Abstract
p27Kip1 is a cyclin-dependent kinase inhibitor that regulates the G1/S transition. Increased degradation of p27Kip1 is associated with cellular transformation. Previous work demonstrated that the ubiquitin ligases KPC1/KPC2 and SCFSkp2 ubiquitinate p27Kip1 in G1 and early S, respectively. The regulation of these ligases remains unclear. We report here that the USP19 deubiquitinating enzyme interacts with and stabilizes KPC1, thereby modulating p27Kip1 levels and cell proliferation. Cells depleted of USP19 by RNA interference exhibited an inhibition of cell proliferation, progressing more slowly from G0/G1 to S phase, and accumulated p27Kip1. This increase in p27Kip1 was associated with normal levels of Skp2 but reduced levels of KPC1. The overexpression of KPC1 or the use of p27−/− cells inhibited significantly the growth defect observed upon USP19 depletion. KPC1 was ubiquitinated in vivo and stabilized by proteasome inhibitors and by overexpression of USP19, and it also coimmunoprecipitated with USP19. Our results identify USP19 as the first deubiquitinating enzyme that regulates the stability of a cyclin-dependent kinase inhibitor and demonstrate that progression through G1 to S phase is, like the metaphase-anaphase transition, controlled in a hierarchical, multilayered fashion.
The ubiquitin proteasome pathway plays essential roles in regulating the cell cycle. The best-defined functions of this pathway in cell cycle regulation are those mediated by the multisubunit ubiquitin protein ligases SKP1-CUL1-F-box (SCF) and the anaphase-promoting complex/cyclosome (APC) (reviewed in reference 25). The functions of the APC in the cell cycle are predominant at the mitosis-anaphase transition, while the activities of SCF-type ubiquitin protein ligase complexes are involved at various steps of the cycle (reviewed in references 21 and 25). One of the best-defined functions of the SCF is mediated by SCFskp2, which plays a vital role in regulating the G1-S transition by ubiquitinating the cyclin-dependent kinase inhibitor p27Kip1, thereby targeting it for degradation by the proteasome (3, 31, 32).
The central role of p27Kip1 in restricting cell proliferation is demonstrated by the fact that mice lacking the p27Kip1 gene manifest increased body and organ weights and develop pituitary adenomas (6, 13, 18). In addition, the results of clinical studies suggest that low p27Kip1 levels are associated with increased aggressivity of tumors (1, 28). Unlike the case with the p53 or Rb tumor suppressors, mutation or deletion of p27Kip1 in tumors is rare. Rather, its deregulation in human tumors is due mainly to reduced protein levels, mediated in large part by increased proteolysis (reviewed in reference 21). In support of this, the low p27Kip1 levels seen in tumors are associated with increased levels of Skp2, the substrate recognition subunit of the SCFskp2 ligase. The loss of Skp2 in mice results in p27Kip1 accumulation, and cells from Skp2−/− animals contain enlarged nuclei with polyploidy and multiple centrosomes. They also show a reduced growth rate and increased apoptosis (19). Many of the cellular phenotypes observed in Skp2−/− mice disappear in Skp2−/− p27−/− double-mutant mice (14, 20). Thus, the oncogenic nature of Skp2 is largely due to its ability to mediate p27Kip1 degradation.
In spite of the clear role of SCFskp2 in mediating the ubiquitination and degradation of p27Kip1, the downregulation of this cyclin-dependent kinase inhibitor proceeds normally in lymphocytes isolated from Skp2−/− mice (8, 21). In addition, in the normal cell cycle, p27Kip1 is also degraded in G1, before the expression of Skp2, which occurs in early S phase (8, 9, 36). Also, p27Kip1 is exported from the nucleus to the cytoplasm in G1, whereas Skp2 is localized in the nucleus (9, 26). These observations suggest the existence of another pathway for the degradation of p27Kip1. Indeed, KPC (Kip1 ubiquitination-promoting complex) was subsequently identified as a novel cytoplasmic ligase complex that interacts with and ubiquitinates p27Kip1 (10). KPC consists of two subunits: KPC1, a 140-kDa RING-finger domain-containing protein, and KPC2, a 50-kDa protein containing a ubiquitin-like domain and two ubiquitin-associated domains.
Although the role of ubiquitin protein ligases in the cell cycle has received considerable attention, fewer data are available regarding the roles of deubiquitinating enzymes in the cell cycle (22). We recently described USP19 as a deubiquitinating enzyme that is induced in skeletal muscle atrophying in response to numerous catabolic stimuli. To study its function, we used RNA interference to explore the consequences of depletion of this enzyme in cultured muscle cells. Our early studies indicated that the loss of USP19 interfered with the growth of L6 myoblasts. We have observed similar effects in FR3T3 fibroblasts and have explored the underlying mechanisms of this growth defect.
MATERIALS AND METHODS
Reagents.
Cell culture media and Lipofectamine were obtained from Invitrogen (Burlington, Ontario, Canada). Propidium iodide, RNase A, anti-Flag, antitubulin, and antihemagglutinin epitope antibodies were obtained from Sigma, and anti-p27 antibodies from Cell Signaling. Anti-p21, p16, and Skp2 antibodies were from Santa Cruz Biotechnology; anti-histone H2A was from Upstate Biotechnologies; and annexin V-Flous from Roche (Laval, Quebec, Canada). Anti-USP19 antibodies were generated by immunizing rabbits with a fragment of USP19 containing the first 129 amino acids of USP19. The antibodies were affinity purified from the serum by using agarose beads coupled to glutathione-S-transferase fused to the first 44 amino acids of USP19.
Cell culture and transfection.
Rat L6 myoblasts, FR3T3, and mouse embryonic fibroblasts (MEF) were cultured in Dulbecco's modified Eagle's medium (DMEM; high glucose) supplemented with 10% fetal bovine serum (FBS) and antibiotic-antimycotic preparations (Invitrogen), unless otherwise indicated. For typical transfection of small interference RNA (siRNA) oligonucleotides, 50,000 FR3T3 cells or 20,000 MEF cells were seeded into six-well plates. Transfection of siRNA oligonucleotides was carried out the next day when cells were ∼30% confluent by incubating oligonucleotides with cells in the presence of Opti-MEM1 medium and Lipofectamine for 5 h. siRNA oligonucleotides were designed against the following USP19 coding sequences: rat siRNA, no. 1, AAAGTGCAGACTCACAAGGGT; no. 7, AAGGGTGGTCTTCTACAGTTG; no. 8, AAACCTCTAGGGACCCAAGAA; no. 43, ACCAAGTCATTGATCTTGTCACGCCAA; no. 44, CATACGAATCATTCTTGACAAATGCCA; and no. 47, GAACTCTTGGGCATCATGCTGTGCGTA, and mouse siRNA, no. m2, CCAGGTTCACCATAGACTCTGGTTGGT, and no. m4, CAGGTCATAGCTAGGCAGCTGCTCCTC. The sequences used for designing the nonspecific control and the scrambled version of no. 7 control oligonucleotides were GGTTCATGGTCTGACTGGT and ACTCTATCTGCACGCTGACTT, respectively. The siRNA oligonucleotides were from Dharmacon (Chicago, IL) or Integrated DNA Technologies (Coralville, IA). Pilot studies established that oligonucleotides at a concentration of 25 nM exerted the maximal effect in reducing USP19 levels without any detectable nonspecific toxicity.
For transient transfections, plasmids expressing hemagglutinin (HA)-tagged KPC1, Flag-USP19, or His6-ubiquitin were transfected into FR3T3 cells at ∼50% confluence. After 48 h, cells were lysed and protein analyzed by immunoblotting with the indicated antibodies. For detection of ubiquitinated KPC1, transfected FR3T3 cells in 10-cm-diameter plates were incubated with 10 μM MG132 6 h prior to lysis. Cells were lysed in 1 ml buffer A (6 M guanidine-HCl, 0.1 M Na2 HPO4-NaH2PO4, 10 mM imidazole, pH 8.0). Following sonication of the cell suspension, the lysate was mixed with 50 μl of Ni-nitrilotriacetic acid (NTA)-agarose beads (50% slurry) (Qiagen) for 3 h at room temperature. The Ni-NTA-agarose beads were washed twice with 1 ml of buffer A and twice with 1 ml of buffer A-TI (1 volume buffer A and 3 volumes buffer TI [25 mM Tris-HCl, 20 mM imidazole, pH 6.8]). Proteins were eluted with 6× Laemmli sample buffer, boiled for 10 min, and analyzed by immunoblotting.
To create stable cell lines, retroviruses were produced by transfecting 293VSV cells with either empty pLXSN plasmid (Clontech, Mountain View, CA) or the same plasmid encoding N-terminally Flag3-tagged USP19 or N-terminally HA-tagged KPC1. Silent mutations were also introduced into USP19 in the region targeted by siRNA oligonucleotide no. 7 (nucleotides 355 to 375) to render the derived mRNA transcripts resistant to this siRNA. Following the transduction of FR3T3 cells with viruses encoding the above proteins, stable cell lines were selected by growing the cells in G418 (400 μg/ml). To measure half-lives of KPC1 or p27, cells were incubated with cycloheximide (100 μg/ml). At various times, cells were harvested, protein prepared from the lysates, and the levels of KPC1 or p27 quantitated by immunoblotting.
Cell cycle synchronization and analysis of apoptosis.
FR3T3 cells were cultured in DMEM for 72 h in the absence of serum to synchronize cells in G0. Thirty-six hours after the start of serum deprivation, cells were transfected with USP19 or control siRNA oligonucleotides. Thirty-six hours later, cells were stimulated to reenter cell cycle by replacing the medium with DMEM supplemented with 20% FBS and harvested at various times. The cell profile was then analyzed by fluorescence-activated cell sorting (FACS) (FACSCalibur; BD Bioscience), and the acquired data analyzed by using Cellquant software. Apoptotic cells were identified by staining cells with propidium iodide fluorescently labeled annexin V (Roche) according to the manufacturer's instructions.
Cell fractionation.
To separate cells into nuclear and cytoplasmic fractions, cell pellets were resuspended in 1 ml of ice-cold 10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.5% NP-40. The cell lysates were incubated on ice for 5 min with brief vortexing at low speed each minute. After centrifugation in a microfuge at 2,500 rpm for 3 min at 4°C, the supernatant containing cytosol was transferred to a new tube and the pellet containing nuclei was resuspended in 300 μl of ice-cold 50 mM Tris, pH 7.4, 5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, and 40% (wt/vol) glycerol for subsequent immunoblotting.
Northern blot analysis.
RNA was isolated from FR3T3 cells by using Trizol reagent (Invitrogen). RNA (15 μg) was electrophoresed on a 1% agarose gel, transferred to a nylon membrane, and hybridized with KPC1 or 18S rRNA probes.
Immunoblotting and IP.
For immunoblotting, cells were harvested in a denaturing buffer (50 mM Tris, pH 7.5, 2.5% sodium dodecyl sulfate [SDS], 1 mM dithiothreitol). Following determination of protein concentration with a bicinchoninic acid protein assay kit (Pierce), equal amounts of proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were transferred onto polyvinylidene difluoride or nitrocellulose membranes, which were then probed with the indicated antibodies. Immunoblots were visualized by chemiluminescence using an ECL detection system (Amersham) and quantitated by using a cooled digital camera and Quantity One software (Bio-Rad). For immunoprecipitation (IP), anti-Flag (anti-M2, 2.5 μg) antibody was incubated with 100 μl of 50% protein G beads (Santa Cruz) and 1 μg of anti-HA antibody or 3 μg of anti-KPC1 antibody was incubated with 100 μl of 50% protein A beads (GE Healthcare) in 0.5 ml IP buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% NP-40) overnight at 4°C. Cell extracts (0.5 to 1 mg of protein lysed in IP buffer) were then incubated with the antibody-protein A/G beads and ubiquitin aldehyde (10 μg/ml) in a 0.5-ml total volume for 3 h at 4°C. The inclusion of ubiquitin aldehyde was found to stabilize the USP19 during the IP. After five washes of the beads with 0.5 ml of cold IP buffer, the bound proteins were eluted with 6× Laemmli sample buffer, boiled, and processed for immunoblotting.
Analysis of immunoisolated KPC1 by mass spectrometry.
FR3T3 cells were transfected with plasmid expressing HA-KPC1 or empty vector plasmid. Forty-three hours later, 15 μM MG132 was added to the medium for 5 h to accumulate ubiquitinated proteins. Cells were harvested by scraping, lysed with two pellet volumes of 2% SDS in 50 mM Tris (pH 8.0), and boiled for 10 min. DNA was sheared by passing the lysate through a syringe. After the addition of 10 volumes of 1% Triton X-100 in 50 mM Tris (pH 8.0) to lower the concentration of SDS, 34 mg of lysate were cleared by ultracentrifugation at 100,000 × g for 1 h at 4°C. The supernatant was precleared with rabbit immunoglobulin G (15 μg) preincubated with 400 μl of 25% protein A-agarose beads. HA-KPC1 was then immunoprecipitated by using 15 μg of anti-HA antibody-protein A beads for 5.5 h at 4°C. After the beads were washed five times with normal IP buffer, the samples were eluted in 140 μl of loading buffer containing 1% SDS and concentrated to 70 μl by using a Speed-Vac concentrator. The eluates from cells transfected with KPC1 expressing plasmid or empty vector control were electrophoresed on a 9% SDS gel and stained with Coomassie blue G-250. Three gel bands were excised from each gel lane (∼100 to 200 kDa, 200 to 300 kDa, and >300 kDa), followed by in-gel trypsin digestion. The tryptic peptides were extracted and analyzed by liquid chromatography coupled with tandem mass spectrometry using an LTQ-Orbitrap mass spectrometer (Thermo Finnigan, San Jose, CA) as previously described (27). We only accepted proteins identified by at least two peptides after database search and stringent filtering. The relative abundance of the proteins was evaluated by assigned spectral counts (the number of spectra matched to the proteins) after normalization by protein size (abundance index = [spectral counts/protein mass] × 50 kDa, assuming the average protein size is 50 kDa) (23).
RESULTS
Cells depleted of USP19 have reduced rates of growth.
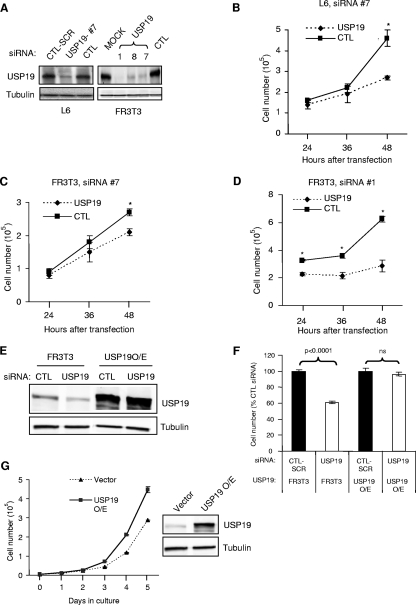
To obtain insights into the physiological functions of USP19, we depleted L6 muscle cells of this enzyme by using siRNA oligonucleotides. Forty-eight hours after transfection, the USP19 protein levels were suppressed by ∼80% in L6 cells transfected with USP19 siRNA oligonucleotide no. 7 (Fig. 1A). In these USP19-depleted cells, a marked reduction in cell number was observed compared to the growth of cells transfected with a nonspecific control siRNA that is predicted not to target any rodent gene (Fig. 1B). To determine whether these effects on cell growth of silencing USP19 can be observed in nonmuscle cells, rat FR3T3 fibroblasts were similarly treated with USP19 siRNA. Reduced cell growth was also seen in cells depleted of USP19 but not in cells treated with nonspecific or scrambled siRNA control oligonucleotides (Fig. 1A and C).
FIG. 1.
Depletion of USP19 leads to reduced cell proliferation. (A) Effectiveness of USP19 siRNA oligonucleotides in depleting the enzyme. L6 myoblasts and FR3T3 fibroblasts were transfected with USP19 or scrambled (CTL-SCR) or nonspecific control siRNA oligonucleotides (CTL) or transfected without siRNA oligonucleotides (MOCK). Forty-eight hours after transfection, cells were harvested and the lysate subjected to immunoblot analysis with antibody against either USP19 or γ-tubulin. #7, 1, 8, and 7, USP19 siRNA oligonucleotides. (B to D) Cells depleted of USP19 show reduced proliferation. L6 myoblasts (B) or FR3T3 fibroblasts (C, D) were transfected with USP19 siRNA oligonucleotide no. 7 (#7) (B and C) or no. 1 (#1) (D) or nonspecific or scrambled control (CTL) siRNA oligonucleotides. Cell numbers were determined at the indicated times after transfection. Shown are means ± standard deviations of the results. *, significantly different from results for control oligonucleotide transfection (P < 0.001). (E) Normal FR3T3 cells or those stably overexpressing (O/E) USP19 bearing mutations that do not change the coding sequence but render the mRNA resistant to siRNA-mediated degradation were transfected with USP19 siRNA or scrambled siRNA (CTL). After 48 h, cells were harvested and lysates prepared and analyzed by immunoblotting. (F) Normal proliferation in cells expressing USP19 resistant to siRNA. Cells were treated as described for panel E and counted at 48 h after transfection with the indicated siRNA oligonucleotides. Shown are means ± standard errors of the results. (G) Expression of exogenous USP19 increases the rate of cell proliferation. FR3T3 cells stably overexpressing Flag-tagged USP19 were cultured for the indicated number of days. Vector-transfected FR3T3 cells were used as control. Shown are means ± standard errors of the results.
To minimize the likelihood that our observations were due to off-target effects, we designed additional siRNA oligonucleotides (no. 1 and no. 8) and showed that these siRNAs, like siRNA no. 7, were effective in reducing USP19 protein levels (Fig. 1A) and also led to reductions in cell numbers (Fig. 1D and data not shown). The levels of USP19 were most effectively depleted by oligonucleotide no. 1, and this oligonucleotide also had the most-dramatic effect on cell number (Fig. 1D). Finally, the stable expression of a form of USP19 in which the sequence targeted by the siRNA oligonucleotide was mutated with bases that do not alter the coding sequence resulted in a cell line in which USP19 levels did not fall (Fig. 1E) and in which growth was no longer inhibited upon siRNA depletion (Fig. 1F), confirming that these effects were specific to USP19 depletion. Since depletion of USP19 inhibited cell growth, we determined whether overexpression of USP19 would enhance cell growth. Indeed, clones that overexpressed USP19 showed increased rates of cell proliferation compared to the rates of cell proliferation of FR3T3 cells transfected with empty vector (Fig. 1G). These results confirmed the ability of USP19 to modulate cell growth.
Depletion of USP19 results in defects in cell cycle progression.
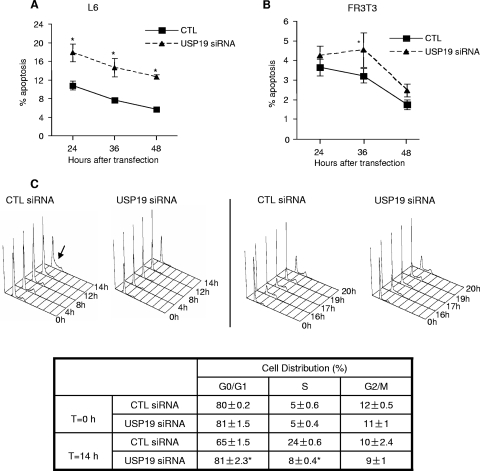
The reduction in cell number in response to USP19 depletion may be due to defects in cell cycle and/or increased rates of apoptosis. We therefore examined whether USP19 depletion led to increased apoptosis by quantifying the cells that stained with labeled annexin V. The percentage of apoptotic cells was higher in USP19-depleted cells, but overall, the rates of apoptosis were small, particularly for FR3T3 cells (Fig. 2A and B). Consistent with the effect on apoptosis being minor, there were no detectable effects of USP19 depletion on the levels of other markers of apoptosis, such as poly(ADP-ribose) polymerase or activated caspase-3 (data not shown). Neither the low basal rate of apoptosis nor the increase in apoptotic cells can explain the up-to-twofold difference in cell number observed 48 h after transfection (Fig. 1B to D). We conclude that upon USP19 depletion, apoptosis plays a minor role in the reduction in cell growth.
FIG. 2.
Cells depleted of USP19 show only a small increase in apoptosis but impaired progression from G0 to S phase. (A, B) Apoptosis is mildly increased in USP19-depleted L6 myoblasts or FR3T3 fibroblasts. Cells were treated with the indicated siRNA oligonucleotides. At the indicated times, cells were harvested and analyzed by FACS to quantify apoptotic cells (identified as annexin V positive, propidium iodide negative). *, significantly different from results for control at the indicated time points (P < 0.001). (C) FR3T3 cells depleted of USP19 showed delayed entry into S phase. Cells were serum starved for 36 h. They were then transfected with scrambled control (CTL) or USP19 siRNA oligonucleotides. Following another 36 h of serum starvation, cells were stimulated to reenter cell cycle with DMEM containing 20% FBS. Cells were harvested at the indicated times following serum stimulation and subjected to FACS analysis. Arrow indicates entry into S phase seen in CTL cells at 14 h (left) and at ∼17 h in cells transfected with USP19 siRNA oligonucleotides (right). At 17 h, CTL cells were starting to reenter G1. Table shows percentages of cells in different phases of the cell cycle before and 14 h after release from G0. *, significantly different (P < 0.001) from results for CTL. (D) p27Kip1 accumulates in USP19-depleted cells. Equal amounts of protein from lysates (from experiment described in panel C) were analyzed by immunoblotting with the indicated antibodies. Shown are a representative blot and quantitation of results for multiple samples. Values for USP19 siRNA-treated cells are significantly different from those for CTL-treated cells. #, P < 0.001; two-way analysis of variance. (E) FR3T3 cells overexpressing USP19 enter S phase earlier. Cells stably transfected with plasmid overexpressing USP19 or an empty vector (control) were starved of serum for 72 h and then stimulated to reenter cell cycle with DMEM containing 20% FBS. Cells were harvested at the indicated times following serum stimulation and subjected to FACS analysis as described for panel C above. Means ± standard errors of the results are shown. Asterisks indicate means that are significantly different from those for control cells (*, P < 0.01; **, P < 0.001). WT, wild type.
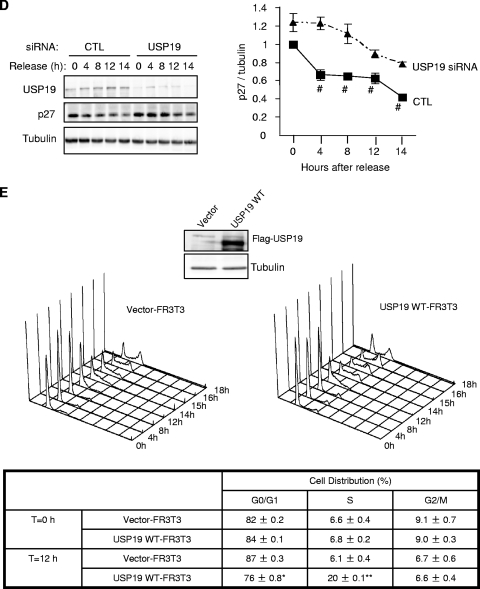
We therefore determined if USP19 had any effect in regulating the cell cycle. FACS analysis of unsynchronized USP19-depleted cells did not reveal any significant changes in the distribution of cells among the various stages of the cell cycle (data not shown). To examine more precisely the role of this enzyme in cell cycle progression, we synchronized FR3T3 cells in G0 by serum starvation and concomitantly transfected them with control or USP19 siRNA oligonucleotides. Both USP19-depleted cells and those that were not depleted exited cell cycle and synchronized in G0 at similar rates (Fig. 2C and data not shown). Cells were stimulated with serum to reenter the cell cycle and harvested at various times for analysis. In cells transfected with control siRNA oligonucleotides, cells entered into S phase beginning at 12 to 14 h (Fig. 2C) and showed the expected decreases in p27Kip1 levels (Fig. 2D). However, in USP19-depleted cells, the p27Kip1 levels were significantly elevated compared to the levels in cells that were not depleted (Fig. 2D), and there was a delay in entry into S phase until 17 h (Fig. 2C). At 17 h, cells transfected with control siRNA oligonucleotides had largely completed the cycle and reentered G1 (Fig. 2C). We then tested whether overexpression of USP19 would result in enhanced entry into S phase. Cells stably overexpressing USP19 (Fig. 1G) or vector-transfected control cells were synchronized by serum starvation and stimulated to reenter the cell cycle as described above. Indeed, the USP19-overexpressing cells entered S phase at 12 h, earlier than control cells which entered S phase at 14 h (Fig. 2E).
p27Kip1 accumulates specifically in USP19-depleted cells.
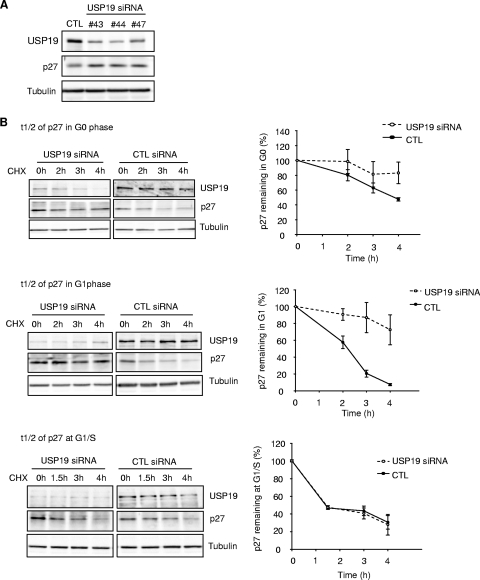
We observed that p27Kip1 levels were increased even in asynchronously dividing cells depleted of USP19. This effect was seen with multiple independent oligonucleotides, making it unlikely that this was due to an off-target effect (Fig. 3A). To confirm that the increase in p27Kip1 levels was due to a decrease in the rate of degradation, the rate of disappearance of p27Kip1 was measured in USP19-depleted cells treated with cycloheximide. Since p27Kip1 can be degraded during both G1 and early S phase, but by different mechanisms, the half-lives of the protein were measured in cells that were synchronized in G0, G1, and at the G1/S transition (Fig. 3B). In cells transfected with control oligonucleotides, p27Kip1 was degraded relatively slowly in G0, with a half-life of approximately 4 h. In G1, the rate of degradation was accelerated, and it was further accelerated at the G1/S transition. Cells transfected with USP19 siRNA showed a mild stabilization of p27Kip1 in G0 and more-marked stabilization in G1 but showed no effect on the rate of degradation in G1/S. This indicates that USP19 acts on the process responsible for p27Kip1 degradation in G0 and G1 but not on the process involved at the G1/S transition. To determine whether this effect was specific to p27Kip1, we tested the effect of USP19 depletion on the levels of two other cyclin-dependent kinase inhibitors, p21 and p16. Neither p21 nor p16 was affected, suggesting a specific effect on p27Kip1 (Fig. 3C). Since SCFSkp2 is the best-known ubiquitin protein ligase for p27Kip1, we tested whether depletion of USP19 decreased Skp2 levels. No changes were observed in Skp2 expression (Fig. 3C). This and the fact that SCFSkp2 acts primarily on p27Kip1 in early S phase, at which time silencing USP19 did not result in stabilization of p27Kip1, suggested that the stabilization of p27Kip1 in USP19-depleted cells was due to a factor(s) other than Skp2.
FIG. 3.
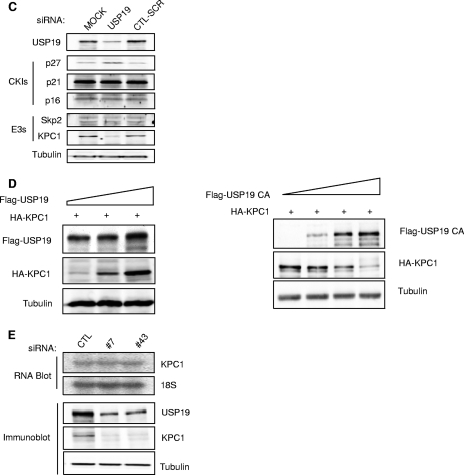
Levels of the cyclin-dependent kinase inhibitor (CKI) p27Kip1 and the ubiquitin protein ligase (E3) KPC1 are modulated by USP19. (A) Multiple independent USP19 siRNA oligonucleotides (#43, no.43; #44, no.44; #47, no. 47) increase p27Kip1 levels in asynchronous FR3T3 cells. (B) Depletion of USP19 decreases the rate of degradation of p27Kip1 in G0 and G1 phases but not at the G1/S transition. FR3T3 cells transfected with USP19 no. 7 siRNA or scrambled control oligonucleotides were synchronized in G0 phase as described for Fig. 2. Cells were stimulated to reenter the cell cycle with DMEM medium containing 10% FBS and 2 mM thymidine (to accumulate cells at G1/S). At 7 h (G1) or 12.5 h (G1/S, 15 h for USP19 siRNA-transfected cells which progressed more slowly) of stimulation (Fig. 2C) (aliquots of cells were analyzed by FACS to confirm positions in cell cycle), the cells were incubated with cycloheximide to block protein synthesis and measure the rate of degradation of p27Kip1. At the indicated times, cell protein was analyzed by immunoblotting with the indicated antibodies. Shown are representative immunoblots and quantitation of p27Kip1 levels from triplicate samples. Rates of degradation of p27Kip1 were significantly inhibited in USP19 siRNA-treated cells in G0 (P < 0.05) and G1 phases (P < 0.001) but not in S phase (not significant) (all analyses were by two-way analysis of variance). t1/2, half-life; CHX, cycloheximide. (C) Levels of KPC1 but not Skp2 are decreased in USP19-depleted cells. Proliferating FR3T3 cells were treated with scrambled control or USP19 siRNA oligonucleotides. Cells were harvested 48 h later, and lysates subjected to immunoblot analysis with the indicated antibodies. Levels of p27Kip1 but not p21 or p16 are increased upon USP19 depletion. Levels of KPC1 but not Skp2 are decreased upon USP19 depletion. (D) USP19 modulates levels of KPC1. FR3T3 cells were transfected with plasmid expressing HA-KPC1 (+) and increasing amounts of a plasmid expressing Flag-USP19 (left) or catalytically inactive Flag-USP19 C545A mutant and His-tagged ubiquitin (right). Proteins from cell extracts were analyzed by immunoblotting with anti-Flag (USP19), anti-HA (KPC1), and anti-γ-tubulin antibodies. (E) Silencing of USP19 does not lower KPC1 levels by lowering mRNA levels. FR3T3 cells were transfected with USP19 siRNA oligonucleotides no. 7 (#7) or no. 43 (#43) or control oligonucleotides (CTL). Two days later, RNA or protein was isolated and analyzed by RNA blot analysis (top) or by immunoblotting (below), respectively.
USP19 stabilizes KPC1 and, like KPC1, is localized in the cytosol.
KPC has been shown to be another ligase for p27 and responsible for targeting cytoplasmic p27Kip1 for degradation in G1 (10). Therefore, we tested if the effects of depletion of USP19 on p27Kip1 might be due to modulation of KPC1 levels. Indeed, USP19 depletion led to a marked reduction in KPC1 levels (Fig. 3C). Conversely, we also observed that cells transfected with increasing amounts of USP19-expressing plasmid were able to stabilize cotransfected KPC1 in a dose-dependent manner (Fig. 3D). This stabilization required USP19 enzymatic activity, as transfection with plasmid expressing a catalytically inactive USP19 mutant, in which the active-site cysteine had been mutated to alanine, had the opposite effect and destabilized KPC1 (Fig. 3D). Thus, USP19 can regulate the level of KPC1. In contrast, depletion of USP19 did not alter the levels of KPC2, the noncatalytic subunit of the KPC1/KPC2 ligase complex (data not shown). The decrease in KPC1 induced by USP19 depletion did not appear to be due to a decrease in gene transcription, as KPC1 mRNA levels did not change (Fig. 3E).
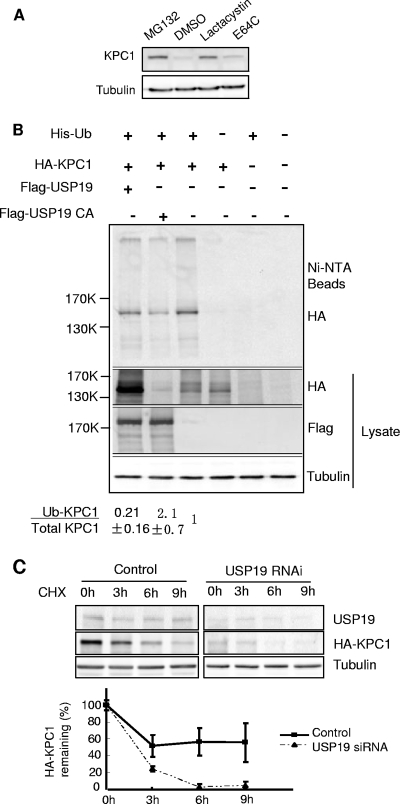
The simplest model to explain these observations would be that USP19 stabilizes KPC1 by deubiquitinating it and protecting it from ubiquitin-proteasome-dependent degradation. Therefore, we examined whether KPC1 is degraded by the ubiquitin-proteasome pathway by treating cells with proteasome inhibitors. Indeed, KPC1 accumulated in the presence of the proteasome inhibitors lactacystin and MG132 but not in cells treated with the cysteine protease inhibitor E64c (Fig. 4A). We then tested whether KPC1 is ubiquitinated in vivo. Following the overexpression of His-tagged ubiquitin and HA-tagged KPC1, polyubiquitinated KPC1 was detected among the ubiquitinated proteins isolated by Ni+2 affinity chromatography (Fig. 4B). Furthermore, when USP19 was simultaneously overexpressed, there was a decrease in the proportion of KPC1 that was ubiquitinated and an overall stabilization of KPC1. Conversely, the simultaneous overexpression of a catalytically inactive form of USP19, in which the active-site cysteine was mutated to alanine, resulted in destabilization of KPC1 but an increase in the proportion of KPC1 that was ubiquitinated. To confirm these findings, we immunoisolated KPC1 under denaturing conditions from FR3T3 cells transiently transfected with plasmid expressing KPC1, resolved the immunoprecipitate by SDS-PAGE, and subjected the gel lane to mass spectrometric analysis (Table 1). The same immunopurification and analysis was applied to cells transfected with empty vector as a negative control. In the control sample, we identified a few nonspecific proteins. In the immunoisolation from KPC1-expressing cells, we found that KPC1 was the dominant species distributed in all three gel bands, with high levels in the 100-to-200-kDa band (corresponding to its unmodified form and possibly monoubiquitinated species) and in the top gel band (more than ∼300 kDa). Ubiquitin was also identified in all three gel bands at a high level that exceeded the abundance of KPC1. All other proteins were present at very low levels and were mostly high-molecular-mass proteins, indicating that these were contaminants. The copresence of KPC1 and ubiquitin as the only abundant proteins in regions of the gel much above their native molecular mass clearly confirm that the KPC1 molecules are indeed ubiquitinated. However, we were not able to detect the ubiquitinated site(s) on KPC1, because the entire sequencing coverage of KPC1 in the liquid chromatography-tandem mass spectrometry analysis is low (∼20%) due to limited amounts of KPC1 in the initial sample.
FIG. 4.
KPC1 is a substrate of the ubiquitin-proteasome pathway and is stabilized by USP19. (A) KPC1 is stabilized by proteasome inhibitors. FR3T3 cells were treated with a proteasome inhibitor, either MG132 (10 μΜ) or lactacystin (5 μΜ), or with cysteine protease inhibitor E64C (10 μΜ) or vehicle (DMSO [dimethyl sulfoxide]) for 6 h. Cells were then harvested and blotted with anti-KPC1 antibody. (B) KPC1 is polyubiquitinated in vivo, and the ubiquitination is modulated by USP19. FR3T3 cells were transfected with the indicated plasmids. Cells were treated with MG132 for 6 h before lysis to accumulate ubiquitinated proteins. Lysate was either analyzed by immunoblotting with the indicated antibodies or incubated with Ni+-agarose beads. Bound ubiquitinated proteins were washed extensively and then eluted with Laemmli buffer and analyzed by immunoblotting with anti-HA antibody. Quantitation and ratios of the levels of ubiquitinated KPC1 (anti-HA blot of proteins bound to Ni+-NTA beads) to total KPC1 (anti-HA blot of lysate) (means ± standard errors) are indicated below. Means for cells overexpressing wild-type USP19 or the USP19CA mutant are significantly different (P value of <0.025 by one-way analysis of variance) from that for non-USP19-overexpressing cells, which was set at 1. Ub, ubiquitin; +, present; −, absent. (C) Silencing of USP19 destabilizes KPC1. FR3T3 cells stably overexpressing KPC1 were transfected with USP19 siRNA or control oligonucleotides. Forty-eight hours later, the cells were incubated with cycloheximide. At the indicated times, cell protein was analyzed by immunoblotting with the indicated antibodies. Shown are representative immunoblots and quantitation of KPC1 levels from triplicate samples. Error bars show standard errors. CHX, cycloheximide; RNAi, RNA interference.
TABLE 1.
Mass spectrometric analysis of KPC1 shows that it is ubiquitinated in FR3T3 cellsa
| Accession no. | Results for protein in indicated molecular mass range (kDa) of gel sample
|
MW | Annotation | |||||
|---|---|---|---|---|---|---|---|---|
| 100-200
|
200-300
|
>300
|
||||||
| No. of pept. | AI | No. of pept. | AI | No. of pept. | AI | |||
| KPC1-expressing cells | ||||||||
| NP_001029102.1 | 3 | 25 | 4 | 25 | 5 | 56 | 8 | Ubiquitin |
| NP_071347.2 | 28 | 17 | 20 | 11 | 26 | 17 | 148 | Ring finger protein 123 (KPC1) |
| NP_001605.1 | 0 | 0 | 0 | 0 | 3 | 3.6 | 42 | Actin, gamma 1 propeptide |
| NP_001393.1 | 0 | 0 | 3 | 3 | 3 | 3 | 50 | Eukaryotic translation elongation factor 1 alpha 1 |
| NP_001093241.1 | 0 | 0 | 0 | 0 | 2 | 0.8 | 121 | Protein encoded on chromosome 2, expressed in prostate, ovary, and testis |
| NP_001077007.1 | 0 | 0 | 0 | 0 | 2 | 0.8 | 121 | Protein expressed in prostate, ovary, testis, and placenta 2 |
| NP_001120959.1 | 0 | 0 | 2 | 0.3 | 3 | 0.5 | 287 | Gamma filamin isoform b |
| NP_001449.3 | 0 | 0 | 2 | 0.3 | 3 | 0.5 | 291 | Gamma filamin isoform a |
| NP_001448.2 | 0 | 0 | 0 | 0 | 2 | 0.4 | 278 | Filamin B, beta (actin binding protein 278) |
| NP_005518.3 | 0 | 0 | 2 | 1.4 | 0 | 0 | 70 | Heat shock 70 kDa protein 1-like |
| NP_000468.1 | 0 | 0 | 2 | 1.4 | 0 | 0 | 69 | Albumin preproprotein |
| NP_002464.1 | 0 | 0 | 5 | 1.1 | 0 | 0 | 226 | Myosin, heavy polypeptide 9, nonmuscle |
| NP_001933.2 | 2 | 0.9 | 0 | 0 | 0 | 0 | 114 | Desmoglein 1 preproprotein |
| Control cells | ||||||||
| XP_001721581.1 | 2 | 0.5 | 0 | 0 | 0 | 0 | 190 | Predicted to be similar to hornerin |
| NP_001014364.1 | 2 | 0.4 | 0 | 0 | 0 | 0 | 248 | Filaggrin family member 2 |
| NP_001009931.1 | 2 | 0.4 | 0 | 0 | 0 | 0 | 282 | Hornerin |
FR3T3 cells were transfected with plasmid expressing HA-tagged KPC1 or empty vector. Lysates were prepared under denaturing conditions and then subjected to immunoprecipitation with anti-HA antibodies. The eluates from the pellets were electrophoresed on 9% SDS-PAGE gels, and the indicated gel slices subjected to proteomic analysis as described in Materials and Methods. Shown are the number of peptides (pept.) identified for each protein and the abundance index (AI), which estimates the relative abundance by normalizing the number of spectra assigned to the protein to its molecular mass (see Materials and Methods for details).
Finally, we tested whether USP19 depletion would increase the rate of degradation of KPC1. Indeed, USP19 siRNA resulted in a more rapid rate of disappearance of KPC1 following the inhibition of protein synthesis with cycloheximide (Fig. 4C). Taken together, the results of these studies argue strongly that KPC1 is a substrate of USP19.
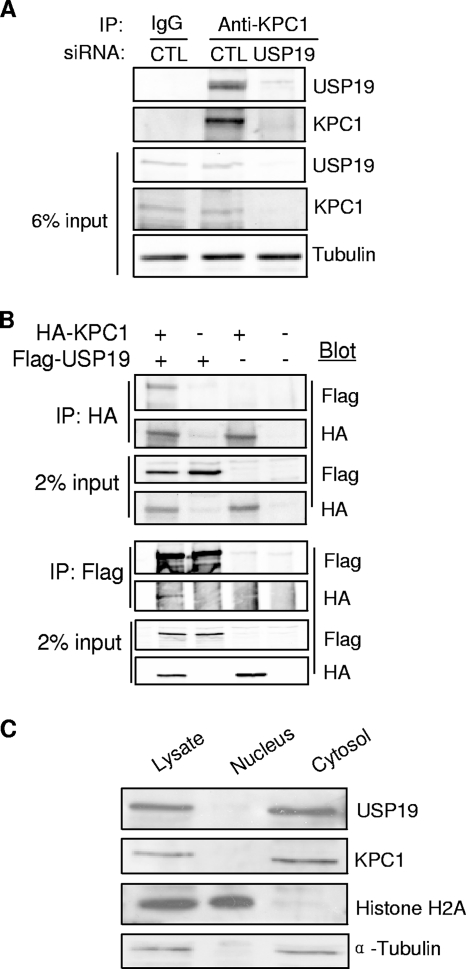
To further test this model, we verified whether the two proteins interact (Fig. 5A). USP19 could be coimmunoprecipitated with KPC1 when anti-KPC1 antibodies were used to immunoprecipitate the endogenous protein from FR3T3 lysates. This interaction was not detected in cells transfected with USP19 siRNA, confirming the identity of the bands detected in the immunoprecipitates. This interaction was confirmed in cells cotransfected with USP19- and KPC1-expressing plasmids. KPC1 and USP19 were detected in the pellets of immunoprecipitates of the reciprocal protein (Fig. 5B).
FIG. 5.
USP19 interacts with KPC1 and, like KPC1, is localized to the cytosol. (A) Interaction between endogenous KPC1 and USP19. FR3T3 cell lysates were subjected to IP using anti-KPC1 antibody, followed by immunoblot analysis with anti-USP19 or anti-KPC1 antibodies. As a control (CTL) for specificity of the antibodies, the experiment was also conducted in cells in which USP19 had been depleted by RNA interference. IgG, immunoglobulin G. (B) FR3T3 cells were transfected with the indicated combinations of plasmids expressing Flag-USP19 and HA-KPC1. Extracts of the transfected cells were subjected to IP using anti-Flag or anti-HA antibody. The immunoprecipitates were blotted with the indicated antibodies. +, present; −, absent. (C) Subcellular fractions of lysate from FR3T3 cells were subjected to immunoblotting with anti-USP19, anti-KPC1, anti-histone H2A (nuclear marker), and anti-α-tubulin (cytoplasmic marker) antibodies.
Previous work has shown that KPC is localized in the cytosol (10). Therefore, we tested whether USP19 is also located in the cytoplasm by subjecting FR3T3 cells to subcellular fractionation. Analysis by immunoblotting revealed that USP19, like KPC1, is located in the cytoplasmic fraction, supporting its ability to modulate KPC1 (Fig. 5C).
USP19 exerts its effects on cell proliferation via KPC1/P27-dependent and -independent pathways.
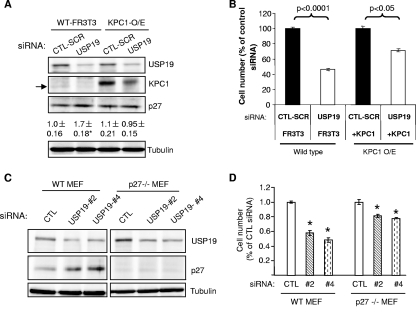
If proliferation defects in USP19-depleted cells were due to destabilization of KPC1, these defects should be prevented by overexpressing KPC1. Therefore, we further verified the model by testing the effects of siRNA-mediated depletion of USP19 on cell growth in FR3T3 cells stably overexpressing KPC1. Lowering USP19 levels by using siRNA lowered KPC1 levels in wild-type or KPC1-overexpressing cells, but the remaining levels in KPC1-overexpressing cells were greater than those seen in control wild-type FR3T3 cells, confirming the ability of the overexpression to restore KPC1 levels (Fig. 6A). USP19 depletion increased the levels of p27Kip1 in wild-type FR3T3 cells, as previously seen, but not in KPC1-overexpressing cells (Fig. 6A). These data are consistent with USP19 exerting its effects on p27 levels through its modulation of KPC1 levels. As seen before, depletion of USP19 in wild-type cells led to a marked reduction (∼50%) in cell number after 2 days (Fig. 6B). However, in cells overexpressing KPC1, a similar depletion of USP19 was much less effective in inhibiting cell proliferation, reducing the cell number at 48 h by only ∼25% (Fig. 6B). Thus, USP19 does mediate some of its effects on cell growth via KPC1. However, since overexpression of USP19 completely reverses the siRNA-mediated inhibition of cell growth (Fig. 1F) but overexpression of KPC1 does so incompletely, this argues that USP19 likely has additional substrates/interacting proteins that are involved in modulating cell growth. Since our model proposes that USP19 exerts its effects on cell growth through KPC1 modulation of p27, we tested it further by determining whether USP19 depletion would be ineffective in cells lacking p27. Wild-type mouse embryonic fibroblasts responded to the lowering of USP19 levels as expected, with an increase in p27 levels and an approximately 50% reduction in cell proliferation (Fig. 6C). USP19 depletion was indeed less effective in p27−/− cells, which showed only an ∼20% reduction in cell proliferation (Fig. 6D). These findings confirm that USP19 exerts its effects on cell proliferation through both KPC1/p27-dependent and -independent pathways.
FIG. 6.
USP19 modulates cell growth through KPC1/p27Kip1-dependent and -independent pathways. (A, B) KPC1 overexpression normalizes p27Kip1 levels and partially rescues growth defects of USP19-depleted cells. Wild-type (WT) or FR3T3 cells stably overexpressing (O/E) KPC1 were transfected with USP19 siRNA or scrambled control oligonucleotide (CTL-SCR). After 48 h, the cells were counted or harvested in a denaturing buffer. (A) Proteins from cells exposed to the indicated siRNA oligonucleotides were analyzed by immunoblotting with the indicated antibodies (arrow indicates endogenous KPC1, which migrates faster than overexpressed epitope-tagged KPC1). Shown below the p27Kip1 blot is quantitation of p27Kip1 protein as means ± standard errors. *, P value of <0.025 compared to result for CTL-SCR. (B) Cell numbers in the different treatment groups. Shown are means ± standard errors. (C, D) Ability of USP19 depletion to inhibit cell growth is partially blocked in MEF lacking p27Kip1. Wild-type or p27Kip1−/− MEF were transfected with USP19 siRNA (#2, no. 2; #4, no. 4) or nonspecific control oligonucleotide (CTL). After 48 h, the cells were counted or harvested in a denaturing buffer. (C) Proteins from cells exposed to the indicated siRNA oligonucleotides were analyzed by immunoblotting with the indicated antibodies. (D) Cell numbers in the different treatment groups. Shown are means ± standard errors. *, P value of <0.001 compared to result for CTL.
DISCUSSION
The requirement for fidelity and directionality in the execution of the cell cycle demands that the levels and activities of the various regulators of the cycle be precisely controlled. This is achieved, at least in part, by hierarchical and multilayered control at different steps of the cycle. For example, exit from mitosis and entry into anaphase require the degradation of securin and mitotic cyclins, respectively, by the APC (5, 21, 25, 30). The APC is in turn activated at anaphase by binding of the substrate-specific activator CDC20. CDC20 in turn is usually sequestered by the mitotic checkpoint complex and is only released and made available to the APC when the kinetochores are attached to the spindles at the metaphase-anaphase transition.
At the G1-S transition, p27Kip1 is ubiquitinated and degraded in a Skp2-dependent manner. To prevent precocious entry into the S phase, Skp2 is degraded and its levels kept low by APCcdh1 (reviewed in references 21 and 25). In this paper, we provide evidence for the first time for multilayered control of p27Kip1 levels in G1 phase. During G1, p27Kip1 levels fall, and this decrease is due significantly to ubiquitination by the KPC ubiquitin protein ligase complex. Here we have shown that the KPC1 catalytic subunit is in turn stabilized by the USP19 deubiquitinating enzyme. Thus, through its stabilizing effects on KPC1, USP19 indirectly promotes the degradation of p27Kip1 in the cytoplasm during G1. This degradation of cytoplasmic p27Kip1 may indirectly decrease nuclear p27Kip1 levels by decreasing the pool of p27Kip1 available for shuttling between the nuclear and cytoplasmic compartments and thereby promote cell cycle progression. Alternatively, the altered cytoplasmic p27Kip1 may modulate the cell cycle through yet-undefined cytoplasmic pathways. Depletion of USP19 did not affect the levels of KPC2, the noncatalytic subunit of the KPC complex. This, in conjunction with previous data showing that the association of KPC2 with KPC1 stabilizes the latter (7), argues that that the mechanism of stabilization of KPC1 by USP19 is largely independent of KPC2.
The role of ubiquitin protein ligases in the cell cycle has received considerable attention and is now well established. However, there is very limited literature on the role of deubiquitinating enzymes in regulating cell growth. The classic tumor suppressor protein p53 is a substrate of the deubiquitinating enzyme HAUSP (USP7). Overexpression of this deubiquitinating enzyme stabilizes p53 and strongly inhibits the growth of human carcinoma cells expressing wild-type p53 (15). Also, deubiquitinating enzyme 3, a member of the USP17 family of related genes, was described as a cytokine-inducible enzyme that blocks proliferation in diverse cell lines (2). Finally, USP44 has recently been shown to participate in the maintenance of the spindle checkpoint by promoting the deubiquitination of cdc20, which when ubiquitinated is not degraded but releases the bound Mad2 checkpoint protein and inhibitor (24, 29). Although these studies demonstrate roles for deubiquitinating enzymes in regulating cell proliferation, our results identify USP19 as the first deubiquitinating enzyme implicated in regulating the stability of a specific modulator of the G0/G1-to-S phase transition.
The overexpression of KPC1 in USP19-depleted cells did increase cell growth and so supports our model of action of USP19. However, it did so only partially, despite the expression of KPC1 to levels that exceeded endogenous levels of KPC1. Similarly, cells lacking p27 were only partly resistant to the effects of USP19 depletion on cell proliferation. These findings argue that USP19 has additional effects on cell growth that are independent of KPC1 and p27. These effects would presumably be mediated through other substrates of USP19 as, in contrast to the partial reversal with KPC1 overexpression and genetic inactivation of p27, overexpression of USP19 completely reversed the effects of the siRNA. That USP19 would have additional substrates is not surprising in view of the current data that indicate that deubiquitinating enzymes are many fewer in number than ubiquitin-protein ligases. The identification of such substrates is clearly an important question to address.
Our data showing the ability of the KPC ubiquitin protein-ligase and the USP19 deubiquitinating enzyme to bind and interact also illustrate the growing phenomenon of having opposing enzymatic activities within the same protein or protein complex. This appears to be an emerging mechanism of metabolic regulation that appears, at least so far, to be unique to the ubiquitin system. The ligase and deubiquitinating enzyme activity may reside within the same protein, as has been demonstrated for UCH-L1 (16) and A20 (33), or may be part of oligounit complexes (as shown here for USP19/KPC1) or in a megacomplex, as is the case with the Hul5 ubiquitin protein ligase and the Ubp6/USP14 deubiquitinating enzyme associated with the proteasome (4). Based on described functional consequences of such interactions, the deubiquitinating enzyme in the complex may serve to rescue the substrates of the ligase, as in the case of RSP5-Ubp2 (11, 12), or may serve to stabilize the ligase itself, as with KPC1/USP19 (this work), Itch/FAM/USP9X (17), cullin ring E3s/CSN-USP15 (34), and Nrdp1/USP8 (35). This complexing of enzymes of opposing activities represents an emerging method of enzyme regulation in the ubiquitin pathway, conferring a local and specific regulation of the level and activity of a subunit of the complex.
In summary, we have identified USP19 as a deubiquitinating enzyme that interacts with the KPC-ubiquitin ligase. This deubiquitinating enzyme regulates cell growth and does so at least partly through indirect control of p27Kip1 degradation in the G1 phase of the cell cycle. Since a number of cancers are associated with enhanced degradation of p27Kip1, the overexpression of USP19 may also be a pathogenetic mechanism of the disordered cell growth in cancer and a potential novel therapeutic target for its treatment.
Acknowledgments
This work was supported by grants from the Canadian Institutes of Health Research (MOP14700 and MT12121) and the National Cancer Institute of Canada to S.S.W. and from the National Institutes of Health (AG025688) to J.P. O.A.J.A. is a recipient of postdoctoral fellowships from the Canadian Diabetes Association and Royal Victoria Hospital Research Institute of McGill University. S.S.W. is the recipient of a Chercheur National salary award from the Fonds de la recherche en santé du Québec.
Footnotes
Published ahead of print on 17 November 2008.
REFERENCES
- 1.Bloom, J., and M. Pagano. 2003. Deregulated degradation of the cdk inhibitor p27 and malignant transformation. Semin. Cancer Biol. 1341-47. [DOI] [PubMed] [Google Scholar]
- 2.Burrows, J. F., M. J. McGrattan, A. Rascle, M. Humbert, K. H. Baek, and J. A. Johnston. 2004. DUB-3, a cytokine-inducible deubiquitinating enzyme that blocks proliferation. J. Biol. Chem. 27913993-14000. [DOI] [PubMed] [Google Scholar]
- 3.Carrano, A. C., E. Eytan, A. Hershko, and M. Pagano. 1999. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1193-199. [DOI] [PubMed] [Google Scholar]
- 4.Crosas, B., J. Hanna, D. S. Kirkpatrick, D. P. Zhang, Y. Tone, N. A. Hathaway, C. Buecker, D. S. Leggett, M. Schmidt, R. W. King, S. P. Gygi, and D. Finley. 2006. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 1271401-1413. [DOI] [PubMed] [Google Scholar]
- 5.Deng, C., P. Zhang, J. W. Harper, S. J. Elledge, and P. Leder. 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82675-684. [DOI] [PubMed] [Google Scholar]
- 6.Fero, M. L., M. Rivkin, M. Tasch, P. Porter, C. E. Carow, E. Firpo, K. Polyak, L. H. Tsai, V. Broudy, R. M. Perlmutter, K. Kaushansky, and J. M. Roberts. 1996. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell 85733-744. [DOI] [PubMed] [Google Scholar]
- 7.Hara, T., T. Kamura, S. Kotoshiba, H. Takahashi, K. Fujiwara, I. Onoyama, M. Shirakawa, N. Mizushima, and K. I. Nakayama. 2005. Role of the UBL-UBA protein KPC2 in degradation of p27 at G1 phase of the cell cycle. Mol. Cell. Biol. 259292-9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hara, T., T. Kamura, K. Nakayama, K. Oshikawa, S. Hatakeyama, and K. Nakayama. 2001. Degradation of p27(Kip1) at the G(0)-G(1) transition mediated by a Skp2-independent ubiquitination pathway. J. Biol. Chem. 27648937-48943. [DOI] [PubMed] [Google Scholar]
- 9.Ishida, N., T. Hara, T. Kamura, M. Yoshida, K. Nakayama, and K. I. Nakayama. 2002. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J. Biol. Chem. 27714355-14358. [DOI] [PubMed] [Google Scholar]
- 10.Kamura, T., T. Hara, M. Matsumoto, N. Ishida, F. Okumura, S. Hatakeyama, M. Yoshida, K. Nakayama, and K. I. Nakayama. 2004. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 61229-1235. [DOI] [PubMed] [Google Scholar]
- 11.Kee, Y., N. Lyon, and J. M. Huibregtse. 2005. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 242414-2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kee, Y., W. Munoz, N. Lyon, and J. M. Huibregtse. 2006. The Ubp2 deubiquitinating enzyme modulates Rsp5-dependent K63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J. Biol. Chem. 28136724-36731. [DOI] [PubMed] [Google Scholar]
- 13.Kiyokawa, H., R. D. Kineman, K. O. Manova-Todorova, V. C. Soares, E. S. Hoffman, M. Ono, D. Khanam, A. C. Hayday, L. A. Frohman, and A. Koff. 1996. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1). Cell 85721-732. [DOI] [PubMed] [Google Scholar]
- 14.Kossatz, U., N. Dietrich, L. Zender, J. Buer, M. P. Manns, and N. P. Malek. 2004. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 182602-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, M., D. Chen, A. Shiloh, J. Luo, A. Y. Nikolaev, J. Qin, and W. Gu. 2002. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature 416648-653. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y., L. Fallon, H. A. Lashuel, Z. Liu, and P. T. Lansbury, Jr. 2002. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell 111209-218. [DOI] [PubMed] [Google Scholar]
- 17.Mouchantaf, R., B. A. Azakir, P. S. McPherson, S. M. Millard, S. A. Wood, and A. Angers. 2006. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J. Biol. Chem. 28138738-38747. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama, K., N. Ishida, M. Shirane, A. Inomata, T. Inoue, N. Shishido, I. Horii, and D. Y. Loh. 1996. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell 85707-720. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama, K., H. Nagahama, Y. A. Minamishima, M. Matsumoto, I. Nakamichi, K. Kitagawa, M. Shirane, R. Tsunematsu, T. Tsukiyama, N. Ishida, M. Kitagawa, and S. Hatakeyama. 2000. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 192069-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakayama, K., H. Nagahama, Y. A. Minamishima, S. Miyake, N. Ishida, S. Hatakeyama, M. Kitagawa, S. Iemura, T. Natsume, and K. I. Nakayama. 2004. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell 6661-672. [DOI] [PubMed] [Google Scholar]
- 21.Nakayama, K. I., and K. Nakayama. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6369-381. [DOI] [PubMed] [Google Scholar]
- 22.Nijman, S. M., M. P. Luna-Vargas, A. Velds, T. R. Brummelkamp, A. M. Dirac, T. K. Sixma, and R. Bernards. 2005. A genomic and functional inventory of deubiquitinating enzymes. Cell 123773-786. [DOI] [PubMed] [Google Scholar]
- 23.Peng, J., M. J. Kim, D. Cheng, D. M. Duong, S. P. Gygi, and M. Sheng. 2004. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 27921003-21011. [DOI] [PubMed] [Google Scholar]
- 24.Reddy, S. K., M. Rape, W. A. Margansky, and M. W. Kirschner. 2007. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446921-925. [DOI] [PubMed] [Google Scholar]
- 25.Reed, S. I. 2003. Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4855-864. [DOI] [PubMed] [Google Scholar]
- 26.Rodier, G., A. Montagnoli, L. Di Marcotullio, P. Coulombe, G. F. Draetta, M. Pagano, and S. Meloche. 2001. p27 cytoplasmic localization is regulated by phosphorylation on Ser10 and is not a prerequisite for its proteolysis. EMBO J. 206672-6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seyfried, N. T., P. Xu, D. M. Duong, D. Cheng, J. Hanfelt, and J. Peng. 2008. Systematic approach for validating the ubiquitinated proteome. Anal. Chem. 804161-4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Signoretti, S., L. Di Marcotullio, A. Richardson, S. Ramaswamy, B. Isaac, M. Rue, F. Monti, M. Loda, and M. Pagano. 2002. Oncogenic role of the ubiquitin ligase subunit Skp2 in human breast cancer. J. Clin. Investig. 110633-641. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Stegmeier, F., M. Rape, V. M. Draviam, G. Nalepa, M. E. Sowa, X. L. Ang, E. R. McDonald III, M. Z. Li, G. J. Hannon, P. K. Sorger, M. W. Kirschner, J. W. Harper, and S. J. Elledge. 2007. Anaphase initiation is regulated by antagonistic ubiquitination and deubiquitination activities. Nature 446876-881. [DOI] [PubMed] [Google Scholar]
- 30.Sudakin, V., D. Ganoth, A. Dahan, H. Heller, J. Hershko, F. C. Luca, J. V. Ruderman, and A. Hershko. 1995. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol. Biol. Cell 6185-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sutterluty, H., E. Chatelain, A. Marti, C. Wirbelauer, M. Senften, U. Muller, and W. Krek. 1999. p45SKP2 promotes p27Kip1 degradation and induces S phase in quiescent cells. Nat. Cell Biol. 1207-214. [DOI] [PubMed] [Google Scholar]
- 32.Tsvetkov, L. M., K. H. Yeh, S. J. Lee, H. Sun, and H. Zhang. 1999. p27(Kip1) ubiquitination and degradation is regulated by the SCF(Skp2) complex through phosphorylated Thr187 in p27. Curr. Biol. 9661-664. [DOI] [PubMed] [Google Scholar]
- 33.Wertz, I. E., K. M. O'Rourke, H. Zhou, M. Eby, L. Aravind, S. Seshagiri, P. Wu, C. Wiesmann, R. Baker, D. L. Boone, A. Ma, E. V. Koonin, and V. M. Dixit. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430694-699. [DOI] [PubMed] [Google Scholar]
- 34.Wu, J. T., Y. R. Chan, and C. T. Chien. 2006. Protection of cullin-RING E3 ligases by CSN-UBP12. Trends Cell Biol. 16362-369. [DOI] [PubMed] [Google Scholar]
- 35.Wu, X., L. Yen, L. Irwin, C. Sweeney, and K. L. Carraway III. 2004. Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 247748-7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, H., R. Kobayashi, K. Galaktionov, and D. Beach. 1995. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell 82915-925. [DOI] [PubMed] [Google Scholar]