkinje cells of the cerebellum. In contrast, Rad50 was required for viability of proliferating tissue culture and bone marrow cells. Rad50-deficient hepatocytes that were induced to divide via hepatectomy were able to achieve limited division and survived despite the presence of DNA damage that persisted long after the bulk of regeneration was complete. Rad50-deficient cells did not exhibit overtly dysfunctional telomeres, suggesting that their loss of viability was not due to acute telomere failure. These data indicate that the Mre11 complex and, by extension, HR may be dispensable in postmitotic cells and are consistent with the interpretation that the replication-associated functions of the Mre11 complex account for its essential nature.
MATERIALS AND METHODS
Mice.
Mice were housed in ventilated rack caging in a pathogen-free facility. The Institutional Animal Care and Use Committee of Memorial Sloan-Kettering Cancer Center approved animal use protocols. Mice were maintained on a mixed C57BL/6 and 129Sv background. For mice carrying an inducible Rad50 allele (Rad50ind), genotypes were determined using the primers TGTTCATGATCCCAAGGTAATGGTGTCT (sense), and TCAGAGAACTCATTGTGGAGTCAATTCT (antisense). Rad50− genotypes were determined using CGCTGTTAAACAGTACTGTCCG (sense) and the antisense primer from the Rad50ind PCR above. All other genotype strategies were described previously (3, 29, 37).
Rad50ind targeting.
Details of the Rad50ind targeting construct will be provided upon request. Targeting and Southern blot analyses of embryonic stem cell clones were carried out using previously described methods (37).
Ear fibroblast derivation.
Mice were anesthetized with isoflurane, and ear tissue was collected using sterile scissors. Ear fragments were rinsed twice each in 70% ethanol and phosphate-buffered saline (PBS) supplemented with 100 μg/ml kanamycin. Tissue was transferred into 0.3 ml of protease solution (4 mg/ml each of collagenase D and dispase in Dulbecco's modified Eagle's medium [DMEM]; filter sterilized), cut into pieces, and incubated at 37° for 45 min. DMEM (1.5 ml) containing 10% fetal bovine serum (Gemini), 1× glutamine, and 5× antibiotic-antimycotic solution was added, and samples were incubated at 37° overnight. Cells were dissociated by pipetting, passed through a 40-μm-pore-size cell strainer, and plated in DMEM as above except using 1× antibiotic-antimycotic solution. Cells were passaged upon reaching confluence and immortalized via transfection with a plasmid expressing simian virus 40 (SV40) large T-antigen. Transformed cells were grown in DMEM plus 10% cosmic calf serum (CCS; HyClone).
Rad50 deletion in cultured cells.
Adenovirus was commercially obtained (University of Iowa Gene Transfer Vector Core), and cells were infected in suspension at 2.5 × 106 cells/ml at a multiplicity of infection of 50 under low-serum conditions (2% CCS in DMEM) with 5 μg/ml Polybrene for 2 h at 37° on a rotating shaker.
Lentiviral production, concentration, and determination of titers were carried out using established methods (12, 36). For infections using a lentivirus-Cre-green fluorescent protein vector, cells were infected in suspension at 1 × 106 cells/ml at a multiplicity of infection of 10 in DMEM plus 10% CCS with 5 μg/ml polybrene. Tubes were spun at 1,900 rpm (∼600 × g) for 90 min, with occasional stops to manually resuspend cells. After viral infection, cells were plated and grown in DMEM containing 10% CCS.
Cellular assays.
Western blotting was carried out on 40 μg of protein lysates prepared by subjecting cells to three freeze-thaw cycles in NETN buffer (20 mM Tris, pH 8, 150 mM NaCl, 1 mM EDTA, and 0.5% NP-40 plus protease inhibitors). Binding and washing steps were done in 5% milk and PBS-Tween 20 buffer. Rabbit anti-Rad50 polyclonal (1:5,000; custom Petrini lab antibody m84-7) and antiactin mouse monoclonal (1:1,000; AC-40; Sigma) primary antibodies and species-specific secondary antibodies (Pierce) were used, and horseradish peroxidase signal was detected with ECL Plus reagent (Amersham).
For analysis of nuclear aberrations and γ-H2AX foci, cells were seeded onto multiwell slides (Erie Scientific), fixed with 4% paraformaldehyde (PFA), permeabilized (50 mM NaCl, 3 mM MgCl2, 200 mM sucrose, 10 mM HEPES, 0.5% Triton X-100), blocked with 10% fetal bovine serum in PBS, incubated with rabbit anti-γ-H2AX polyclonal antibody (1:300; Upstate) for 2 h, washed, incubated with secondary antibody for 1 h, washed, and stained with DAPI (4′,6′-diamidino-2-phenylindole; Sigma). Slides for this and all subsequent experiments described were scored blind as to the status of genotype. Images were captured on a Zeiss Axiovert microscope using a charge-coupled-device camera (Hammamatsu) and Volocity software (Improvision). For each sample, >100 nuclei were scored.
Metaphases were prepared from cultures treated with colcemid (2 × 10−7 M) for 1 h. Cells were trypsinized, hypotonically swelled (0.075 M KCl) for 7 min at 37°, fixed, washed in ice-cold methanol-acetic acid (3:1), and dropped on slides. Slides were stained with Giemsa (Sigma) for 10 min and rinsed with distilled water, and coverslips were mounted with Permount (Fisher). For each sample, >40 spreads were scored.
For fluorescence-activated cell sorter analysis of the sub-G1 population, cells were harvested, fixed in 70% ethanol, and stained with propidium iodide (Sigma). For each sample, >10,000 events were collected. Data were analyzed using FlowJo software.
Rad50 cDNA-expressing cultures were derived for colony assays via transfection and stable selection of cells bearing an Rad50 expression vector. The assay was done in duplicate using virus-Cre-infected and uninfected control cultures. Plates were stained with crystal violet, and colonies were tabulated after 18 days.
Telomere fluorescence in situ hybridization (FISH) staining was performed on metaphase spreads prepared as outlined above. Slides were then rehydrated in PBS, treated with 0.5 mg/ml RNase A in PBS at 37° for 10 min, rinsed in PBS, dehydrated through a series of ethanol washes (70%, 90%, and 100%), and air dried. A hybridization mixture (10 mM Tris-Cl, pH 7.2, 70% formamide 0.5% block reagent [Dupont NEN], 1:1,000 TelG-Cy3 peptide nucleic acid probe [Applied Biosystems]) was added, coverslips were applied to the slides, and DNA was denatured at 80° for 3 min. Probes were hybridized at room temperature for 2 h in a dark, humid chamber; slides were washed two times for 15 min in wash I (70% formamide, 10 mM Tris-Cl, pH 7.2, 0.1% bovine serum albumin), two times for 5 min in wash II (100 mM Tris-Cl, pH 7.2, 150 mM NaCl, 0.08% Tween, with DAPI added to the second wash), dehydrated in ethanol as above, and dried; coverslips were mounted. A total of >1,000 metaphase chromosomes from a minimum of 15 spreads were analyzed.
For immunofluorescence-FISH, cells were seeded on multiwell slides, fixed for 15 min in 2% PFA, washed in PBS, and blocked for 30 min (1 mg/ml bovine serum albumin, 3% goat serum, 0.1% Triton X-100, 1 mM EDTA). Slides were incubated at room temperature for 3 h with 53BP1 antibody (1:1,000 diluted in blocking solution; Novus), washed, incubated with secondary antibody for 2 h, fixed for 5 min in 2% PFA, and washed in PBS. Slides were dehydrated and denatured for 5 min each under conditions as described for telomere FISH. Slides were hybridized overnight at room temperature using a TelC-fluorescein isothiocyanate probe (1:1,000; Applied Biosystems). Slides were washed two times for 15 min in FISH wash (70% formamide, 10 mM Tris-Cl, pH 7.2) and three times in PBS, with DAPI added to the second wash. For IR-treated samples >1,000 53BP1 foci from at least 35 cells were analyzed for colocalization with telomeres; for Cre-treated Trf2ind/ind (Trf2-deficient) and Rad50Δ/ind (Rad50-deficient) samples, >200 foci from at least 18 cells were analyzed; for untreated samples and Rad50+/ind (Rad50-proficient) Cre-treated samples, 53BP1 focus-positive cells were rare, but >65 foci from at least 11 cells were analyzed.
Partial hepatectomy (PH) and liver regeneration analyses.
Mx-Cre-mediated Rad50 deletion was induced by injecting 6- to 12-week-old mice twice with 400 mg of poly(I)·poly(C) ([pI-pC] Sigma) intraperitoneally at 48-h intervals. PHs were performed after a wait of >28 days for bone marrow recovery. Mice were anesthetized with isoflurane, an incision was made above the abdomen, the large left and two median lobes of the liver were extruded, and lobes were ligated with silk suture and excised. Incisions were cleaned and stitched, and analgesic was administered. Studies conducted on mice at 2, 3, 4, and 8 days post-PH were performed in triplicate; studies at days 1 and 6 post-PH were performed in duplicate. For proliferative indices, bromodeoxyuridine (BrdU) injections were administered at 24-h intervals (50 mg/g of body weight) up to 3 days post-PH. Livers were used for calculation of regeneration kinetics, genotyping, and histological preparations.
Histological sample preparation and staining.
Tissue samples for histological analyses were fixed overnight at 4°C in 4% PFA, stored at 4°C in 70% ethanol, and then processed for paraffin embedding. Sections (8 μm) were prepared, and slides were processed and stained at the Memorial Sloan-Kettering molecular cytogenetics core for γ-H2AX, phospho-H3, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay, and Rad50. p16 and p19 staining was carried out by the Memorial Sloan-Kettering pathology core. For γ-H2AX and TUNEL analyses, >200 cells per sample were scored. For phospho-H3 analyses >250 cells per sample were scored for determining mitotic indices, and >45 cells per sample were scored for metaphase distributions. Phospo-H3-positive nuclei with chromatin beginning to condense were classified as prophase; those with condensed unaligned chromosomes were scored as prometaphase; aligned, unsegregated chromosomes were scored as metaphase; condensed chromosomes undergoing segregation were scored as anaphase; and decondensed sister nuclei were scored as telophase.
Behavioral analyses.
To assess gait abnormalities in Pcp2-Cre experiments, mice were conditioned to walk down a corridor, and gaits were recorded by dipping front paws in nontoxic red paint and hind paws in blue paint. Measurements from three to four runs per mouse were averaged. At 4 months, two controls and five mutants were analyzed; at 16 months three controls and four mutants were analyzed. To assess balance, studies were conducted as described previously with two controls and four mutant mice at 16 months (22).
RESULTS
Rad50 gene targeting and deletion in cultured cells.
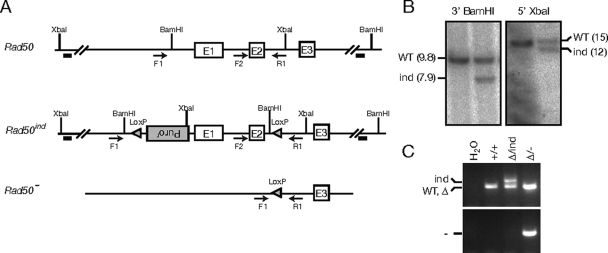
We generated mice carrying an inducible allele of Rad50 (Fig. 1A and B) to investigate the consequences of Rad50 deficiency in vivo. These mice were bred with the Rad50+/Δ strain (37) to produce Rad50Δ/ind mice. A PCR strategy was devised to determine the status of the inducible allele (Fig. 1A), and in conjunction with the Rad50Δ-specific PCR (37), final genotypes were determined.
FIG. 1.
Rad50-inducible construction and deletion. (A) The germ line region targeted for conditional deletion is shown as Rad50. The targeting vector was constructed to contain LoxP sites and a puromycin resistance (Puror) marker flanking exons 1 and 2. Conventional gene targeting methods were used to generate embryonic stem cells harboring the conditional allele (Rad50ind). Expression of Cre recombinase in cells or mice harboring this allele results in generation of the Rad50− allele. (B) Integration of the targeting construct was confirmed via Southern blotting using BamHI- and XbaI-digested genomic DNA with 3′- and 5′-specific probes, respectively (depicted as black bars in panel A). (C) Genotype PCR strategies were devised to differentiate the Rad50 loci. Primers F2 and R1 (see panel A for primer diagram) amplify Rad50 (or Rad50Δ), a larger fragment from Rad50ind, and no product from Rad50−. Primers F1 and R1 amplify a Rad50−-specific band. WT, wild type.
SV40-immortalized ear fibroblast cultures were derived and used to assess the effects of Rad50 deletion in vitro. Adenovirus- or lentivirus-mediated delivery of Cre resulted in deletion of the Rad50ind allele and production of the Rad50− allele (Fig. 1A and C). Time course analysis indicated that Cre-mediated recombination was complete at 2 days postinfection (data not shown). At 4 days after lentivirus-Cre infection, Rad50 protein was undetectable (Fig. 2A), and the levels of the remaining complex members were diminished (data not shown). Rad50 depletion was faster in adenovirus-infected cultures, and a moderate increase in phenotypic severity was noted, most likely due to Cre-independent effects of adenovirus. Because of this, adenovirus experiments were terminated at 4 days whereas lentivirus experiments could be carried to 6 days postinfection.
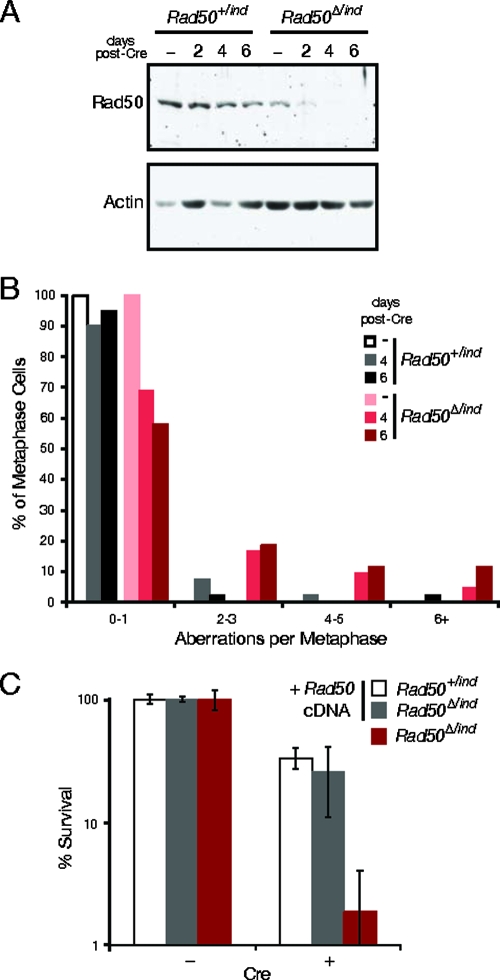
FIG. 2.
Rad50 deletion in cultured cells results in genomic instability. (A) Rad50 levels from lentivirus-Cre-infected SV40-transformed ear fibroblasts were detected by Western blotting. Actin is shown as a loading control. (B) The frequency of metaphase aberrations was determined for control (Rad50+/ind) and Rad50-deficient (Rad50Δ/ind) cells, as indicated on the graph, in uninfected cells (−) and cells at 4 and 6 days postdeletion. (C) Colony formation was assessed in control cells (white bars), inducible cells transfected with a vector expressing Rad50 cDNA (gray bars), and Rad50-deficient cells (red bars).
Following Rad50 deletion, cells were examined for indices of spontaneous DNA damage. Four days after adenovirus-Cre infection, 61% of Rad50Δ/− cells contained two or more γ-H2AX foci, whereas 18% of Rad50+/− control cells exhibited this staining pattern (χ2 = 39; P = 3.4 × 10−10). Analysis of metaphase spreads indicated that 42% of Rad50Δ/− cells contained two or more aberrations 6 days after lentivirus-Cre infection versus 5% in controls (Fig. 2B) (χ2 = 16; P = 0.0013). At this time point, cells primarily exhibited chromatid breaks (55.8% versus 10% in controls), but fragments, chromosome breaks, fusions, and tri-/quadra-radial exchange structures were also frequently observed (Table 1 and Fig. 3A; see also Fig. S1A in the supplemental material). Endoreduplicated cells containing four sister chromatids were occasionally seen, as previously noted in Nbs1-deficient B cells (45). Nuclear aberrations were also observed in Rad50-deficient cells including micronuclei, telophase bridges, and fragmented nuclei (see Fig. S1B in the supplemental material). These aberrations were likely the by-product of cell division in the presence of acentric or dicentric chromosomes.
TABLE 1.
Percentage of metaphase cells containing the indicated aberrations
| Days postinfection | Genotype | Frequency of aberrations in metaphase cells (%)
|
|||
|---|---|---|---|---|---|
| Chromatid breaks | Fragments | Chromosome breaks | Exchange structures | ||
| 4 | Rad50+/− | 10 | 10 | 2.5 | 2.5 |
| Rad50Δ/− | 14.3 | 31 | 4.8 | 0 | |
| 6 | Rad50+/− | 10 | 10 | 2.5 | 0 |
| Rad50Δ/− | 55.8 | 16.3 | 14 | 14 | |
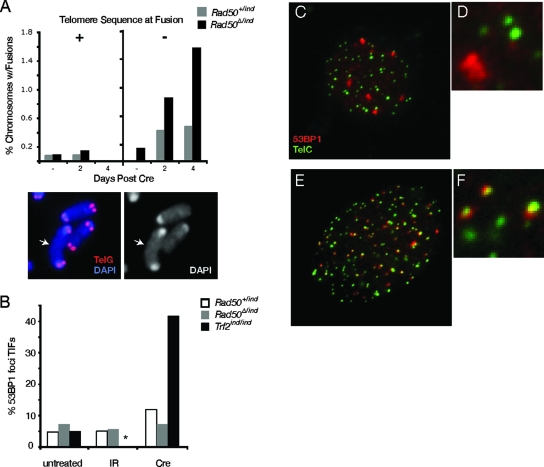
FIG. 3.
Rad50-deficient cells do not exhibit telomere defects. (A) Telomere FISH-stained metaphase spreads were analyzed for the presence of fused chromosomes. Fusions with and without telomere sequence at junctions were tabulated. An example of a fusion lacking telomere sequence is shown from a Rad50Δ/− spread. (B) Spontaneous and IR-induced 53BP1 foci in Rad50 control (white bars), Rad50-deficient (gray bars), and Trf2-deficient (black bars) cells were analyzed for colocalization with telomeres. Values represent percentages of 53BP1 foci exhibiting TIFs (*, IR-treated Trf2 deficient cells were not analyzed). Examples of 53BP1 (red)- and TelC (green)-stained cells from Rad50-deficient (C and D) and Trf2-deficient (E and F) cultures.
Consistent with the DNA damage observed, Rad50-deficient cells lost viability rapidly. Fluorescence-activated cell sorter analysis at 4 days postinfection revealed a sevenfold increase in the percentage of sub-G1 events in Rad50Δ/− cells, indicative of cell death, and colony formation was dramatically reduced. Rad50Δ/− colonies were never isolated (surviving clones retained the functional, undeleted allele), whereas colony-forming ability was restored by expression of a Rad50 cDNA (Fig. 2C). Collectively these results indicate that genomic instability is likely to account for cell death in the absence of Rad50.
To address whether Rad50 deficiency was associated with telomere dysfunction, we asked whether the chromosome fusions observed occurred within telomeric sequences. This would be expected if Rad50 deficiency were to phenocopy loss of Trf2. Southern blot analysis of telomeric DNA from Rad50Δ/− cells at 2 and 4 days postdeletion did not reveal the presence of aberrantly large telomeric DNA fragments (see Fig. S1C in the supplemental material), as would be expected to arise from telomere fusions and which are seen upon expression of a dominant negative Trf2 (Trf2ΔBΔM) (55). Southern blots also did not reveal precipitous telomere shortening, and despite an increase in the frequency of chromosome fusions, telomere sequence was rarely observed at fusion sites in metaphase spreads examined by telomere FISH (Fig. 3A). Additionally, in contrast to Trf2-deficient cells that exhibit telomere dysfunction-induced DNA damage foci (TIFs) (50), no increase in TIFs was observed in Rad50-deficient cells (Fig. 3B to F). These data indicate that Mre11 complex deficiency does not result in telomere uncapping and argue against telomere dysfunction as the underlying basis of lethality in Rad50Δ/− cells.
Rad50 deletion in vivo.
To query the role of the Mre11 complex in proliferative and postmitotic settings in vivo, the Mx-Cre transgene was crossed into Rad50Δ/ind mice. In Mx-Cre-containing mice, injection with double-stranded pI-pC induces Cre expression in both proliferative and nonproliferative tissues, including bone marrow, thymus, and liver (10, 29). Cre expression in the liver enabled analysis of Rad50 deficiency under both quiescent and proliferating conditions since quiescent hepatocytes can be stimulated to proliferate by resection of the liver mass.
The floxed Rad50 allele was inactivated in adult Mx-Cre transgenic mice, and bone marrow samples were analyzed. Genotype PCR results indicated that deletion of the Rad50ind allele occurred within 6 days of pI-pC administration (data not shown). Since all mice were alive at 4 weeks and showed no signs of anemia, we hypothesized that deletion did not occur in 100% of hematopoietic stem cells, permitting repopulation of the bone marrow with hematopoietic cells that had escaped deletion. We confirmed this at 7 weeks after pI-pC injection, when loss of the Rad50-inducible allele was detected in liver and bone marrow of Rad50+/ind controls as well as in the liver of Rad50Δ/− animals. At this time, the Rad50ind (i.e., functional) allele was still present in bone marrow from Rad50Δ/ind mice (Fig. 4A). These data indicate that the viable bone marrow cells detected were those that had escaped Rad50 deletion. This demonstrates that Rad50 is required for the viability of bone marrow cells, consistent with observations made in Nbs1-deficient mice (10).
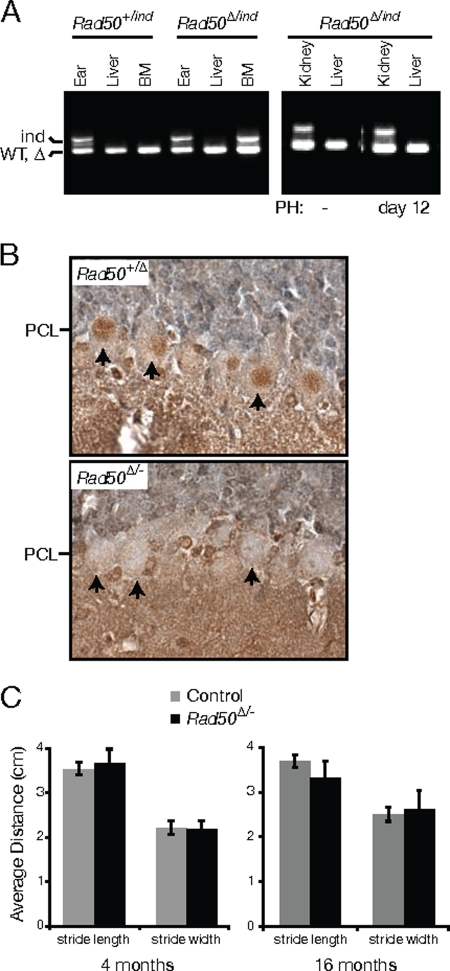
FIG. 4.
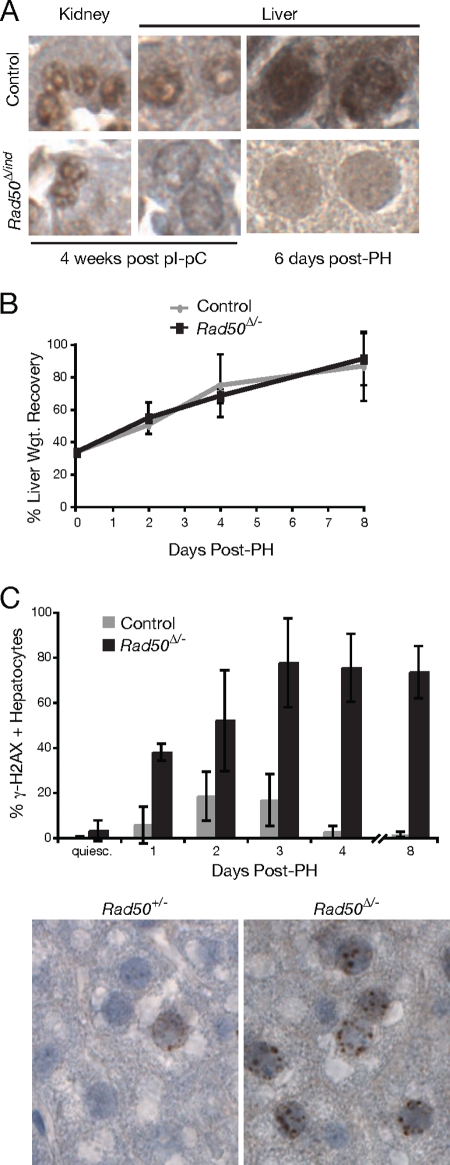
Rad50 deletion in bone marrow, quiescent liver, and postmitotic Purkinje cells. (A) Ear, liver, and bone marrow from adult Rad50+/ind and Rad50Δ/ind Mx-Cre+ mice were analyzed for deletion of the Rad50ind allele at 7 weeks after pI-pC injection; kidney and liver from pI-pC-injected Rad50Δ/ind Mx-Cre+ mice were analyzed before (−) and 12 days after PH. (B) Immunohistochemical staining of cerebellar sections from Pcp2-Cre+ control (Rad50+/Δ) and mutant (Rad50Δ/−) mice with Rad50 antibody. Loss of nuclear Rad50 staining in the Purkinje cell layer (PCL) of mutant mice is evident (arrows). (C) Gaits were recorded at 4 and 16 months, and average stride lengths and widths were calculated for control and Rad50Δ/− Pcp2-Cre+ mice.
To assess whether Rad50 deficiency was lethal in quiescent cells, we examined the livers of Rad50Δ/− Mx-Cre mice after administration of pI-pC. Hepatocytes of the adult liver are largely quiescent unless liver injury or resection induces regeneration. Rad50 deletion in the liver (Fig. 4A and 5A) resulted in a slight increase in levels of spontaneous DNA damage as assessed by γ-H2AX labeling (Fig. 5C). Nevertheless, liver function, as inferred from serum levels of liver enzymes, was unaffected up to 36 weeks postdeletion (see Fig. S2A in the supplemental material). Additionally, survival up to 40 weeks postdeletion was not significantly compromised, and no liver-associated pathology was observed. These results indicate that, in contrast to hematopoietic cells, quiescent Rad50-deficient hepatocytes are viable and functional.
FIG. 5.
Liver regeneration is normal in Rad50-deficient cells, but hepatocytes exhibit indices of DNA damage. (A) Kidney and liver sections from control and Rad50Δ/ind Mx-Cre+ mice were immunohistochemically stained with Rad50 antibody. Examples from mice sacrificed 4 weeks after pI-pC injection and 6 days post-PH are shown. (B) Percent liver weight recovery was calculated, and results were graphed for control and mutant mice up to 8 days post-PH. (C) Immunohistochemical staining of liver sections prepared from control and mutant mice were used to determine the percent γ-H2AX-positive hepatocytes before and after PH. Examples of γ-H2AX-labeled sections from Rad50+/− and Rad50Δ/− livers at 2 days post-PH are shown. quiesc, quiescent.
We examined a second tissue in which DNA replication is rare, terminally differentiated neurons. The Pcp2-Cre mouse line (3) was used to test the requirement for Rad50 in maintenance of postmitotic Purkinje cells. In this strain, Cre expression is activated postnatally under the control of the Purkinje cell-specific Pcp2 promoter. Although loss of Rad50 in Pcp2-Cre mice was evident in postmitotic Purkinje cells of adults (Fig. 4B), no alteration in cerebellar size or architecture was noted, and no increase in spontaneous γ-H2AX foci or pyknotic nuclei was observed in Purkinje cells of the mutants. Furthermore, mice lacking Rad50 did not show signs of akinesis or balance abnormalities up to 16 months postdeletion (χ2 = 0.037; P = 0.85), and only minor gait abnormalities were observed (Fig. 4C) (for stride length at 4 months, Wilcoxan rank sum [W] = 20,586 and P [two-sided] = 0.28; for width, W = 19,638 and P [two-sided] = 0.32; for stride length at 16 months, W = 11,591 and P [two-sided] = 8.7 × 10−4; for stride width, W = 14,206 and P [two-sided] = 0.16). These data support the interpretation that the Mre11 complex is dispensable for the viability of nondividing cells.
Rad50 deletion in proliferating hepatocytes.
Having established that Rad50 was dispensable in quiescent hepatocytes and postmitotic Purkinje cells, we induced proliferation in Rad50-deficient liver to determine whether proliferating hepatocytes would be similarly robust. The Rad50ind locus was deleted in Mx-Cre transgenic mice at least 4 weeks prior to regeneration studies to allow for bone marrow recovery. Mutant and control mice then underwent PH to remove two-thirds of the liver to induce regeneration. In contrast to the bone marrow, which relies upon the stem cell pool for repopulation, the proliferation of hepatocytes accounts for liver repopulation; hence, approximately two cell divisions per hepatocyte are required for full recovery of the liver mass (14). Mean survival after this procedure is approximately 5 days when regeneration is impaired (for examples, see references 4 and 60).
Livers from Rad50Δ/ind mice lacked detectable signal from the inducible locus when assayed by genotype PCR, both before and after regeneration (Fig. 4A). Furthermore, immunohistochemical staining of kidney and liver sections indicated that Rad50 was present in kidney cell nuclei of all mice, whereas Rad50-deficient hepatocytes exhibited only background levels of staining in comparison to controls (Fig. 5A). These data indicate that regeneration occurred in the absence of Rad50. Rad50Δ/− liver regeneration (as assessed by calculation of liver weight recovery) did not significantly differ from controls (Fig. 5B) (χ2 = 0.58; P = 0.90). Furthermore, liver function (see Fig. S2B in the supplemental material) was uncompromised at 2 and 16 weeks post-PH, and survival up to 36 weeks was comparable to controls.
Although liver function appeared normal, staining with γ-H2AX antibody revealed a marked increase in the frequency of hepatocytes exhibiting DNA damage foci in Rad50Δ/− livers following regeneration (Fig. 5C) (χ2 = 30.8; P = 1 × 10−5). Damage appeared within 24 h of surgery and peaked at 3 days post-PH when 79% of hepatocytes exhibited two or more foci. Damage persisted at essentially the same level up to 8 days. In contrast, hepatocytes from control livers peaked at 20% γ-H2AX positive and declined to 1% by 8 days post-PH.
Given the evidence for DNA damage in repopulating hepatocytes, we assessed DNA damage checkpoint activation in that setting. Hepatectomized mice were injected with BrdU immediately after surgery and at 24-h intervals thereafter, a procedure that labels a subset of replicating cells. The cumulative percentage of BrdU-positive hepatocytes at 3 days post-PH, the time frame in which the bulk of regeneration occurs (14), was significantly higher in controls than in mutant liver sections (38% ± 7.7% versus 22% ± 7.7%, respectively; χ2 = 51; P = 8.8 × 10−13), suggesting that DNA damage accumulating during repopulation impaired the proliferation of Rad50-deficient hepatocytes. This interpretation predicts that the mitotic index of Rad50Δ/− hepatocytes at time points during repopulation would be reduced relative to controls.
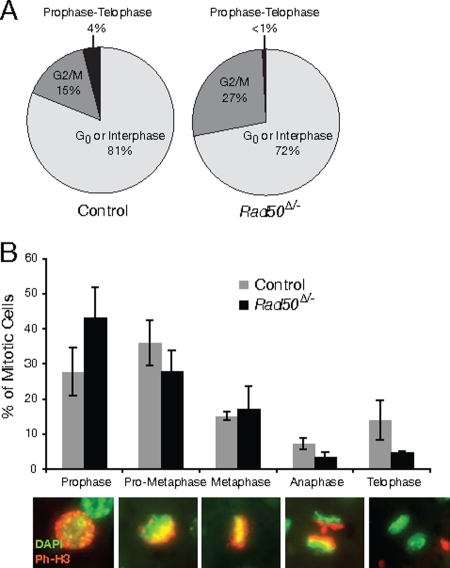
To assess cell division, liver sections were labeled with the mitotic marker phospho-histone H3. Mitotic cells were observed beginning 2 days posthepatectomy. In control livers, 15% (standard deviation, ±11%) of hepatocytes had the speckled phospho-H3 pattern of cells at the G2/M border (21) and 4% (±3%) were present between prophase and telophase of mitosis. The remaining cells were either in G0 or interphase (Fig. 6A; see Fig. S2C in the supplemental material for examples of G2/M and G0/interphase nuclei). In Rad50-deficient livers 27% (±23%) appeared to be at the G2/M boundary and less than 1% (±0.6%) were in mitosis (χ2 = 79; P = 6.14 × 10−18). At 3 days post-PH, mitotic indices had decreased in all mice, but the pattern was similar to that observed at day 2: G2/M levels were approximately threefold higher in mutants, but mitotic cells were 20-fold less frequent than in controls (χ2 = 158; P = 5.6 × 10−35). At 4 days post-PH, mitotic ratios were less than 0.1% in all mice. These results show that mitotic progression is impaired in Rad50-deficient hepatocytes. A similar increase in cells at the G2/M boundary was observed in Nbs1-deficient B cells (45).
FIG. 6.
Rad50-deficient hepatocytes divide at a reduced rate. The mitotic index at 2 days post-PH was determined by immunofluorescent labeling with phospho-H3 and DAPI staining of sections from control and Rad50Δ/− livers (A). (B) Phospho-H3-positive cells at 2 days post-PH were classified according to the indicated stages, and the percentages in each class were graphed for control and mutant mice. Representative images from the various stages are shown. Red, phospho-H3 (Ph-H3); pseudo-green, DAPI.
To determine whether Rad50-deficient cells that enter mitosis ultimately divide, mitotic cells were classified according to their mitotic progress through prophase, prometaphase, metaphase, anaphase, and telophase. These cells were identified on the basis of their morphological and staining patterns as described in Materials and Methods (Fig. 6B). There was a significant difference between mutant and control distributions at 2 days post-PH (Fig. 6B) (χ2 = 24; P = 7.01 × 10−5). This difference was due to a reduction in anaphase (7.1% in controls versus 3.2% in mutants) and telophase cells (13.9% in controls versus 4.7% in mutants). At 3 days post-PH, the distributions did not significantly differ (χ2 = 1.2; P = 0.87), and anaphase and telophase frequencies were comparable (6% and 14%, respectively, in controls versus 5% and 13% in mutants). These data indicate that despite high levels of damage observed in Rad50-deficient cells, a subset of cells are able to enter mitosis and divide either by resolving damage before mitotic entry or by dividing in the presence of damage.
The reduction in proliferation and impaired mitotic progression in Rad50Δ/− mice was associated with a decrease in liver cellularity. A reduction in the average number of cells per field (59 in controls versus 41 in mutants at 8 days post-PH; W = 489 and P [two-sided] = 9.92 × 10−5) and an increase in the average nuclear area per hepatocyte (3,251 pixels in controls versus 4113 pixels in mutants; W = 26,076 and P [two-sided] = 3.8 × 10−8) suggest that the cellularity of Rad50-deficient livers is reduced and, further, that Rad50Δ/− hepatocytes may undergo endoreduplication.
Since γ-H2AX foci persisted several days after regeneration, we analyzed hepatocytes for markers of apoptosis and senescence. A TUNEL assay of liver sections indicated that there was no increase in apoptotic cells from either mutant or control mice up to 12 days post-PH. Similarly, markers of senescence such as p16 and p19 were absent from repopulating hepatocytes. These data suggest that, as in other proliferating cells examined, Rad50 deficiency has a profound effect on genome integrity in proliferating hepatocytes. The apparent persistence of DNA damage notwithstanding, liver function did not appear to be grossly impaired.
DISCUSSION
DNA repair in postmitotic tissues.
In terminally differentiated tissues, the rates of cellular turnover can range from days to months or even decades. In the case of murine Purkinje cells, proliferation ceases at day 14 of embryogenesis (20), and the cells persist over the organism's life span without dividing. In these tissues, DNA replication-associated chromosome breakage, the most common source of spontaneous DSBs, will be commensurately rare. Spontaneous chromosome breakage has also been attributed to oxidative damage induced by reactive oxygen species and free radical by-products of oxidative metabolism. Nondividing cells, predominantly in G1, lack a sister chromatid and rely upon NHEJ for DSB repair. This may account for the observation that DNA ligase IV-deficient mice exhibit widespread apoptosis in postmitotic neurons (31) and that disruption of HR in the developing nervous system primarily affects proliferating neural precursors (41). These data predict that Mre11 complex-dependent homologous recombination, which promotes the sister chromatid as a repair template (6, 26), may not be required for maintenance of postmitotic cells. To test this hypothesis, we analyzed the effects of Rad50 deletion in postmitotic tissues of the mouse.
As expected from previous studies in proliferating cells (45), Rad50 deficiency led to profound chromosome instability and death within 4 to 6 days. In contrast, that condition was well tolerated in quiescent hepatocytes and postmitotic Purkinje cells, and no overt pathology was observed. These findings support the possibility that the Mre11 complex and, by extension, Mre11 complex-dependent HR are dispensable in nonproliferative tissues.
Implicitly, these data also suggest that DSBs that arise in nondividing cells—some of which likely result from oxidative damage—are not repaired in an Mre11 complex-dependent manner. An alternative possibility is that such DSBs are sufficiently rare that they do not deleteriously affect postmitotic tissue homeostasis. The phenotypic outcomes of NHEJ-deficient mice, in which postmitotic neurons are severely affected (2, 15, 16, 18, 41), argue against the latter possibility. A parsimonious interpretation is that the Mre11 complex does not play a significant role in mammalian NHEJ, and so Rad50 deficiency does not affect the repair of such lesions.
The data also suggest that mitigation of other consequences of oxidative damage such as base modifications is not heavily dependent on the Mre11 complex. It is clear from a number of studies that certain endogenous DNA lesions such as abasic sites can trap topoisomerase I-DNA covalent complexes (43). Recent studies suggest that the Mre11 complex may contribute to the resolution of such intermediates (11, 35, 40, 54) although TDP1 (13, 35) may play a more significant role in this regard. Furthermore, the most common form of oxidative DNA damage in mammalian cells, 7,8-dihydro-8-oxoguanine, stabilizes the noncovalently DNA-bound form of topoisomerase I (32, 44) and thus is not likely to be removed by either TDP1 or the Mre11 complex.
Essential functions of the Mre11 complex.
The Mre11 complex's effects on chromosome metabolism are diverse; however, several lines of evidence suggest that its functions are acutely required during DNA replication. The complex associates with chromatin during S phase in undamaged cells and forms foci that colocalize with PCNA (38, 39). Treatment of cells with agents that cause replication fork stalling or collapse results in additional recruitment of the Mre11 complex (34, 39, 56). Recruitment of the Mre11 complex in either circumstance may reflect a role in stabilization, processing, or repair of damaged replication forks.
Furthermore, the major recombinational repair function of the complex is the promotion of DNA damage-induced sister chromatid recombination (6, 26). A primary determinant of this function is the zinc hook domain of Rad50, in which two cysteine residues from each of two protomers coordinate a zinc atom to establish a dimerization interface. This mode of interaction underlies molecular bridging by the Mre11 complex, which we have proposed enforces the physical proximity of participants in the DSB repair reaction (8, 24, 58).
The chromosomal outcomes observed in Rad50Δ/− fibroblasts, Mre11-deficient DT40 cells (61), and Nbs1-deficient B cells (45) support the interpretation that the essential function of the Mre11 complex in vertebrates is the metabolism of DNA replication-associated damage. The effects of Rad50 depletion on hepatocyte repopulation are also consistent with the view that proliferation is severely impaired by Rad50 deficiency and suggest that the consequences of Rad50 loss are manifest within the first passage through S phase.
On the other hand, the data do not support the possibility that the complex's role at the telomere underlies its essential nature as depletion of Rad50 did not result in acute telomere dysfunction. No telomere fusions—which would indicate telomere uncapping—were observed in metaphase cells upon Rad50 deletion, and telomere dysfunction-induced foci (50) were not evident in interphase nuclei (Fig. 3B to F).
The deletion of Rad50 in postmitotic Purkinje cells did not phenocopy the mild ataxia seen in Atm−/− mice (1), even at relatively advanced ages (>12 months) (Fig. 4C and data not shown). This raises the possibility that events during neuronal development may contribute to Purkinje cell loss in AT and AT-like disorder patients. This interpretation is consistent with the report of ataxia in Nestin-Cre mice that lose Nbs1 during development of the nervous system (17) as well as studies implicating ATM (23, 46) and HR (41) during proliferation of precursors rather than maintenance of postmitotic Purkinje cells and neurons.
It remains an open question as to whether other essential members of the DDR pathway are dispensable for the maintenance of postmitotic or quiescent cells. Resolution of this issue is required to fully understand the role of the DDR in vivo.
Supplementary Material
Acknowledgments
This work was supported by grants GM56888 and GM59413 and the Joel and Joan Smilow Initiative.
We thank E. Lazzerini-Denchi and G. Plitas for providing hepatectomy training, C. Attwooll for providing immunofluorescence-FISH-labeled Trf2 samples, E. Brown for lentivirus vectors, K. Manova and S. Couto for cytology and pathology input and services, and members of our laboratory for insightful discussion.
Footnotes
Published ahead of print on 10 November 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Barlow, C., S. Hirotsune, R. Paylor, M. Liyanage, M. Eckhaus, F. Collins, Y. Shiloh, J. N. Crawley, T. Ried, D. Tagle, and A. Wynshaw-Boris. 1996. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 86159-171. [DOI] [PubMed] [Google Scholar]
- 2.Barnes, D. E., G. Stamp, I. Rosewell, A. Denzel, and T. Lindahl. 1998. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 81395-1398. [DOI] [PubMed] [Google Scholar]
- 3.Barski, J. J., K. Dethleffsen, and M. Meyer. 2000. Cre recombinase expression in cerebellar Purkinje cells. Genesis 2893-98. [PubMed] [Google Scholar]
- 4.Blindenbacher, A., X. Wang, I. Langer, R. Savino, L. Terracciano, and M. H. Heim. 2003. Interleukin 6 is important for survival after partial hepatectomy in mice. Hepatology 38674-682. [DOI] [PubMed] [Google Scholar]
- 5.Boulton, S. J., and S. P. Jackson. 1998. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 171819-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bressan, D. A., B. K. Baxter, and J. H. Petrini. 1999. The Mre11-Rad50-Xrs2 protein complex facilitates homologous recombination-based double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 197681-7687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Celli, G. B., and T. de Lange. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat. Cell Biol. 7712-718. [DOI] [PubMed] [Google Scholar]
- 8.de Jager, M., J. van Noort, D. C. van Gent, C. Dekker, R. Kanaar, and C. Wyman. 2001. Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol. Cell 81129-1135. [DOI] [PubMed] [Google Scholar]
- 9.de Lange, T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 192100-2110. [DOI] [PubMed] [Google Scholar]
- 10.Demuth, I., P. O. Frappart, G. Hildebrand, A. Melchers, S. Lobitz, L. Stockl, R. Varon, Z. Herceg, K. Sperling, Z. Q. Wang, and M. Digweed. 2004. An inducible null mutant murine model of Nijmegen breakage syndrome proves the essential function of NBS1 in chromosomal stability and cell viability. Hum. Mol. Genet. 132385-2397. [DOI] [PubMed] [Google Scholar]
- 11.Deng, C., J. A. Brown, D. You, and J. M. Brown. 2005. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics 170591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dull, T., R. Zufferey, M. Kelly, R. J. Mandel, M. Nguyen, D. Trono, and L. Naldini. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 728463-8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Khamisy, S. F., G. M. Saifi, M. Weinfeld, F. Johansson, T. Helleday, J. R. Lupski, and K. W. Caldecott. 2005. Defective DNA single-strand break repair in spinocerebellar ataxia with axonal neuropathy-1. Nature 434108-113. [DOI] [PubMed] [Google Scholar]
- 14.Fausto, N., and J. S. Campbell. 2003. The role of hepatocytes and oval cells in liver regeneration and repopulation. Mech. Dev. 120117-130. [DOI] [PubMed] [Google Scholar]
- 15.Frank, K. M., J. M. Sekiguchi, K. J. Seidl, W. Swat, G. A. Rathbun, H. L. Cheng, L. Davidson, L. Kangaloo, and F. W. Alt. 1998. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature 396173-177. [DOI] [PubMed] [Google Scholar]
- 16.Frank, K. M., N. E. Sharpless, Y. Gao, J. M. Sekiguchi, D. O. Ferguson, C. Zhu, J. P. Manis, J. Horner, R. A. DePinho, and F. W. Alt. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5993-1002. [DOI] [PubMed] [Google Scholar]
- 17.Frappart, P. O., W. M. Tong, I. Demuth, I. Radovanovic, Z. Herceg, A. Aguzzi, M. Digweed, and Z. Q. Wang. 2005. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat. Med. 11538-544. [DOI] [PubMed] [Google Scholar]
- 18.Gao, Y., Y. Sun, K. M. Frank, P. Dikkes, Y. Fujiwara, K. J. Seidl, J. M. Sekiguchi, G. A. Rathbun, W. Swat, J. Wang, R. T. Bronson, B. A. Malynn, M. Bryans, C. Zhu, J. Chaudhuri, L. Davidson, R. Ferrini, T. Stamato, S. H. Orkin, M. E. Greenberg, and F. W. Alt. 1998. A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95891-902. [DOI] [PubMed] [Google Scholar]
- 19.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95583-586. [DOI] [PubMed] [Google Scholar]
- 20.Hatten, M. E., and N. Heintz. 1995. Mechanisms of neural patterning and specification in the developing cerebellum. Annu. Rev. Neurosci. 18385-408. [DOI] [PubMed] [Google Scholar]
- 21.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106348-360. [DOI] [PubMed] [Google Scholar]
- 22.Herson, P. S., M. Virk, N. R. Rustay, C. T. Bond, J. C. Crabbe, J. P. Adelman, and J. Maylie. 2003. A mouse model of episodic ataxia type-1. Nat. Neurosci. 6378-383. [DOI] [PubMed] [Google Scholar]
- 23.Herzog, K. H., M. J. Chong, M. Kapsetaki, J. I. Morgan, and P. J. McKinnon. 1998. Requirement for Atm in ionizing radiation-induced cell death in the developing central nervous system. Science 2801089-1091. [DOI] [PubMed] [Google Scholar]
- 24.Hopfner, K. P., L. Craig, G. Moncalian, R. A. Zinkel, T. Usui, B. A. Owen, A. Karcher, B. Henderson, J. L. Bodmer, C. T. McMurray, J. P. Carney, J. H. Petrini, and J. A. Tainer. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418562-566. [DOI] [PubMed] [Google Scholar]
- 25.International Nijmegen Breakage Syndrome Study Group. 2000. Nijmegen breakage syndrome. Arch. Dis. Child. 82400-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov, E. L., V. G. Korolev, and F. Fabre. 1992. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics 132651-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kang, J., R. T. Bronson, and Y. Xu. 2002. Targeted disruption of NBS1 reveals its roles in mouse development and DNA repair. EMBO J. 211447-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kironmai, K. M., and K. Muniyappa. 1997. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 2443-455. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn, R., F. Schwenk, M. Aguet, and K. Rajewsky. 1995. Inducible gene targeting in mice. Science 2691427-1429. [DOI] [PubMed] [Google Scholar]
- 30.Lavin, M. F. 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 267749-7758. [DOI] [PubMed] [Google Scholar]
- 31.Lee, Y., D. E. Barnes, T. Lindahl, and P. J. McKinnon. 2000. Defective neurogenesis resulting from DNA ligase IV deficiency requires Atm. Genes Dev. 142576-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesher, D. T., Y. Pommier, L. Stewart, and M. R. Redinbo. 2002. 8-Oxoguanine rearranges the active site of human topoisomerase I. Proc. Natl. Acad. Sci. USA 9912102-12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim, D. S., and P. Hasty. 1996. A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 167133-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Limoli, C. L., E. Giedzinski, W. M. Bonner, and J. E. Cleaver. 2002. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma-H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. USA 99233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, C., J. J. Pouliot, and H. A. Nash. 2002. Repair of topoisomerase I covalent complexes in the absence of the tyrosyl-DNA phosphodiesterase Tdp1. Proc. Natl. Acad. Sci. USA 9914970-14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lois, C., E. J. Hong, S. Pease, E. J. Brown, and D. Baltimore. 2002. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 295868-872. [DOI] [PubMed] [Google Scholar]
- 37.Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl, A. Bradley, and J. H. Petrini. 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 967376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maser, R. S., O. K. Mirzoeva, J. Wells, H. Olivares, B. R. Williams, R. Zinkel, P. J. Farnham, and J. H. J. Petrini. 2001. The MRE11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol. Cell. Biol. 216006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mirzoeva, O. K., and J. H. Petrini. 2003. DNA replication-dependent nuclear dynamics of the Mre11 complex. Mol. Cancer Res. 1207-218. [PubMed] [Google Scholar]
- 40.Morales, M., Y. Liu, E. C. Laiakis, W. F. Morgan, S. D. Nimer, and J. H. Petrini. 2008. DNA damage signaling in hematopoietic cells: a role for Mre11 complex repair of topoisomerase lesions. Cancer Res. 682186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orii, K. E., Y. Lee, N. Kondo, and P. J. McKinnon. 2006. Selective utilization of nonhomologous end-joining and homologous recombination DNA repair pathways during nervous system development. Proc. Natl. Acad. Sci. USA 10310017-10022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petrini, J. H., and T. H. Stracker. 2003. The cellular response to DNA double-strand breaks: defining the sensors and mediators. Trends Cell Biol. 13458-462. [DOI] [PubMed] [Google Scholar]
- 43.Pommier, Y. 2006. Topoisomerase I inhibitors: camptothecins and beyond. Nat. Rev. Cancer. 6789-802. [DOI] [PubMed] [Google Scholar]
- 44.Pourquier, P., L. M. Ueng, J. Fertala, D. Wang, H. J. Park, J. M. Essigmann, M. A. Bjornsti, and Y. Pommier. 1999. Induction of reversible complexes between eukaryotic DNA topoisomerase I and DNA-containing oxidative base damages. 7, 8-dihydro-8-oxoguanine and 5-hydroxycytosine. J. Biol. Chem. 2748516-8523. [DOI] [PubMed] [Google Scholar]
- 45.Reina-San-Martin, B., M. C. Nussenzweig, A. Nussenzweig, and S. Difilippantonio. 2005. Genomic instability, endoreduplication, and diminished Ig class-switch recombination in B cells lacking Nbs1. Proc. Natl. Acad. Sci. USA 1021590-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soares, H. D., J. I. Morgan, and P. J. McKinnon. 1998. Atm expression patterns suggest a contribution from the peripheral nervous system to the phenotype of ataxia-telangiectasia. Neuroscience 861045-1054. [DOI] [PubMed] [Google Scholar]
- 47.Sonoda, E., M. S. Sasaki, J. M. Buerstedde, O. Bezzubova, A. Shinohara, H. Ogawa, M. Takata, Y. Yamaguchi-Iwai, and S. Takeda. 1998. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 17598-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, G. S., R. S. Maser, T. Stankovic, D. A. Bressan, M. I. Kaplan, N. G. Jaspers, A. Raams, P. J. Byrd, J. H. Petrini, and A. M. Taylor. 1999. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 99577-587. [DOI] [PubMed] [Google Scholar]
- 49.Stohr, B. A., and K. N. Kreuzer. 2001. Repair of topoisomerase-mediated DNA damage in bacteriophage T4. Genetics 15819-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takai, H., A. Smogorzewska, and T. de Lange. 2003. DNA damage foci at dysfunctional telomeres. Curr. Biol. 131549-1556. [DOI] [PubMed] [Google Scholar]
- 51.Tauchi, H., J. Kobayashi, K. Morishima, D. C. van Gent, T. Shiraishi, N. S. Verkaik, D. vanHeems, E. Ito, A. Nakamura, E. Sonoda, M. Takata, S. Takeda, S. Matsuura, and K. Komatsu. 2002. Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 42093-98. [DOI] [PubMed] [Google Scholar]
- 52.Theunissen, J. W., M. I. Kaplan, P. A. Hunt, B. R. Williams, D. O. Ferguson, F. W. Alt, and J. H. Petrini. 2003. Checkpoint failure and chromosomal instability without lymphomagenesis in Mre11(ATLD1/ATLD1) mice. Mol. Cell 121511-1523. [DOI] [PubMed] [Google Scholar]
- 53.Tsuzuki, T., Y. Fujii, K. Sakumi, Y. Tominaga, K. Nakao, M. Sekiguchi, A. Matsushiro, Y. Yoshimura, and T. Morita. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc. Natl. Acad. Sci. USA 936236-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vance, J. R., and T. E. Wilson. 2002. Yeast Tdp1 and Rad1-Rad10 function as redundant pathways for repairing Top1 replicative damage. Proc. Natl. Acad. Sci. USA 9913669-13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Steensel, B., A. Smogorzewska, and T. de Lange. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92401-413. [DOI] [PubMed] [Google Scholar]
- 56.Wang, Y., D. Cortez, P. Yazdi, N. Neff, S. J. Elledge, and J. Qin. 2000. BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev. 14927-939. [PMC free article] [PubMed] [Google Scholar]
- 57.Williams, B. R., O. K. Mirzoeva, W. F. Morgan, J. Lin, W. Dunnick, and J. H. Petrini. 2002. A murine model of Nijmegen breakage syndrome. Curr. Biol. 12648-653. [DOI] [PubMed] [Google Scholar]
- 58.Wiltzius, J. J., M. Hohl, J. C. Fleming, and J. H. Petrini. 2005. The Rad50 hook domain is a critical determinant of Mre11 complex functions. Nat. Struct. Mol. Biol. 12403-407. [DOI] [PubMed] [Google Scholar]
- 59.Xiao, Y., and D. T. Weaver. 1997. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 252985-2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamada, Y., I. Kirillova, J. J. Peschon, and N. Fausto. 1997. Initiation of liver growth by tumor necrosis factor: deficient liver regeneration in mice lacking type I tumor necrosis factor receptor. Proc. Natl. Acad. Sci. USA 941441-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamaguchi-Iwai, Y., E. Sonoda, M. S. Sasaki, C. Morrison, T. Haraguchi, Y. Hiraoka, Y. M. Yamashita, T. Yagi, M. Takata, C. Price, N. Kakazu, and S. Takeda. 1999. Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J. 186619-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang, Y. G., A. Saidi, P. O. Frappart, W. Min, C. Barrucand, V. Dumon-Jones, J. Michelon, Z. Herceg, and Z. Q. Wang. 2006. Conditional deletion of Nbs1 in murine cells reveals its role in branching repair pathways of DNA double-strand breaks. EMBO J. 255527-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25347-352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.