Abstract
The infant leukemia-associated gene Ott1 (Rbm15) has broad regulatory effects within murine hematopoiesis. However, germ line Ott1 deletion results in fetal demise prior to embryonic day 10.5, indicating additional developmental requirements for Ott1. The spen gene family, to which Ott1 belongs, has a transcriptional activation/repression domain and RNA recognition motifs and has a significant role in the development of the head and thorax in Drosophila melanogaster. Early Ott1-deficient embryos show growth retardation and incomplete closure of the notochord. Further analysis demonstrated placental defects in the spongiotrophoblast and syncytiotrophoblast layers, resulting in an arrest of vascular branching morphogenesis. The rescue of the placental defect using a conditional allele with a trophoblast-sparing cre transgene allowed embryos to form a normal placenta and survive gestation. This outcome showed that the process of vascular branching morphogenesis in Ott1-deficient animals was regulated by the trophoblast compartment rather than the fetal vasculature. Mice surviving to term manifested hyposplenia and abnormal cardiac development. Analysis of global gene expression of Ott1-deficient embryonic hearts showed an enrichment of hypoxia-related genes and a significant alteration of several candidate genes critical for cardiac development. Thus, Ott1-dependent pathways, in addition to being implicated in leukemogenesis, may also be important for the pathogenesis of placental insufficiency and cardiac malformations.
spen (split ends) family proteins have a significant role in development. In Drosophila, spen was isolated from screens searching for modifiers of egfr (Epidermal Growth Factor Receptor)/ras (Rat Sarcoma Virus)/mapk (Mitogen-Activated Protein Kinase) pathways (32, 49). In addition, spen has been implicated in downstream hox (homeobox) gene pathways responsible for head and thorax development (66). The murine homolog, Mint (Msx-Interacting Nuclear Target), suppresses marginal zone B-cell development while promoting follicular B-cell differentiation in the spleen, purportedly through an inhibition of Notch2 signaling (33). The deletion of Mint is embryonic lethal at embryonic day 14.5 (E14.5) and has a phenotype including defects in cardiac, pancreatic, and skin cells as well as a quantitative loss of fetal liver cells (33).
The spen family member OTT1 (One-Twenty Two 1) (RBM15 [RNA Binding Motif 15]) was originally isolated as a fusion partner in t(1;22)(p13;q13), associated with infant acute megakaryocytic leukemia (39, 41), but its function is not well understood. In vitro experiments showed a cell context-dependent stimulation or inhibition of Notch-dependent transcription in myeloid cell lines and nonhematopoietic lines, respectively (38). An additional function in RNA transport and splicing has been described; however, thus far, only viral transcripts rather than physiological mRNAs have been identified (27, 37).
We previously reported that the germ line deletion of Ott1 is embryonic lethal beyond E9.5, whereas conditional deletion within the hematopoietic compartment results in a block in B-cell development at the pre-B stage and increases in hematopoietic progenitor, myeloid, and megakaryocytic populations (48). Because Ott1 has a significant role in multiple aspects of hematopoiesis and is widely expressed in multiple tissue types, additional roles of Ott1 in development were investigated. Analysis of Ott1 homozygous null embryos revealed a specific defect in placental trophoblast development and placental vascular branching morphogenesis that is responsible for the demise of the early embryos. Using a conditional deletion of Ott1 that spared trophoblastic lineages, the early placental phenotype was rescued, allowing for the delineation of additional requirements of Ott1 during development, including roles in cardiac and splenic organogenesis.
MATERIALS AND METHODS
Mice.
Mice containing the Ott1flox and Ott1null alleles were generated as previously reported (48). Transgenic Sox2-cre animals were obtained from Jackson Labs (Bar Harbor, ME). Genotyping was performed by PCR analysis of murine or embryonic tissue (48). Timed matings were determined by the presence of a vaginal plug at E0.5. Animals were housed in microisolator cages, and all experiments were performed with the approval of the Institutional Animal Care and Use Committee.
Histology.
Mouse litters were sacrificed at the appropriate gestational age. For routine histology and light microscopy, tissues were fixed overnight in 10% formalin or Bouin's solution, processed, paraffin embedded, sectioned (5-μm thickness), and stained with hematoxylin and eosin (H&E). E8.5 and E.9.5 embryos were kept within deciduas and sectioned along the embryonic/abembryonic axis. For detailed analysis of cardiac phenotype, thoraces from E18.5 embryos were embedded and serially sectioned transversely at a 10-μm thickness. Immunohistochemical staining for CD34 was performed with a rat monoclonal antibody, MEC14.7 (Abcam, Cambridge, MA).
In situ hybridization.
Plasmids to generate in situ probes for Tpbp/4311, Gcm1, and Vegf were generously provided by J. Rossant (Toronto, Ontario, Canada). The Ott1 probe was generated by cloning the first 1,000 bp of exon 1 of Ott1 from murine genomic DNA using PCR amplification with primers 5′-ATGAGGTCTGCGGGGCGGGAG-3′ and 5′-ACGGATAGTCTCGTTCTCTTTCCA-3′ into pCR4-TOPO (Invitrogen, Carlsbad, CA). A sense Ott1 probe was used as a control for the antisense probe. Linearized plasmids were transcribed using T7 polymerase. 33P-radiolabeled in situ hybridization of E9.5 sections was performed. After deparaffinization, the slides were fixed in 4% paraformaldehyde for 10 min and then treated with proteinase K for 10 min at 37°C. The slides were hybridized overnight at 60°C with labeled riboprobe diluted in hybridization buffer. The slides were washed at 65°C for 2 h in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), immersed in Kodak NTB emulsion, and exposed for 1 to 4 weeks at 4°C. Following exposure, the slides were developed and counterstained with hematoxylin.
Gene expression analysis.
RNA was extracted from three hearts each from either Ott1null/null Sox2-cre or Ott1null/wt Sox2-cre E18.5 embryos using the RNeasy microkit (Qiagen, Valencia, CA) and treated with RNase-free DNase (Qiagen, Valencia, CA). Fifty nanograms of purified total RNA was linearly amplified using the Ovation RNA amplification system, V2 (Nugen, San Carlos, CA), and biotinylated with the FL-Ovation Biotin Module, V2 (Nugen, San Carlos, CA), according to the supplied protocol. cDNA was then hybridized to Affymetrix mouse expression array 430A2.0 chips by the Dana Farber Microarray Core Facility (Boston, MA). The raw gene expression values were preprocessed with a robust multiarray analysis algorithm (28) using BioConductor software (21). To find differentially expressed genes, the preprocessed gene expression values were then filtered to remove genes with relatively constant expression. The filter kept genes that had at least a threefold change and an absolute change of 100 gene expression units. On this filtered set, differential expression was measured with the Comparative Marker Selection algorithm (50) using GenePattern software (50). The signal-to-noise ratio was used as the test statistic to rank order the genes for permutation testing. In addition to finding differentially expressed genes, we also considered gene sets. Gene sets were tested for enrichment with gene set enrichment analysis (GSEA) (61). We tested all 1,837 gene sets in the Molecular Signatures Database (http://www.broad.mit.edu/gsea/msigdb/index.jsp).
Microarray data accession number.
Array data are listed in the Gene Expression Omnibus repository under accession number GSE12628.
RESULTS
Deletion of Ott1 results in numerous developmental defects prior to fetal demise at E10.5.
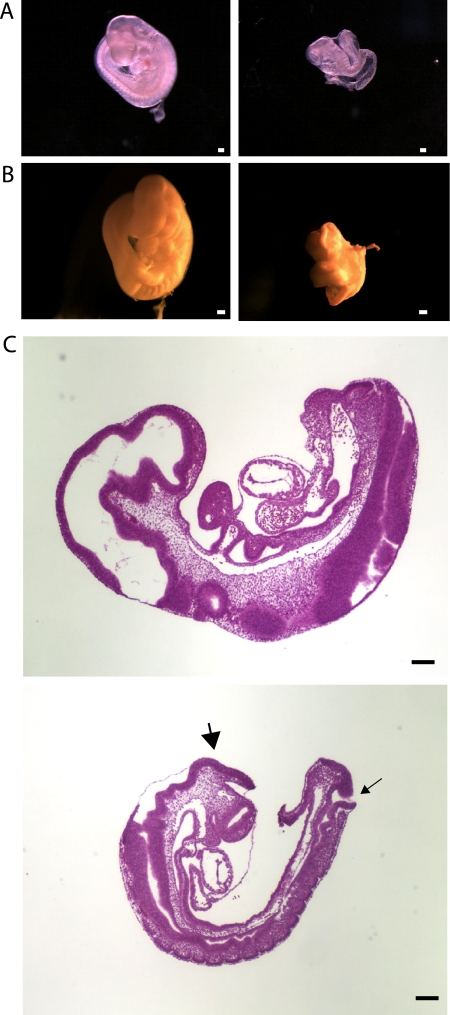
Analysis of embryos resulting from timed matings of Ott1null/+ mice shows that Ott1null/null embryos are dead or undergoing degeneration by E10.5 (48). Further comparison with wild-type or heterozygous littermates showed severe growth retardation, delayed turning, and incomplete closure of the neural tube in Ott1 homozygous null embryos (Fig. 1A and B). Blood was present and the heart was visibly pumping in the Ott1null/null embryos (data not shown). Histological cross sections of E9.5 Ott1null/null embryos showed that the neural tube defect was manifested by exencephaly and spina bifida (Fig. 1C). Somitogenesis did not appear disorganized but reflected an otherwise delayed state. No major structures were missing, and no specific tissues were observed to undergo degeneration.
FIG. 1.
Gross histopathology of E9.5 Ottnull/null and wild-type embryos. (A and B) Micrographs of whole E9.5 embryos (A) and embryos fixed in Bouin's solution (B). (Left) Wild-type embryo; (right) Ott1null/null embryo. (C) H&E-stained cross section. Exencephaly and spina bifida are indicated by large and small arrows, respectively. (Top) Wild-type embryo; (bottom) Ott1null/null embryo. Magnification bars represent 200 μm.
Placentas of Ott1null/null embryos have defective vascularization.
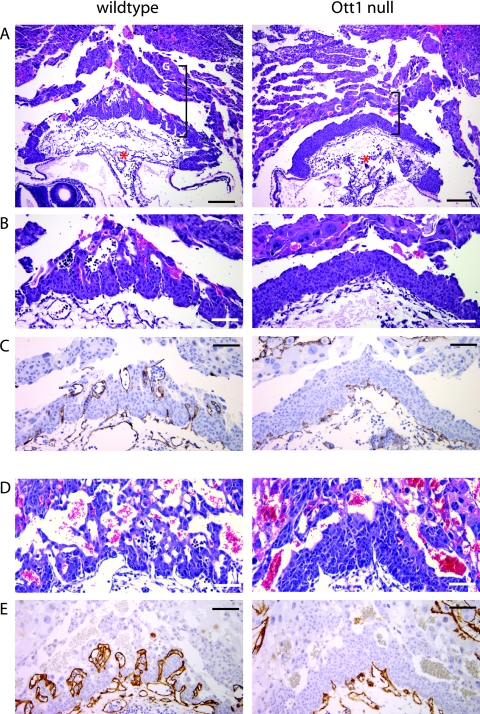
Numerous gene deletions in mice with similar growth delay phenotypes due to inadequate placentation, thus limiting fetal oxygen exchange and resulting in fetal death, have been described (53). Because the embryonic defects in the mutant embryos were similar to one of global growth retardation and occurred at a point requiring placental function, we studied placenta formation in Ott1null/null deciduas. At E7.5, the allantois was seen in both wild-type and mutant embryos (not shown), and by E8.5, chorioallantoic fusion had occurred in both genotypes (Fig. 2A). Histologically, beginning at E8.5 and progressing at E9.5, Ott1-null embryos exhibited a smaller, thinner placenta. The outermost placental layer consisting of trophoblast giant cells appeared unchanged in size and number (Fig. 2A). However, the region consisting of the spongiotrophoblast and the labyrinthine syncytiotrophoblast layers was markedly reduced and did not show a vascular network that was present in the control littermate embryos (Fig. 2B). To examine this vascular phenotype further, immunoperoxidase staining with the endothelial marker CD34 was performed. CD34 staining showed a nearly complete lack of vascularization of the syncytiotrophoblast layer by fetal vessels (Fig. 2C). Penetration of the placenta by fetal vessels was observed; however, there was no expansion, branching, or development of adjacent maternal sinuses. While an increase in vascular branching was seen in control E9.5 embryos, no developmental progression was seen in mutants (Fig. 2D and E). Collectively, these findings are consistent with an arrest of fetal vascular branching morphogenesis in Ott1null/null animals.
FIG. 2.
Ott1null/null placentas have impaired fetal vascularization. Shown are cross sections through E8.5 (A to C) and E9.5 (D and E) placenta and decidual maternal tissues. Littermate control placentas are depicted on the left, and Ott1null/null placentas are depicted on the right. (A and B) H&E stain. Brackets indicate the layers comprising the placenta. G, giant cell trophoblast layer; S, spongiotrophoblast layer; L, labyrinthine syncytiotrophoblast layer; asterisk, allantoic stalk. (B) Higher magnification of placenta. (C) Immunostaining of the same embryo with endothelial marker CD34 to highlight fetal vasculature. (D and E) E9.5 placentas showing a highly vascularized labyrinth in controls but not mutants. (E) CD34 stain. Magnification bars represent 200 μm (A) and 100 μm (B to E).
Loss of Ott1 causes a decrease in the syncytiotrophoblast and spongiotrophoblast layers.
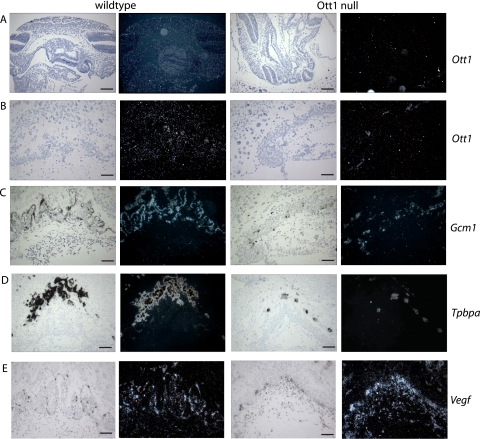
To ascertain the involvement and tissue specificity of Ott1 in the embryo and placenta, in situ hybridization of Ott1 RNA expression was performed with Ott1null/null mice and their wild-type littermates. Ott1 was widely expressed throughout the embryo, similar to the nearly ubiquitous expression in adult tissues (Fig. 3A) (39, 41). The neural tube, however, did not show a significant expression of Ott1, providing additional evidence that the observed defects were likely to be secondary rather than primary effects of the loss of Ott1 within the neural tube. Within the placenta, Ott1 was observed to have a diffuse expression pattern and was present in all three trophoblast layers (Fig. 3B). As expected, Ott1 null homozygous embryos lacked expression of Ott1 within both the embryo proper and placenta (Fig. 3A and B).
FIG. 3.
In situ gene expression in E9.5 Ott1null/null embryos and placentas. (A) In situ hybridization of E9.5 embryos with the Ott1 probe. (B to E) In situ hybridization of E9.5 placentas with Ott1 (B), Gcm1 (C), Tpbpa (4311) (D), and Vegf (E) probes. (Left panes) bright field; (right panes) dark field. Magnification bars, 200 μm (A and D) and 100 μm (B, C, and E).
To further define the involvement of the syncytiotrophoblast and spongiotrophoblast layers in Ott1null/null animals, in situ hybridization for Gcm1 (Glial Cell Missing 1) and Tpbpa (Trophoblast-Specific Protein α/4311) expression was performed. The lineage-specific Gcm1 is expressed on syncytiotrophoblast cells, specifically on cells adjacent to fetal vessels (2). In situ analysis of the expression of Gcm1 in Ott1-deleted E9.5 placentas showed markedly decreased cell numbers (Fig. 3C). Punctate clusters of Gcm1-positive cells were observed abutting narrow, nonbranching fetal vessels. In situ hybridization for Tpbpa that identified the spongiotrophoblast cells (7) demonstrated a dramatic reduction in the spongiotrophoblast layer in the Ott1null/null placentas.
The Vegf (Vascular Endothelial Growth Factor) pathway plays a pivotal role in fetal vascular branching morphogenesis (51). To determine whether an Ott1 deficiency causes a loss of Vegf induction, in situ hybridization for Vegf was performed. Vegf expression was observed within the syncytiotrophoblast layer in wild-type littermates, as previously noted (17). The Ott1-deleted placentas still demonstrated expression of Vegf; however, and similar to Gcm1 expression, it was confined to punctuate areas adjacent to fetal vessels. These data indicate that Ott1-mediated fetal vascular branching morphogenesis occurs either downstream or independent of Vegf expression. In summary, the deletion of Ott1 results in the hypoplasia of the spongiotrophoblast and syncytiotrophoblast layers and an arrest of fetal vascular branching morphogenesis within the placenta independent of Vegf expression.
Ott1 is required solely in extraembryonic tissue for normal placental development and vascular branching morphogenesis.
The formation of a vascular network within the placenta is thought to be driven primarily by trophoblast cells rather than the penetrating fetal vessels (16). To determine whether placental development and vascular branching morphogenesis required Ott1 expression within the trophoblast compartment and/or the fetal vasculature, we utilized the Sox2 (SRY-containing gene 2)-cre transgene. Sox2-cre expression is confined to the epiblast and has a high excision efficiency for alleles flanked by loxP (floxed) sites (24). This results in a complete cre-mediated excision of floxed loci within the embryo proper including the yolk sac but leaves the trophoectoderm, which gives rise to the placental trophoblast lineages, intact. Male Ott1null/wt Sox2-cre mice were bred with female Ott1flox/flox mice in timed matings (Table 1). Ott1null/null embryos, the result of the excision of the original Ott1null/flox Sox2-cre genotype, were identified with near the expected Mendelian ratio of 1:3 throughout gestation, in contrast to the nonconditional Ott1null/null embryos, which died beyond E9.5. Genotyping of Ott1null/flox Sox2-cre embryos by PCR demonstrated a complete conversion of the floxed Ott1 alleles to the null allele, and residual floxed alleles were undetectable (data not shown).
TABLE 1.
Genotypes of embryos from Ottnull/wt transgenic Sox2-cre × Ottflox/flox mice
| DPCa | No. of litters | No. of embryos | No. of flox/wt or flox/null Sox2-cre embryos (%) | No. of null/wt Sox2-cre embryos (%) | No. of null/null Sox2-cre embryos (%) |
|---|---|---|---|---|---|
| E9.5 | 2 | 21 | 12 (57) | 6 (29) | 3 (14) |
| E10.5 | 1 | 12 | 7 (58) | 3 (25) | 2 (17) |
| E14.5 | 7 | 55 | 26 (47) | 15 (27) | 14 (26) |
| E15.5 | 13 | 74 | 38 (51) | 16 (22) | 20 (27) |
| E16.5 | 3 | 21 | 14 (67) | 1 (5) | 6 (29) |
| E17.5 | 1 | 7 | 1 (14) | 4 (57) | 2 (29) |
| E18.5 | 7 | 44 | 25 (58) | 12 (27) | 7 (16) |
| E19.5/P0 | 5 | 34 | 17 (50) | 11 (32) | 6 (18)b |
Days postconception. The day of the vaginal plug is counted as E0.5. P0 is postpartum day 0.
All pups were found dead or dying <5 h after birth.
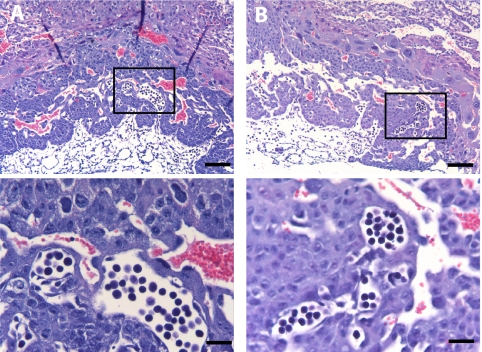
The histology of Ott1null/flox Sox2-cre E9.5 placentas was indistinguishable from that of littermate placentas with an intact Ott1 allele (Fig. 4A and B), with the preservation of the spongiotrophoblast and syncytiotrophoblast layers. Therefore, defects in spongiotrophoblast and syncytiotrophoblast development caused by a loss of Ott1 are intrinsic to Ott1 function within the trophoblast cell compartment alone. Placental vascular branching morphogenesis was present, and Ott1null/flox Sox2-cre placentas had a normal juxtaposition of fetal vasculature and maternal sinuses (Fig. 4A). Therefore, in the context of Ott1-dependent mechanisms, placental vascular branching morphogenesis is mediated via the trophoblasts rather than by the invading embryonic fetal vessels.
FIG. 4.
Normal E9.5 placental vascularization after Sox2-cre-mediated Ott1 deletion, sparing extraembryonic tissue. Shown are H&E-stained cross sections of Ott1null/null (A) and wild-type E9.5 (B) placentas. Bottom panels represent enlargements of the indicated areas. Magnification bars, 200 μm (top) and 25 μm (bottom).
Rescue of the placental defect in Ott1-deleted embryos enables peripartum survival and reveals defects in cardiac and splenic organogenesis.
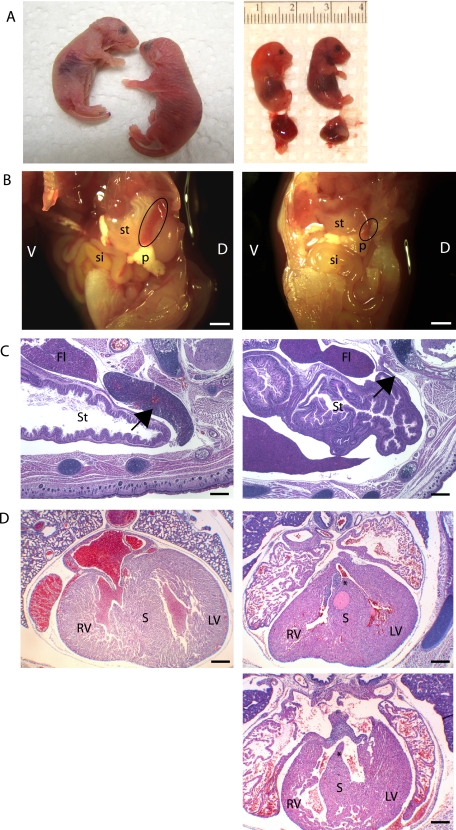
Timed matings of Ott1null/wt Sox2-cre and Ott1flox/flox mice demonstrated viable embryos at near-Mendelian ratios throughout gestation (Table 1). However, Ott1null/null pups, the product of the Sox2-cre Ott1flox/null genotype, were often found dead after birth. Occasional live mutant pups were observed, but they were runted and unable to feed and died within a few hours peripartum (Fig. 5A, left). Necropsies at E18.5 or postpartum day 0 were performed to assess the peripartum lethal phenotype. Examination of the lungs showed no histologic abnormalities, with the exception of a lack of lung inflation in mutant pups. However, pericardial effusion was often observed, and histological evaluation of the heart revealed a membranous ventricular septal defect in the majority of mutant embryos (Fig. 5D). No atrial defects or defects in the endocardial cushions, valves, or great vessels were seen. Ott1null/null embryos harvested during late gestation (E18.5) were often edematous, consistent with heart failure. In the absence of other major organ defects, this was a likely cause of peripartum demise (Fig. 5A, right). Interestingly, gross dissection also revealed hyposplenism with variable penetrance (Fig. 5B). Histological analysis showed a small remnant of splenic tissue attached to the splenic ligament (Fig. 5C).
FIG. 5.
Ott1null/null Sox2-cre mice are runted and hyposplenic and have a cardiac ventricular septal defect. (A, left) Photograph of newborn pups. (Right) Photograph of E18.5 embryos. Ott1null/null embryos are shown at left, and wild-type embryos are shown at right. (B) Gross photograph of dissected left flank/abdomen of E18.5 embryos. The black circle overlays the spleen. The fetal liver has been removed. V, ventral; D, dorsal; si, small intestine; st, stomach; p, pancreas. (C) H&E-stained cross section of E18.5 embryos. The arrow points to the spleen. St, stomach; Fl, fetal liver. (D) H&E-stained cross section of E18.5 embryo through the cardiac ventricles. RV, right ventricle; S, ventricular septum; LV, left ventricle. Asterisks indicate the location of the septal defect. (Left) Wild-type embryo; (right) two representative Ott1 null embryos. Magnification bars represent 1,000 μm (B) and 200 μm (C and D).
Cardiac gene expression in E18.5 embryos.
To gain insight into the pathways affected by the loss of Ott1, total RNA was isolated from E18.5 Ott1null/null Sox2-cre and littermate hearts to analyze global changes in gene expression. Total RNA was linearly amplified and then hybridized to Affymetrix 430 chips. Cardiac development in the embryo has overlapping gene expression requirements with other organs, including spleen and placenta. A hierarchy of gene expression in the developing spleen was previously determined and involves Pbx1 (Pre-B-cell leukemia transcription factor 1), Tlx1 (T-cell leukemia homeobox 1), Nkx2.5 (NK2 transcription factor related, locus 5), and Wt1 (Wilms’ Tumor 1), each of which has also been implicated in cardiogenesis (6, 10, 13). Sox11 (SRY-box-containing gene 11), a high-mobility-group protein, and Acvr2b (Activin Receptor type IIB) were also shown to be required for both cardiac and splenic development (47, 59). Of the above-mentioned genes, Acvr2b was significantly altered, with a threefold upregulation (P = 0.001) (Table 2). Other genes implicated in cardiac development that were found to have significantly upregulated expression in Ott1null/null hearts included Gnb1l (guanine nucleotide binding protein, beta-1-like) (twofold; P < 0.001), a gene deleted in DiGeorge syndrome (23), and E2f3 (E2F transcription factor 3) (twofold; P = 0.05). Significantly downegulated cardiac-associated genes in Ott1null/null embryo hearts included Il18r1 (Interleukin 18 receptor 1) (twofold; P < 0.004).
TABLE 2.
Changes in gene expression among selected genes in Ott1-deleted E18.5 heartsc
| Gene | Fold differencea | Signal-to-noise ratiob | P value |
|---|---|---|---|
| Pbx1 | 1.16 | 0.59 | 0.23 |
| Tlx1 | −1.12 | −0.50 | 0.39 |
| Nkx2.5 | −1.07 | −0.29 | 0.53 |
| Wt1 | 1.06 | 0.21 | 0.63 |
| Sox11 | 1.06 | 0.27 | 0.62 |
| Acvr2b | 3.02 | 7.35 | 0.001 |
| Hif1α | 1.26 | 0.68 | 0.17 |
| Cited2 | 1.11 | 0.39 | 0.41 |
| Gnb1l | 1.95 | 4.43 | <0.001 |
| E2f3 | 2.03 | 2.05 | 0.05 |
| Il18r1 | −1.91 | −2.94 | 0.004 |
| Hes1 | −1.06 | −.028 | 0.53 |
| EphrinB2 | 1.15 | 0.37 | 0.52 |
| Hey1 | 1.11 | −0.35 | 0.48 |
| Hey2 | 1.06 | 0.79 | 0.21 |
Mean change of Ott1null/null compared to Ott1null/wt genotypes (n = 3).
Signal-to-noise ratio of gene expression values.
Values in boldface type indicate significant changes.
Ott1 and the family member Mint have been implicated in Notch signaling. In vitro studies of Ott1 showed a cell context-dependent inhibition or stimulation of Notch-activated pathways (38). Notch signaling is also known to play a crucial role in cardiac and trophoblast development (26, 35, 45).The expression levels of downstream Notch targets, including Hes1 (hairy and enhancer of split 1), EphrinB2 (eph-related receptor tyrosine kinase ligand 5), Hey1 (hairy/enhancer-of-split related with YRPW motif 1), and Hey2 (hairy/enhancer-of-split related with YRPW motif 2) were not significantly altered between the Ott1-deleted and wild-type embryos, arguing against a significant change in Notch pathway activation (Table 2).
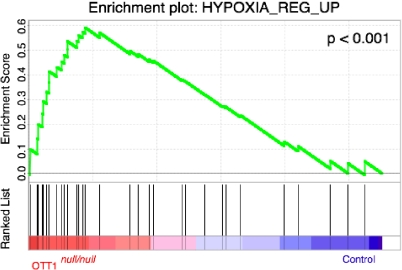
The hypoxia-inducible genes Hif1α (hypoxia-inducible factor 1 α) and Cited2 (Cbp/p300-interacting transactivator, with Glu/Asp-rich C-terminal domain, 2) have essential roles in cardiac development and in placental vascular branching morphogenesis (1, 4, 14, 58, 67). We examined expression levels of these genes between Ott1null/null embryos and littermates, but no significant differences were observed. To further understand global changes resulting from the deletion of Ott1 in embryonic hearts, the expression array data were used for GSEA to compare to 1,837 curated gene sets in the Molecular Signature Database (61). Interestingly, the Ott1null/null set was significantly enriched (normalized enrichment score of 1.98; P < 0.001) in comparison to a gene set composed of genes upregulated in response to hypoxia (Fig. 6) (34). However, a gene set specifically composed of HIF-1α targets was not enriched (normalized enrichment score of 1.12; P = 0.12) (57). In addition, Notch pathway gene sets were also not enriched (3, 44).
FIG. 6.
Enrichment plot for genes upregulated in hypoxia. Shown are a profile of the running enrichment score and positions of the gene set members on the rank order list. Ott1null/null embryonic heart gene expression signatures are compared to those of Ott1null/wt embryonic hearts at E18.5 (n = 3).
DISCUSSION
The Ott1 gene is widely expressed in adult and fetal tissues including the placenta. Early embryonic lethality of Ott1null/null embryos at E9.5 to E10.5 can be avoided through the use of the epiblast-specific Sox2-cre transgene, which spares the extraembryonic tissue that develops into the placenta. Therefore, the early stages of embryogenesis such as gastrulation are independent of Ott1. Although germ line Ott1null/null embryos have incomplete closure of the neural tube, the Sox2-cre deletion of Ott1 confined to the embryo proper lacks this defect. Taken together with the dearth of Ott1 expression seen in wild-type neural tube tissue, the neural tube defect in germ line Ott1null/null embryos must be a consequence of placental insufficiency rather than a primary consequence of the loss of Ott1.
Ott1 is required for the vascular branching morphogenesis of fetal vessels into the placental labyrinth. The disruption of this process by the deletion of Ott1 results in fetal death at E9.5 to 10.5, as diffusion alone is unable to supply the oxygen requirements of the embryo beyond this stage (52). Both the spongiotrophoblast and syncytiotrophoblast layers are required for vascular branching morphogenesis (53). Identification of the individual trophoblast layers showed a significant reduction in the sizes of both the spongiotrophoblast and syncytiotrophoblast layers but a preservation of the giant cell trophoblast layer. The formation of the syncytiotrophoblast layer into the labyrinth is in part dependent on the process of vascular branching morphogenesis; therefore, the defect in Ott1null/null placentas could be either intrinsic to syncytiotrophoblasts or a non-cell-autonomous effect resulting from the failure of vascular branching morphogenesis (16). In contrast, the development of the spongiotrophoblast layer was previously observed in the absence of an intact labyrinth in murine knockouts of the Hai-1 (hepatocyte growth factor activator inhibitor 1) and Fosl1 (fos-like antigen 1) genes, suggesting that the loss of Ott1 causes a primary defect in spongiotrophoblasts (55, 62). The role of the spongiotrophoblast layer is not completely understood; however, it is thought to be required for the proper establishment of the fetal and maternal portions of the labyrinth through the production of vasoactive factors (15, 63).
The expansion and branching of the fetal vessels are presumed to be guided through growth factors and contact originating from the trophoblast tissues rather than directed by the fetal vasculature itself (15). Evidence for this include the observations that (i) there is expression of vascular remodeling genes such as Vegf in the trophoblast cells but not the fetal vessels and (ii) Vegf expression is essential for vascular branching morphogenesis (8, 19, 64). The deletion of Tfeb (transcription factor EB) causes a loss of vascular branching morphogenesis as well and specifically abolishes Vegf expression within the syncytiotrophoblast cells (60). Vegf expression can still be induced in the Ott1null/null placenta; therefore, Ott1-mediated vascular branching morphogenesis is either independent or downstream of Vegf signaling. In addition, the excision of Ott1 by Sox2-cre in the embryo, including the fetal vasculature, demonstrates that vascular branching morphogenesis, at least in the context of Ott1, is a trophoblast-driven process.
The extraembryonic-sparing Sox2-cre excision demonstrated that Ott1 was required for spleen and cardiac development in the later stages of embryonic development. Interestingly, splenic, cardiac, and hematopoietic tissues are all derived from mesoderm, suggesting that despite the nearly ubiquitous expression of Ott1, mesodermal tissue has specifically dependent or nonredundant requirements. Asplenia or hyposplenia has been associated with the deletion of a relatively small set of genes including Tlx1, Pbx1, Nkx2.3 (NK2 transcription factor related, locus 3), Nkx3.2 (NK3 homeobox 2), Tcf21 (transcription factor 21), Sox11, Wt-1, Acvrb2, and Cited2 (4, 6, 47). A transcriptional hierarchy among several of these genes has been established, placing Wt-1 downstream of the homeobox genes Tlx1, Pbx1, and Nkx3.2 (6). Nkx2.5 marks the spleen primordium and is downstream of Tcf21 and Pbx1 (6). Spen/Mint was previously implicated in Hox gene regulation but not specifically with any of the above-mentioned genes or their homologs (43, 66).
Sox11, Pbx1, Wt-1, Cited2, and Acvr2b are known to be involved in both cardiac and splenic development (25, 47, 56, 59, 65). Of these genes, evaluation of cardiac gene expression in Ott1 knockout embryos revealed a threefold increase in levels of Acvr2b. Deletion mutants of Acvr2b in mice demonstrate a complex phenotype including cardiac atrial and ventricular septal defects, hyposplenism, right isomerization of the lung, and vertebral defects (46, 47) Mutations of ACVR2B in humans have been observed in relation to cardiac defects including ventricular septal defects, left-right axis abnormalities, and splenic malposition (31). Although no placental abnormalities were noted in the knockout model, activin and activin receptors are believed to be important regulators of human trophoblast differentiation (29). No studies have yet examined the effects of an overexpression of Acvr2b; however, gene dosage appears to be an important factor in phenotype and penetrance in mice (46).
Twofold overexpression was also observed for E2f3 and Gnb1l. An E2f3 deletion results in congestive heart failure in adult mice and causes apoptosis in cardiomyocytes when overexpressed (11, 18). GNB1L is among the deleted genes in human DiGeorge syndrome (del22q11), which manifests cardiac abnormalities including ventricular septal defects; however, it is believed that another gene, Tbx1 (T-box 1), is responsible for the cardiac phenotype (36, 42). A threefold underexpression of Il18r1 was seen in the Ott1 knockout hearts, and Il-18 has been shown to be important for cardiac remodeling and the hypertrophic response (12, 40).
The expression levels of Hif1α, a regulator of the hypoxia response, and its downstream target Cited2 have been shown to contribute to cardiac, splenic, and placental development. (4, 5). Therefore, it was important to exclude a perturbation of gene expression within this set as the basis for the Ott1 knockout phenotype. GSEA showed a significant enrichment of hypoxia-related genes without a similar enrichment of a specific Hif1α target set. It is unclear whether changes in hypoxia-related gene expression may be primary or secondary to inadequate cardiac function in the Ott1null/null embryos. Likewise, alterations in the expressions of Acvr2b, E2f3, Gnb1l, and Il18r1 will need further investigation to determine whether Ott1 directly affects expression.
The Notch pathway is also critical for embryonic heart development. Ott1 can inhibit or activate the Notch-responsive gene Hes1 in vitro depending on cell context (38). The family member Mint has already been implicated in the suppression of Notch2 signaling via RbpJκ (recombination signal binding protein for immunoglobulin kappa J region) binding. Although Ott1 lacks the domain identified for this function, an alternative RbpJκ binding site in Ott1 has been identified (38). In murine gene knockout experiments, the Notch target genes Hes1, EphrinB2, Hey1, and Hey2 have each been shown to be required for cardiac development and manifest phenotypes including ventricular septal defects (20, 22, 30). Surprisingly, the expression of these Notch targets did not change in Ott1null/null embryonic hearts compared to littermate controls, suggesting that the generation of the Ott1null/null cardiac phenotype is not mediated via these Notch pathway signaling intermediates and that an alternative pathway exists.
In summary, Ott1 has essential roles in hematopoietic, trophoblast, cardiac, and splenic development. These findings implicate the spen family and homologs as significant global regulators of embryonic and adult hematopoietic development. It is possible that Ott1 has a more expansive role than Mint by virtue of its wider expression pattern. The deletion of the family member Mint in mice results in a cardiac septal defect as well; however, Ott1 must fulfill a nonredundant role. The deletion of Ott1 causes a defect within the syncytiotrophoblast and spongiotrophoblast cell layers responsible for placental vascular branching morphogenesis, as confirmed by embryo-specific cre. The diverse affected tissues in Ott1-deleted mice suggest that Ott1 or Ott1-dependent pathways may play an important part in human pathophysiology, specifically, placental insufficiency and cardiac malformation. Of note, certain leukemia translocation-associated genes such as Cdx2 (caudal-type homeobox 2) and Rxr-α (retinoid X receptor alpha) have significant roles in development and particularly in trophoblast development (9, 54). Further studies into the Ott1-dependent mechanisms essential for development, including the putative targets Acvr2b, E2f3, Gnb1l, and Il18r1, may also yield significant insight into pathways coopted by OTT1-MAL in t(1;22)-associated leukemogenesis.
Acknowledgments
We thank T. Mercher for initial work on the Ott1 knockout construct and review of the manuscript, J. Rossant for supplying valuable reagents, Samuel Perry and Li Zhang in the DF/HCC Rodent Histopathology Core for expert histology assistance, and Yu Yang for assistance with in situ hybridization.
This work was supported in part by National Institutes of Health grants K08 CA111399 (G.D.R.) and P01 CA66996 (D.G.G.) and the Leukemia and Lymphoma Society. D.G.G. is an Investigator for the Howard Hughes Medical Institute. We report no relevant conflicts of interest.
Footnotes
Published ahead of print on 3 November 2008.
REFERENCES
- 1.Adelman, D. M., M. Gertsenstein, A. Nagy, M. C. Simon, and E. Maltepe. 2000. Placental cell fates are regulated in vivo by HIF-mediated hypoxia responses. Genes Dev. 143191-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anson-Cartwright, L., K. Dawson, D. Holmyard, S. J. Fisher, R. A. Lazzarini, and J. C. Cross. 2000. The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25311-314. [DOI] [PubMed] [Google Scholar]
- 3.Ashburner, M., C. A. Ball, J. A. Blake, D. Botstein, H. Butler, J. M. Cherry, A. P. Davis, K. Dolinski, S. S. Dwight, J. T. Eppig, M. A. Harris, D. P. Hill, L. Issel-Tarver, A. Kasarskis, S. Lewis, J. C. Matese, J. E. Richardson, M. Ringwald, G. M. Rubin, G. Sherlock, et al. 2000. Gene Ontology: tool for the unification of biology. Nat. Genet. 2525-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamforth, S. D., J. Braganca, J. J. Eloranta, J. N. Murdoch, F. I. Marques, K. R. Kranc, H. Farza, D. J. Henderson, H. C. Hurst, and S. Bhattacharya. 2001. Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat. Genet. 29469-474. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharya, S., C. L. Michels, M. K. Leung, Z. P. Arany, A. L. Kung, and D. M. Livingston. 1999. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1364-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brendolan, A., E. Ferretti, V. Salsi, K. Moses, S. Quaggin, F. Blasi, M. L. Cleary, and L. Selleri. 2005. A Pbx1-dependent genetic and transcriptional network regulates spleen ontogeny. Development 1323113-3126. [DOI] [PubMed] [Google Scholar]
- 7.Calzonetti, T., L. Stevenson, and J. Rossant. 1995. A novel regulatory region is required for trophoblast-specific transcription in transgenic mice. Dev. Biol. 171615-626. [DOI] [PubMed] [Google Scholar]
- 8.Carmeliet, P., V. Ferreira, G. Breier, S. Pollefeyt, L. Kieckens, M. Gertsenstein, M. Fahrig, A. Vandenhoeck, K. Harpal, C. Eberhardt, C. Declercq, J. Pawling, L. Moons, D. Collen, W. Risau, and A. Nagy. 1996. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature 380435-439. [DOI] [PubMed] [Google Scholar]
- 9.Chawengsaksophak, K., R. James, V. E. Hammond, F. Kontgen, and F. Beck. 1997. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature 38684-87. [DOI] [PubMed] [Google Scholar]
- 10.Clark, K. L., K. E. Yutzey, and D. W. Benson. 2006. Transcription factors and congenital heart defects. Annu. Rev. Physiol. 6897-121. [DOI] [PubMed] [Google Scholar]
- 11.Cloud, J. E., C. Rogers, T. L. Reza, U. Ziebold, J. R. Stone, M. H. Picard, A. M. Caron, R. T. Bronson, and J. A. Lees. 2002. Mutant mouse models reveal the relative roles of E2F1 and E2F3 in vivo. Mol. Cell. Biol. 222663-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colston, J. T., W. H. Boylston, M. D. Feldman, C. P. Jenkinson, S. D. de la Rosa, A. Barton, R. J. Trevino, G. L. Freeman, and B. Chandrasekar. 2007. Interleukin-18 knockout mice display maladaptive cardiac hypertrophy in response to pressure overload. Biochem. Biophys. Res. Commun. 354552-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conway, S. J., A. Kruzynska-Frejtag, P. L. Kneer, M. Machnicki, and S. V. Koushik. 2003. What cardiovascular defect does my prenatal mouse mutant have, and why? Genesis 351-21. [DOI] [PubMed] [Google Scholar]
- 14.Cowden Dahl, K. D., B. H. Fryer, F. A. Mack, V. Compernolle, E. Maltepe, D. M. Adelman, P. Carmeliet, and M. C. Simon. 2005. Hypoxia-inducible factors 1α and 2α regulate trophoblast differentiation. Mol. Cell. Biol. 2510479-10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross, J. C., M. Hemberger, Y. Lu, T. Nozaki, K. Whiteley, M. Masutani, and S. L. Adamson. 2002. Trophoblast functions, angiogenesis and remodeling of the maternal vasculature in the placenta. Mol. Cell. Endocrinol. 187207-212. [DOI] [PubMed] [Google Scholar]
- 16.Cross, J. C., H. Nakano, D. R. Natale, D. G. Simmons, and E. D. Watson. 2006. Branching morphogenesis during development of placental villi. Differentiation 74393-401. [DOI] [PubMed] [Google Scholar]
- 17.Dumont, D. J., G. H. Fong, M. C. Puri, G. Gradwohl, K. Alitalo, and M. L. Breitman. 1995. Vascularization of the mouse embryo: a study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev. Dyn. 20380-92. [DOI] [PubMed] [Google Scholar]
- 18.Ebelt, H., N. Hufnagel, P. Neuhaus, H. Neuhaus, P. Gajawada, A. Simm, U. Muller-Werdan, K. Werdan, and T. Braun. 2005. Divergent siblings: E2F2 and E2F4 but not E2F1 and E2F3 induce DNA synthesis in cardiomyocytes without activation of apoptosis. Circ. Res. 96509-517. [DOI] [PubMed] [Google Scholar]
- 19.Ferrara, N., K. Carver-Moore, H. Chen, M. Dowd, L. Lu, K. S. O'Shea, L. Powell-Braxton, K. J. Hillan, and M. W. Moore. 1996. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature 380439-442. [DOI] [PubMed] [Google Scholar]
- 20.Fischer, A., N. Schumacher, M. Maier, M. Sendtner, and M. Gessler. 2004. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 18901-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentleman, R. C., V. J. Carey, D. M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, K. Hornik, T. Hothorn, W. Huber, S. Iacus, R. Irizarry, F. Leisch, C. Li, M. Maechler, A. J. Rossini, G. Sawitzki, C. Smith, G. Smyth, L. Tierney, J. Y. Yang, and J. Zhang. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerety, S. S., and D. J. Anderson. 2002. Cardiovascular ephrinB2 function is essential for embryonic angiogenesis. Development 1291397-1410. [DOI] [PubMed] [Google Scholar]
- 23.Gong, L., M. Liu, J. Jen, and E. T. Yeh. 2000. GNB1L, a gene deleted in the critical region for DiGeorge syndrome on 22q11, encodes a G-protein beta-subunit-like polypeptide. Biochim. Biophys. Acta 1494185-188. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, S., P. Lewis, L. Pevny, and A. P. McMahon. 2002. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev. 119(Suppl. 1)S97-S101. [DOI] [PubMed] [Google Scholar]
- 25.Herzer, U., A. Crocoll, D. Barton, N. Howells, and C. Englert. 1999. The Wilms tumor suppressor gene wt1 is required for development of the spleen. Curr. Biol. 9837-840. [DOI] [PubMed] [Google Scholar]
- 26.High, F. A., and J. A. Epstein. 2008. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet. 949-61. [DOI] [PubMed] [Google Scholar]
- 27.Hiriart, E., H. Gruffat, M. Buisson, I. Mikaelian, S. Keppler, P. Meresse, T. Mercher, O. A. Bernard, A. Sergeant, and E. Manet. 2005. Interaction of the Epstein-Barr virus mRNA export factor EB2 with human Spen proteins SHARP, OTT1, and a novel member of the family, OTT3, links Spen proteins with splicing regulation and mRNA export. J. Biol. Chem. 28036935-36945. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry, R. A., B. Hobbs, F. Collin, Y. D. Beazer-Barclay, K. J. Antonellis, U. Scherf, and T. P. Speed. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4249-264. [DOI] [PubMed] [Google Scholar]
- 29.Jones, R. L., L. A. Salamonsen, and J. K. Findlay. 2002. Potential roles for endometrial inhibins, activins and follistatin during human embryo implantation and early pregnancy. Trends Endocrinol. Metab. 13144-150. [DOI] [PubMed] [Google Scholar]
- 30.Kokubo, H., S. Miyagawa-Tomita, M. Nakazawa, Y. Saga, and R. L. Johnson. 2005. Mouse hesr1 and hesr2 genes are redundantly required to mediate Notch signaling in the developing cardiovascular system. Dev. Biol. 278301-309. [DOI] [PubMed] [Google Scholar]
- 31.Kosaki, R., M. Gebbia, K. Kosaki, M. Lewin, P. Bowers, J. A. Towbin, and B. Casey. 1999. Left-right axis malformations associated with mutations in ACVR2B, the gene for human activin receptor type IIB. Am. J. Med. Genet. 8270-76. [DOI] [PubMed] [Google Scholar]
- 32.Kuang, B., S. C. Wu, Y. Shin, L. Luo, and P. Kolodziej. 2000. Split ends encodes large nuclear proteins that regulate neuronal cell fate and axon extension in the Drosophila embryo. Development 1271517-1529. [DOI] [PubMed] [Google Scholar]
- 33.Kuroda, K., H. Han, S. Tani, K. Tanigaki, T. Tun, T. Furukawa, Y. Taniguchi, H. Kurooka, Y. Hamada, S. Toyokuni, and T. Honjo. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J. signaling pathway. Immunity 18301-312. [DOI] [PubMed] [Google Scholar]
- 34.Leonard, M. O., D. C. Cottell, C. Godson, H. R. Brady, and C. T. Taylor. 2003. The role of HIF-1 alpha in transcriptional regulation of the proximal tubular epithelial cell response to hypoxia. J. Biol. Chem. 27840296-40304. [DOI] [PubMed] [Google Scholar]
- 35.Li, L., I. D. Krantz, Y. Deng, A. Genin, A. B. Banta, C. C. Collins, M. Qi, B. J. Trask, W. L. Kuo, J. Cochran, T. Costa, M. E. Pierpont, E. B. Rand, D. A. Piccoli, L. Hood, and N. B. Spinner. 1997. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat. Genet. 16243-251. [DOI] [PubMed] [Google Scholar]
- 36.Lindsay, E. A., F. Vitelli, H. Su, M. Morishima, T. Huynh, T. Pramparo, V. Jurecic, G. Ogunrinu, H. F. Sutherland, P. J. Scambler, A. Bradley, and A. Baldini. 2001. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 41097-101. [DOI] [PubMed] [Google Scholar]
- 37.Lindtner, S., A. S. Zolotukhin, H. Uranishi, J. Bear, V. Kulkarni, S. Smulevitch, M. Samiotaki, G. Panayotou, B. K. Felber, and G. N. Pavlakis. 2006. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 28136915-36928. [DOI] [PubMed] [Google Scholar]
- 38.Ma, X., M. J. Renda, L. Wang, E. C. Cheng, C. Niu, S. W. Morris, A. S. Chi, and D. S. Krause. 2007. Rbm15 modulates Notch-induced transcriptional activation and affects myeloid differentiation. Mol. Cell. Biol. 273056-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma, Z., S. W. Morris, V. Valentine, M. Li, J. A. Herbrick, X. Cui, D. Bouman, Y. Li, P. K. Mehta, D. Nizetic, Y. Kaneko, G. C. Chan, L. C. Chan, J. Squire, S. W. Scherer, and J. K. Hitzler. 2001. Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13) of acute megakaryoblastic leukemia. Nat. Genet. 28220-221. [DOI] [PubMed] [Google Scholar]
- 40.Mallat, Z., C. Heymes, A. Corbaz, D. Logeart, S. Alouani, A. Cohen-Solal, T. Seidler, G. Hasenfuss, Y. Chvatchko, A. M. Shah, and A. Tedgui. 2004. Evidence for altered interleukin 18 (IL)-18 pathway in human heart failure. FASEB J. 181752-1754. [DOI] [PubMed] [Google Scholar]
- 41.Mercher, T., M. B. Coniat, R. Monni, M. Mauchauffe, F. N. Khac, L. Gressin, F. Mugneret, T. Leblanc, N. Dastugue, R. Berger, and O. A. Bernard. 2001. Involvement of a human gene related to the Drosophila spen gene in the recurrent t(1;22) translocation of acute megakaryocytic leukemia. Proc. Natl. Acad. Sci. USA 985776-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Merscher, S., B. Funke, J. A. Epstein, J. Heyer, A. Puech, M. M. Lu, R. J. Xavier, M. B. Demay, R. G. Russell, S. Factor, K. Tokooya, B. S. Jore, M. Lopez, R. K. Pandita, M. Lia, D. Carrion, H. Xu, H. Schorle, J. B. Kobler, P. Scambler, A. Wynshaw-Boris, A. I. Skoultchi, B. E. Morrow, and R. Kucherlapati. 2001. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell 104619-629. [DOI] [PubMed] [Google Scholar]
- 43.Newberry, E. P., T. Latifi, and D. A. Towler. 1999. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 3810678-10690. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen, B. C., K. Lefort, A. Mandinova, D. Antonini, V. Devgan, G. Della Gatta, M. I. Koster, Z. Zhang, J. Wang, A. Tommasi di Vignano, J. Kitajewski, G. Chiorino, D. R. Roop, C. Missero, and G. P. Dotto. 2006. Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation. Genes Dev. 201028-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oda, T., A. G. Elkahloun, P. S. Meltzer, and S. C. Chandrasekharappa. 1997. Identification and cloning of the human homolog (JAG1) of the rat Jagged1 gene from the Alagille syndrome critical region at 20p12. Genomics 43376-379. [DOI] [PubMed] [Google Scholar]
- 46.Oh, S. P., and E. Li. 2002. Gene-dosage-sensitive genetic interactions between inversus viscerum (iv), nodal, and activin type IIB receptor (ActRIIB) genes in asymmetrical patterning of the visceral organs along the left-right axis. Dev. Dyn. 224279-290. [DOI] [PubMed] [Google Scholar]
- 47.Oh, S. P., and E. Li. 1997. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes Dev. 111812-1826. [DOI] [PubMed] [Google Scholar]
- 48.Raffel, G. D., T. Mercher, H. Shigematsu, I. R. Williams, D. E. Cullen, K. Akashi, O. A. Bernard, and D. G. Gilliland. 2007. Ott1(Rbm15) has pleiotropic roles in hematopoietic development. Proc. Natl. Acad. Sci. USA 1046001-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rebay, I., F. Chen, F. Hsiao, P. A. Kolodziej, B. H. Kuang, T. Laverty, C. Suh, M. Voas, A. Williams, and G. M. Rubin. 2000. A genetic screen for novel components of the Ras/mitogen-activated protein kinase signaling pathway that interact with the yan gene of Drosophila identifies split ends, a new RNA recognition motif-containing protein. Genetics 154695-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reich, M., T. Liefeld, J. Gould, J. Lerner, P. Tamayo, and J. P. Mesirov. 2006. GenePattern 2.0. Nat. Genet. 38500-501. [DOI] [PubMed] [Google Scholar]
- 51.Reynolds, L. P., P. P. Borowicz, K. A. Vonnahme, M. L. Johnson, A. T. Grazul-Bilska, J. M. Wallace, J. S. Caton, and D. A. Redmer. 2005. Animal models of placental angiogenesis. Placenta 26689-708. [DOI] [PubMed] [Google Scholar]
- 52.Rinkenberger, J. L., J. C. Cross, and Z. Werb. 1997. Molecular genetics of implantation in the mouse. Dev. Genet. 216-20. [DOI] [PubMed] [Google Scholar]
- 53.Rossant, J., and J. C. Cross. 2001. Placental development: lessons from mouse mutants. Nat. Rev. Genet. 2538-548. [DOI] [PubMed] [Google Scholar]
- 54.Sapin, V., P. Dolle, C. Hindelang, P. Kastner, and P. Chambon. 1997. Defects of the chorioallantoic placenta in mouse RXRalpha null fetuses. Dev. Biol. 19129-41. [DOI] [PubMed] [Google Scholar]
- 55.Schreiber, M., Z. Q. Wang, W. Jochum, I. Fetka, C. Elliott, and E. F. Wagner. 2000. Placental vascularisation requires the AP-1 component fra1. Development 1274937-4948. [DOI] [PubMed] [Google Scholar]
- 56.Selleri, L., M. J. Depew, Y. Jacobs, S. K. Chanda, K. Y. Tsang, K. S. Cheah, J. L. Rubenstein, S. O'Gorman, and M. L. Cleary. 2001. Requirement for Pbx1 in skeletal patterning and programming chondrocyte proliferation and differentiation. Development 1283543-3557. [DOI] [PubMed] [Google Scholar]
- 57.Semenza, G. L. 2001. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7345-350. [DOI] [PubMed] [Google Scholar]
- 58.Semenza, G. L., F. Agani, N. Iyer, L. Kotch, E. Laughner, S. Leung, and A. Yu. 1999. Regulation of cardiovascular development and physiology by hypoxia-inducible factor 1. Ann. N. Y. Acad. Sci. 874262-268. [DOI] [PubMed] [Google Scholar]
- 59.Sock, E., S. D. Rettig, J. Enderich, M. R. Bosl, E. R. Tamm, and M. Wegner. 2004. Gene targeting reveals a widespread role for the high-mobility-group transcription factor Sox11 in tissue remodeling. Mol. Cell. Biol. 246635-6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steingrimsson, E., L. Tessarollo, S. W. Reid, N. A. Jenkins, and N. G. Copeland. 1998. The bHLH-Zip transcription factor Tfeb is essential for placental vascularization. Development 1254607-4616. [DOI] [PubMed] [Google Scholar]
- 61.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 10215545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka, H., K. Nagaike, N. Takeda, H. Itoh, K. Kohama, T. Fukushima, S. Miyata, S. Uchiyama, S. Uchinokura, T. Shimomura, K. Miyazawa, N. Kitamura, G. Yamada, and H. Kataoka. 2005. Hepatocyte growth factor activator inhibitor type 1 (HAI-1) is required for branching morphogenesis in the chorioallantoic placenta. Mol. Cell. Biol. 255687-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tanaka, M., M. Gertsenstein, J. Rossant, and A. Nagy. 1997. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev. Biol. 19055-65. [DOI] [PubMed] [Google Scholar]
- 64.Vuorela, P., E. Hatva, A. Lymboussaki, A. Kaipainen, V. Joukov, M. G. Persico, K. Alitalo, and E. Halmesmaki. 1997. Expression of vascular endothelial growth factor and placenta growth factor in human placenta. Biol. Reprod. 56489-494. [DOI] [PubMed] [Google Scholar]
- 65.Weninger, W. J., K. Lopes Floro, M. B. Bennett, S. L. Withington, J. I. Preis, J. P. Barbera, T. J. Mohun, and S. L. Dunwoodie. 2005. Cited2 is required both for heart morphogenesis and establishment of the left-right axis in mouse development. Development 1321337-1348. [DOI] [PubMed] [Google Scholar]
- 66.Wiellette, E. L., K. W. Harding, K. A. Mace, M. R. Ronshaugen, F. Y. Wang, and W. McGinnis. 1999. spen encodes an RNP motif protein that interacts with Hox pathways to repress the development of head-like sclerites in the Drosophila trunk. Development 1265373-5385. [DOI] [PubMed] [Google Scholar]
- 67.Withington, S. L., A. N. Scott, D. N. Saunders, K. Lopes Floro, J. I. Preis, J. Michalicek, K. Maclean, D. B. Sparrow, J. P. Barbera, and S. L. Dunwoodie. 2006. Loss of Cited2 affects trophoblast formation and vascularization of the mouse placenta. Dev. Biol. 29467-82. [DOI] [PubMed] [Google Scholar]