Abstract
We previously showed that ribosomal protein L13a is required for translational silencing of gamma interferon (IFN-γ)-induced ceruloplasmin (Cp) synthesis in monocytes. This silencing also requires the presence of the GAIT (IFN-gamma activated inhibitor of translation) element in the 3′ untranslated region (UTR) of Cp mRNA. Considering that Cp is an inflammatory protein, we hypothesized that this mechanism may have evolved to silence a family of proinflammatory proteins, of which Cp is just one member. To identify the other mRNAs that are targets for this silencing, we performed a genome-wide analysis of the polysome-profiled mRNAs by using an Affymetrix GeneChip and an inflammation-responsive gene array. A cluster of mRNAs encoding different chemokines and their receptors was identified as common hits in the two approaches and validated by real-time PCR. In silico predicted GAIT hairpins in the 3′ UTRs of the target mRNAs were confirmed as functional cis-acting elements for translational silencing by luciferase reporter assays. Consistent with Cp, the newly identified target mRNAs also required L13a for silencing. Our studies have identified a new inflammation-responsive posttranscriptional operon that can be regulated directly at the level of translation in IFN-γ-activated monocytes. This regulation of a cohort of mRNAs encoding inflammatory proteins may be important to resolve inflammation.
Emerging evidence suggests that the assessment of mRNA expression level in characterizing a cellular phenotype is only partially valid because the transcriptome does not accurately represent the proteome (31). As the cellular phenotype is ultimately governed by the protein(s) expressed from mRNAs, expression profiling data would be more realistic if mRNA samples were enriched by the mRNAs that are actively involved in translation since, by definition, they are associated with multiple ribosomes, which form a polysome. The polysome-bound mRNA pool is significantly heavier than the translationally inactive mRNAs. The inactive mRNAs are sequestered in messenger ribonucleoprotein (mRNP) particles or can be associated with a single ribosome, called a monosome. Thus, the actively translated pool of polysome-bound mRNAs can be readily separated from the pool of nontranslated/undertranslated mRNAs by sucrose gradient centrifugation, allowing a functional distinction between the two. Through microarray analysis of these two pools of mRNA, several groups have identified target mRNAs for translational control on a genome-wide scale (1, 2, 15, 43).
We previously showed that gamma interferon (IFN-γ) induces ceruloplasmin (Cp) mRNA and protein in U937 monocytic cells and peripheral blood monocytes (23). However, IFN-γ-induced Cp protein synthesis ceases after about 16 h, even in the presence of abundant Cp mRNA, due to uncoupling of the Cp transcript from the polyribosome as a result of inhibition of translation initiation. Detailed analysis revealed that cytosolic extracts made from U937 cells treated with IFN-γ for 16 h contain a protein complex, called IFN-gamma-activated inhibitor of translation (GAIT), that binds to the 29-nucleotide (nt)-long GAIT element present in the Cp (3′ untranslated region [UTR]) and specifically silences Cp mRNA translation (22, 33). We showed that the inhibition depends on the poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G (eIF4G), thereby documenting a requirement for end-to-end circularization of the Cp transcript (25). We also identified the large ribosomal subunit protein L13a as a component of the GAIT complex and the key molecular switch for Cp silencing (24). Our studies revealed that IFN-γ treatment results in the phosphorylation of L13a and its release from the 60S ribosome. Phospho-L13a (P∼L13a) then combines with the three other protein constituents of the GAIT complex: glutamyl-prolyl-tRNA synthetase (EPRS), NS1-associated protein-1, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH). This complex binds to the GAIT element in the 3′ UTR of Cp mRNA and silences its translation (34). Knockdown of L13a by RNA silencing abrogates IFN-γ-mediated inhibition of Cp mRNA translation (4). However, the global translational activity of L13a-depleted ribosomes remains unchanged, pointing to a dispensable extraribosomal function of L13a (4). Recent results from our laboratory have shown that, as a part of the GAIT complex, P∼L13a can bind the translation initiation factor eIF4G and prevent the formation of 48S preinitiation complex. The prevention of 48S complex formation relies on the direct competition between GAIT element-bound P∼L13a and 43S-bound eIF3 for binding to cap-bound eIF4G (16), which blocks the translation initiation of GAIT element-containing mRNAs.
It should be mentioned that Cp is an inflammatory protein. We therefore hypothesized that the translational silencing mechanism described above may have evolved to target not only one particular protein (i.e., Cp) but also a set of proteins associated with the particular function (e.g., inflammation). This hypothesis is based on the high abundance of GAIT complex components and stoichiometric release of L13a and EPRS from their parental complexes (24, 34). We envisioned that the simultaneous control of a cluster of inflammatory proteins by this mechanism might be an important cellular strategy to resolve inflammation.
To this end, we specifically searched for mRNAs that would follow the same signature of regulation as Cp mRNA, i.e., enrichment in the polyribosome within 4 h of IFN-γ treatment and disappearance from polyribosome but appearance in free fractions at 18 h of IFN-γ treatment. Thus, our interest was specifically focused on those mRNAs that show a pattern of a reduced ratio of polysomal to nonpolysomal abundance upon 18 h of IFN-γ treatment compared to 4 h of treatment. Using genome-wide analysis of the polysome-bound and unbound mRNA pools, we identified a cohort of mRNAs that follow the same pattern of regulation as Cp. Real-time PCR analysis and translation of reporter mRNA constructs carrying fragments of the 3′ UTR sequences of the target mRNAs functionally validated the results from genome-wide studies. Interestingly, these genes appeared to code for inflammatory proteins, such as many chemokine and chemokine receptors and molecules important in cytokine response and signaling. Thus, this cohort of mRNAs may represent a new inflammation-responsive posttranscriptional operon regulated directly at the level of translation in order to control inflammation in a temporal and expedited fashion.
MATERIALS AND METHODS
Cells and culture conditions.
Human monocytic U937 cells were cultured in RPMI 1640 medium with 10% heat-inactivated fetal bovine serum, 2 mM glutamine, and 100 U/ml of penicillin and streptomycin at 37°C and 5% CO2. U937 cells were treated with 500 U/ml of human IFN-γ (R&D Systems, Minneapolis, MN) for up to 18 h. U937 cells depleted of L13a were generated by lentivirus-mediated stable expression of short hairpin RNA (shRNA) against L13a, as described in one of our previous publications (4).
Isolation of translationally active and inactive pool of mRNAs by polysome fraction analysis.
Polysomes were analyzed from cytoplasmic extracts. Cytoplasmic extracts from exponentially growing cells were prepared following a method described previously (4). Twenty-five optical density units of cytoplasmic extract was carefully layered over 5 to 50% linear sucrose gradients in polysome buffer (10 mM HEPES, pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol, 50 U of recombinant RNasin [Promega, Madison, WI], and 0.1% Igepal-CA630 [Sigma, St. Louis, MO]) and centrifuged at 17,000 rpm in a Beckman SW32.1 Ti rotor for 18 h at 4°C. Gradients were fractionated using an ISCO gradient fractionation system equipped with a UA-6 detector following an upward displacement method. Light RNP fractions, 40S, 60S, 80S, and heavy polysome fractions were monitored by continuous UV absorption profiles at A264, and 22 tubes of 750-μl fractions were collected. The fractions collected in the first six tubes, representing light RNP and free ribosomes, were used to isolate a translationally inactive pool of mRNAs, and fractions numbered 7 to 22, representing polysomes, were used as a source of translationally active mRNAs. Total RNA was isolated from these fractions by extraction with Trizol (Invitrogen, Carlsbad, CA) and purified by an RNeasy minikit (Qiagen, Valencia, CA), following the manufacturer's suggested procedure. The RNA was quantitated and checked for purity by agarose formaldehyde gel and used for the Affymetrix GeneChip, an inflammation-responsive array, and real-time PCR analysis.
Microarray analysis using Affymetrix GeneChip.
Ten micrograms of RNA was subjected to labeling reaction using a one-cycle target labeling kit (Affymetrix, Santa Clara, CA). This procedure is a combination of cDNA synthesis, cleanup, and synthesis of biotin-labeled antisense cRNAs using a biotinylated ribonucleotide analog and in vitro transcription. The biotinylated cRNAs were subjected to cleanup and fragmentation using a cleanup module (Affymetrix, Santa Clara, CA) according to the manufacturer's suggested procedure. The fragmented and biotinylated cRNAs were subjected to hybridization with a Human Genome U133 Plus 2.0 chip (Affymetrix, Santa Clara, CA) using a hybridization control kit (Affymetrix, Santa Clara, CA). The chips were washed and stained with streptavidin-phycoerythrin biotinylated antistreptavidin antibody (Affymetrix, Santa Clara, CA). The washing and staining were performed using Affymetrix Fluidic Station 400. The stained chips were scanned using a GeneChip scanner 3000 (Affymetrix, Santa Clara, CA).
Data analysis.
Affymetrix data were analyzed by using the Affymetrix GeneChip Operating Software (GCOS) and Data Mining Tools software packages (Affymetrix, Santa Clara, CA). Signal values for all genes from each sample were imported directly from GCOS into Data Mining Tools. The SOM (for self-organizing map) clustering algorithm (38) of the Data Mining Tools software package was used at default filtering values, with parameters set to identify the 25 most abundant gene clusters. After clustering, results were screened to eliminate genes that failed to attain a “present” call in any of the samples (e.g., to eliminate genes scored as “absent” in all samples), and then primary clusters of interest were reanalyzed using the Data Mining Tools software to identify more refined clusters. Genes exhibiting expression patterns of interest in these smaller clusters (for example, genes induced by IFN-γ in total RNA at 18 h but suppressed in the polysome fraction at 18 h) were selected from these targeted clusters for verification by real-time PCR. In parallel, pairwise comparison data were exported from GCOS into Data Mining Tools and then carried through multiparametric comparisons to identify genes that were scored as “increased” (P < 0.005) in expression at 18 h and were scored as either “not changed” or “decreased” in expression in the polysome samples at this same time point, relative to untreated controls. The ratios of the raw signal intensity values of the polysomal and nonpolysomal mRNAs of the selected genes were determined. Heat maps were generated from gene expression data for selected genes by using the programs Cluster, version 2.11 (6), and TreeView, version 1.6 (6). For the unfractionated RNA, the raw signal intensity values of the total mRNA obtained from different times of IFN-γ treatment were used for the heat maps.
Microarray analysis using inflammatory response pathway array.
RNAs from translationally active and inactive pools were isolated by polysome fractionation following the method described above. Four micrograms of RNA was subjected to a labeling reaction using a TrueLabeling-AMP, version 2.0, kit (Superarray Inc., Frederick, MD). This process is a combination of cDNA synthesis followed by synthesis of the biotinylated cRNA by in vitro transcription in the presence of a biotinylated ribonucleotide analog. The labeled cRNAs were used to hybridize the Oligo GEArray Human Inflammatory Response and Autoimmunity microarray (Superarray Inc., Frederick, MD) at 60°C using GEAhyb hybridization solution (Superarray Inc., Frederick, MD). After 18 h of hybridization, the hybridized arrays were washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) plus 1% sodium dodecyl sulfate (SDS) and with 0.1× SSC plus 0.5% SDS at 60°C. The washed arrays were subjected to chemiluminescent detection using a Chemiluminescent Detection kit (Superarray Inc., Frederick, MD). This procedure relies on the binding of alkaline phosphatase-conjugated streptavidin with the hybridized biotinylated cRNAs, followed by incubation with the chemiluminescent substrate CDP-Star (Superarray Inc., Frederick, MD). All the steps of prehybridization, hybridization, washing, and chemiluminescent detection were performed using the suggested procedure of the Oligo GEArray System manual (37). The arrays were exposed to Clear Blue X-ray film (Pierce, Rockford, IL) in order to capture the luminescence. The image was scanned. Each spot representing one mRNA was identified and quantified using a web-based platform (GEArray Expression Analysis Suite, version 2.0; http://geasuite.superarray.com) of Superarray Inc. (Frederick, MD). The normalization of the data is based on the signal intensities of several spots representing housekeeping genes, e.g., GAPDH, β-actin, RPS27A, and β2-microglobulin. Signal intensities from each mRNA present in polysomal and nonpolysomal pools were retrieved and used to determine the ratio of the translationally active to the inactive pool of a particular mRNA.
Real-time PCR analysis.
Real-time PCR analysis was performed to validate the microarray results described above. Five micrograms of the total RNA from translationally active and inactive pools was subjected to reverse transcription using a TaqMan reverse transcription reagent kit (Applied Biosystems, Foster City, CA). PCR amplification was carried out using Sybr Green PCR Master Mix (Applied Biosystems, Foster City, CA) and an ABI Thermo Cycler (ABI Prism 7000 SDS). Primers were designed so as to produce 100 to 200 bp of amplified product. Before being used in real-time PCR, each primer pair was authenticated by standard reverse transcription-PCR and by sequencing the amplification product of the expected size. The results were expressed as the relative amount of each mRNA after normalization with GAPDH. The primer sequences for the different genes are as follows: APOL2 forward (F), 5′-AGC CTG GAA TGG ATT CGT GG-3′; APOL2 reverse (R), 5′-GTG GCG GTT TTT GTC CTT CA-3′; CCL22 (F), 5′-TCC TTG TGC TCC CAA AGT GC-3′; CCL22 (R), 5′-AGG AAA TGT AAG CAG ACA CG-3′; CXCL13 (F), 5′-GTT CCA GTT GAT GGG CAA GT-3′; CXCL13 (R), 5′-TCT GGC ACC TGT ATT CAC CA-3′; CCR3 (F), 5′-TTG GAG AGA GGT TCC GGA AG-3′; CCR3 (R), 5′-AGG GTA CAT GCA TCT ACT GC-3′; CCR4 (F), 5′-AGA AGG CAT CAA GGC ATT TG-3′; CCR4 (R), 5′-AGC CCA CCA AGT ACA TCC AG-3′; CCR6 (F), 5′-AGT CTC CCT AAG GCA TGT GT-3′; and CCR6 (R), 5′-ATG CTA CCT ATC AGA GAC CA-3′.
Bioinformatic prediction of GAIT-like elements.
The 3′ UTR sequence of the candidate mRNA was examined for structural similarities with the Cp GAIT element using the Foldalign program on a web-based server (http://foldalign.ku.dk/). This procedure allows pairwise local structural alignment of RNA sequences where no significant sequence similarity exists between the query and the reference. In addition, the program allows multiple structural alignments and clustering of RNA sequences (12, 40). Various lengths of the identified sequences were then folded using the Vienna RNAfold secondary structure prediction program, version 2.0 (14) (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). The hairpins that best resembled the Cp GAIT (33) by visual selection were further screened by structure-based homology with the Cp GAIT hairpin using the Foldalign program, version 2.0.3 (13).
Construction of CMV-Luc-GAIT plasmids.
The following genes were selected for the verification of translational inactivation by reporter gene assay: APOL2 (NM_030882), CCL22 (NM_002990), and CCR4 (NM_005508). This list represents only the common genes obtained by polysomal profiling using both the pathway-specific array (see Table S2 in the supplemental material) and the Affymetrix GeneChip (see Fig. 2). Based on the bioinformatics analysis described above, the following sequences were selected for cloning (the numbers refer to nucleotide positions in the 3′ UTR, i.e., starting the count after the stop codon) APOL2, nt 928 to 958; CCL22, nt 433 to 462; and CCR4, nt 280 to 307. For each sequence, sense and antisense strands were synthesized with SpeI and HindIII half-sites at the termini for cloning into the same sites of pMIR-reporter vector (Ambion, Austin, TX). In addition, we added an ApaI site (GGGCCC) immediately after the SpeI half-site for easy screening of the clones as this would be the unique ApaI site in the construct. The resultant reporter constructs had the following structure: cytomegalovirus(CMV) promoter-luciferase (Luc)-3′ UTR (test element).
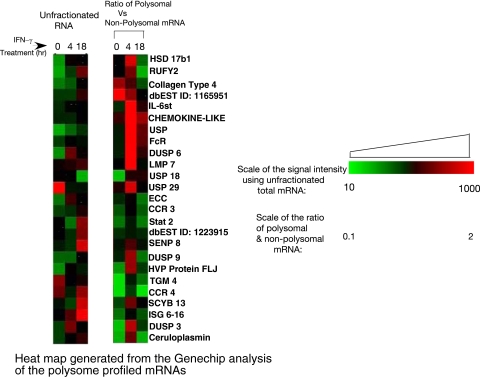
FIG. 2.
Heat map generated from the GeneChip analysis of translationally active and inactive pools of mRNAs. Raw expression data were exported from GCOS (Affymetrix, Santa Clara, CA) to the Data Mining Tools program (Affymetrix, Santa Clara, CA) and subjected to SOM cluster analysis to identify groups of genes with expression patterns similar to the Cp pattern, i.e., induction in response to IFN-γ, greater induction in polysome fractions and less in nonpolysome fractions in 4 h, and the reverse at 18 h after IFN-γ treatment. The ratios of the raw signal intensity values of the polysomal to nonpolysomal mRNAs of the selected genes were determined. Heat maps were generated using gene expression data for selected genes using the programs Cluster, version 2.11 (6), and TreeView, version 1.6 (6). For the unfractionated RNA, the raw signal intensity values of the total mRNA obtained from different times of IFN-γ treatment were used. The color progression scale represents the relationship between different colors and relative quantities of a particular mRNA. IL-6st, interleukin-6 signal transducer; USP, ubiquitin-specific protease; HVP, herpesvirus papio.
Transfection of U937 cells with chimeric Luc-GAIT reporters.
To measure the translation of the chimeric reporter Luc, U937 cells (106 cells) were transiently transfected with 10 μg of CMV-firefly Luc-GAIT in 0.2 ml of RPMI 1640 medium containing 10 mM HEPES following a previously published method (33). Transfection efficiency of the cells was monitored by cotransfection of 1.0 μg of CMV-Renilla Luc plasmid (CMV-ph RL; Promega, Madison,WI). The cells were electroporated (Amaxa, Gaithersburg, MD), allowed to recover for 16 h in RPMI 1640 medium containing 10% serum, and then treated with IFN-γ (500 U/ml) in medium containing 2% serum. At 4 h and 18 h after the addition of IFN-γ, the treated cells were washed twice in cold phosphate-buffered saline and lysed, and firefly and Renilla Luc activities were measured by luminescence using a Dual Luciferase Reporter assay system (Promega, Madison,WI) and 20/20n Luminometer (Turner BioSystems, Sunnyvale, CA). Firefly luciferase activity was normalized to Renilla luciferase activity. Each assay was done in triplicate, and the mean and standard error were determined.
In the second set of experiments, U937 cells stably expressing shRNA against L13a and control shRNA (4) were transfected with the above constructs and treated with IFN-γ in an identical manner, and Luc activity was measured.
RNA gel shift analysis.
Synthetic RNA was end labeled in the presence of [γ-32P]ATP (7,000 Ci/mmol) and T4 polynucleotide kinase using a KinaseMax kit (Ambion, Austin, TX). Unincorporated radioactivity was removed by a NucAway spin column (Ambion, Austin, TX). The radiolabeled RNA (50 fmol) was incubated for 30 min at room temperature with U937 cell extract (20 μg of protein) in 20 μl of reaction buffer containing 25 mM HEPES (pH 8.0), 15 mM KCl, 0.25 mM dithiothreitol, 5 mM MgCl2, 0.1 mM phenylmethylsulfonyl fluoride, 200 μg of Saccharomyces cerevisiae tRNA per ml, 40 U of RNasin, and 10% glycerol. RNA-protein complexes were resolved by native gel electrophoresis (5% polyacrylamide in 0.5× Tris-buffered EDTA). The gel was dried, and the retarded probe was detected by autoradiography.
RESULTS
Identification of potential target mRNAs for IFN-γ-mediated delayed translational silencing using human GeneChip analysis.
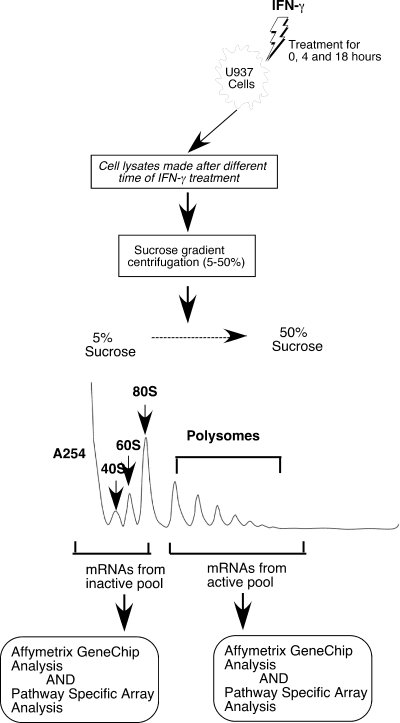
Our earlier studies showed that GAIT complex-mediated translational silencing in U937 monocytic cells is active only after 16 h of IFN-γ treatment. Using sucrose gradient centrifugation analysis (following the scheme shown in Fig. 1), we have isolated the total pool of mRNAs from the active polysome fractions and from the inactive free mRNP/monosome fractions after treatment of U937 cells with IFN-γ for 0, 4, and 18 h. The different pools of mRNAs were subjected to microarray analysis using GeneChip Human Genome U133 Plus 2.0 (Affymetrix, Santa Clara, CA). The mRNAs that showed (i) enhanced association with active polyribosome and less with inactive free fractions at 4 h of IFN-γ treatment and (ii) less association with polyribosome and enhanced association with free fractions at 18 h of treatment were identified by cluster analysis (see Materials and Methods for details). The ratios of the signal intensities of polysomal (active) to nonpolysomal (inactive) association of each mRNA were determined at each time point, i.e., 0, 4, and 18 h of IFN-γ treatment. These ratios represent the translational status of an mRNA and are shown as a heat plot (Fig. 2). A complete list of the different target mRNAs and the raw data of the signal intensities representing the polysomal and nonpolysomal associations and their ratios are presented as supplemental data (see Table S1 in the supplemental material). This approach identified 27 mRNAs including Cp mRNA, thus confirming that the approach is valid. Interestingly, several of the identified mRNAs have functions potentially related to Cp; i.e., they have a role in inflammation. For example, we found chemokine and chemokine receptors, e.g., CCL11, CCL22, CCL25, CCR3, CCR4, and CXCL13 (SCYB13); molecules important in cytokine response and signaling, e.g., IFN-stimulated gene 6-16, Stat2, interleukin-6 signal transducer gp130; human-specific (27) and IFN-activated (11) acute phase protein (8) APOL2 (Fig. 2; see Table S1 in the supplemental material).
FIG. 1.
Flow chart of the isolation of translationally active and inactive mRNAs from IFN-γ treated U937 cells for different time.
Identification of the common target mRNAs of translational silencing using GeneChip and an inflammatory response pathway microarray.
Expression profiling studies using polysome-bound mRNAs described above identified several mRNAs coding for proteins important in inflammation. Using a more focused approach, we next conducted one more round of expression profiling studies using an inflammatory response pathway microarray (Super Array Inc., Frederick, MD), in which 440 genes involved in cellular inflammation were prespotted. The functional groupings of these genes include chemokines and chemokine receptors; cytokines and cytokine receptors; genes involved in cytokine metabolism, cytokine production, and cytokine-receptor interaction; and acute phase, inflammatory, and humoral immune response genes. The rationale for this strategy is based on our expectation to identify common hits between GeneChip and the pathway-specific approach.
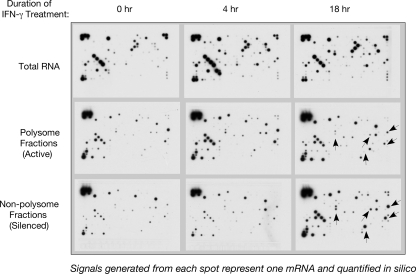
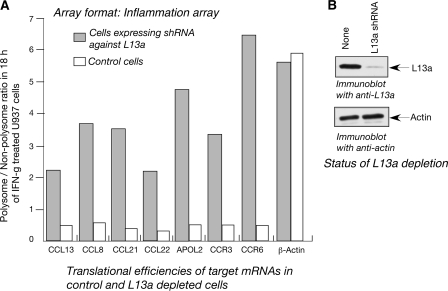
mRNAs from the translationally active (polysome) and silenced pools (mRNP/monosome) were isolated from IFN-γ-treated U937 cells following the same scheme as above (Fig. 1), followed by hybridization with the human inflammatory response microarray and subsequent processing (see Materials and Methods for details). The raw image of the array is shown in Fig. 3. Enrichment of some representative spots hybridized with the inactive mRNA pool isolated from 18 h of IFN-γ-treated cells is marked by arrowheads (Fig. 3). The ratio of the translationally active versus the inactive (polysomal versus nonpolysomal) pool of a particular mRNA was determined from the signal intensities. The mRNAs that showed significant reductions in this ratio following 18 h of IFN-γ treatment were identified. Table S2 in the supplemental material shows the complete list of such mRNAs. Previously, we discovered the requirement of L13a in the IFN-γ-mediated delayed translational silencing of Cp mRNA in U937 monocytic cells (4, 24). In addition, we generated a permanent line of U937 cells stably expressing shRNA of L13a showing abrogation of L13a protein; these cells failed to show translational silencing of Cp mRNA (4). Following the Cp paradigm, we have used these L13a knockdown cells to ask directly whether the translational silencing of these clusters of mRNAs is also L13a dependent. In contrast to the wild-type cells (see Table S2 in the supplemental material), the majority of the mRNAs of this cluster exhibited reversal of the ratio of the polysomal to nonpolysomal association in L13a knockdown cells after 18 h of IFN-γ treatment (see Table S3 in the supplemental material). In contrast, this reversal was not observed in control β-actin mRNA. A direct comparison of the ratios of polysomal to nonpolysomal associations of some representative mRNAs at 18 h of IFN-γ-treated wild-type and L13a-depleted monocytes is shown in Fig. 4A. The extent of L13a depletion was confirmed by immunoblot analysis, and found to be more than 98% (Fig. 4B). These results show that the IFN-γ-mediated translational silencing of these mRNA clusters is L13a dependent.
FIG. 3.
Raw data from the mRNA profiling analysis using an inflammatory response pathway array. mRNAs isolated from the translationally active (polysomal) and silenced pools from IFN-γ-treated U937 cells were subjected to microarray analysis using a human inflammatory response microarray (catalog item OHS-803; SuperArray Inc., Frederick, MD). The raw image was generated after chemiluminescence detection, scanned, and analyzed using the GE Expression Analysis Suite, version 2.0 (http://geasuite.superarray.com). The results of this analysis are presented in Table S2 in the supplemental material.
FIG. 4.
(A) Comparison of the translational efficiencies of the target mRNAs in normal and L13a-depleted cells using an inflammatory response pathway array. The ratios of polysomal to nonpolysomal abundance of the common mRNAs (i.e., found positive both in GeneChip and inflammatory response pathway arrays) was determined in L13a-depleted and normal U937 cells after 18 h of IFN-γ treatment. Ratios were retrieved from data in Tables S1 and S2 in the supplemental material. (B) Extent of L13a depletion. Immunoblot analysis using anti-L13a antibody was carried out using the lysate made from the cells expressing shRNA against L13a and normal cells (upper panel). The same blot was reprobed with antiactin antibody as a control (lower panel).
In summary, both GeneChip analysis and inflammation-responsive gene array analysis (Fig. 2; see Tables S1 and S2 in the supplemental material) using mRNAs from two different experiments have identified several common mRNAs as potential targets for translational silencing. The mRNAs identified as common hits from these two different experimental approaches are APOL2, CCL11, CCL22, CCL25, CCR3, and CCR4. In addition, analysis of the inflammation-responsive array also showed a reduced ratio of the polysomal to nonpolysomal abundance for a number of other chemokine ligands such as CCL1, CCL3, CCL8, CCL20, CCL21, CCL27, and CCL28 and chemokine receptors such as CCR2, CCR5, CCR6, and CCR7 (see Table S2 in the supplemental material). However, these mRNAs were not identified using Affymetrix GeneChip analysis. It was also important to test whether the change in the ratio of the signal intensities of polysomal (active) to nonpolysomal (inactive) mRNAs is a true reflection of translational silencing or the consequence of the change of ribosomal components by IFN-γ. To address this concern, we performed polysome profile analysis from the U937 cells treated with IFN-γ for different periods of time. These studies showed that there is no significant difference in the levels of 40S, 60S, and 80S ribosomal components or in the extent of translationally active polysome formation between cells treated with IFN-γ for different timeperiods (data not shown).
Validation of selected common hits from the GeneChip and inflammation-responsive array.
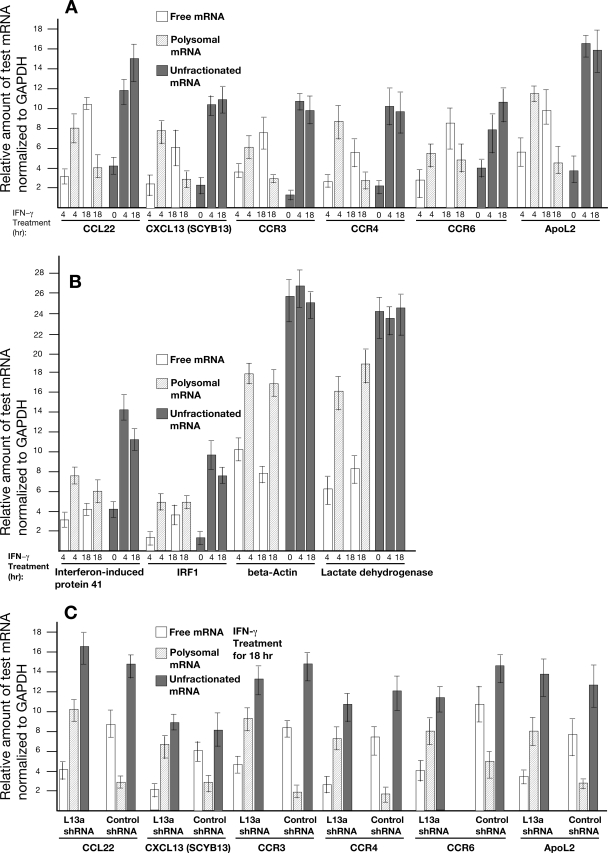
In an independent experiment we isolated mRNAs from the translationally active polysome pool and the inactive free or nonpolysomal pool from IFN-γ-treated U937 cells following the scheme shown in Fig. 1. The presence of CCL22, CCR3, CCR4, CCR6, CXCL13, and APOL2 mRNAs in each pool was determined by quantitative real-time PCR. Results presented in Fig. 5A show that the expression of these mRNAs paralleled the results obtained from the GeneChip and inflammation-responsive array. In other words, for each mRNA greater association with active polysome and less association with inactive free or nonpolysomal fractions was found at 4 h after IFN-γ treatment. However, the reduced association with the active polysome and concomitant elevated levels in the inactive free fractions were found at 18 h of IFN-γ treatment. Importantly, 18 h of IFN-γ treatment caused translational silencing of these mRNAs even under conditions where they were induced. This is identical to the profile of previously reported translational silencing of Cp in response to IFN-γ (22). As an additional control, we also determined the polysomal and nonpolysomal abundances of two IFN-induced mRNAs, IFN-induced protein 41 and IRF1, and two noninduced mRNAs, β-actin and lactate dehydrogenase. Results from real-time PCR showed no significant change in the polysomal and nonpolysomal abundances of these mRNAs at different times of IFN-γ treatment (Fig. 5b).
FIG. 5.
Validation of the potential target mRNAs by real-time PCR analysis. (A) Total RNAs were isolated from the translationally active polysomal fraction and inactive free fraction after 4 and 18 h of IFN-γ treatment of normal U937 cells. The total mRNAs from unfractionated cell lysates were also isolated at 0, 4, and 18 h of IFN-γ treatment to check the steady-state levels. Total RNAs were subjected to reverse transcription using a TaqMan Reverse Transcription Reagent Kit (Applied Biosystem, Foster City, CA). PCR amplification was carried out using Sybr Green PCR Master Mix (Applied Biosystem, Foster City, CA) and an ABI Thermo Cycler (ABI Prism 7000 SDS). In addition, the authenticity of each primer pair used in real-time PCR was confirmed by classical reverse transcription-PCR and sequencing of the amplified products (100 to 200 bp). The value shown is the relative amount of each mRNA after normalization with GAPDH from triplicates. (B) Status of polysomal and nonpolysomal abundances of two IFN-γ-induced and two noninduced mRNAs upon IFN-γ treatment as a negative control. Total RNAs were isolated from the translationally active polysomal fraction and inactive free fraction after 4 and 18 h of IFN-γ treatment of normal U937 cells. The total mRNAs from unfractionated cell lysates were also isolated at 0, 4, and 18 h of IFN-γ treatment to check the steady-state levels. Total RNAs were subjected to real-time PCR using a TaqMan Reverse Transcription Reagent Kit, Sybr Green PCR Master Mix, and an ABI Thermo Cycler (ABI Prism 7000 SDS). The value shown is the relative amount of each mRNA after normalization with GAPDH from triplicates. (C) The experiments described in panel A above were carried out using L13a-depleted cells expressing shRNA against L13a and control shRNA.
In order to check whether the reduced association of these mRNAs with polysomes at 18 h of IFN-γ treatment requires L13a, we performed the same experiment using cells expressing shRNA against L13a and control shRNA. Results presented in Fig. 5C show that the reduced polysomal association of the mRNA compared to free fractions was not observed in L13a-depleted cells. On the other hand, cells expressing control shRNA continued to show reduced polysomal association of the target mRNAs upon 18 h of IFN-γ treatment (Fig. 5c).
In silico analysis of the 3′ UTRs of potential target mRNAs for translational silencing.
As mentioned before, the 29-nt GAIT element within the 3′ UTR of Cp mRNA that was responsible for IFN-γ-mediated translational silencing has been previously identified (33). The structural signature of the GAIT element appears to consist of a 6-bp helical stem, an asymmetric internal bulge, a distal and weak 3-bp helix, and a 5-nt terminal loop. However, the exact nucleotide length or sequence requirement of the different segments and the structural footprinting of the GAIT element by chemical or enzymatic probing remain unknown. Therefore, it is possible that different segments of an active GAIT element can accommodate a range of nucleotide lengths and sequences.
To identify functional GAIT elements in our newly discovered candidate genes, we first narrowed our selection to the mRNAs that were identified as common hits from both the human GeneChip (Fig. 2; see Table S1 in the supplemental material) and the inflammation-responsive pathway-specific array (see Table S2 in the supplemental material). These mRNAs are the following: APOL2, CCL11, CCL22, CCL25, CCR3, CCR4, and CCR6. Six of these mRNAs including APOL2, CCL22, CCR3, CCR4, CCR6, and CXCL13 were validated by real-time PCR (Fig. 5). The prospective GAIT elements in the 3′ UTR of these mRNAs were then examined for structural similarities with the Cp GAIT element using the Foldalign program and for potential hairpins using the Vienna RNA secondary structure prediction program, as detailed in Materials and Methods. The resultant structures, found in the 3′ UTRs of CCL22, CCL11, CCR3, CCR4, CCR6, and APOL2, appeared to contain significant folding similarities to the Cp GAIT element (see Fig. S1 in the supplemental material). In contrast, Foldalign failed to identify any significant folding similarity between the Cp GAIT element and the 3′ UTRs of CCL25 and CXCL13 mRNAs. We hypothesize that these novel Cp GAIT-like elements may be functional targets for IFN-γ mediated translational silencing.
Functional testing of the translational silencing response driven by the 3′ UTRs of the target mRNAs.
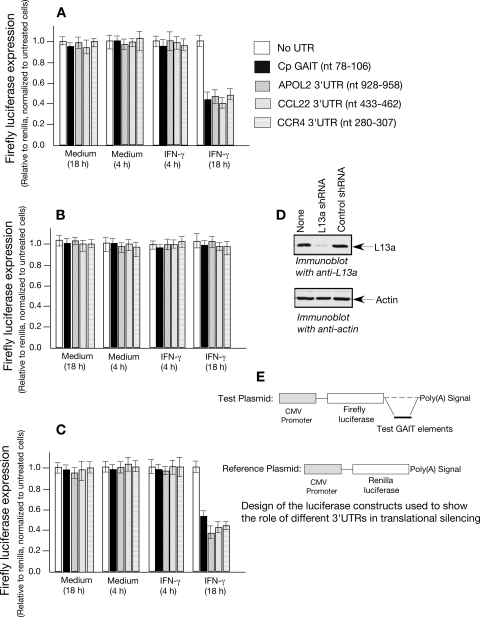
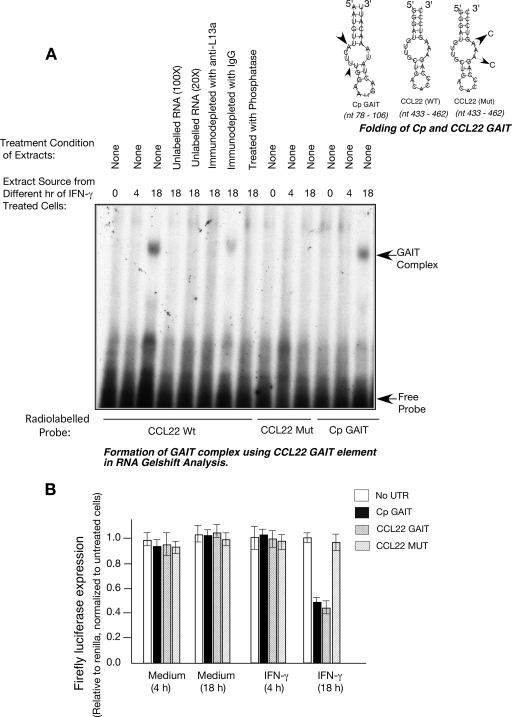
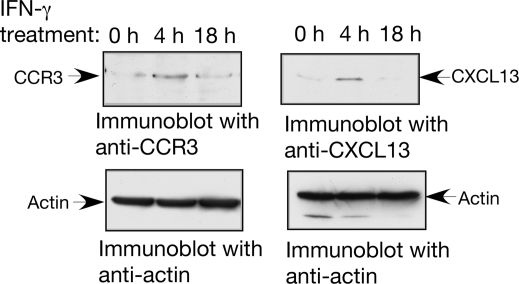
We constructed translational reporter plasmids in which the bioinformatically identified candidate GAIT elements described above were cloned downstream of a Luc reporter gene (see Materials and Methods for details). The ability of the predicted elements to confer the translational silencing of Luc was tested upon transfection in U937 cells. Note that the 3-h half-life of Luc in mammalian cells permits its appropriate use as a reporter of protein synthesis in mammalian cells (39). U937 cells were cotransfected by electroporation (Amaxa, Gaithersburg, MD) with chimeric CMV-Luc and with CMV-Renilla Luc to correct for transfection efficiency. As shown (Fig. 6), incubation of cells with IFN-γ for 18 h (but not 4 h) inhibited the expression of Luc from the plasmids containing the predicted GAIT elements. No such inhibition was observed in the control plasmid lacking the 3′ UTR (Fig. 6A). We then addressed whether the translational silencing of the novel GAIT elements is L13a dependent by testing for silencing in cells depleted of L13a. In contrast to wild-type cells (Fig. 6A), the L13a-depleted cells failed to promote the translational silencing of the same constructs (Fig. 6B). In cells expressing negative control shRNA used previously (4), translational silencing was unaffected (Fig. 6C). Western analysis of L13a confirmed the efficacy of the L13a shRNA (Fig. 6D). The stability of firefly Luc mRNA in transfected U937 cells was tested by real-time PCR using primers for firefly Luc and GAPDH (as a control). Consistent with our earlier observation (33), Luc mRNA levels were found to be similar in all treatments (data not shown). These results demonstrate that the segments of 3′ UTRs from APOL2 (nt 928 to 958), CCL22 (nt 433 to 462), and CCR4 (nt 280 to 307) harboring the folding similarities to the Cp GAIT element are effective for translational silencing, and, like Cp GAIT, require L13a for their silencing activity in response to IFN-γ. In agreement with these results, we found that the CCL22 GAIT element (nt 433 to 462) can support GAIT complex formation in RNA gel shift experiments. The authenticity of the GAIT complex formed on the CCL22 GAIT element was confirmed by the time dependence of IFN-γ treatment and its dependence on L13a since immunodepletion of L13a from the 18-h extract could abrogate the RNA protein complex formation (Fig. 7A). To better understand the critical residues of the potential new GAIT elements, we compared the internal loops of CCL22 (nt 433 to 462) and Cp (nt 78 to 106) GAIT elements. Previously, we had shown the importance of this internal loop in Cp GAIT-mediated translational silencing. These studies showed that changing U to C at positions 84 and 87 caused inactivation of this element (33). In accordance with the Cp GAIT, mutation of the A to C at positions 453 and 455 in the same internal loop of the CCL22 GAIT inactivated the element both in terms of GAIT complex formation (Fig. 7A) and translational silencing of the reporter gene (Fig. 7B). Finally, we examined the abundance of the proteins encoded by the target mRNAs of this regulatory mechanism. Our results obtained with the use of immunoblot analysis show a significant reduction of the steady-state levels of CCR3 and CXCL13 proteins after 18 h compared to 4 h of IFN-γ treatment (Fig. 8).
FIG. 6.
Abilities of the 3′ UTRs of target mRNAs to confer translational silencing to reporter mRNAs in response to IFN-γ treatment. (A) Normal U937 cells were electroporated using the construct harboring the CMV promoter-driven firefly Luc upstream of the different 3′ UTR elements under test and CMV-Renilla Luc construct as a control for transfection efficiencies. After the recovery period of the electroporated cells, they were treated with IFN-γ (500 U/ml) for 4 and 18 h. Chemiluminescence was measured to quantify the firefly and Renilla Luc activity. Renilla Luc expression was used to normalize the firefly Luc and then expressed as a percentage compared to untreated control cells. The data shown represent the means and standard errors from three independent experiments. (B) The experiment described above was conducted using L13a-depleted cells by stable expression of shRNA against L13a. (C) The experiment described above was conducted using cells expressing control shRNA. (D) Status of L13a depletion of the U937 cells used in the experiment. (E) Design of the reporter gene constructs harboring the elements from different 3′ UTRs at the end of the reporter Luc.
FIG. 7.
The asymmetric bulge or internal loop of CCL22 GAIT element is important for its activity. (A) Synthetic RNAs from the CCL22 wild-type (Wt) GAIT element (nt 433 to 462), CCL22 mutant (Mut) GAIT (A453 455C), and wild-type Cp GAIT (nt 78 to 106) were synthesized (Dharmacon) and 32P labeled by T4 polynucleotide kinase. The cell extracts from IFN-γ-treated U937 cells were incubated with 32P-labeled RNA. In competition experiments, the extracts were first incubated with 100- and 20-fold molar excesses of the unlabeled CCL22 wild-type GAIT element before the extract was added to the labeled probe. To test the requirement of L13a in the RNA protein complex formation, the extract was immunodepleted with L13a and control antibody before being added to the labeled probe. RNA protein complexes were resolved by electrophoresis on a nondenaturing 5% polyacrylamide gel and detected by autoradiography. (B) Reporter gene constructs were made harboring either the CCL22 wild-type GAIT element (nt 433 to 462) or mutant (MUT) GAIT element (A453 455C) at the end of the reporter Luc. These constructs were transfected in to U937 cells alongside the reporter construct harboring Cp GAIT (nt 78 to 106) as a positive control, followed by IFN-γ treatment for 4 and 18 h (for details, see the legend of Fig. 6A).
FIG. 8.
Detection of the steady-state levels of CCR3 and CXCL13 in IFN-γ-treated monocytes. U937 cells were treated with IFN-γ for different times. The cells were lysed and then subjected to by SDS-polyacrylamide gel electrophoresis analysis and immunoblotting using anti-CCR3 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-CXCL13 (R&D Systems, Minneapolis, MN).
DISCUSSION
We have used a combination of genome-wide polysomal profiling, bioinformatics analysis of RNA folding, and translational silencing assays of reporter genes to identify a family of translationally controlled mRNAs in IFN-γ-activated monocytes. Our confidence in these target mRNAs identified by the genome-wide approach is based on the following observations: (i) identification of translational silencing of Cp mRNA by the Affymetrix GeneChip (Fig. 2) shows proof of principle; (ii) identification of the same cohort of target mRNAs using two independent experiments e.g., GeneChip (Fig. 2; also Table S1 in the supplemental material) and inflammation-responsive gene array analysis (see Table S2 in the supplemental material); (iii) independent validation of mRNAs by real-time PCR analysis of the polysomal and nonpolysomal fractions isolated from IFN-γ-treated cells (Fig. 5); (iv) recognition of folding similarities between the segments of 3′ UTRs of some mRNAs (e.g., CCL22, APOL2, CCL11, CCR4, CCR3, and CCR6) and the Cp GAIT element (see Fig. S1 in the supplemental material); and, finally, (iv) the ability of the segments of the 3′ UTRs present in APOL2, CCL22, and CCR4 to confer the translational silencing and its dependence on L13a (Fig. 6A and B) in heterologous reporter gene constructs. On the other hand, this approach may have some limitations as it requires that the level of expression of a particular mRNA should be well above the cutoff limit of detection, with a high signal-to-noise ratio. For example, this approach identified Cp mRNA, a well-characterized target for a GAIT-mediated silencing mechanism (22, 24, 25, 33). However, another previously identified target, e.g., vascular endothelial growth factor mRNA (32), remained unidentified due to its level of expression, with a low signal-to-noise ratio.
Extracellular signal-dependent release of EPRS and L13a from their parental complexes, i.e., multisynthetase complex and 60S ribosome, respectively, is critical for the assembly of a silencing-competent GAIT complex (24, 34). The assembly process relies on a highly concerted two-stage mechanism, driven by the phosphorylation of both proteins. It is reasonable to believe that evolution of such a complex translational silencing mechanism would be strategically more advantageous if it could simultaneously target a cluster of mRNAs instead of a single Cp mRNA. It is now increasingly appreciated that regulation of functionally related proteins can be orchestrated as a posttranscriptional operon in which expression from their cognate mRNAs can be regulated though a RNP-driven mechanism (18). Consistent with this idea, we hypothesize that this silencing may have evolved to target not only a particular protein (i.e., Cp) but also a function associated with this protein, i.e., inflammation. A rational way to achieve this physiological outcome may rely on simultaneous silencing of a cohort of other inflammatory proteins. Here, we report a novel inflammation-responsive posttranscriptional operon composed of mRNAs for several chemokines and their cognate receptors. Our report also shows that delayed translational silencing activity induced by IFN-γ also targets this new posttranscriptional operon in addition to Cp mRNA, and this mechanism may be important to resolve chronic inflammation.
The family of chemokines and their cognate receptors regulate the activation-directed migration of leukocytes to the sites of inflammation during immune surveillance. The integrated signals initiated by various chemokines offer combinatorial control of leukocyte response (41). Uncontrolled synthesis of chemokines and their receptors by the mononuclear cells is now recognized as a major player in the pathogenesis of inflammatory conditions such as atherosclerosis (19, 20, 29) and neoplastic, infectious, allergic, and autoimmune diseases (9). Our report shows that the mechanism for delayed translational silencing of Cp can also target a cohort of chemokines and their specific receptors such as CCL-1/CCR8, CCL-3/CCR5, CCL8/CCR3, CCL11/CCR3, CCL20/CCR6, and CCL21/CCR7 (7, 42). The simultaneous silencing of these chemokine ligands and their specific receptors may provide a highly efficient mechanism to limit leukocyte trafficking and resolve inflammation. Cumulative evidence suggests that resolution of inflammation is an active process involving dedicated pathways and mechanisms that initiate in the first few hours after an inflammatory challenge (10, 35, 36). The initiation of this translational silencing mechanism involving the release of EPRS from multisynthetase complex within 4 h of IFN-γ treatment (34) is highly consistent with this idea. Another interesting feature of our report is the regulation of a diverse group of mRNAs by IFN-γ-mediated translational silencing.
It is to be noted that our selection of the prospective GAIT hairpins was somewhat subjective, albeit aided by two bioinformatic searches, including structure-based homology. We subsequently tested them by a functional gain-of-silencing assay in translational reporter plasmids. Our positive results warrant a more comprehensive analysis in the future in which we plan to clone the full 3′ UTR of the candidate gene(s), followed by systematic deletion analysis for functional loss of silencing. It will be interesting to see whether the latter approach identifies the same hairpins or perhaps detects additional sequences of similar or stronger silencing activity. All hairpins proposed in Fig. S1 in the supplemental material have a loop and a stem with a predominantly asymmetric bulge; however, these features show considerable diversity, which is in part due to the somewhat arbitrary nature of RNA structure prediction itself. The CCL11 hairpin, for example, has a long U-A stem that can be denatured to the desired extent to increase the size of the U-C bulge. In addition, there are some obvious differences between the folding units of these RNAs, e.g., the thermodynamic stability of terminal stem, asymmetric bulge or internal loop, internal stem, and terminal loop. Nonetheless, the diversity of GAIT elements may be explained by the flexibility of the GAIT hairpin, the heterogeneous nature of the GAIT complex itself in terms of exact subunit composition, and the diversity of the RNA-binding surface of the GAIT component proteins. Indeed, an induced fit conformational change in the GAIT element during the interaction with RNA-binding proteins has been proposed (33). Furthermore, in a recent report the PUF protein has been shown to recognize diverse RNA targets by using its flexible RNA-binding surface (26).
Translational regulation is critical for controlling the expression of a variety of proinflammatory proteins synthesized by macrophages. For example, tumor necrosis factor alpha mRNA can be translationally controlled by a number of RNA binding proteins that bind to the 3′ UTR, e.g., hnRNPA1 (3), Hur (17), and TIA-1 (30). Enhanced inflammation and significant pathological outcomes were reported in TIA-1 knockout mice (30). Another inflammatory protein, Cox-2, could be translationally controlled by binding of CUGB2 (28), TIA-1 (5), and Hur (17) in the 3′ UTR. A genome-wide analysis of TIA-1-bound transcripts also identified a number of mRNAs coding for inflammatory proteins (21). A broad array of chemokines are regulated at the posttranscriptional level in response to a variety of inflammatory stimuli; however, this mechanism primarily depends on alteration of mRNA stability (7). Currently, there is no report of translational regulation of a cohort of chemokine ligands and their cognate receptors by a single stimulus. Our report, therefore, shows the existence of a novel inflammation-responsive posttranscriptional operon in the IFN-γ-activated monocytes. Coordinated translation regulation of this posttranscriptional operon by a ribonucleoprotein complex-driven mechanism such as the GAIT complex may have an important physiological role in resolving inflammation. A model representation of this concept is presented in Fig. 9. The simultaneous translational silencing of a cluster of inflammatory proteins by the GAIT complex may also offer new targets for future anti-inflammatory therapeutics. Our results show the dependence of the translational silencing of this newly identified posttranscriptional operon on L13a. These findings also motivate future experiments using a myeloid cell-specific L13a-knockout mouse that might bring significant new insight into the physiological consequences of this translational silencing.
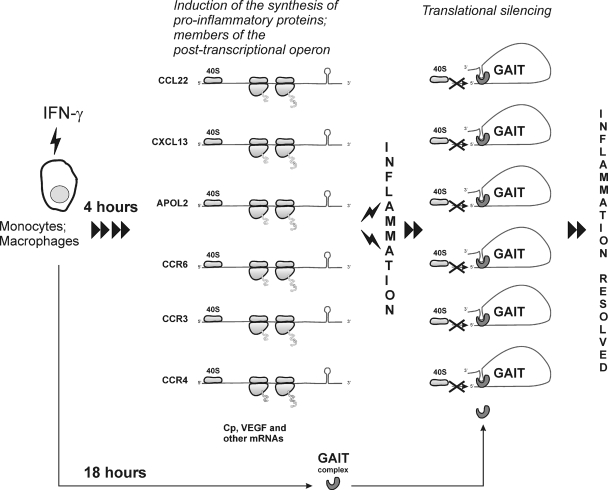
FIG. 9.
GAIT complex-mediated translational silencing of the posttranscriptional operon controls inflammation in monocytes/macrophages. IFN-γ induces a cluster of mRNAs and synthesis of the encoded proteins involved in the inflammatory response; this process is proinflammatory. However, at 18 h posttreatment the formation of active GAIT complex causes the simultaneous translational silencing of this cluster by binding to the GAIT elements present in the 3′ UTRs of the target mRNAs. Using Cp mRNA, the first discovered member of this posttranscriptional operon, we have shown that the GAIT complex blocks the recruitment of the 40S ribosomal subunit (16). This process may resolve inflammation.
Supplementary Material
Acknowledgments
This work was supported by PHS grant HL79164 from the NIH and grant-in-aid 0855555D from American Heart Association Great Rivers Affiliate (to B.M.), PHS grants AI068133 (to D.L.) and AI059267 (to S.B.) from the NIH, and National American Heart Association grant 0730120N (to A.A.K.). S.C. was supported by postdoctoral fellowship grant 0725628B from the American Heart Association Great Rivers Affiliate.
We acknowledge Joseph D. Fontes (University of Kansas School of Medicine) for helpful comments on the manuscript.
Footnotes
Published ahead of print on 10 November 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Arava, Y., Y. Wang, J. D. Storey, C. L. Liu, P. O. Brown, and D. Herschlag. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1003889-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown, V., P. Jin, S. Ceman, J. C. Darnell, W. T. O'Donnell, S. A. Tenenbaum, X. Jin, Y. Feng, K. D. Wilkinson, J. D. Keene, R. B. Darnell, and S. T. Warren. 2001. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell 107477-487. [DOI] [PubMed] [Google Scholar]
- 3.Buxade, M., J. L. Parra, S. Rousseau, N. Shpiro, R. Marquez, N. Morrice, J. Bain, E. Espel, and C. G. Proud. 2005. The Mnks are novel components in the control of TNF alpha biosynthesis and phosphorylate and regulate hnRNP A1. Immunity 23177-189. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhuri, S., K. Vyas, P. Kapasi, A. A. Komar, J. D. Dinman, S. Barik, and B. Mazumder. 2007. Human ribosomal protein L13a is dispensable for canonical ribosome function but indispensable for efficient rRNA methylation. RNA 132224-2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon, D. A., G. C. Balch, N. Kedersha, P. Anderson, G. A. Zimmerman, R. D. Beauchamp, and S. M. Prescott. 2003. Regulation of cyclooxygenase-2 expression by the translational silencer TIA-1. J. Exp. Med. 198475-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 9514863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan, J., N. M. Heller, M. Gorospe, U. Atasoy, and C. Stellato. 2005. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur. Respir. J. 26933-947. [DOI] [PubMed] [Google Scholar]
- 8.Gabay, C., and I. Kushner. 1999. Acute-phase proteins and other systemic responses to inflammation. N. Engl. J. Med. 340448-454. [DOI] [PubMed] [Google Scholar]
- 9.Gerard, C., and B. J. Rollins. 2001. Chemokines and disease. Nat. Immunol. 2108-115. [DOI] [PubMed] [Google Scholar]
- 10.Han, J., and R. J. Ulevitch. 2005. Limiting inflammatory responses during activation of innate immunity. Nat. Immunol. 61198-1205. [DOI] [PubMed] [Google Scholar]
- 11.Hartman, S. E., P. Bertone, A. K. Nath, T. E. Royce, M. Gerstein, S. Weissman, and M. Snyder. 2005. Global changes in STAT target selection and transcription regulation upon interferon treatments. Genes Dev. 192953-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Havgaard, J. H., R. B. Lyngso, G. D. Stormo, and J. Gorodkin. 2005. Pairwise local structural alignment of RNA sequences with sequence similarity less than 40%. Bioinformatics 211815-1824. [DOI] [PubMed] [Google Scholar]
- 13.Havgaard, J. H., E. Torarinsson, and J. Gorodkin. 2007. Fast pairwise structural RNA alignments by pruning of the dynamical programming matrix. PLoS Comput. Biol. 31896-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofacker, I. L. 2003. Vienna RNA secondary structure server. Nucleic Acids Res. 313429-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johannes, G., M. S. Carter, M. B. Eisen, P. O. Brown, and P. Sarnow. 1999. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc. Natl. Acad. Sci. USA 9613118-13123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapasi, P., S. Chaudhuri, K. Vyas, D. Baus, A. A. Komar, P. L. Fox, W. C. Merrick, and B. Mazumder. 2007. L13a blocks 48S assembly: role of a general initiation factor in mRNA-specific translational control. Mol. Cell 25113-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katsanou, V., O. Papadaki, S. Milatos, P. J. Blackshear, P. Anderson, G. Kollias, and D. L. Kontoyiannis. 2005. HuR as a negative posttranscriptional modulator in inflammation. Mol. Cell 19777-789. [DOI] [PubMed] [Google Scholar]
- 18.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8533-543. [DOI] [PubMed] [Google Scholar]
- 19.Lambert, M. P., B. S. Sachais, and M. A. Kowalska. 2007. Chemokines and thrombogenicity. Thromb. Haemost. 97722-729. [DOI] [PubMed] [Google Scholar]
- 20.Libby, P. 2002. Inflammation in atherosclerosis. Nature 420868-874. [DOI] [PubMed] [Google Scholar]
- 21.Lopez de Silanes, I., S. Galban, J. L. Martindale, X. Yang, K. Mazan-Mamczarz, F. E. Indig, G. Falco, M. Zhan, and M. Gorospe. 2005. Identification and functional outcome of mRNAs associated with RNA-binding protein TIA-1. Mol. Cell. Biol. 259520-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mazumder, B., and P. L. Fox. 1999. Delayed translational silencing of ceruloplasmin transcript in gamma interferon-activated U937 monocytic cells: role of the 3′ untranslated region. Mol. Cell. Biol. 196898-6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazumder, B., C. K. Mukhopadhyay, A. Prok, M. K. Cathcart, and P. L. Fox. 1997. Induction of ceruloplasmin synthesis by IFN-gamma in human monocytic cells. J. Immunol. 1591938-1944. [PubMed] [Google Scholar]
- 24.Mazumder, B., P. Sampath, V. Seshadri, R. K. Maitra, P. E. DiCorleto, and P. L. Fox. 2003. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 115187-198. [DOI] [PubMed] [Google Scholar]
- 25.Mazumder, B., V. Seshadri, H. Imataka, N. Sonenberg, and P. L. Fox. 2001. Translational silencing of ceruloplasmin requires the essential elements of mRNA circularization: poly(A) tail, poly(A)-binding protein, and eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 216440-6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller, M. T., J. J. Higgin, and T. M. Tanaka Hall. 2008. Basis of altered RNA-binding specificity by PUF proteins revealed by crystal structures of yeast Puf4p. Nat. Struct. Mol. Biol. 15397-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monajemi, H., R. D. Fontijn, H. Pannekoek, and A. J. Horrevoets. 2002. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 79539-546. [DOI] [PubMed] [Google Scholar]
- 28.Mukhopadhyay, D., C. W. Houchen, S. Kennedy, B. K. Dieckgraefe, and S. Anant. 2003. Coupled mRNA stabilization and translational silencing of cyclooxygenase-2 by a novel RNA binding protein, CUGBP2. Mol. Cell 11113-126. [DOI] [PubMed] [Google Scholar]
- 29.Osterud, B., and E. Bjorklid. 2003. Role of monocytes in atherogenesis. Physiol. Rev. 831069-1112. [DOI] [PubMed] [Google Scholar]
- 30.Piecyk, M., S. Wax, A. R. Beck, N. Kedersha, M. Gupta, B. Maritim, S. Chen, C. Gueydan, V. Kruys, M. Streuli, and P. Anderson. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 194154-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pradet-Balade, B., F. Boulme, H. Beug, E. W. Mullner, and J. A. Garcia-Sanz. 2001. Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci. 26225-229. [DOI] [PubMed] [Google Scholar]
- 32.Ray, P. S., and P. L. Fox. 2007. A post-transcriptional pathway represses monocyte VEGF-A expression and angiogenic activity. EMBO J. 263360-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampath, P., B. Mazumder, V. Seshadri, and P. L. Fox. 2003. Transcript-selective translational silencing by gamma interferon is directed by a novel structural element in the ceruloplasmin mRNA 3′ untranslated region. Mol. Cell. Biol. 231509-1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sampath, P., B. Mazumder, V. Seshadri, C. A. Gerber, L. Chavatte, M. Kinter, S. M. Ting, J. D. Dignam, S. Kim, D. M. Driscoll, and P. L. Fox. 2004. Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell 119195-208. [DOI] [PubMed] [Google Scholar]
- 35.Serhan, C. N., N. Chiang, and T. E. Van Dyke. 2008. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 8349-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serhan, C. N., and J. Savill. 2005. Resolution of inflammation: the beginning programs the end. Nat. Immunol. 61191-1197. [DOI] [PubMed] [Google Scholar]
- 37.Superarray, Inc. 2007. Oligo GEArray system manual, version 3.2. Superarray Inc., Frederick, MD.
- 38.Tamayo, P., D. Slonim, J. Mesirov, Q. Zhu, S. Kitareewan, E. Dmitrovsky, E. S. Lander, and T. R. Golub. 1999. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc. Natl. Acad. Sci. USA 962907-2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson, J. F., L. S. Hayes, and D. B. Lloyd. 1991. Modulation of firefly luciferase stability and impact on studies of gene regulation. Gene 103171-177. [DOI] [PubMed] [Google Scholar]
- 40.Torarinsson, E., J. H. Havgaard, and J. Gorodkin. 2007. Multiple structural alignment and clustering of RNA sequences. Bioinformatics 23926-932. [DOI] [PubMed] [Google Scholar]
- 41.Weber, C., and R. R. Koenen. 2006. Fine-tuning leukocyte responses: towards a chemokine “interactome”. Trends Immunol. 27268-273. [DOI] [PubMed] [Google Scholar]
- 42.Zlotnik, A., O. Yoshie, and H. Nomiyama. 2006. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome Biol. 7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zong, Q., M. Schummer, L. Hood, and D. R. Morris. 1999. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc. Natl. Acad. Sci. USA 9610632-10636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.