Abstract
Individual cells of the budding yeast, Saccharomyces cerevisiae, have a limited life span and undergo a form of senescence termed replicative aging. Replicative life span is defined as the number of daughter cells produced by a yeast mother cell before she ceases dividing. Replicative aging is asymmetric: a mother cell ages but the age of her daughter cells is “reset” to zero. Thus, one or more senescence factors have been proposed to accumulate asymmetrically between mother and daughter yeast cells and lead to mother-specific replicative senescence once a critical threshold has been reached. Here we evaluate potential candidates for senescence factors and age-associated phenotypes and discuss potential mechanisms underlying the asymmetry of replicative aging in budding yeast.
Introduction
Asymmetric cell divisions are the foundation for generating cells with distinct phenotypes from a common genetic blueprint. Asymmetric cell divisions lead to the diversity of tissues and organs in metazoans, the creation of spores and fruiting bodies in microorganisms, and are required for the preservation of a stem cell’s qualities while permitting it to produce progeny that differentiate into another cell type [reviewed in 1,2,3]. A number of mechanisms for generating asymmetry have been documented, but the common objective they all accomplish is differential segregation of biomolecules (e.g. protein, RNA) between two cells that ultimately determines their distinct fates [reviewed in 4].
Studies of metazoan development have defined and addressed many of the questions about asymmetric cell division. However, the simple budding yeast, Saccharomyces cerevisiae, has also served as a model system to analyze asymmetry. Discoveries in S. cerevisiae have contributed to understanding how cell polarity is established, how different genetic programs are established within one cell division, and how molecules are asymmetrically segregated between dividing cells [reviewed in 5,6].
S. cerevisiae undergoes a stereotypical pattern of growth and asymmetric division [reviewed in 6]. Once a site for bud (daughter cell) emergence is chosen in a mother cell, an axis of polarity is established and is built upon an actin cytoskeletal framework. The cytoskeleton facilitates targeted secretion at the nascent bud site to allow for bud growth. After bud emergence, subsequent cell growth is restricted to the bud as the cell duplicates and segregates its organelles and bud-specific molecules. Typically, when the bud reaches a slightly smaller size than the mother, mitosis and cytokinesis commence and a septum is built between the mother and daughter cells to complete their separation.
A Finite Number of Asymmetric Cell Divisions
While there are a number of fundamental questions to be asked about asymmetric cell divisions, one was asked nearly 50 years ago: how many divisions can one cell go through, continuously serving as a “mother”? This question was steeped in a rather fundamental issue of the times. Budding yeast cultures seemed to be immortal; the culture would continue to divide as long as there were nutrients. This “immortality” was measured by monitoring the population of yeast cells, but was it based upon immortal individuals? Taking advantage of the ability to visually distinguish between mother and daughter yeast cells and to micromanipulate single cells, Mortimer and Johnston discovered that mother cells have a finite replicative capacity [7]. The replicative life span (RLS) of a mother yeast cell is defined as the number of daughter cells she can produce before she ceases to divide and ultimately lyses.
One obvious question from the yeast life span results is, what caused the mother cells to stop dividing and die? Even though it has been nearly 50 years since the question was posed, there is no clear answer. However, this has become an active area of research and a number of advances have been made [reviewed in 8].
While the answer to how mother yeast cells die remains elusive, these earlier studies have also spawned an interest in understanding the aging process in S. cerevisiae. Identifying and characterizing age-associated phenotypes may ultimately provide a key insight that explains the limited life span potential of a mother cell and gets to the root of the aging process. However, there is also an appreciation for studying age-associated phenotypes in their own right.
Age-Associated Phenotypes Observed in Asymmetric Cell Divisions
Some of the age-associated phenotypes of mother yeast cells were akin to noting wrinkled skin on older people; mother yeast cells of advanced replicative age become enlarged and have wrinkled surfaces, and the time between cell divisions becomes greatly extended during their last few cell divisions [7]. But other discoveries have laid an intellectual groundwork that has guided recent research.
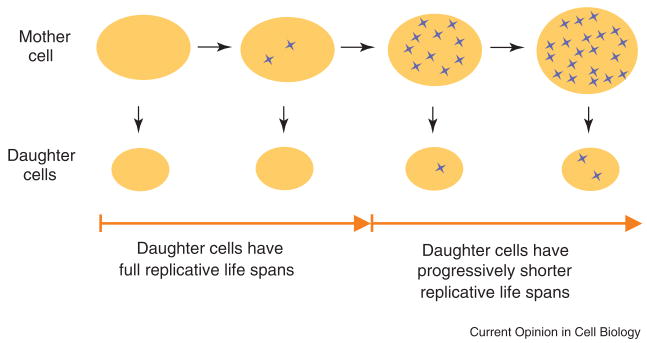
For the first half of their lives, yeast mother cells produce daughter cells capable of full replicative life spans (see Figure). The replicative age of mother cells is not normally transferred to her daughters; the age of daughter cells appears to be “reset” to zero. However, this “age-asymmetry” between mother and daughter deteriorates later in life [9]. The closer a mother cell is to the end of her life, the shorter the life span of the daughter cells she produces. At the extreme, daughter cells produced by very old mother cells have life spans only 25% the length of the mother cell’s life span.
Figure. Model for the Molecular Basis of Replicative Age-Asymmetry.
A single yeast mother cell (top) undergoes replicative aging as she produces successive smaller daughter cells (below the mother cell). One or more senescence factors accumulate in the mother cell (blue Xs) and cause senescence or an age-associated phenotype once a critical threshold is reached. The senescence factor(s) is asymmetrically distributed between the mother cell and the daughter cells for most of the mother cell life span. Likewise, for most of the mother cell’s life span, replicative aging is asymmetric. Daughter cells do not inherit their mother’s age – the replicative age of the daughter cells is “reset” to zero. In the last half of a mother cell’s life span, there may be a complete breakdown of senescence factor asymmetry. Alternatively, senescence factor asymmetry could still exist but so much senescence factor may have accumulated in the mother cell that even the small proportion of the mother cell’s senescence factor acquired by the daughter cell has a deleterious affect on replicative life span. Once daughter cells start to acquire sufficient levels of senescence factor, they are born with progressively decreasing replicative life spans.
These and other studies led to the proposal of a “senescence factor” that accumulates in mother cells as they age and causes senescence once it reaches a critical threshold [9,10]. The senescence factor appears to acts dominantly; when old and young cells are mated, the life span of the resulting diploid most closely resembles the remaining life span of the old cell [11]. These results and interpretations have defined the thinking about the aging process in yeast, which is largely built upon this age-asymmetry.
Criteria for a Replicative Life Span Senescence Factor
Based on the behavior of replicatively aged yeast cells, four criteria can be assigned to a putative senescence factor. 1. The senescence factor should be more abundant in old mother cells than in young cells. 2. Introduction of high levels of a putative senescence factor into young cells should limit their RLS. 3. Reduction in the amount of senescence factor in old cells should extend RLS. 4. The abundance of a senescence factor should be asymmetric between aged mother cells and their daughter cells for most of the RLS of the mother cell.
Not only are these criteria applicable to the cause of a mother cell’s death, but they can also be adapted to explain age-associated phenotypes. Here we will use the criteria as a guide for evaluating a few studies of age-associated phenotypes and senescence.
Extrachromosomal rDNA Circles Fit the Criteria for a Senescence Factor
Extrachromosomal rDNA circles (ERCs) are a well-characterized age-associated phenotype in budding yeast [12]. The yeast rDNA locus consists of 100–200 copies of a directly repeated unit containing a sequence that can act as an origin of replication. Because of the structure of the rDNA locus, recombination can generate extrachromosomal circles. ERCs accumulate in mother cells with replicative age because they are replicated and experience mother-biased segregation similar to circular ARS-containing plasmids without centromeres [12, 13]. Thus, ERCs are much more abundant in replicatively old mother cells than young mother cells and are asymmetrically distributed between mother and daughter cells. Moreover, generating high levels of ERCs in young cells limits their life span [12], and limiting ERC formation extends life span [14], strongly supporting the idea that ERC accumulation has a causal role in replicative aging. Thus, ERCs fulfill all of the expectations of a senescence factor and are likely to have a causal role in replicative aging. Although ERCs were identified as a causal agent in replicative aging more than 10 years ago, the mechanism by which ERCs limit a mother cell’s life span has not been elucidated and remains an important future challenge.
However, there may be a reason to look more carefully before accepting ERCs as the senescence factor in replicatively aged yeast cells. When yeast cells enter stationary phase for a time and are then re-introduced to nutrients, the mother cells have a reduced RLS compared to those that never entered stationary phase. This reduction in RLS occurs even though ERC levels are not increased during this period. This suggests that dependent upon growth conditions, there is at least one other senescence factor in addition to ERCs [15]. Results like this, and the fact that there is no convincing evidence that ERCs are a senescence factor in other organisms, have motivated a continued hunt for senescence factors and age-associated phenotypes in the asymmetric divisions of yeast.
Is Reactive Oxygen-Mediated Damage to Proteins a Senescence Factor?
More than 50 years ago, reactive oxygen species were proposed as causative agents in aging. The oxidative damage to diverse cellular components that reactive oxygen species inflict is, in fact, correlated with age in many organisms [16]. This prompted the investigation of oxidative damage of protein in budding yeast during replicative aging. Protein carbonylation is considered an irreversible form of oxidative damage proposed to disrupt protein function and its presence correlates with advanced age in many organisms [reviewed in 17]. Thus, it is intellectually pleasing that carbonylated proteins accumulate in yeast mother cells with increasing replicative age [18]. Furthermore, whereas normal cellular proteins diffuse freely between mother cells and their buds [19], carbonylated proteins are asymmetrically distributed between young mother cells and their buds. These observations led to the proposal that protein carbonyls are a senescence factor [18]. Currently, however, protein carbonyls only fulfill two of the four criteria we defined for a senescence factor. Importantly, there is not yet evidence that decreasing the levels of protein carbonyls in cells extends RLS. Thus, while a correlation exists between increased carbonyls and replicative age, it is not obvious that protein carbonyls are a causative agent in replicative senescence. Furthermore, there are some caveats to using protein carbonyls as general indicator of oxidative damage. First, carbonylated proteins only represent a small subset of the possible oxidative modifications possible to proteins, not to mention other macromolecules [reviewed in 20,21]. Second, detection of protein carbonylation monitors bulk carbonylation [20] and perhaps carbonylation of only one or a few proteins acts as a senescence factor. Thus, while it is not yet clear whether protein carbonylation is a senescence factor, this does not exclude other forms of oxidative damage or a subset of protein carbonyls from being senescence factors. Nevertheless, protein carbonyls continue to be studied, based on the belief that they may cause age-associated phenotypes even if they do not cause senescence.
Protein Aggregation Increases with Replicative Age in Budding Yeast
Because oxidative damage to proteins can cause them to aggregate [22], Erjavec et al. examined whether protein aggregates were asymmetrically distributed between aging mother cells and their daughters [23]. They used Hsp104 as a marker of protein aggregation because of its activity as a cellular “disaggregase” that interacts with aggregated proteins [24]. Hsp104-GFP aggregates increased as a function of replicative age of the mother cell and partially co-localized with carbonyls [23]. Furthermore, just like protein carbonyls, Hsp104-GFP aggregates generated in young cells are asymmetrically distributed between mother cells and their buds. However, like carbonylated proteins, it is not clear that general protein aggregation is a senescence factor – there have not been experiments that eliminate or reduce protein aggregates and show an extension of life span. Attempts to change the amount of aggregates in a yeast cell by deleting or overexpressing HSP104 had no significant effect on life span in otherwise wild-type cells [23,25]. Nevertheless, it remains possible that a subset of unknown aggregated proteins act as a senescence factor or cause age-associated phenotypes.
Senescence Factors: Everything and Nothing
Much of the work to identify senescence factors in budding yeast has focused on particular classes of macromolecules. However, these age-associated phenomena may just be the tip of the iceberg. Potential senescence factors could include forms of protein modification in addition to carbonylation [reviewed in 21], and could include other forms of reactive oxygen-mediated damage besides damage to protein [reviewed in 20]. Furthermore, dysfunctional membranes or organelles and/or aberrant forms of RNA or other cofactors could be senescence factors [26].
It is also possible that there is no senescence factor, in the sense that there is no single molecule, or group of molecules that leads to cellular aging and senescence. Rather it may be that the maintenance of protein homeostasis begins to break down over time [27]. There are a myriad of quality control and repair processes, beginning with DNA replication and transcription and including protein synthesis, degradation, complex assembly and cellular targeting, that are engaged to maintain this homeostasis. If defects begin to amass at one or more places in this process, then there may be a crisis cascade that uncovers the “weak link” in the process, which ultimately leads to catastrophe (cell death/senescence). The initiating molecules may be random stochastic defects that simply accumulate with time and overtax the components of quality control. The defective molecules could be anything, but their effect would be to permit other proteins that are normally handled by quality control systems to escape detection. Thus, senescence factors may be random molecules which escaped detection but eventually accumulate and cause a system-wide failure. For instance, if a chaperone protein such as Hsp90, which serves hundreds of client proteins, becomes overwhelmed by an accumulation of randomly generated aberrant proteins, then client proteins that most need Hsp90 for maintenance will become non-functional [28]. The demise of this important hypothetical client protein was not the result of a specific defect in the protein, or Hsp90, but rather accumulation of too many proteins that required Hsp90 assistance.
Possible Sources of Age-Asymmetry
Despite the paucity of identified senescence factors in budding yeast, some recent findings suggest that the actin cytoskeleton is part of the mechanism maintaining age-asymmetry [23]. A clever experiment was performed to investigate the role of the actin cytoskeleton in generating age-asymmetry. Middle-aged budded mother cells were transiently treated with latrunculin, a reagent that prevents actin polymerization and disrupts the cytoskeleton. The subsequent withdrawal of latrunculin allowed a second bud to form on the mother cell and both buds completed cytokinesis nearly simultaneously – they were twins. Intriguingly, the RLS of the daughter cell that formed when the actin cytoskeleton was intact was normal. However, the RLS of the daughter that formed when the cytoskeleton was perturbed was reduced by 20%. Disruption of the actin cytoskeleton may thus have caused the daughter cell to inherit a portion of the mother cell’s senescence factor(s). Therefore, this “twin experiment” supports the idea that the actin cytoskeleton is part of the mechanism for maintaining senescence factor asymmetry, and thereby age-asymmetry.
More recent work has uncovered the likely mechanism leading to mother-daughter asymmetry of ERCs [29]. A septin-mediated diffusion barrier restricts the movement of outer-membrane proteins of the nuclear envelope from the mother cell into the bud. Thus, the mother cell preferentially retains pre-existing nuclear pores and they are instead assembled de novo in daughter cells. In addition, non-centromeric episomes similar to ERCs (and likely ERCs as well) associate with the nuclear pore and appear to be anchored there. Thus, this septin-mediated diffusion barrier to the nuclear pore can explain the previously observed mother cell bias in ERC accumulation. Importantly, mutations that allow daughter cells to inherit nuclear pores from mother cells increase ERC inheritance by daughters and disrupt mother-daughter age-asymmetry. Thus, retention of nuclear pores by mother cells helps to explain the mother-cell bias of ERC inheritance and how age-asymmetry may be generated.
We hypothesize that any molecular asymmetry between mother and daughter cells could potentially contribute to age-asymmetry. For instance, like the diffusion barrier to the outer-membrane proteins of the nuclear envelope, budding yeast also have distinct plasma membrane compartments created by this septin-mediated diffusion barrier at the mother-bud neck [30,31]. The diffusion barrier prevents translocation of transmembrane proteins in the plasma membrane from the mother cell past the bud neck and also affects the cortical ER [32]. Thus, age-asymmetry could be established by the requirement for daughter cells to synthesize new plasma membrane and cortical ER proteins. Furthermore, the composition of the plasma membrane itself is thought to be asymmetric [33] and could provide an additional unexplored source of age-asymmetry between mother and daughter cells. Moreover, bud-enrichment of several mRNAs has been identified [reviewed in 5] and could provide daughter cells with an abundance of factors required to repair aberrant molecules inherited by the mother cell. Finally, most organelles are not generated de novo in daughter cells and are inherited from mother cells [34]. If organelles experienced dysfunction with replicative age but segregation of their dysfunctional components to daughter cells were prevented, age-asymmetry could result.
While it is interesting to speculate how age-asymmetry could be established and maintained, further progress in this area will likely depend on a clearer understanding of what molecules or changes to cellular processes cause senescence.
Are Age-Associated Phenotypes in Budding Yeast Common to other Organisms?
Increased oxidative damage to many macromolecules increases nearly universally with age and is potentially a conserved senescence factor. However, a clear causal relationship between increased oxidative damage and aging has still not been established [reviewed in 16]. The discovery that at least one form of oxidative damage increases with replicative age in budding yeast [18] brings continued hope that cause and effect for age-associated phenotypes will be distinguished due to the tractability of S. cerevisiae as a model system.
Even though protein aggregates are not necessarily senescence factors in budding yeast, it is striking that asymmetric inheritance of aberrant proteins has been demonstrated in other organisms. For example, protein aggregates localize asymmetrically during the division of D. melanogaster neural precursor cells and in the epithelial crypts of humans with a protein folding disease [35]. Protein aggregates also localize asymmetrically during E. coli cell division [36]. Thus, in several model systems, aggregated proteins are asymmetrically distributed so that they do not accumulate in the longest-lived daughter cell.
Thus, a conserved senescence factor that causes senescence in all organisms remains elusive – as has been suggested, there may not be a single molecule or molecular event that is a senescence factor [reviewed in 37]. Nevertheless, the commonality of age-associated changes at the molecular level in diverse organisms is suggestive of a common molecular basis for age-related phenotypes. Furthermore, the observation that molecular changes that extend life span are common between divergent species [38] supports this idea.
Conclusions
Recent work in budding yeast has described age-associated changes in addition to the accumulation of ERCs and has begun to identify the requirements for their asymmetry. As discussed, it is likely that only a small subset of age-associated changes have been discovered. Although extensive analyses to identify loss-of-function mutations that extend RLS have been performed in several organisms [reviewed in 39], these studies can only reveal the pathways whose modulation extends RLS. This will reveal which pathways modulate the abundance of molecules that end life, but will not necessarily lead to identification of senescence factors. We believe that more careful identification and analysis of age-associated phenotypes will augment and extend research in the field of budding yeast replicative aging, because it will provide a wider foundation for understanding the replicative aging process. Unfortunately, it has been technically difficult to collect large numbers of old mother yeast cells to readily discover age-associated changes in an unbiased manner. Thus, progress in this aspect of the study of replicative aging in yeast has been rather slow. With the advent of new technologies to isolate replicatively old budding yeast cells, progress in identifying senescence factors, age-associated changes, and the mechanisms maintaining their asymmetry will accelerate.
Footnotes
Papers of Special Interest:
Lindner et al., 2008
Although the daughter cells of E. coli appear identical, the cell that inherits the old pole after division grows more slowly and divides less frequently, which is suggestive of aging. Here, the authors demonstrate that the cell that acquires the old pole is more likely to inherit spontaneous protein aggregates at cell division.
Erjavec et al., 2007
Because protein carbonyls seemed to form aggregates, the authors examined colocalization of protein carbonyls in replicatively old cells with Hsp104-GFP, a marker for general protein aggregation. Like protein carbonyls, protein aggregates accumulated as a function of mother cell replicative cell age and were asymmetrically distributed between mother cells and their buds. In addition, the authors demonstrated that the actin cytoskeleton is likely a component of the mechanism of age-asymmetry.
Papers of Outstanding Interest:
Shcheprova et al., 2008
Although ERCs were identified as a cause of replicative aging 10 years ago, both the mechanism by which they cause replicative aging and the mechanism accounting for their asymmetric distribution between mother and daughter cells were unknown. Here, the authors identify a septin-mediated barrier to diffusion between mother and daughter cells of outer-membrane proteins of the nuclear membrane. This barrier prevents nuclear pores and nuclear pore-associated non-centromeric plasmids similar to ERCs from entering daughter cells. Disruption of the septin-mediated diffusion barrier both increases the number of daughter cells that inherit ERCs and disrupts mother daughter-age asymmetry. Thus, retention of nuclear pores by the mother cell offers an explanation for the mother-cell bias of ERC accumulation and for how age-asymmetry may be generated.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Kroos L. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu Rev Genet. 2007;41:13–39. doi: 10.1146/annurev.genet.41.110306.130400. [DOI] [PubMed] [Google Scholar]
- 3.Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068–1074. doi: 10.1038/nature04956. [DOI] [PubMed] [Google Scholar]
- 4.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 5.Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 7.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 8.Kaeberlein M, Burtner CR, Kennedy BK. Recent developments in yeast aging. PLoS Genet. 2007;3:e84. doi: 10.1371/journal.pgen.0030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy BK, Austriaco NR, Jr, Guarente L. Daughter cells of Saccharomyces cerevisiae from old mothers display a reduced life span. J Cell Biol. 1994;127:1985–1993. doi: 10.1083/jcb.127.6.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egilmez NK, Jazwinski SM. Evidence for the involvement of a cytoplasmic factor in the aging of the yeast Saccharomyces cerevisiae. J Bacteriol. 1989;171:37–42. doi: 10.1128/jb.171.1.37-42.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller I. Parental age and the life-span of zygotes of Saccharomyces cerevisiae. Antonie Van Leeuwenhoek. 1985;51:1–10. doi: 10.1007/BF00444223. [DOI] [PubMed] [Google Scholar]
- 12.Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- 13.Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- 14.Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- 15.Ashrafi K, Sinclair D, Gordon JI, Guarente L. Passage through stationary phase advances replicative aging in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:9100–9105. doi: 10.1073/pnas.96.16.9100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bokov A, Chaudhuri A, Richardson A. The role of oxidative damage and stress in aging. Mech Ageing Dev. 2004;125:811–826. doi: 10.1016/j.mad.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aguilaniu H, Gustafsson L, Rigoulet M, Nystrom T. Asymmetric inheritance of oxidatively damaged proteins during cytokinesis. Science. 2003;299:1751–1753. doi: 10.1126/science.1080418. [DOI] [PubMed] [Google Scholar]
- 19.Dobbelaere J, Barral Y. Spatial coordination of cytokinetic events by compartmentalization of the cell cortex. Science. 2004;305:393–396. doi: 10.1126/science.1099892. [DOI] [PubMed] [Google Scholar]
- 20.Levine RL, Stadtman ER. Oxidative modification of proteins during aging. Exp Gerontol. 2001;36:1495–1502. doi: 10.1016/s0531-5565(01)00135-8. [DOI] [PubMed] [Google Scholar]
- 21.Schoneich C. Protein modification in aging: an update. Exp Gerontol. 2006;41:807–812. doi: 10.1016/j.exger.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Bota DA, Davies KJ. Lon protease preferentially degrades oxidized mitochondrial aconitase by an ATP-stimulated mechanism. Nat Cell Biol. 2002;4:674–680. doi: 10.1038/ncb836. [DOI] [PubMed] [Google Scholar]
- 23.Erjavec N, Larsson L, Grantham J, Nystrom T. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 2007;21:2410–2421. doi: 10.1101/gad.439307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: a novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 25.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev. 2005;126:491–504. doi: 10.1016/j.mad.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 26.Lai CY, Jaruga E, Borghouts C, Jazwinski SM. A mutation in the ATP2 gene abrogates the age asymmetry between mother and daughter cells of the yeast Saccharomyces cerevisiae. Genetics. 2002;162:73–87. doi: 10.1093/genetics/162.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- 28.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396:336–342. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 29.Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- 30.Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- 31.Barral Y, Mermall V, Mooseker MS, Snyder M. Compartmentalization of the cell cortex by septins is required for maintenance of cell polarity in yeast. Mol Cell. 2000;5:841–851. doi: 10.1016/s1097-2765(00)80324-x. [DOI] [PubMed] [Google Scholar]
- 32.Luedeke C, Frei SB, Sbalzarini I, Schwarz H, Spang A, Barral Y. Septin-dependent compartmentalization of the endoplasmic reticulum during yeast polarized growth. J Cell Biol. 2005;169:897–908. doi: 10.1083/jcb.200412143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beh CT, Rine J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci. 2004;117:2983–2996. doi: 10.1242/jcs.01157. [DOI] [PubMed] [Google Scholar]
- 34.Fagarasanu A, Rachubinski RA. Orchestrating organelle inheritance in Saccharomyces cerevisiae. Curr Opin Microbiol. 2007;10:528–538. doi: 10.1016/j.mib.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 35.Rujano MA, Bosveld F, Salomons FA, Dijk F, van Waarde MA, van der Want JJ, de Vos RA, Brunt ER, Sibon OC, Kampinga HH. Polarised asymmetric inheritance of accumulated protein damage in higher eukaryotes. PLoS Biol. 2006;4:e417. doi: 10.1371/journal.pbio.0040417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindner AB, Madden R, Demarez A, Stewart EJ, Taddei F. Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc Natl Acad Sci U S A. 2008;105:3076–3081. doi: 10.1073/pnas.0708931105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirkwood TB. A systematic look at an old problem. Nature. 2008;451:644–647. doi: 10.1038/451644a. [DOI] [PubMed] [Google Scholar]
- 38.Smith ED, Tsuchiya M, Fox LA, Dang N, Hu D, Kerr EO, Johnston ED, Tchao BN, Pak DN, Welton KL, et al. Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res. 2008;18:564–570. doi: 10.1101/gr.074724.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy BK. The genetics of ageing: insight from genome-wide approaches in invertebrate model organisms. J Intern Med. 2008;263:142–152. doi: 10.1111/j.1365-2796.2007.01903.x. [DOI] [PubMed] [Google Scholar]