Abstract
Cyclic AMP response element-binding protein (CREB) is a widely expressed transcription factor whose role in neuronal protection is now well established. Here we report that CREB is present in the mitochondrial matrix of neurons and that it binds directly to cyclic AMP response elements (CREs) found within the mitochondrial genome. Disruption of CREB activity in the mitochondria decreases the expression of a subset of mitochondrial genes, including the ND5 subunit of complex I, down-regulates complex I-dependent mitochondrial respiration, and increases susceptibility to 3-nitropropionic acid, a mitochondrial toxin that induces a clinical and pathological phenotype similar to Huntington disease. These results demonstrate that regulation of mitochondrial gene expression by mitochondrial CREB, in part, underlies the protective effects of CREB and raise the possibility that decreased mitochondrial CREB activity contributes to the mitochondrial dysfunction and neuronal loss associated with neurodegenerative disorders.
The cAMP response element-binding protein (CREB)3 is a transcription factor known to mediate stimulus-dependent expression of genes critical for the plasticity, growth, and survival of neurons (1). A variety of stimuli alter levels of intracellular second messengers in neurons, such as cAMP and calcium, and activate CREB by leading to phosphorylation at its critical regulatory site, serine 133 (2, 3). Overexpression of constitutively active CREB prevents cell death induced by growth factor deprivation, while expression of a dominant negative form of CREB leads to apoptosis in both sympathetic neurons and cerebellar granule cells (4, 5). A recent report that CREB is present in the mitochondria raises the possibility that CREB could mediate mitochondrial gene expression (6). Nonetheless, the function of mitochondrial CREB is not known. The present study confirms the presence of CREB in the mitochondria and addresses the role of CREB in mitochondrial gene expression and neuronal survival. The results raise the possibility of a novel mechanism for CREB dysfunction in the pathogenesis of neurodegenerative disorders.
MATERIALS AND METHODS
Isolation of Mitochondria
Mitochondria were isolated from primary cultured cortical neurons and adult rat brains by sucrose density gradient centrifugation (6).
Confocal Microscopy
Indirect labeling methods were used to determine the levels of CREB, phosphorylated CREB (pCREB), and neurofilament (200 kDa) in cortical neuronal cultures and human and rat brain tissues as described previously (7).
Immunogold Labeling and Electron Microscopy
Frozen samples were sectioned at −120 °C, and the sections were transferred to Formvar/carbon-coated copper grids. Samples were incubated with antibody in 1% bovine serum albumin for 30 min. After rinsing the samples four times with PBS, protein A-gold (10 nm) in 1% bovine serum albumin was added for 20 min. Contrasting stain procedures were carried out using 2% methyl cellulose: 3% uranyl acetate (9:1) for 10 min on ice. To dry the samples, grids were picked up with a loop and excess liquid was removed using filter paper.
DNase I Footprinting Analysis
The mitochondrial DNA fragment encompassing 15858/16063 bp (GenBank™ accession number J01420) was prepared by PCR and used as a probe in the DNase I footprinting experiment (8).
Electrophoretic Mobility Shift Assay (EMSA)
We performed EMSAs on mitochondrial extracts from rat brain tissues and cortical neurons using a 32P-labeled oligonucleotide containing a wild-type CREB-binding site as described previously (7). Mitochondrial D-loop CRE oligonucleotides were designed from the CRE I–III sequences shown in Fig. 2B. Supershifts were performed with pCREB-specific antibody for the Ser-133 residue (Upstate Biotechnology Inc., Lake Placid, NY), ATF-1/CREB (25C10G; Santa Cruz Biotechnology), or CREB-1 (24H4B and 240; Santa Cruz Biotechnology).
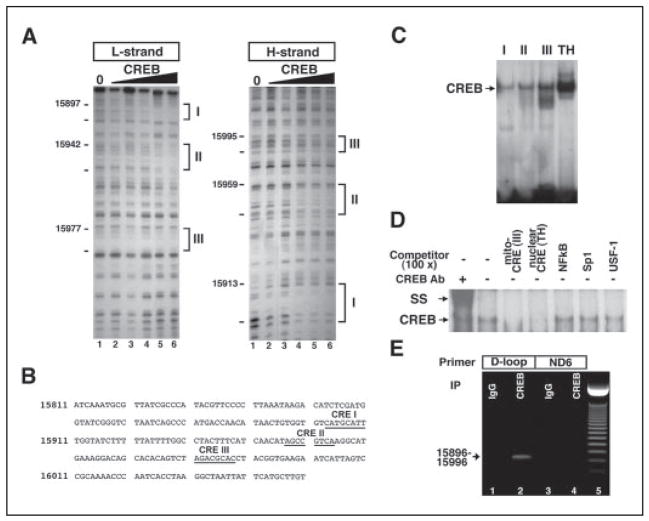
FIGURE 2. CREB binds to the non-coding region of mitochondrial DNA.
A, in vitro footprinting analysis reveals three putative CRE-like sites in the D-loop of mouse mitochondrial DNA. B, sequences of CRE-like sites in the D-loop are presented. Three CRE-like sequences (I–III) as underlined are found in the D-loop. Arrows indicate the direction of CREB binding to the putative CRE. C, EMSA shows that CREB binds to mitochondrial CRE-like sites (I–III). A canonical CRE site in a nuclear gene, tyrosine hydroxylase (TH), promoter was used as a standard of CREB binding activity to mitochondrial DNA. D, nonspecific competitor analysis confirmed that CREB is specifically associated with mitochondrial CRE-like sequences. SS, supershift analysis. E, CREB association with mitochondrial DNA was further examined using DNA-protein cross-linking, immunoprecipitation of pCREB, and PCR methods (see supplemental methods). The mitochondria pellet was cross-linked by 1% formaldehyde, sonicated, and immunoprecipitated. Immuoprecipitated and eluted mitochondrial DNA with CREB antibody and IgG were amplified with primers designed for CRE sites in D-loop sequences and non-CRE sites in ND6 sequences. Lanes 1 and 3, IgG immunoprecipitation; lanes 2 and 4, CREB antibody immunoprecipitation; lane 5, 50-bp molecular marker. The arrow indicates an amplified signal with CRE site primers (15896 –15996).
Mitochondrial DNA and Protein Cross-linking and Immunoprecipitation
Mitochondrial DNA and protein cross-linking and immunoprecipitation analysis for CREB binding to mitochondrial DNA was performed using a chromatin immunoprecipitation assay kit (Upstate Biotechnology Inc.). Mitochondrial fraction pellets or HT-22 cells transfected with pDs-Red2-Mito empty vector, pDs-Red2-Mito-wt-CREB, and pDsRed2-A-CREB for 24 h were cross-linked with 1% formaldehyde for 20 min at room temperature. PCR amplification was carried out for 35 cycles, and PCR products were separated on 2% agarose gels. Three primers were used to amplify the segment flanking the three or two CRE-like sites in the D-loop of mitochondria. The forward primers were 5′-GTGGTGTCATGCATTTGGTATCT-3′ and 5′-ATCAACATAGCCGTCAAGGCATG-3′, and the reverse primer was 5′-TCACCGTAGGTGCGTCTAGACTGT-3′. Normal rabbit IgG served as a negative control.
Construction of Plasmids
To generate mitochondrially targeted fusion proteins, wt-CREB and A-CREB (9) were subcloned into pECFP-Mito and pDs-Red2 Mito vector (CLONTECH Laboratories, Inc., Palo Alto, CA).
Real-time PCR and Conventional RT-PCR
To quantify the copy number of mRNA of the ND5 and ND6 genes, real-time PCR was performed using a DNA Engine Opticon System (MJ Research Inc., Las Vegas, NV). For the detection of mitochondria-encoded gene expression, total cellular RNA digested with RNase-free DNase was reverse-transcribed with SuperScript RT-PCR kit (Invitrogen). The probe and primers designed to amplify mitochondrial transcripts were as follows: human ND2, 4704–5103; human ND4, 11479–11929; human ND5, 13569–13917, human cytochrome b, 15494–15748; human ATPase 6, 8854–9087; human complex IV, 6188–6377, human mitochondrial 12 S rRNA, 576–422; human 18 S RNA, 5′-CCGAGATTGAGCAATAACAGG-3′ (forward) and 5′-AGTTCGACCGTCTTCTCAGG-3′ (reverse).
Measurement of Mitochondrial Enzyme Activities
Respiratory activities were measured polargraphically as described previously (10).
Western Blot Analysis
Western blot was performed using subcellular fractions and cell or tissue lysates as described previously (7).
RESULTS AND DISCUSSION
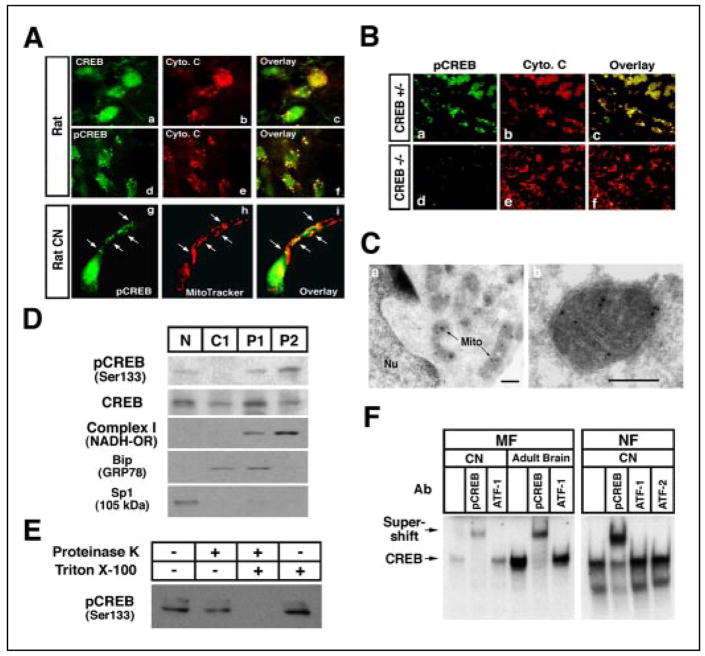
In the neurons of the adult rat cortex, we found that CREB and pCREB (Ser-133) were partially extranuclear. Furthermore, the extranuclear CREB colocalized with the mitochondrial marker cytochrome c (Fig. 1A, panels a–f). In cultured immature embryonic cortical neurons, CREB and pCREB partially colocalized with the mitochondrial marker MitoTracker and were found within the proximal segments of dendrites (Fig. 1A, panels g–i). CREB staining was most prominent in neurofilament-200 (NF-200)-positive cells (data not shown). Moreover, both nuclear and extranuclear CREB and pCREB immunoreactivity was absent in neurons derived from CREB null mice (Fig. 1B). In addition, we found that CREB-binding protein is not localized in the mitochondria (supplemental Fig. 1). To pinpoint the subcellular localization of extranuclear CREB, we performed immuno-electron microscopy using cultured cortical neurons. This analysis demonstrated that pCREB is present within both the nucleus and the mitochondrial matrix of these neurons (Fig. 1C). We further verified the localization of CREB to the mitochondrial fraction using subcellular fractionation of adult rat brain tissue by sucrose density centrifugation (Fig. 1D). The presence of complex I (NADH oxidoreductase) immunoreactivity, together with the absence of immunoreactivity of the endoplasmic reticulum marker Bip (GRP78) or the nuclear marker Sp1, confirmed that the fractions contained only mitochondria. Mitochondrial pCREB was resistant to proteinase K treatment in the absence of detergent, suggesting that pCREB is located within the mitochondrial matrix (Fig. 1, C and E). We employed EMSA to determine whether mitochondrial CREB is capable of binding to a canonical CRE DNA sequence (5′-TGACGTCA-3′). Consistent with the immunoblot and immunolocalization experiments, mitochondrial extracts from rat embryonic cortical neurons and adult rat brain tissue revealed the presence of CRE-binding activities (Fig. 1F).
FIGURE 1. Identification of mitochondria as a site of CREB localization in neurons.
A, both CREB (panels a– c) and pCREB (Ser-133) (panels d–i) colocalize with mitochondria in the cerebral cortex of rat adult brain in situ (panels a–f) and primary cultured cortical neurons (panels g–i). White arrows indicate colocalization with punctate staining of pCREB and mitochondria in a single cortical neuron. B, pCREB immunoreactivity (panels a and d) with cytochrome c (Cyto. c) (panels b and e) is impaired in dorsal root ganglia in CREB null (−/−) mice (panels a– c) but not in CREB+/− (panels d–f). C, immunogold electron microscopy shows pCREB in the mitochondrial matrix of cortical neurons. Mito, mitochondria; Nu, nucleus. Scale bar: 200 nm. D, CREB is found in the mitochondrial subcellular fraction. pCREB was detected as a 43-kDa band. The same blot was then stripped and reblotted with anti-complex I (mitochondrial NADH oxidoreductase (OR), 37-kDa subuint) (mitochondrial marker), Bip (GRP78) (endoplasmic reticulum marker), and Sp1 (nucleus marker), each of which were also detected. N, nuclear fraction; C1, cytoplasmic fraction 1; P1, pellet 1 mitochondria-rich fraction; P2, pellet 2 mitochondria fraction. E, proteinase K assay using the mitochondrial fraction shows that pCREB is localized to the mitochondrial matrix. F, mitochondrial fraction (MF) of cortical neurons (CN) and adult rat brain show specific CREB DNA binding activity by EMSA. EMSA was performed with mitochondrial and nuclear extracts using 32P-labeled CRE oligonucleotides. Supershift analysis for identification of CREB complex used antibodies (Ab) for pCREB (Ser-133), ATF-1, and ATF-2. NF, nuclear fraction.
Although CREB lacks a classical mitochondrial targeting sequence, alternative pathways exist for targeting proteins to the mitochondria (11, 12). To determine whether chaperone molecules may play a role in the mitochondrial targeting of CREB, we performed a coimmunoprecipitation assay using lysates from the mitochondrial fraction to assess the degree of association of CREB and mitochondria (mt) HSP70 (GRP 75) (11, 12). We found that CREB coprecipitated with mtHSP70 in human and rat brain tissues and in embryonic neurons (supplemental Fig. 2). The data support the possibility that chaperone molecules are involved in delivering CREB to the mitochondria.
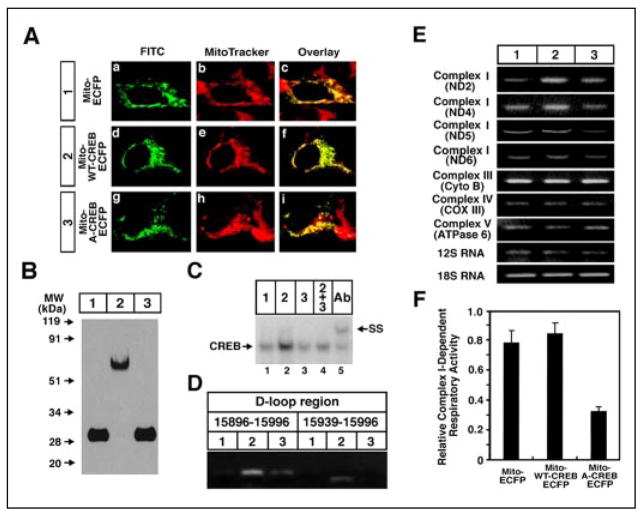
The D-loop is the control site for both transcription and DNA replication in the mitochondrial genome. Within this region, we identified variant CRE-like sequences by in vitro footprinting analysis (Fig. 2, A and B) and observed that these sites formed specific DNA-protein complexes with CREB (Fig. 2C) and that complex formation by mitochondrial CRE elements did not compete with NFκB, Sp1, or USF-1 cis-elements (Fig. 2D). Furthermore, we performed chromatin immunoprecipitation with a CREB antibody to demonstrate that CREB is bound to these mitochondrial DNA sites in the intact cell (Fig. 2E) (supplemental Fig. 3). To evaluate the role of CREB in mitochondrial gene expression and survival, we expressed a mitochondrially targeted form of either CREB (mito-wt-CREB-ECFP) or its dominant negative (mito-A-CREB). Transiently expressed mito-wt-CREB-ECFP protein colocalized with mitotracker staining, confirming that the heterologous protein is localized in the mitochondria (Fig. 3A). Immunoblotting verified that mito-wt-CREB (~80 kDa), mito-A-CREB (~30 kDa), and mito-ECFP (~28 kDa) fusion proteins were expressed at similar levels (Fig. 3B). EMSA analysis demonstrated that the dominant negative mito-A-CREB disrupts the mitochondrial DNA-binding activity of CREB (Fig. 3, C and D). To more definitively address the function of CREB targeted to mitochondria, we prepared cell lines that stably express either mito-wt-CREB or mito-A-CREB. We used real-time PCR and RT-PCR to determine to what extent mitochondrial gene expression was altered in the stable cell lines (Fig. 3E and supplemental Table 1). We found that mito-wt-CREB and mito-A-CREB inversely regulate the expression of some mitochondrial genes. Mito-wt-CREB increased levels of transcripts of the ND2, ND4, and ND5 mitochondrial genes, while mito-A-CREB decreased them. Interestingly, ND5 expression was significantly reduced in mito-A-CREB cells. Consistent with reduced expression of ND5 (a complex I subunit), we also observed a relative reduction of complex I activity in mito-A-CREB cells (Fig. 3F). We monitored levels of the c-fos gene, a transcript regulated by nuclear CREB levels, to verify that mito-A-CREB does not affect nuclear CREB activity. As expected, we found that neither mito-wt-CREB or A-CREB influences c-fos expression as compared with control (supplemental Fig. 4). Our results that mito-A-CREB down-regulates several of the mitochondrial genes, in part, likely reflect diminished mito-CREB transcriptional activity. However, the failure to detect a decrease in levels of some mitochondrial genes, such as the cytochrome b or ATPase 6 genes that are also encoded on the H strand, could be due to other factors, such as differences in mRNA stability. Indeed, mutations in the mitochondrial RNA binding protein, LRPPRC (leucine-rich pentatricopeptide repeat cassette) are responsible for a French Canadian form of Leigh’s syndrome. In this syndrome, cytochrome c oxidase mRNAs are selectively decreased as compared with other mRNAs encoded in the mitochondrial H-strand (13).
FIGURE 3. Mitochondrially targeted CREB directly regulates the expression of mitochondrial genes.
A, confocal microscopy showed the expression of mito-wt-CREB-ECFP vector (panel a; green) and mitotracker (panel b; red) staining and overlaid images (panel c) in SH-SY5Y cells. ECFP fusion proteins were visualized by indirect labeling using mouse anti-ECFP antibody and goat anti-mouse IgG antibody conjugated with fluorescein isothiocyanate. B, Western blot analysis of mito-ECFP (lane 1), mito-wt-CREB-ECFP (lane 2), and mito-A-CREB-ECFP fusion protein (lane 3). C, A-CREB inhibits CREB DNA binding to the mitochondrial EMSA of mitochondrial extracts from mito-ECFP (lane 1), mito-wt-CREB-ECFP (lane 2), and mito-A-CREB-ECFP (lane 3) or mito-wt-CREB-ECFP and mito-A-CREB-ECFP co-transfected (lane 4) cells. Anti-CREB antibody (Ab) was used for supershift analysis (lane 5). D, mitochondrially targeted A-CREB inhibits mito-wt-CREB association with mitochondrial D-loop CRE sequences. HT-22 cells were transfected with mito-Red2 (lane 1), mito-wt-CREB-Red2 (lane 2), and mito-A-CREB-Red2 and mito-wt-CREB-Red2 (lane 3) for 24 h. E, RT-PCR analysis of the expression of mitochondrial DNA-encoded genes in mito-ECFP (lane 1), mito-wt-CREB-ECFP (lane 2), and mito-A-CREB-ECFP (lane 3) in SH-SY5Y cell lines. Complex I (ND2, ND4, and ND5), complex III (cytochrome b (Cyto B)), complex IV (cytochrome c oxidase subunit III (COXIII)), complex V (ATPase 6), and 12 S RNA gene fragments were examined for mitochondrial gene expression. 18 S RNA was used as a nuclear-encoded gene control. The representative data are shown from three separate experiments (see numerical and real-time PCR data in the supplemental Table 1 and Fig. 4). F, the ratio of complex I-dependent respiration on glutamate/malate compared with complex II-dependent respiration on succinate (plus rotenone) was significantly decreased in permeabilized mito-A-CREB-ECFP cells compared with mito-ECFP and mito-wt-CREB-ECFP cells. Data are expressed as the mean ± S.E. of three to five separate experiments.
Previous studies have established that decreasing ND5 expression corresponds with decreasing complex I-dependent respiration, suggesting that ND5 transcript levels may tightly control mitochondrial respiration rate (14). Indeed, mitochondrial DNA mutations in the genes encoding the ND5 subunit of complex I are associated with mitochondrial myopathy and Lebers hereditary optic neuropathy, which show defects in complex I activity (14–16). Thus, CREB regulation of expression of the ND5 and/or ND6 subunit may influence mitochondrial respiration (17). Interestingly, however, intracellular ATP levels were not decreased by mito-A-CREB expression (supplemental Table 2).
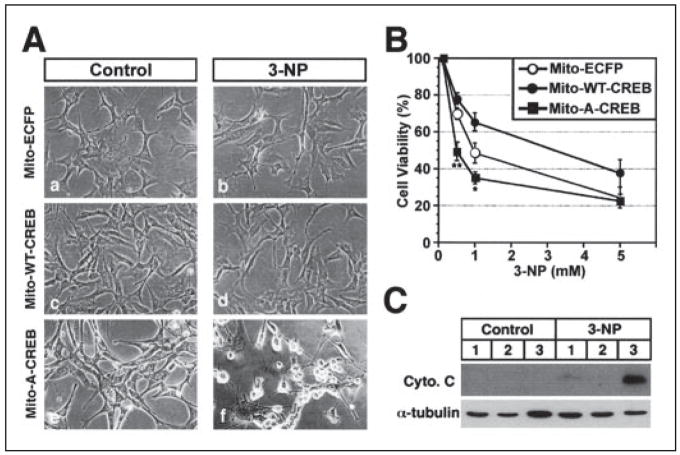
There are a number of mitochondrial inhibitors that affect complexes of the electron transport chain by reducing cellular levels of ATP, resulting in energy deficiency and mimicking HD pathogenesis (18–20). One such naturally occurring plant toxin, 3-nitropropionic acid (3-NP), is an irreversible inhibitor of succinate dehydrogenase and both the Krebs cycle and complex II activity of the electron transport chain (19). 3-NP is associated with HD-like symptoms in both humans and animals and, as such, has been used as an experimental model for HD (20). We further hypothesized that mitochondria may be more susceptible to mitochondrial toxins in the presence of a dominant negative or mutant mitochondrial CREB. To begin to address this possibility, we examined the effect of 3-NP on cell lines stably expressing either mito-wt-CREB or mito-A-CREB. Mito-wt-CREB cells were relatively resistant to 3-NP-induced cytotoxicity, whereas mito-A-CREB cells were more susceptible (Fig. 4, A and B). Increased 3-NP cytotoxicity in mito-A-CREB cells was associated with an increase of cytochrome c release compared with mito-wt-CREB and mito-ECFP cells (Fig. 4C). These results support the hypothesis that defects in mitochondrial transcription are associated with increased vulnerability to mitochondrial toxins.
FIGURE 4. Mitochondrial CREB is involved in the neuronal survival.
A, mito-A-CREB cells (panels e and f) were highly susceptible to 3-NP-induced cytotoxicity compared with mito-ECFP (panels a and b) and mito-wt-CREB cells (panels c and d). Cells were treated with vehicle (control) and 3-NP (0.5–5 mM) for 72 h. B, cell viability was measured by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. Data are the average ± S.E. of three separate experiments: significant at p < 0.05 (*) and p < 0.01 (**). C, cytochrome c release is higher in mito-A-CREB cells (lane 3) compared with mito-ECFP (lane 1) and mito-wt-CREB cells (lane 2) in response to 3-NP. Cytosol fractions were prepared from cells treated with vehicle (control) or 3-NP (0.5 mM) for 72 h.
We further found atrophy of striatal neurons in the CREB−/− mice measured by Nissl staining (1). The data directly support a survival and/or trophic role for CREB in striatal neurons (supplemental Fig. 5A) (21). Interestingly, a decreased level of mitochondrial CREB proteins was found to correlate with the shrinkage of the striatum in R6/2 mice (supplemental Fig. 5B). Because the striatal phenotype of CREB−/− mice is reminiscent of HD (21), our data are consistent with a loss of function of mitochondrial CREB as a determinant of striatal neuron atrophy and/or loss in HD.
To determine whether there is an alteration of mitochondrial transcription in a transgenic mouse HD model, we examined mitochondrial transcript levels in R6/2 mice. Mitochondrial ND5 and ND6 mRNA were significantly decreased in R6/2 mice, similar to the changes induced by a dominant negative CREB targeted to the mitochondria (supplemental Fig. 6A and B, and Fig. 3). Furthermore, striatal cell damage and neuronal loss in R6/2 mice were greater than in control mice in response to 3-NP injection (supplemental Fig. 6C). These results support the notion that defects in mitochondrial transcription are associated with increased vulnerability to mitochondrial toxins in vivo as well as in vitro (Fig. 4). In concert with previous studies demonstrating defective mitochondrial function in HD patients and transgenic mice (19, 22–24), our findings suggest that loss of mitochondrial CREB may play a role in the pathophysiology of HD.
It has been hypothesized that mitochondrial dysfunction plays a role in aging and in neurodegenerative diseases such as HD, Alzheimer disease, and Parkinson disease (18–20, 22–26). Our data demonstrate that CREB is present within the mitochondria and functions to regulate mitochondrial gene expression in neurons. Other transcription factors, such as p53 and the orphan receptor TR3, localize, in part, to mitochondria and exert pro-apoptotic effects with or without affecting the expression of the mitochondrial genes (27, 28). In the present study, mitochondrial CREB appears to regulate pro-survival effects, at least in part, by regulating mitochondrial genes whose expression may be controlled by CRE-like elements within the D-loop of the mitochondrial genome (29, 30). Our data also suggest that CREB-mediated modulation of mitochondrial gene expression plays an important role in determining vulnerability to cell death upon expressing a defective CREB (A-CREB) and a mitochondrial toxin known to induce a phenotype similar to HD. This is in agreement with previous studies showing that overexpression of a dominant negative CREB transgene induces apoptosis (4, 5, 31) and that tissue-specific deletion of both CREB and CREM leads to striatal toxicity similar to that observed in HD (21).
However, these previous studies have assumed that the toxicity is explained by a loss of nuclear CREB activity. We show that mutant huntingtin (mtHtt) can bind CREB (supplemental Fig. 7). Thus, mutant huntingtin has the potential to sequester CREB and decrease CREB-mediated mitochondrial transcriptional activity (32–36). In addition to the effects that the loss of CREB may cause in nuclear function, huntingtin proteins may affect the mitochondrial CREB function. Our work suggests the latter function may be equally important in Huntington disease.
Acknowledgments
We thank K. Smith for technical support and Dr. F. Takaku for providing pCG-CREB plasmid.
Footnotes
This work was supported by National Institutes of Health Grants NS52724-01 (to H. R.), P30 AG13846 (to J. L.), MH48866 (to K.-S. K.), NS045242 and NS045806 (to R. J. F.), and NS39170 and NS40591 (to R. R. R.); by the High Q Foundation and the Huntington Disease Society of America (to H. R. and R. J. F.); and by the Veterans Administration (to R. J. F. and J. L.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental methods, references, and Figs. 1–7 and Tables 1 and 2.
The abbreviations used are: CREB, cAMP response element-binding protein; CRE, CREB response element; ATF, activating transcription factor; ND, NADH dehydrogenase; EMSA, electric mobility shift assay; mtHtt, mutant huntingtin; PBS, phosphate-buffered saline; ECFP, enhanced cyan fluorescent protein; RT, reverse transcription; HD, Huntington disease; 3-NP, 3-nitropropionic acid.
References
- 1.Lonze BE, Ginty DD. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 2.Mayr B, Montminy M. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 3.Impey S, Goodman RH. Sci STKE. 2001;82:PE1. doi: 10.1126/stke.2001.82.pe1. [DOI] [PubMed] [Google Scholar]
- 4.Riccio A, Ahn S, Davenport CM, Blendy JA, Ginty DD. Science. 1999;286:2358–2361. doi: 10.1126/science.286.5448.2358. [DOI] [PubMed] [Google Scholar]
- 5.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 6.Cammarota M, Paratcha G, Bevilaqua LR, Levi de Stein M, Lopez M, Pellegrino de Iraldi A, Izquierdo I, Medina JH. J Neurochem. 1999;72:2272–2277. doi: 10.1046/j.1471-4159.1999.0722272.x. [DOI] [PubMed] [Google Scholar]
- 7.Ryu H, Lee J, Olofsson BA, Mwidau A, Dedeoglu A, Escudero M, Flemington E, Azizkhan-Clifford J, Ferrante RJ, Ratan RR. Proc Natl Acad Sci U S A. 2003;100:4281–4286. doi: 10.1073/pnas.0737363100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim CH, Hwang DY, Park JJ, Kim KS. J Neurosci. 2002;22:2579–2589. doi: 10.1523/JNEUROSCI.22-07-02579.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahn S, Olive M, Aggarwal S, Krylov D, Ginty DD, Vinson C. Mol Cell Biol. 1998;18:967–977. doi: 10.1128/mcb.18.2.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jazayeri M, Andreyev A, Will Y, Ward M, Anderson CM, Clevenger W. J Biol Chem. 2003;278:9823–9830. doi: 10.1074/jbc.m211730200. [DOI] [PubMed] [Google Scholar]
- 11.Marchenko ND, Zaika A, Moll UM. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 12.Merrick BA, He C, Witcher LL, Patterson RM, Reid JJ, Pence-Pawlowski PM, Selkirk JK. Biochim Biophys Acta. 1996;1297:57–68. doi: 10.1016/0167-4838(96)00089-1. [DOI] [PubMed] [Google Scholar]
- 13.Xu F, Morin C, Mitchell G, Ackerley C, Robinson BH. Biochem J. 2004;382:331–336. doi: 10.1042/BJ20040469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bai Y, Shakwley RM, Attardi G. Mol Cell Biol. 2000;20:805–815. doi: 10.1128/mcb.20.3.805-815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bentlage HA, Janssen AJ, Chomyn A, Attardi G, Walker JE, Schagger H, Sengers RC, Trijbels FJ. Biochim Biophys Acta. 1995;1234:63–73. doi: 10.1016/0005-2736(94)00288-z. [DOI] [PubMed] [Google Scholar]
- 16.Wissinger B, Besch D, Baumann B, Fauser S, Christ-Adler M, Jurklies B, Zrenner E, Leo-Kottler B. Biochem Biophys Res Commun. 1997;234:511–515. doi: 10.1006/bbrc.1997.6660. [DOI] [PubMed] [Google Scholar]
- 17.Enriquez JA, Fernandez-Silva P, Garrido-Perez N, Lopez-Perez MJ, Perez-Martos A, Montoya J. Mol Cell Biol. 1999;19:657–670. doi: 10.1128/mcb.19.1.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludolph AC, Seelig M, Ludolph AG, Sabri MI, Spencer PS. Ann N Y Acad Sci. 1992;648:300–302. doi: 10.1111/j.1749-6632.1992.tb24562.x. [DOI] [PubMed] [Google Scholar]
- 19.Beal MF, Brouillet E, Jenkins BG, Ferrante RJ, Kowall NW, Miller JM, Storey E, Srivastava R, Rosen BR, Hyman BT. J Neurosci. 1993;13:4181–4192. doi: 10.1523/JNEUROSCI.13-10-04181.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bogdanov MB, Ferrante RJ, Kuemmerle S, Klivenyi P, Beal MF. J Neurochem. 1998;71:2642–2644. doi: 10.1046/j.1471-4159.1998.71062642.x. [DOI] [PubMed] [Google Scholar]
- 21.Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, Otto C, Schmid W, Schutz G. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- 22.Brouillet E, Hantraye P, Ferrante RJ, Dolan R, Leroy-Willig A, Kowall NW, Beal MF. Proc Natl Acad Sci U S A. 1995;92:7105–7109. doi: 10.1073/pnas.92.15.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beal MF. Ann Neurol. 1995;38:357–366. doi: 10.1002/ana.410380304. [DOI] [PubMed] [Google Scholar]
- 24.Borthwick GM, Johnson MA, Ince PG, Shaw PJ, Turnbull DM. Ann Neurol. 1999;46:787–790. doi: 10.1002/1531-8249(199911)46:5<787::aid-ana17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 25.Chandrasekaran K, Giordano T, Brady DR, Stoll J, Martin LJ, Rapoport SI. Brain Res Mol Brain Res. 1994;24:336–340. doi: 10.1016/0169-328x(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 26.Green DR, Reed JC. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 27.Mihara M, Erster S, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM. Mol Cell. 2003;11:577–590. doi: 10.1016/s1097-2765(03)00050-9. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Kolluri SK, Gu J, Dawson MI, Cao X, Hobbs PD, Lin B, Chen G, Lu J, Lin F, Xie Z, Fontana JA, Reed JC, Zhang X. Science. 2000;289:1159–1164. doi: 10.1126/science.289.5482.1159. [DOI] [PubMed] [Google Scholar]
- 29.Casas F, Rochard P, Rodier A, Cassar-Malek I, Marchal-Victorion S, Wiesner RJ, Cabello G, Wrutniak C. Mol Cell Biol. 1999;19:7913–7924. doi: 10.1128/mcb.19.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ryu H, Lee J, Impey S, Ratan RR, Ferrante RJ. Proc Natl Acad Sci U S A. 2005;102:13915–13920. doi: 10.1073/pnas.0502878102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaworski J, Mioduszewska B, Sanchez-Capelo A, Figiel I, Habas A, Gozdz A, Proszynski T, Hetman M, Mallet J, Kaczmarek L. J Neurosci. 2003;23:4519–4526. doi: 10.1523/JNEUROSCI.23-11-04519.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimohata T, Nakajima T, Yamada M, Uchida C, Onodera O, Naruse S, Kimura T, Koide R, Nozaki K, Sano Y, Ishiguro H, Sakoe K, Ooshima T, Sato A, Ikeuchi T, Oyake M, Sato T, Aoyagi Y, Hozumi I, Nagatsu T, Takiyama Y, Nishizawa M, Goto J, Kanazawa I, Davidson I, Tanese N, Takahashi H, Tsuji S. Nat Genet. 2000;26:29–36. doi: 10.1038/79139. [DOI] [PubMed] [Google Scholar]
- 33.Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft TF, Kato K, Rubinsztein DC. Hum Mol Genet. 2001;10:1829–1845. doi: 10.1093/hmg/10.17.1829. [DOI] [PubMed] [Google Scholar]
- 34.Nucifora FC, Jr, Sasaki M, Peters MF, Huang H, Cooper JK, Yamada M, Takahashi H, Tsuji S, Troncoso J, Dawson VL, Dawson TM, Ross CA. Science. 2001;291:2423–2428. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 35.Gines S, Seong IS, Fossale E, Ivanova E, Trettel F, Gusella JF, Wheeler VC, Persichetti F, MacDonald ME. Hum Mol Genet. 2003;12:497–508. doi: 10.1093/hmg/ddg046. [DOI] [PubMed] [Google Scholar]
- 36.Panov AV, Gutekunst CA, Leavitt BR, Hayden MR, Burke JR, Strittmatter WJ, Greenamyre JT. Nat Neurosci. 2002;5:731–736. doi: 10.1038/nn884. [DOI] [PubMed] [Google Scholar]