Abstract
B-cell-activating factor (BAFF) is a survival and maturation factor for B cells belonging to the tumour necrosis factor superfamily. Among three identified functional receptors, the BAFF receptor (BAFF-R) is thought to be responsible for the effect of BAFF on B cells though details of how remain unclear. We determined that a hairy-cell leukaemia line, MLMA, expressed a relatively high level of BAFF-R and was susceptible to apoptosis mediated by either CD20 or B-cell antigen receptor (BCR). Using MLMA cells as an in vitro model of mature B cells, we found that treatment with BAFF could inhibit apoptosis mediated by both CD20 and BCR. We also observed, using immunoblot analysis and microarray analysis, that BAFF treatment induced activation of nuclear factor-κB2 following elevation of the expression level of Bcl-2, which may be involved in the molecular mechanism of BAFF-mediated inhibition of apoptosis. Interestingly, BAFF treatment was also found to induce the expression of a series of genes, such as that for CD40, related to cell survival, suggesting the involvement of a multiple mechanism in the BAFF-mediated anti-apoptotic effect. MLMA cells should provide a model for investigating the molecular basis of the effect of BAFF on B cells in vitro and will help to elucidate how B cells survive in the immune system in which BAFF-mediated signalling is involved.
Keywords: apoptosis, B-cell-activating factor, Bcl-2, B-cell receptor, CD20
Introduction
The immune system comprises a variety of immune effector cells, including T and B lymphocytes and antigen-presenting cells, such as dendritic cells and others; it protects individuals from infections and cancer. To maintain these sophisticated mechanisms, a very subtle balance between the life and death of the immune effector cells must be maintained to eliminate, by apoptosis, potentially harmful self-reactive lymphocytes and only allow the survival, development and activation of safe and protective immune cells. For this purpose, a number of molecules are involved in this regulatory system.1
B-cell-activating factor (BAFF, also termed BlyS, TALL-1, THANK and zTNF4) produced by monocytes, dendritic cells and some T cells is a member of the tumour necrosis factor (TNF) superfamily and is a type 2 transmembrane-bound protein that can also be expressed as a soluble ligand.2 BAFF was first described as a factor that stimulates cell proliferation and the secretion of immunoglobulin in B cells.3–7 Transgenic mice that overexpress BAFF in lymphoid tissues exhibited hyperplasia of the mature B-cell compartment.8–10 In contrast, mice deficient in BAFF showed a deficit in peripheral B lymphocytes10,11 and an almost complete loss of follicular and marginal zone B lymphocytes in secondary lymphoid organs. This suggests an absolute requirement for BAFF in normal B-cell development.10 In contrast, a later examination of immunized BAFF-null mice validated the BAFF-independent nature of germinal centre formation and that antibody responses, including high-affinity responses, were attenuated, indicating that BAFF is required for maintenance, but not initiation, of the germinal centre reaction.12 Based on the above evidence, BAFF is considered to be a survival and maturation factor for B lymphocytes and has emerged as a crucial factor that modulates B-cell tolerance and homeostasis.2,13 However, the precise role of BAFF in B-cell development is still controversial and it has been reported that the capacity of B lymphocytes to bind BAFF is correlated with their maturation state and that the effect of BAFF is dependent on the maturation stage of the B lymphocytes.2,14
Recent studies have further shown that BAFF affects not only B lymphocytes but also T lymphocytes.15,16 The three distinct receptors for BAFF, namely the BAFF receptor (BAFF-R, also termed BR3), the B-cell maturation antigen (BCMA), and the transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI), have been identified and BAFF binds with a similar high affinity to these receptors.7,17–23 Among these receptors, however, BAFF-R is thought to be responsible for the survival and differentiation of B cells,24 whereas the molecular basis of BAFF-mediated signalling remains unclear.
A number of systems inducing apoptosis in B cells are present to eliminate inappropriate clones, such as self-acting B cells. For example, it is reported that stimulation via particular surface molecules, including B-cell receptor antigen (BCR) and CD20, induces apoptosis in cultured B cells.25,26 The balance between apoptosis-inducing systems and survival systems, such as CD20 and BAFF-mediated signalling, would be important for the maintenance of appropriate B-cell development, though details are not known.
To elucidate the molecular basis of the interaction between apoptosis-inducing signals and BAFF-mediated cell survival signals in B cells, we have employed a B-cell line that expresses BAFF-R and is sensitive to CD20-mediated and BCR-mediated apoptosis. In this paper, we present evidence that BAFF-mediated stimulation inhibits the apoptosis induced by both CD20-mediated and BCR-mediated signalling. The possible mechanisms involved in BAFF-mediated cell responses that regulate these apoptotic stimuli are discussed.
Materials and methods
Cells and reagents
The human hairy cell leukaemia cell line MLMA was obtained from the Japanese Cancer Research Resources Bank (JCRB, Tokyo, Japan). Cells were cultured in RPMI-1640 medium supplemented with 10% fetal calf serum at 37° in a humidified 5% CO2 atmosphere.
Recombinant human BAFF and a proliferation-inducing ligand (APRIL) were obtained from R&D Systems, Inc. (Minneapolis, MN), and used at a concentration of 400 ng/ml for cell stimulation unless otherwise described. The mouse monoclonal antibodies (mAbs) used for the immunofluorescence analysis were anti-CD10, anti-CD20, anti-CD21, anti-CD22, anti-CD24, anti-CD40, anti-human leucocyte antigen DR (HLA-DR; Beckman Coulter, Inc., Fullerton, CA); anti-CD19 (Becton Dickinson and Company, BD, Franklin Lakes, NJ); anti-κ, anti-λ, anti-μ, anti-δ, anti-γ (Dako, Denmark A/S); anti-BAFF-R (Santa Cruz Biotechnology, Santa Cruz, CA); and anti-CD45 (American Type Culture Collection, ATCC, Manassas, VA). The rat mAbs against BCMA (Vicky-1) and TACI (1A1) were purchased from Santa Cruz Biotechnology. The mouse mAbs used for the immunochemical analysis were anti-caspase-2, anti-caspase-3 and anti-glycogen synthase kinase-3β (GSK-3β; Becton Dickinson); anti-caspase-9 (Medical & Biological Laboratories Co., Ltd, Nagoya, Japan); anti-nuclear factor-κB (NF-κB) p52 (C-5), anti-Bcl-2 (100) from Santa Cruz; and anti-β-actin (AC-15) from Sigma-Aldrich Co. (St Louis, MO). The rabbit polyclonal antibodies used were anti-cleaved poly ADP-ribose polymerase (PARP), anti-cleaved caspase-3, anti-phospho-GSK-3β (Ser9) and anti-phospho-GSK-3α/β (Ser9, 21) from Cell Signaling Technology, Inc. (Danvers, MA). A goat anti-NF-κB p50 (C-19) from Santa Cruz was also used. Secondary antibodies, including fluorescein isothiocyanate- (FITC) and enzyme-conjugated antibodies, were purchased from either Jackson ImmunoResearch Laboratories, Inc. (West Grove, PA) or Dako. To cross-link BCR, purified anti-μ rabbit polyclonal antibody (10 μg/ml) from Jackson ImmunoResearch Laboratories, Inc. was used. To cross-link CD20, a mouse anti-CD20 mAb from Beckman Coulter and a secondary anti-mouse immunoglobulin antibody from Jackson ImmunoResearch Laboratories, Inc. were used each at a concentration of 5 μg/ml.
Immunofluorescence analysis and detection of apoptosis
Cells were stained with FITC-labelled mAbs and analysed by flow cytometry (EPICS-XL, Beckman Coulter) as described previously.27 To quantify the incidence of apoptosis, cells were incubated with FITC-labelled annexin V using a MEBCYTO-Apoptosis kit (Medical & Biological Laboratories Co., Ltd) and then analysed by flow cytometry according to the manufacturer’s directions. Apoptotic cells were also detected by nuclear-staining with DAPI and examined by confocal microscopy as described previously.28 The enzymatic activity of caspases -2, -3, -9 was assessed by using a colorimetric protease assay kit for each caspase (Medical & Biological Laboratories Co., Ltd) according to the manufacturer’s protocol.
Immunoblotting
Immunoblotting was performed as described previously.29 Briefly, cell lysates were prepared by solubilizing the cells in lysis buffer (containing 20 mm Na2PO4, pH 7·4, 150 mm NaCl, 1% Triton X-100, 1% aprotinin, 1 mm phenylmethylsulphonylfluoride, 100 mm NaF, and 2 mm Na3VO4), and the total protein concentration was determined using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). For each cell lysate, 20 μg was separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis and transferred to a nitrocellulose membrane using a semidry Transblot system (Bio-Rad). After blocking with 3% skimmed milk in phosphate-buffered saline, the membrane was incubated with the appropriate combination of primary and secondary antibodies as indicated, washed intensively, and examined using the enhanced chemiluminescence reagent system (ECL plus; GE Healthcare Bio-Sciences AB, Uppsala, Sweden).
DNA microarray analysis
The DNA microarray analysis was performed using GeneChip (Affymetrix, Santa Clara, CA). Total RNA isolated from MLMA cells treated with and without BAFF for 12 hr was reverse transcribed and labelled using One-Cycle Target Labeling and Control Reagents as instructed by the manufacturer (Affymetrix). The labelled probes were hybridized to Human Genome U133 Plus 2.0 Arrays (Affymetrix). The arrays were analysed using genechip operating Software 1.2 (Affymetrix). Background subtraction and normalization were performed with genespring gx 7.3 software (Agilent Technologies, Santa Clara, CA). Signal intensities were prenormalized based on the median of all measurements on that chip. To account for the difference in detection efficiency between the spots, prenormalized signal intensities on each gene were normalized to the median of prenormalized measurements for that gene. The data were filtered with the following steps. (1) Genes that were scored as absent in both samples were eliminated. (2) Genes with a signal intensity lower than 90 in both samples were eliminated. (3) Performing cluster analysis using filtering genes, genes were selected that exhibited increased expression or decreased expression in BAFF-treated cells.
Results
Immunophenotypic characterization of MLMA cells
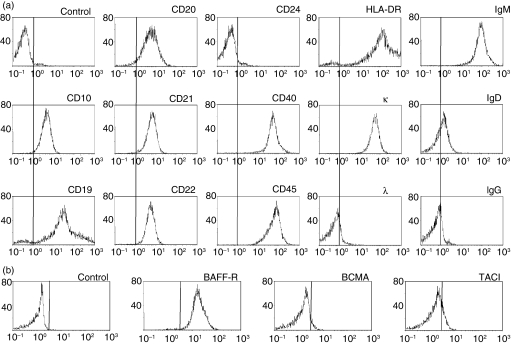
While screening to identify human cell lines expressing BAFF-R, we found that MLMA cells expressed higher levels of BAFF-R than other human B-cell lines. Although the MLMA cell line is known to have been established from a patient with hairy-cell leukaemia, details were not reported. Therefore, we first examined the immunophenotypic characteristics of MLMA cells. Consistent with the JCRB records, flow cytometric analysis revealed that MLMA cells expressed high levels of μ heavy chain and low levels of δ heavy chain with expression of κ light chain (Fig. 1a). In addition to the CD19 and HLA-DR, MLMA cells were found to express mature B-cell antigens, including CD20, CD21, CD22 and CD40, but not CD24. Notably, MLMA cells showed the expression of CD10. When the expression of three types of receptors for BAFF was similarly examined, MLMA cells exhibited apparent expression of BAFF-R, while the levels of BCMA and TACI were found to be quite low (Fig. 1b). The data indicate that MLMA cells exhibit immunophenotypic characteristics of mature B cells expressing BAFF-R.
Figure 1.
Immunophenotypic characterization of MLMA cells. (a) MLMA cells were stained with specific fluorescein isothiocyanate (FITC)-labelled monoclonal antibodies (mAbs) against B-cell differentiation antigens and analysed by flow cytometry. The x-axis represents fluorescence intensity and the y-axis the relative cell number; control was isotype-matched mouse immunoglobulin. (b) The expression of B-cell-activating factor receptor (BAFF-R), transmembrane activator and calcium modulator and cyclophilin ligand interactor (TACI), and B-cell maturation antigen (BCMA) on MLMA cells was also examined as in (a).
Cross-linking of BCR and CD20 induces apoptosis in MLMA cells
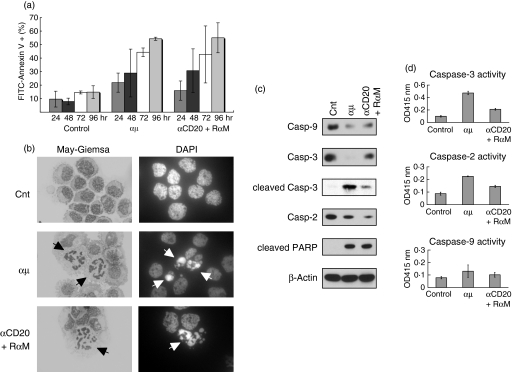
It has been well documented that cross-linking of BCR using anti-μ heavy chain antibodies induces apoptosis in some B cells in vitro.25 Recent studies including our own have also shown that CD20 cross-linking mediates apoptosis in human B-cell lines in a manner involving raft-mediated signalling.26,30 Therefore, we next examined whether cross-linking of either BCR or CD20 mediated apoptosis in MLMA cells. As shown in Fig. 2(a), when anti-μ antibodies were added to the culture, a time-dependent increase in the number of cells bound to annexin V was observed, suggesting the occurrence of apoptosis in MLMA cells after BCR cross-linking. The apoptosis was confirmed by the morphological appearance of nuclear fragmentation, a typical feature of apoptosis, detected by either Giemsa-staining or nuclear-staining with DAPI (Fig. 2b). Immunoblotting revealed the cleavage of caspases -9, -3 and -2 and of PARP after treatment with anti-μ antibodies (Fig. 2c), indicating that caspase activation was involved in the apoptosis. In the case of caspase-3, we also detected a 17 000 molecular weight cleaved fragment by using a specific antibody (Fig. 2c). In addition, elevation of the enzymatic activity of each caspase after cross-linking of BCR was detected by a colorimetric protease assay (Fig. 2d). We also examined the effect of anti-CD20 antibodies and found that CD20 cross-linking signalling induced apoptosis in MLMA cells (Fig. 2).
Figure 2.
Induction of apoptosis in MLMA cells mediated by CD20 and B-cell antigen receptor. (a) MLMA cells were treated with either rabbit anti-μ heavy-chain polyclonal antibody (αμ, 10 μg/ml) or a combination of anti-CD20 monoclonal antibody (mAb; αCD20, 5 μg/ml) and secondary rabbit anti-mouse immunoglobulin antibody (RαM, 5 μg/ml) for 48 hr and binding with fluorescein iosthiocyanate (FITC)-conjugated annexin V was examined by flow cytometry. Each experiment was performed in triplicate and the means + SD are indicated. (b) The same sample preparations as in (a) were cytocentrifuged and morphological appearance was examined by Giemsa-staining and nuclear staining with DAPI, using light microscopy and confocal microscopy, respectively. (c) Cell lysates were obtained from the same sample preparation as in (a) and the proforms of each caspase, cleaved caspase-3 and cleaved PARP were detected by immunoblotting.
BAFF inhibits CD20-mediated and BCR-mediated apoptosis in MLMA cells
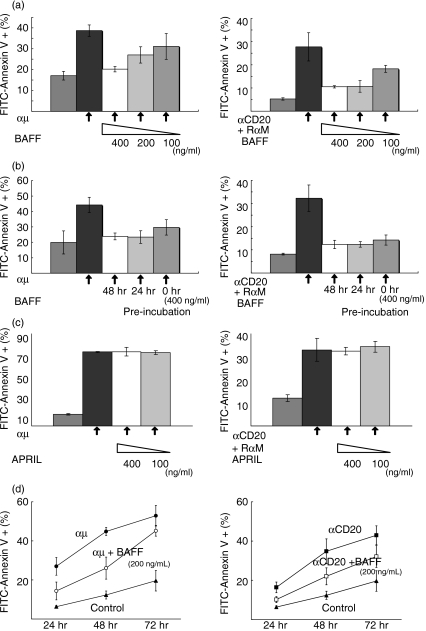
Next, we examined whether BAFF was able to inhibit apoptosis mediated by cross-linking of CD20 and BCR. As shown in Fig. 3(a), when BAFF was added to the culture, the incidence of apoptosis induced by both BCR-mediated and CD20-mediated stimuli was reduced as assessed by annexin V-binding. Although inhibition tended to be more effective with a higher dose of BAFF, the effect was not significant. We also examined the effect of pretreatment with BAFF on the inhibition of apoptosis but found none (Fig. 3b). In contrast, APRIL, another ligand for BCMA and TACI, did not affect apoptosis induced by the BCR-mediated and CD20-mediated stimuli, indicating the specificity of BAFF’s effect (Fig. 3c). Therefore, we concluded that BAFF-mediated stimuli are able to inhibit apoptosis mediated by the cross-linking of either CD20 or BCR and simultaneous treatment with apoptosis-inducing stimuli is almost sufficient to achieve maximum BAFF-mediated inhibition of apoptosis, at least in these cases. However, the inhibitory effect of BAFF against apoptosis mediated by the cross-linking of either CD20 or BCR was only partial and it was more obvious when the inhibition of apoptosis was examined at several different time-points.
Figure 3.
Effect of B-cell-activating factor (BAFF) on B-cell receptor (BCR)-induced and CD20-induced apoptosis in MLMA cells. (a) MLMA cells were treated with either rabbit anti-μ heavy-chain polyclonal antibody (mAb) (αμ, 10 μg/ml, left panel) or a combination of anti-CD20 mAb (αCD20, 5 μg/ml) and secondary rabbit anti-mouse immunoglobulin antibody (RαM, 5 μg/ml) (right panel) for 48 hr in the presence or absence of different concentrations of BAFF as indicated and binding with fluorescein isothiocyanate (FITC)-conjugated annexin V was examined as in Fig. 2(a). (b) MLMA cells preincubated with or without 400 ng/ml of BAFF for the indicated periods were treated with either αμ (left panel) or a combination of αCD20 and RαM (right panel) and examined as in (a). (c) The effect of APRIL on apoptosis induction was also examined as in (a). (d) MLMA cells were treated as in (a) and apoptosis was induced. The inhibitory effect of simultaneous addition of BAFF (200 ng/ml) against apoptosis was examined at different time-points as in (a).
Cellular effect of BAFF involved in the inhibition of apoptosis in MLMA cells
We further examined the molecular basis of the BAFF-mediated inhibition of apoptosis in MLMA cells. First, we tested the effect of BAFF on the growth of MLMA cells. As shown in Fig. 4, when BAFF was added to the culture, the cell proliferation was slightly enhanced, as assessed by cell counting, suggesting that BAFF promotes the growth of MLMA cells.
Figure 4.
Effect of B-cell-activating factor (BAFF) on MLMA cell proliferation. Starting from a cell concentration at 5 × 105/ml, MLMA cells were cultured in the presence (solid line) and absence (dotted line) of 400 ng/ml of BAFF and cell numbers were counted at the time-points indicated. Each experiment was performed in triplicate and the means + SD are indicated.
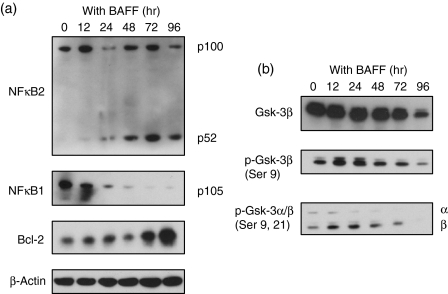
Next, we examined the intracellular signalling induced in MLMA cells by BAFF treatment. As shown in Fig. 5(a), immunoblot analysis revealed cleavage of p100, the precursor of NF-κB2, and a increase in p52, the active form of NF-κB2 after BAFF treatment, suggesting that the activation of NF-κB2 occurred after the treatment. We also observed the cleavage of the precursor of NF-κB1 after BAFF treatment (Fig. 5a). We further examined the activation of other molecules after treatment with BAFF and found that GSK-3β was transiently phosphorylated (Fig. 5b). In addition, we observed an elevation in the level of Bcl-2, an anti-apoptotic protein, after BAFF treatment.
Figure 5.
Intracellular signalling events and induction of Bcl-2 protein by B-cell-activating factor (BAFF). Cell lysates were prepared from MLMA cells treated with 400 ng/ml of BAFF for the periods indicated and an immunoblot analysis was performed using the antibodies indicated.
To investigate the early responses to BAFF in MLMA cells, global screening of candidate genes whose expression is regulated by BAFF was performed by employing a microarray system. First, we selected up-regulated genes that are expressed in MLMA cells treated with BAFF for 12 hr at a level at least 1·5-fold higher than in untreated cells. Under these conditions, 178 probes were selected as up-regulated genes (Table 1). Consistent with the results of the immunoblot analysis presented in Fig. 5(a), the gene expression of Bcl-2 was found to be up-regulated by BAFF treatment (Table 1). Interestingly, the gene expression of CD40, a member of the TNF-receptor family involved in B-cell survival, was also increased after treatment with BAFF. The genes that are known to be involved in anti-apoptotic effect, including Myb, Epstein–Barr virus (EBV)-induced gene 3 (EBI3), and caspase 8 and FADD-like apoptosis regulator (CFLAR), were also up-regulated by BAFF treatment.
Table 1.
Up-regulated genes after BAFF stimulation
| Affy ID | Gene name | Symbol | Fold-change |
|---|---|---|---|
| 204798_at | V-myb myeloblastosis viral oncogene homolog | MYB | 3·6934717 |
| 207861_at | Chemokine (C-C motif) ligand 22 | CCL22 | 3·2195807 |
| 201669_s_at | Myristoylated alanine-rich protein kinase C substrate | MARCKS | 3·0888734 |
| 213138_at | AT rich interactive domain 5A | ARID5A | 2·9730885 |
| 203927_at | IkBe | NFKBIE | 2·6444874 |
| 239412_at | Interferon regulatory factor 5 | IRF5 | 2·6300144 |
| 205173_x_at | CD58 antigen | CD58 | 2·5103657 |
| 230543_at | Similar to Chloride intracellular channel protein 4 | USP9X | 2·4929807 |
| 201932_at | Leucine rich repeat containing 41 | MUF1 | 2·3778253 |
| 203835_at | Leucine rich repeat containing 32 | GARP | 2·3439856 |
| 221912_s_at | Human DNA sequence from clone RP4-622L5 | MGC1203 | 2·296463 |
| 205599_at | TNF receptor-associated factor 1 | TRAF1 | 2·283286 |
| 218470_at | Tyrosyl-tRNA synthetase 2 | CGI-04 | 2·2782216 |
| 203685_at | B-cell CLL/lymphoma 2 | BCL2 | 2·2648098 |
| 202644_s_at | Tumor necrosis factor, alpha-induced protein 3 | TNFAIP3 | 2·2458956 |
| 204897_at | Prostaglandin E receptor 4 (subtype EP4) | PTGER4 | 2·2327275 |
| 217728_at | S100 calcium binding protein A6 | S100A6 | 2·188896 |
| 234339_s_at | Glioma tumor suppressor candidate region gene 2 | GLTSCR2 | 2·1786015 |
| 226354_at | Lactamase, beta | LACTB | 2·1238286 |
| 209680_s_at | Kinesin family member C1 | KIFC1 | 2·1163168 |
| 206508_at | Tumor necrosis factor (ligand) superfamily, member 7 | TNFSF7 | 2·0985012 |
| 223319_at | Gephyrin | GPHN | 2·0934505 |
| 242312_x_at | AV736963 CB | 2·0774355 | |
| 207608_x_at | Cytochrome P450, family 1, subfamily A, polypeptide 2 | CYP1A2 | 2·019507 |
| 229437_at | BIC transcript | BIC | 1·9920377 |
| 224468_s_at | Multidrug resistance-related protein | MGC13170 | 1·9605879 |
| 214101_s_at | Aminopeptidase puromycin sensitive | NPEPPS | 1·949555 |
| 208624_s_at | Eukaryotic translation initiation factor 4 gamma, 1 | EIF4G1 | 1·9305304 |
| 218819_at | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26 | DDX26 | 1·925604 |
| 200648_s_at | Glutamate-ammonia ligase | GLUL | 1·9081395 |
| 210686_x_at | Solute carrier family 25, member 16 | GDA; GDC; ML7; hML7; HGT.1; D10S105E; MGC39851 | 1·903342 |
| 48659_at | Invasion inhibitory protein 45 | FLJ12438 | 1·8990294 |
| 204283_at | Phenylalanine-tRNA synthetase 2 | FARS2 | 1·8852582 |
| 1563796_s_at | KIAA1970 protein | KIAA1970 | 1·8749646 |
| 219424_at | Epstein–Barr virus induced gene 3 | EBI3 | 1·8620349 |
| 213747_at | Antizyme inhibitor 1 | OAZIN | 1·8602847 |
| 205419_at | Epstein–Barr virus induced gene 2 | EBI2 | 1·8580037 |
| 210978_s_at | Transgelin 2 | TAGLN2 | 1·8418014 |
| 201502_s_at | IkBa | NFKBIA | 1·8220907 |
| 207688_s_at | Inhibin, beta C | INHBC | 1·8162365 |
| 208949_s_at | Lectin, galactoside-binding, soluble, 3 | LGALS3 | 1·8159895 |
| 216252_x_at | Fas | FAS | 1·7921783 |
| 203422_at | Polymerase (DNA directed), delta 1 | POLD1 | 1·7856169 |
| 227299_at | Cyclin I | CCNI | 1·7854006 |
| 218872_at | Hypothetical protein FLJ20607 | TSC | 1·7839508 |
| 205749_at | Cytochrome P450, family 1, subfamily A, polypeptide 1 | CYP1A1 | 1·7801132 |
| 210514_x_at | HLA-G histocompatibility antigen, class I, G | HLA-G | 1·780091 |
| 211376_s_at | Chromosome 10 open reading frame 86 | C10orf86 | 1·7799767 |
| 212063_at | CD44 antigen | CD44 | 1·778577 |
| 201404_x_at | Proteasome (prosome, macropain) subunit, beta type, 2 | PSMB2 | 1·774633 |
| 209939_x_at | CASP8 and FADD-like apoptosis regulator | CFLAR | 1·7721851 |
| 225775_at | Tetraspanin 33 | MGC50844 | 1·7708977 |
| 213642_at | Ribosomal protein L27 | RPL27 | 1·7626065 |
| 209100_at | Interferon-related developmental regulator 2 | IFRD2 | 1·7502396 |
| 201572_x_at | DCMP deaminase | DCTD | 1·7400836 |
| 212642_s_at | Human DNA sequence from clone RP1-67K17 | HIVEP2 | 1·7382044 |
| 221866_at | TFEB | 1·7346125 | |
| 204562_at | Interferon regulatory factor 4 | IRF4 | 1·7332152 |
| 230660_at | SERTA domain containing 4 | SERTAD4 | 1·7324636 |
| 229671_s_at | Chromosome 21 open reading frame 45 | C21orf45 | 1·7187352 |
| 201797_s_at | Valyl-tRNA synthetase | VARS2 | 1·7037842 |
| 212857_x_at | Similar to hypothetical protein DKFZp434P0316 | PC4 | 1·7007011 |
| 225360_at | Hypothetical protein PP2447 | PP2447 | 1·6819744 |
| 201565_s_at | Inhibitor of DNA binding 2 | ID2 | 1·6807232 |
| 213113_s_at | Solute carrier family 43, member 3 | SLC43A3 | 1·680492 |
| 212107_s_at | DEAH (Asp-Glu-Ala-His) box polypeptide 9 | DHX9 | 1·6746678 |
| 204211_x_at | Eukaryotic translation initiation factor 2-alpha kinase 2 | EIF2AK2 | 1·6744418 |
| 205153_s_at | CD40 antigen | CD40 | 1·6712306 |
| 214531_s_at | Sorting nexin 1 | SNX1 | 1·6710757 |
| 226853_at | BMP2 inducible kinase | BMP2K | 1·6657684 |
| 217734_s_at | WD repeat domain 6 | WDR6 | 1·6582829 |
| 202418_at | Yip1 interacting factor homolog A | YIF1 | 1·6526384 |
| 203672_x_at | Thiopurine S-methyltransferase | TPMT | 1·6522619 |
| 223287_s_at | Forkhead box P1 | FOXP1 | 1·6494714 |
| 205621_at | AlkB, alkylation repair homolog | ALKBH | 1·6476021 |
| 233310_at | Clone 25119 mRNA sequence | 1·642435 | |
| 201046_s_at | RAD23 homolog A | RAD23A | 1·6409639 |
| 202161_at | Protein kinase N1 | PKN1 | 1·6403072 |
| 219347_at | Nudix-type motif 15 | NUDT15 | 1·6394255 |
| 226099_at | Elongation factor, RNA polymerase II, 2 | ELL2 | 1·6393304 |
| 202715_at | Carbamoyl-phosphate synthetase 2, | CAD | 1·6350683 |
| 214315_x_at | Calreticulin | CALR | 1·6341119 |
| 227371_at | BAI1-associated protein 2-like 1 | LOC55971 | 1·6332316 |
| 37028_at | Protein phosphatase 1, regulatory (inhibitor) subunit 15A | PPP1R15A | 1·6298432 |
| 223079_s_at | Glutaminase | GLS | 1·6280246 |
| 200613_at | Adaptor-related protein complex 2, mu 1 subunit | AP2M1 | 1·6246499 |
| 218611_at | Immediate early response 5 | IER5 | 1·6235775 |
| 228993_s_at | Programmed cell death 4 | PDCD4 | 1·6221018 |
| 224241_s_at | Homo sapiens cDNA clone IMAGE:2820510 | PRO1855 | 1·6194851 |
| 205393_s_at | CHK1 checkpoint homolog | CHEK1 | 1·6177739 |
| 213826_s_at | H3 histone, family 3A | H3F3A | 1·6139177 |
| 202819_s_at | Transcription elongation factor B (SIII), polypeptide 3 | TCEB3 | 1·6136174 |
| 212064_x_at | MYC-associated zinc finger protein | MAZ | 1·613094 |
| 218847_at | IGF-II mRNA-binding protein 2 | IMP-2 | 1·6124035 |
| 201421_s_at | WD repeat domain 77 | MEP50 | 1·6116982 |
| 230509_at | 602041213F1 NCI_CGAP_Brn67 | SNX22 | 1·610629 |
| 214383_x_at | Kelch domain containing 3 | KLHDC3 | 1·604017 |
| 202265_at | Polycomb group ring finger 4 | PCGF4 | 1·6002513 |
| 200894_s_at | FK506 binding protein 4, 59 kDa | FKBP4 | 1·5997517 |
| 202024_at | ArsA arsenite transporter, ATP-binding, homolog 1 | ASNA1 | 1·5976273 |
| 225625_at | Similar to hypothetical protein 9530023G02 | MGC90512 | 1·5957043 |
| 224961_at | SCY1-like 2 | SCYL2 | 1·5939683 |
| 222774_s_at | Neuropilin (NRP) and tolloid (TLL)-like 2 | NETO2 | 1·5898373 |
| 225063_at | Ubiquitin-like 7 (bone marrow stromal cell-derived) | BMSC-UbP | 1·5887783 |
| 218305_at | Importin 4 | IPO4 | 1·5878817 |
| 204228_at | Peptidyl prolyl isomerase H (cyclophilin H) | PPIH | 1·5876329 |
| 224966_s_at | Dihydrouridine synthase 3-like (S. cerevisiae) | LOC56931 | 1·5869961 |
| 239364_at | Ets variant gene 6 (TEL oncogene) | ETV6 | 1·5859513 |
| 206138_s_at | Phosphatidylinositol 4-kinase, catalytic, beta polypeptide | PIK4CB | 1·5855292 |
| 209797_at | Transmembrane protein 4 | TMEM4 | 1·5808252 |
| 204116_at | Interleukin 2 receptor, gamma | IL2RG | 1·5777943 |
| 205965_at | Basic leucine zipper transcription factor, ATF-like | BATF | 1·577366 |
| 201545_s_at | Poly(A) binding protein, nuclear 1 | PABPN1 | 1·5768379 |
| 205235_s_at | M-phase phosphoprotein 1 | MPHOSPH1 | 1·5718083 |
| 220924_s_at | Solute carrier family 38, member 2 | SLC38A2 | 1·5694296 |
| 208858_s_at | Family with sequence similarity 62, member A | MBC2 | 1·5689234 |
| 203235_at | Thimet oligopeptidase 1 | THOP1 | 1·5672989 |
| 215001_s_at | Glutamate-ammonia ligase (glutamine synthetase) | GS; GLNS | 1·5667295 |
| 224571_at | Interferon regulatory factor 2 binding protein 2 | IRF2BP2 | 1·566219 |
| 235759_at | EF-hand calcium binding protein 1 | EFCBP1 | 1·5627139 |
| 218092_s_at | HIV-1 Rev binding protein | HRB | 1·5555688 |
| 244413_at | Dendritic cell-associated lectin-1 | DCAL1 | 1·5547262 |
| 209781_s_at | KH domain containing, RNA binding, signal transduction associated 3 | KHDRBS3 | 1·5527176 |
| 211965_at | Zinc finger protein 36, C3H type-like 1 | ZFP36L1 | 1·5514944 |
| 210740_s_at | Inositol 1,3,4-triphosphate 5/6 kinase | ITPK1 | 1·5482888 |
| 218097_s_at | CUE domain containing 2 | CUEDC2 | 1·5461936 |
| 207618_s_at | BCS1-like | BCS1L | 1·5440258 |
| 200628_s_at | Tryptophanyl-tRNA synthetase | WARS | 1·5433701 |
| 201490_s_at | Peptidylprolyl isomerase F (cyclophilin F) | PPIF | 1·5432048 |
| 222425_s_at | Polymerase (DNA-directed), delta interacting protein 2 | POLDIP2 | 1·5430135 |
| 240277_at | Solute carrier family 30 (zinc transporter), member 7 | SLC30A7 | 1·5403106 |
| 204882_at | Rho GTPase activating protein 25 | ARHGAP25 | 1·5393674 |
| 214784_x_at | Exportin 6 | XPO6 | 1·5390915 |
| 201801_s_at | Solute carrier family 29 (nucleoside transporters), member 1 | SLC29A1 | 1·5378566 |
| 202307_s_at | Transporter 1, ATP-binding cassette, sub-family B | TAP1 | 1·5369159 |
| 224913_s_at | Translocase of inner mitochondrial membrane 50 homolog | TIMM50 | 1·5356098 |
| 226797_at | Mbt domain containing 1 | MBTD1 | 1·5314596 |
| 202887_s_at | DNA-damage-inducible transcript 4 | DDIT4 | 1·53085 |
| 207396_s_at | Asparagine-linked glycosylation 3 homolog | ALG3 | 1·5305443 |
| 228487_s_at | Ras responsive element binding protein 1 | RREB1 | 1·5286149 |
| 201473_at | Jun B proto-oncogene | JUNB | 1·5285981 |
| 222968_at | Chromosome 6 open reading frame 48 | C6orf48 | 1·5267475 |
| 203879_at | Phosphoinositide-3-kinase, catalytic, delta polypeptide | PIK3CD | 1·5265557 |
| 206181_at | Signaling lymphocytic activation molecule family member 1 | SLAMF1 | 1·5258498 |
| 238567_at | Sphingosine-1-phosphate phosphotase 2 | SGPP2 | 1·5248299 |
| 223427_s_at | Erythrocyte membrane protein band 4·1 like 4B | EPB41L4B | 1·5245888 |
| 215450_at | Small nuclear ribonucleoprotein polypeptide E | SNRPE | 1·5239984 |
| 210428_s_at | Hepatocyte growth factor-regulated tyrosine kinase substrate | HGS | 1·5230072 |
| 202968_s_at | Dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 | DYRK2 | 1·5226749 |
| 217289_s_at | glucose-6-phosphatase | G6PT; GSD1a | 1·5194279 |
| 230466_s_at | Mesenchymal stem cell protein DSC96 | 1·5193534 | |
| 229204_at | Heterochromatin protein 1, binding protein 3 | HP1-BP74 | 1·5177599 |
| 223743_s_at | Mitochondrial ribosomal protein L4 | MRPL4 | 1·5151922 |
| 200706_s_at | Lipopolysaccharide-induced TNF factor | LITAF | 1·5070926 |
| 217782_s_at | G protein pathway suppressor 1 | GPS1 | 1·5069752 |
| 219279_at | DOCK10 | 1·5061336 | |
| 215031_x_at | Ring finger protein 126 | RNF126 | 1·5051547 |
| 203466_at | MpV17 transgene, murine homolog, glomerulosclerosis | MPV17 | 1·5044804 |
| 203114_at | Sjogren’s syndrome/scleroderma autoantigen 1 | SSSCA1 | 1·5035471 |
| 222037_at | MCM4 minichromosome maintenance deficient 4 | MCM4 | 1·5032879 |
| 45749_at | Family with sequence similarity 65, member A | FLJ13725 | 1·5015459 |
| 213224_s_at | Hypothetical protein LOC92482 | LOC92482 | 1·5014849 |
| 204171_at | Ribosomal protein S6 kinase, 70 kDa, polypeptide 1 | RPS6KB1 | 1·5013521 |
| 219628_at | P53 target zinc finger protein | WIG1 | 1·5013262 |
We further confirmed the increased CD40 protein expression by flow cytometry (Fig. 6a). Similarly, down-regulated genes that were expressed in BAFF-treated cells at a level at least 0·75-fold lower than in untreated cells were selected. As shown in Table 2, 517 probes were selected as down-regulated genes. The above results of global gene expression profiling suggest that the expression of various types of genes was influenced by BAFF stimulation in MLMA cells.
Figure 6.
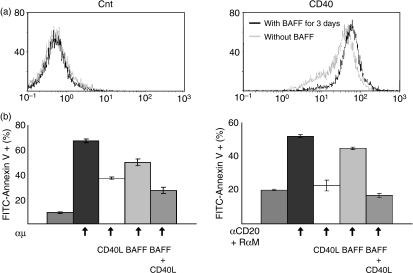
Effect of B-cell-activating factor (BAFF) on CD40 expression in MLMA cells. (a) MLMA cells cultured with or without BAFF for 3 days were stained with fluorescein isothiocyanate (FITC)-labelled monoclonal antibody (mAb) against CD40 and analysed by flow cytometry as in Fig. 1. (b) The inhibitory effect of CD40 stimulation on apoptosis induction was examined. MLMA cells were treated with 500 ng/ml of CD40-ligand in the presence of 2·5 ng/ml of interleukin-4 to stimulate CD40. The effects of either stimulation of CD40 alone or simultaneous stimulation of CD40 and BAFF receptor on apoptosis similarly induced as in Fig. 2 were examined.
Table 2.
Down-regulated genes after BAFF stimulation
| Affy ID | Gene name | Symbol | Fold-change |
|---|---|---|---|
| 223697_x_at | Chromosome 9 open reading frame 64 | C9orf64 | 0·18046121 |
| 237461_at | NACHT, leucine rich repeat and PYD containing 7 | NALP7 | 0·1989845 |
| 243808_at | Cyclin-dependent kinase 6 | CDK6 | 0·25961372 |
| 212027_at | wn02f07.x1 NCI_CGAP_Ut2 | RBM25 | 0·27985397 |
| 211352_s_at | Similar to Hin-2 | CAGH16 | 0·2816477 |
| 225569_at | Eukaryotic translation initiation factor 2C, 2 | EIF2C2 | 0·3363239 |
| 230128_at | Homo sapiens cDNA: FLJ21578 fis, clone COL06726 | 0·33926746 | |
| 204698_at | Interferon stimulated exonuclease gene 20 kDa | ISG20 | 0·33949938 |
| 222633_at | Transducin (beta)-like 1X-linked receptor 1 | TBL1XR1 | 0·34330967 |
| 242601_at | Hypothetical protein LOC253012 | LOC253012 | 0·35196936 |
| 207677_s_at | Neutrophil cytosolic factor 4, 40 kDa | NCF4 | 0·35400128 |
| 1553499_s_at | Serpin peptidase inhibitor, clade A, member 9 | SERPINA9 | 0·37200212 |
| 212028_at | RNA binding motif protein 25 | RBM25 | 0·38822186 |
| 240798_at | Cut-like 1, CCAAT displacement protein | CUTL1 | 0·39412326 |
| 202205_at | Vasodilator-stimulated phosphoprotein | VASP | 0·39878875 |
| 212794_s_at | KIAA1033 | KIAA1033 | 0·39967155 |
| 230831_at | FERM domain containing 5 | MGC14161 | 0·41499138 |
| 222158_s_at | Chromosome 1 open reading frame 121 | PNAS-4 | 0·41996363 |
| 213577_at | Squalene epoxidase | SQLE | 0·42782143 |
| 1557953_at | Transcribed locus | ZNF36 | 0·43064588 |
| 1558678_s_at | Metastasis associated lung adenocarcinoma transcript 1 | MALAT1 | 0·4366038 |
| 201291_s_at | Topoisomerase (DNA) II alpha 170 kDa | TOP2A | 0·43767774 |
| 222186_at | Zinc finger, A20 domain containing 3 | ZA20D3 | 0·43876088 |
| 225937_at | FP6778 | 0·44128555 | |
| 205967_at | Histone 1, H4c | HIST1H4C | 0·44577146 |
| 212592_at | Immunoglobulin J polypeptide | IGJ | 0·45744386 |
| 213850_s_at | Splicing factor, arginine/serine-rich 2, interacting protein | SFRS2IP | 0·4588066 |
| 201678_s_at | DC12 protein | DC12 | 0·46194622 |
| 225640_at | Hypothetical gene supported by AK091718 | 0·4679212 | |
| AFFX-M27830_M_at | 0·4755289 | ||
| 225219_at | 602071082F1 NCI_CGAP_Brn64 | SMAD5 | 0·47642624 |
| 207057_at | Solute carrier family 16, member 7 | SLC16A7 | 0·48003742 |
| 1569344_a_at | Homo sapiens, clone IMAGE:4044872, mRNA | 0·48249447 | |
| 223217_s_at | IkBz | NFKBIZ | 0·48446876 |
| 201454_s_at | Aminopeptidase puromycin sensitive | NPEPPS | 0·48679113 |
| 213906_at | V-myb myeloblastosis viral oncogene homolog-like 1 | MYBL1 | 0·4908449 |
| 210970_s_at | Inhibitor of Bruton agammaglobulinemia tyrosine kinase | IBTK | 0·49121413 |
| 1255_g_at | guanylate cyclase activator 1A | GCAP | 0·49559578 |
| 222858_s_at | Dual adaptor of phosphotyrosine and 3-phosphoinositides | DAPP1 | 0·4963491 |
| 204730_at | Regulating synaptic membrane exocytosis 3 | RIMS3 | 0·5001501 |
| 207826_s_at | Inhibitor of DNA binding 3 | ID3 | 0·50062287 |
| 33304_at | Interferon stimulated exonuclease gene 20 kDa | ISG20 | 0·50399923 |
| 211928_at | Dynein, cytoplasmic 1, heavy chain 1 | DNCH1 | 0·5087544 |
| 242195_x_at | Numb homolog (Drosophila)-like | NUMBL | 0·5093039 |
| 209257_s_at | Chondroitin sulfate proteoglycan 6 | CSPG6 | 0·51205146 |
| 215990_s_at | B-cell CLL/lymphoma 6 | BCL5 | 0·51233035 |
| 227396_at | Protein tyrosine phosphatase, receptor type, J | PTPRJ | 0·5157389 |
| 228056_s_at | Napsin B aspartic peptidase pseudogene | NAPSB | 0·5182605 |
| 228787_s_at | Breast carcinoma amplified sequence 4 | BCAS4 | 0·51919734 |
| 225327_at | KIAA1370 | FLJ10980 | 0·5194209 |
| 212368_at | Zinc finger protein 292 | ZNF292 | 0·5197308 |
| 228343_at | POU domain, class 2, transcription factor 2 | POU2F2 | 0·5240128 |
| 209138_x_at | IGLJ3 | 0·52593404 | |
| 214016_s_at | Splicing factor proline/glutamine-rich | SFPQ | 0·5273748 |
| 201236_s_at | BTG family, member 2 | BTG2 | 0·5273795 |
| 202033_s_at | RB1-inducible coiled-coil 1 | RB1CC1 | 0·5290374 |
| 219911_s_at | Solute carrier organic anion transporter family, member 4A1 | SLCO4A1 | 0·5337818 |
| 204867_at | GTP cyclohydrolase I feedback regulator | GCHFR | 0·53428966 |
| 209579_s_at | Methyl-CpG binding domain protein 4 | MBD4 | 0·5378972 |
| 207761_s_at | DKFZP586A0522 protein | DKFZP586A0522 | 0·53842366 |
| 219517_at | Elongation factor RNA polymerase II-like 3 | ELL3 | 0·53930795 |
| 207339_s_at | Lymphotoxin beta | LTB | 0·53969586 |
| 228031_at | Hypothetical protein LOC149705 | C20orf121 | 0·5410639 |
| 206219_s_at | Vav 1 oncogene | VAV1 | 0·54299057 |
| 231716_at | Membrane associated DNA binding protein | MNAB | 0·5447174 |
| 213036_x_at | ATPase, Ca++ transporting, ubiquitous | SERCA3 | 0·5453293 |
| 230740_at | Transcribed locus | EHD3 | 0·5472777 |
| 230777_s_at | PR domain containing 15 | PRDM15 | 0·54828 |
| 210679_x_at | Homo sapiens cDNA clone MGC:3878 IMAGE:3609162 | BCL7A | 0·55017775 |
| 229147_at | Ras association (RalGDS/AF-6) domain family 6 | RASSF6 | 0·5539612 |
| 233746_x_at | Huntingtin interacting protein K | HYPK | 0·55896664 |
| 224616_at | Dynein, cytoplasmic 1, light intermediate chain 2 | DNCLI2 | 0·5595679 |
| 212677_s_at | RAB1A, member RAS oncogene family | RAB1A | 0·5602901 |
| 213016_at | Bobby sox homolog | BBX | 0·56184375 |
| 211383_s_at | WD repeat domain 37 | WDR37 | 0·56346995 |
| 215504_x_at | Ankyrin repeat domain 10 | ANKRD10 | 0·563754 |
| 212119_at | 602149641F1 NIH_MGC_81 | RHOQ | 0·5639594 |
| 203819_s_at | IGF-II mRNA-binding protein 3 | IMP-3 | 0·5641141 |
| 205124_at | MADS box transcription enhancer factor 2, polypeptide B | MEF2B | 0·56775457 |
| 220071_x_at | Centrosomal protein 27 kDa | C15orf25 | 0·56932724 |
| 219396_s_at | Nei endonuclease VIII-like 1 | NEIL1 | 0·5707603 |
| 226372_at | Carbohydrate (chondroitin 4) sulfotransferase 11 | CHST11 | 0·57161444 |
| 232266_x_at | Homo sapiens cDNA FLJ14317 fis, clone PLACE3000401. | CDC2L5 | 0·57188374 |
| 211445_x_at | Nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 | FKSG17 | 0·5727707 |
| 224829_at | Cytoplasmic polyadenylation element binding protein 4 | CPEB4 | 0·57316726 |
| 229353_s_at | Nuclear casein kinase and cyclin-dependent kinase substrate 1 | NUCKS | 0·5739766 |
| 215457_at | Actin related protein 2/3 complex, subunit 1A, 41 kDa | ARPC1A | 0·5758453 |
| 1569594_a_at | Serologically defined colon cancer antigen 1 | SDCCAG1 | 0·5770196 |
| 200596_s_at | Eukaryotic translation initiation factor 3, subunit 10 theta, 150/170 kDa | EIF3S10 | 0·5771801 |
| 209023_s_at | Stromal antigen 2 | STAG2 | 0·57891893 |
| 203140_at | B-cell CLL/lymphoma 6 | BCL6 | 0·5807235 |
| 217862_at | Protein inhibitor of activated STAT, 1 | PIAS1 | 0·5808474 |
| 227748_at | RNA binding motif protein, X-linked-like 1 | KAT3 | 0·5812906 |
| 229429_x_at | LOC440667 | 0·582004 | |
| 227740_at | U2AF homology motif (UHM) kinase 1 | UHMK1 | 0·5825476 |
| 208615_s_at | Protein tyrosine phosphatase type IVA, member 2 | PTP4A2 | 0·58321685 |
| 209360_s_at | Runt-related transcription factor 1 | RUNX1 | 0·58370924 |
| 213734_at | WD repeat and SOCS box-containing 2 | WSB2 | 0·5844417 |
| 202996_at | Polymerase (DNA-directed), delta 4 | POLD4 | 0·58979243 |
| 212047_s_at | Ring finger protein 167 | RNF167 | 0·59118843 |
| 218886_at | PAK1 interacting protein 1 | PAK1IP1 | 0·59172606 |
| 215179_x_at | Placental growth factor | PGF | 0·59284484 |
| 204141_at | Tubulin, beta 2A | TUBB2 | 0·5932073 |
| 212810_s_at | Solute carrier family 1, member 4 | SLC1A4 | 0·59446627 |
| 201193_at | Isocitrate dehydrogenase 1 (NADP+), soluble | IDH1 | 0·59564257 |
| 228153_at | IBR domain containing 2 | IBRDC2 | 0·59580594 |
| 64418_at | AP1 gamma subunit binding protein 1 | AP1GBP1 | 0·5966325 |
| 203143_s_at | Transcribed locus | KIAA0040 | 0·5968683 |
| 209076_s_at | WDR45-like | WDR45L | 0·5985668 |
| 204076_at | Ectonucleoside triphosphate diphosphohydrolase 4 | ENTPD4 | 0·59857833 |
| 1558080_s_at | Hypothetical protein LOC144871 | DNAJC3 | 0·5994704 |
| 203044_at | Carbohydrate (chondroitin) synthase 1 | CHSY1 | 0·60100013 |
| 234762_x_at | Neurolysin (metallopeptidase M3 family) | NLN | 0·6012079 |
| 207124_s_at | Guanine nucleotide binding protein (G protein), beta 5 | GNB5 | 0·6012481 |
| 204449_at | Phosducin-like | PDCL | 0·6038013 |
| 226508_at | Polyhomeotic like 3 (Drosophila) | PHC3 | 0·60685164 |
| 204681_s_at | Rap guanine nucleotide exchange factor (GEF) 5 | RAPGEF5 | 0·60706514 |
| 203346_s_at | Metal response element binding transcription factor 2 | M96 | 0·6077221 |
| 200998_s_at | Cytoskeleton-associated protein 4 | CKAP4 | 0·608657 |
| 222816_s_at | Zinc finger, CCHC domain containing 2 | ZCCHC2 | 0·6087468 |
| 219158_s_at | Synonyms: Ga19, NAT1, NATH, TBDN100 | NARG1 | 0·6090036 |
| 201901_s_at | YY1 transcription factor | YY1 | 0·6101737 |
| 229072_at | RAB30, member RAS oncogene family | RAB30 | 0·6116257 |
| 212604_at | Mitochondrial ribosomal protein S31 | MRPS31 | 0·61232865 |
| 214352_s_at | V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | KRAS2 | 0·6126175 |
| 231825_x_at | Activating transcription factor 7 interacting protein | ATF7IP | 0·6132226 |
| 225204_at | T-cell activation protein phosphatase 2C | TA-PP2C | 0·6143749 |
| 202379_s_at | Natural killer-tumor recognition sequence | NKTR | 0·61503386 |
| 204853_at | Origin recognition complex, subunit 2-like | ORC2L | 0·6158861 |
| 201138_s_at | Sjogren syndrome antigen B | SSB | 0·6166977 |
| 225136_at | Pleckstrin homology domain containing, family A, member 2 | PLEKHA2 | 0·6169427 |
| 201384_s_at | Neighbor of BRCA1 gene 1 | M17S2 | 0·61722594 |
| 213620_s_at | Intercellular adhesion molecule 2 | ICAM2 | 0·61761326 |
| 226158_at | Kelch-like 24 | DRE1 | 0·6183106 |
| 205383_s_at | Zinc finger and BTB domain containing 20 | ZBTB20 | 0·62210584 |
| 219148_at | PDZ binding kinase | PBK | 0·6229036 |
| 227402_s_at | Chromosome 8 open reading frame 53 | MGC14595 | 0·6240476 |
| 206641_at | Tumor necrosis factor receptor superfamily, member 17 | TNFRSF17 | 0·62757266 |
| 202412_s_at | Ubiquitin specific peptidase 1 | USP1 | 0·6286277 |
| 227224_at | Ral GEF with PH domain and SH3 binding motif 2 | RALGPS2 | 0·62964123 |
| 231809_x_at | EST365840 MAGE resequences, MAGC | PDCD7 | 0·6322018 |
| 200797_s_at | Myeloid cell leukemia sequence 1 | MCL1 | 0·6327875 |
| 204912_at | Interleukin 10 receptor, alpha | IL10RA | 0·63407373 |
| 210754_s_at | V-yes-1 Yamaguchi sarcoma viral related oncogene homolog | LYN | 0·6343251 |
| 235469_at | Similar to RIKEN cDNA 5830415L20 | MGC40405 | 0·6344833 |
| 208415_x_at | Inhibitor of growth family, member 1 | ING1 | 0·6353071 |
| 229656_s_at | Similar to echinoderm microtubule associated protein like 5 | 0·6356807 | |
| 224875_at | Hypothetical protein FLJ37562 | FLJ37562 | 0·6361133 |
| 230110_at | Mucolipin 2 | MCOLN2 | 0·6363045 |
| 213502_x_at | Similar to bK246H3·1 | LOC91316 | 0·63717055 |
| 209272_at | NGFI-A binding protein 1 | NAB1 | 0·6376612 |
| 214677_x_at | Immunoglobulin lambda joining 3 | IGLC2 | 0·6377649 |
| 220999_s_at | synonym: PIR121; p53 inducible protein | CYFIP2 | 0·6382754 |
| 201320_at | SWI/SNF related, matrix associated, actin dependent regulator of chromatin | SMARCC2 | 0·63914317 |
| 223553_s_at | Docking protein 3 | DOK3 | 0·63973147 |
| 214730_s_at | Golgi apparatus protein 1 | GLG1 | 0·64044565 |
| 1555989_at | Dishevelled associated activator of morphogenesis 1 | DAAM1 | 0·641099 |
| 210142_x_at | Flotillin 1 | FLOT1 | 0·64127034 |
| 228098_s_at | Myosin regulatory light chain interacting protein | MYLIP | 0·6423752 |
| 226464_at | Hypothetical protein MGC33365 | MGC33365 | 0·64265686 |
| 227189_at | Copine V | CPNE5 | 0·64277476 |
| 228910_at | CD82 antigen | KAI1 | 0·643213 |
| 208246_x_at | hypothetical protein FLJ20006 | FLJ20006 | 0·64384246 |
| 1565627_a_at | Leucine-rich repeat kinase 1 | LRRK1 | 0·6438542 |
| 208070_s_at | REV3-like, catalytic subunit of DNA polymerase zeta | REV3L | 0·64388376 |
| 226779_at | LMBR1 domain containing 2 | DKFZp434H2226 | 0·64402026 |
| 212760_at | Ubiquitin protein ligase E3 component n-recognin 2 | UBR2 | 0·6447363 |
| 232644_x_at | OCIA domain containing 1 | OCIAD1 | 0·6447408 |
| 205922_at | Vanin 2 | VNN2 | 0·6448793 |
| 209062_x_at | Nuclear receptor coactivator 3 | NCOA3 | 0·64496595 |
| 200842_s_at | Glutamyl-prolyl-tRNA synthetase | EPRS | 0·645458 |
| 212733_at | KIAA0226 | KIAA0226 | 0·64567864 |
| 244887_at | Regulator of G-protein signalling 13 | RGS13 | 0·6465569 |
| 205370_x_at | Dihydrolipoamide branched chain transacylase E2 | DBT | 0·6465768 |
| 219812_at | Stromal antigen 3 | MGC2463 | 0·64680105 |
| 202378_s_at | Leptin receptor | LEPR | 0·6468874 |
| 204285_s_at | Phorbol-12-myristate-13-acetate-induced protein 1 | PMAIP1 | 0·6495961 |
| 228151_at | Transcribed locus | 0·6497464 | |
| 213007_at | KIAA1794 | FLJ10719 | 0·6498637 |
| 222891_s_at | B-cell CLL/lymphoma 11A | BCL11A | 0·64996225 |
| 220085_at | Helicase, lymphoid-specific | HELLS | 0·6502575 |
| 220746_s_at | Receptor associated protein 80 | RAP80 | 0·65041715 |
| 213111_at | Phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III | PIP5K3 | 0·65109867 |
| 203318_s_at | Zinc finger protein 148 | ZNF148 | 0·6520053 |
| 202655_at | Arginine-rich, mutated in early stage tumors | ARMET | 0·6522758 |
| 225273_at | KIAA1280 protein | KIAA1280 | 0·6529812 |
| 204709_s_at | Kinesin family member 23 | KIF23 | 0·65339863 |
| 218358_at | Cysteine-rich with EGF-like domains 2 | MGC11256 | 0·6534136 |
| 201917_s_at | Solute carrier family 25, member 36 | FLJ10618 | 0·65368974 |
| 220933_s_at | Zinc finger, CCHC domain containing 6 | ZCCHC6 | 0·6548457 |
| 212588_at | Protein tyrosine phosphatase, receptor type, C | PTPRC | 0·6555439 |
| 215780_s_at | Human DNA sequence from clone RP1-30P20 | SET | 0·65566504 |
| 239748_x_at | yl95h12.s1 Soares infant brain 1NIB | 0·6571978 | |
| 209780_at | Putative homeodomain transcription factor 2 | PHTF2 | 0·65742826 |
| 211040_x_at | G-2 and S-phase expressed 1 | GTSE1 | 0·659053 |
| 206150_at | TAP binding protein-like | TNFRSF7 | 0·6612941 |
| 209049_s_at | PRKCBP1 | 0·66141385 | |
| 217796_s_at | Nuclear protein localization 4 | NPL4 | 0·6618553 |
| 222737_s_at | Bromodomain containing 7 | BRD7 | 0·6626274 |
| 218306_s_at | Hect domain and RCC1 (CHC1)-like domain (RLD) 1 | HERC1 | 0·662763 |
| 222420_s_at | Ubiquitin-conjugating enzyme E2H | UBE2H | 0·66321975 |
| 210962_s_at | A kinase (PRKA) anchor protein (yotiao) 9 | AKAP9 | 0·6633441 |
| 1555275_a_at | Kelch-like 6 (Drosophila) | KLHL6 | 0·6639439 |
| 218348_s_at | Zinc finger CCCH-type containing 7A | ZC3HDC7 | 0·6641935 |
| 233329_s_at | Hypothetical protein LOC51315 | LOC51315 | 0·66421455 |
| 225232_at | Myotubularin related protein 12 | PIP3AP | 0·66434306 |
| 230917_at | CDNA FLJ45450 fis, clone BRSTN2002691 | 0·664679 | |
| 202181_at | KIAA0247 | KIAA0247 | 0·66494757 |
| 210561_s_at | WD repeat and SOCS box-containing 1 | WSB1 | 0·66532546 |
| 206272_at | S-phase response (cyclin-related) | SPHAR | 0·66562426 |
| 201498_at | Ubiquitin specific peptidase 7 (herpes virus-associated) | USP7 | 0·6661961 |
| 235661_at | ye65a03.r1 Soares fetal liver spleen 1NFLS | 0·666862 | |
| 228087_at | LOC90693 protein | LOC90693 | 0·6674958 |
| 218150_at | ADP-ribosylation factor-like 5A | ARL5 | 0·6685538 |
| 203608_at | ALDH5A1 | 0·6689315 | |
| 213460_x_at | Williams Beuren syndrome chromosome region 20C | WBSCR20C | 0·6701285 |
| 202922_at | Glutamate-cysteine ligase, catalytic subunit | GCLC | 0·67089856 |
| 222408_s_at | Yippee-like 5 | YPEL5 | 0·671024 |
| 223391_at | Sphingosine-1-phosphate phosphatase 1 | SGPP1 | 0·67116857 |
| 213166_x_at | Hypothetical protein FLJ14346 | FLJ14346 | 0·67121977 |
| 219119_at | LSM8 homolog, U6 small nuclear RNA associated | LSM8 | 0·67156994 |
| 203297_s_at | Jumonji, AT rich interactive domain 2 | JARID2 | 0·67171353 |
| 213940_s_at | Formin binding protein 1 | FNBP1 | 0·67211777 |
| 224677_x_at | Chromosome 11 open reading frame 31 | C11orf31 | 0·6722119 |
| 212066_s_at | Ubiquitin specific peptidase 34 | USP34 | 0·67282677 |
| 201779_s_at | Ring finger protein 13 | RNF13 | 0·67283076 |
| 243798_at | B-cell CLL/lymphoma 9-like | BCL9L | 0·6730866 |
| 212023_s_at | Antigen identified by monoclonal antibody Ki-67 | MKI67 | 0·673758 |
| 219502_at | Nei endonuclease VIII-like 3 | FLJ10858 | 0·67435133 |
| 203556_at | Zinc fingers and homeoboxes 2 | ZHX2 | 0·67446464 |
| 237475_x_at | qb48d05.x1 NCI_CGAP_Brn23 | 0·6744719 | |
| 202704_at | Transducer of ERBB2, 1 | TOB1 | 0·67514235 |
| 225701_at | AT-hook transcription factor | AKNA | 0·6767782 |
| 219392_x_at | Proline rich 11 | FLJ11029 | 0·67691547 |
| 205297_s_at | CD79B antigen | CD79B | 0·67695534 |
| 226398_s_at | Chromosome 10 open reading frame 4 | C10orf4 | 0·6774105 |
| 213064_at | Nuclear protein UKp68 | FLJ11806 | 0·6776265 |
| 1559436_x_at | Arrestin, beta 2 | ARRB2 | 0·67766494 |
| 212167_s_at | SWI/SNF related, matrix associated, actin dependent regulator of chromatin | SMARCB1 | 0·6779144 |
| 223268_at | LP4947 | PTD012 | 0·6783802 |
| 217781_s_at | Zinc finger protein 106 homolog | ZFP106 | 0·6795011 |
| 1556059_s_at | Spen homolog, transcriptional regulator | SPEN | 0·6800467 |
| 1552448_a_at | Homo sapiens chromosome 8 open reading frame 12 (C8orf12), mRNA. | C8orf12 | 0·6807321 |
| 224602_at | HCV F-transactivated protein 1 | LOC401152 | 0·6808058 |
| 212944_at | Mitochondrial ribosomal protein S6 | MRPS6 | 0·6811207 |
| 208737_at | ATPase, H+ transporting, lysosomal 13 kDa, V1 subunit G isoform 1 | ATP6V1G1 | 0·6811667 |
| 211997_x_at | H3 histone, family 3B | H3F3B | 0·68119144 |
| 212622_at | Transmembrane protein 41B | KIAA0033 | 0·68157566 |
| 203301_s_at | Cyclin D binding myb-like transcription factor 1 | DMTF1 | 0·68273085 |
| 208899_x_at | ATPase, H+ transporting, lysosomal 34 kDa, V1 subunit D | ATP6V1D | 0·6829173 |
| 202983_at | SWI/SNF related, matrix associated, actin dependent regulator of chromatin | SMARCA3 | 0·6829173 |
| 209250_at | Degenerative spermatocyte homolog 1, lipid desaturase | DEGS1 | 0·6831875 |
| 204581_at | CD22 antigen | CD22 | 0·6840824 |
| 225433_at | General transcription factor IIA, 1, 19/37 kDa | GTF2A1 | 0·685833 |
| 219076_s_at | Peroxisomal membrane protein 2, 22 kDa | PXMP2 | 0·6862011 |
| 208772_at | Ankyrin repeat and KH domain containing 1 | ANKHD1 | 0·68652916 |
| 212571_at | Chromodomain helicase DNA binding protein 8 | CHD8 | 0·68715703 |
| 200920_s_at | B-cell translocation gene 1, anti-proliferative | BTG1 | 0·6878364 |
| 212126_at | Chromobox homolog 5 | CBX5 | 0·687962 |
| 203752_s_at | Jun D proto-oncogene | JUND | 0·68797 |
| 210105_s_at | FYN oncogene related to SRC, FGR, YES | FYN | 0·68825364 |
| 221501_x_at | Hypothetical protein LOC339047 | LOC339047 | 0·6883818 |
| 227696_at | Exosome component 6 | EXOSC6 | 0·688953 |
| 201810_s_at | SH3-domain binding protein 5 | SH3BP5 | 0·6889935 |
| 206513_at | Absent in melanoma 2 | AIM2 | 0·689059 |
| 205484_at | Signaling threshold regulating transmembrane adaptor 1 | SIT | 0·68912697 |
| 225890_at | Chromosome 20 open reading frame 72 | C20orf72 | 0·69149476 |
| 213154_s_at | Bicaudal D homolog 2 | BICD2 | 0·6923916 |
| 217717_s_at | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein | YWHAB | 0·69240123 |
| 212350_at | TBC1 domain family, member 1 | TBC1D1 | 0·69268274 |
| 213128_s_at | Ubiquitin protein ligase E3A | UBE3A | 0·6931148 |
| 212943_at | KIAA0528 gene product | KIAA0528 | 0·6943166 |
| 203434_s_at | Membrane metallo-endopeptidase | MME | 0·69480205 |
| 232515_at | Ankyrin repeat and SOCS box-containing 3 | ASB3 | 0·69493157 |
| 234984_at | Neural precursor cell expressed, developmentally down-regulated 1 | NEDD1 | 0·6951 |
| 204446_s_at | Arachidonate 5-lipoxygenase | ALOX5 | 0·6954707 |
| 221918_at | PCTAIRE protein kinase 2 | PCTK2 | 0·6954857 |
| 218384_at | Calcium regulated heat stable protein 1, 24 kDa | CARHSP1 | 0·6965451 |
| 223022_s_at | Chromosome 6 open reading frame 55 | C6orf55 | 0·69656706 |
| 217249_x_at | Cytochrome c oxidase subunit VIIa pseudogene 2 | COX7A3H | 0·69701755 |
| 243539_at | Ring finger protein 11 | RNF11 | 0·69720584 |
| 210592_s_at | Spermidine/spermine N1-acetyltransferase | SAT | 0·69809985 |
| 217967_s_at | Chromosome 1 open reading frame 24 | C1orf24 | 0·6989367 |
| 228006_at | Phosphatase and tensin homolog | PTEN | 0·6992462 |
| 204710_s_at | WD repeat domain, phosphoinositide interacting 2 | WIPI-2 | 0·699294 |
| 202760_s_at | Paralemmin 2 | PALM2-AKAP2 | 0·6998125 |
| 204872_at | Transducin-like enhancer of split 4 | TLE4 | 0·6999074 |
| 233702_x_at | Homo sapiens cDNA: FLJ20946 fis, clone ADSE01819. | 0·70036465 | |
| 222986_s_at | Scotin | SCOTIN | 0·70078844 |
| 223445_at | Dystrobrevin binding protein 1 | DTNBP1 | 0·7017351 |
| 203007_x_at | Lysophospholipase I | LYPLA1 | 0·7029054 |
| 235333_at | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 | B4GALT6 | 0·7029247 |
| 213729_at | Formin binding protein 3 | FNBP3 | 0·70293915 |
| 227124_at | MRNA full length insert cDNA clone EUROIMAGE 966164 | 0·70326364 | |
| 205733_at | Bloom syndrome | BLM | 0·7036139 |
| 219304_s_at | Platelet derived growth factor D | PDGFD | 0·7036207 |
| 219237_s_at | DnaJ (Hsp40) homolog, subfamily B, member 14 | FLJ14281 | 0·7041835 |
| 209358_at | TAF11 RNA polymerase II | TAF11 | 0·7044149 |
| 225545_at | Eukaryotic elongation factor-2 kinase | EEF2K | 0·70445967 |
| 208146_s_at | Carboxypeptidase, vitellogenic-like | CPVL | 0·7045879 |
| 210972_x_at | T-cell receptor rearranged alpha-chain V-region | TCRA | 0·7055038 |
| 208579_x_at | H2B histone family, member S | H2BFS | 0·7060805 |
| 212263_at | Quaking homolog, KH domain RNA binding | QKI | 0·707485 |
| 202386_s_at | Limkain b1 | LKAP | 0·7075946 |
| 202113_s_at | sorting nexin 2 | TRG-9 | 0·70800734 |
| 206323_x_at | Oligophrenin 1 | OPHN1 | 0·70812446 |
| 202664_at | Wiskott-Aldrich syndrome protein interacting protein | WASPIP | 0·70844746 |
| 233093_s_at | Baculoviral IAP repeat-containing 6 | BIRC6 | 0·7087636 |
| 218662_s_at | Chromosome condensation protein G | HCAP-G | 0·70939666 |
| 214508_x_at | CAMP responsive element modulator | CREM | 0·7095808 |
| 213737_x_at | Transcribed locus | DKFZp434P162 | 0·7103367 |
| 201877_s_at | Protein phosphatase 2, regulatory subunit B (B56), gamma isoform | PPP2R5C | 0·7103621 |
| 212693_at | MDN1, midasin homolog (yeast) | MDN1 | 0·7104688 |
| 212314_at | KIAA0746 protein | KIAA0746 | 0·71076244 |
| 201247_at | Sterol regulatory element binding transcription factor 2 | SREBF2 | 0·7107886 |
| 227993_at | Methionyl aminopeptidase 2 | METAP2 | 0·7108942 |
| 216508_x_at | similar to nonhistone chromosomal protein HMG-1 | WUGSC | 0·71091264 |
| 227833_s_at | Methyl-CpG binding domain protein 6 | MBD6 | 0·71103483 |
| 1566509_s_at | F-box protein 9 | FBX9 | 0·7116769 |
| 203279_at | ER degradation enhancer, mannosidase alpha-like 1 | EDEM1 | 0·7119141 |
| 235372_at | Fc receptor-like and mucin-like 1 | FREB | 0·71208745 |
| 241968_at | Transcribed locus | 0·71281445 | |
| 206296_x_at | Mitogen-activated protein kinase kinase kinase kinase 1 | MAP4K1 | 0·7130861 |
| 212462_at | MYST histone acetyltransferase (monocytic leukemia) 4 | MYST4 | 0·713116 |
| 233665_x_at | Mitochondrial translation optimization 1 homolog | MTO1 | 0·7134478 |
| 212209_at | Thyroid hormone receptor associated protein 2 | THRAP2 | 0·7144091 |
| 235327_x_at | 602284688F1 NIH_MGC_86 | UBXD4 | 0·7144813 |
| 230618_s_at | BAT2 domain containing 1 | XTP2 | 0·71461946 |
| 236641_at | Kinesin family member 14 | KIF14 | 0·71463937 |
| 204224_s_at | GTP cyclohydrolase 1 | GCH1 | 0·7147643 |
| 204531_s_at | Breast cancer 1, early onset | BRCA1 | 0·71507436 |
| 203787_at | Single-stranded DNA binding protein 2 | SSBP2 | 0·7154888 |
| 212492_s_at | Jumonji domain containing 2B | JMJD2B | 0·7159365 |
| 202817_s_at | Synovial sarcoma translocation, chromosome 18 | SS18 | 0·7160071 |
| 212420_at | E74-like factor 1 | ELF1 | 0·71618116 |
| 203338_at | Protein phosphatase 2, regulatory subunit B (B56), epsilon isoform | PPP2R5E | 0·7164281 |
| 207540_s_at | Spleen tyrosine kinase | SYK | 0·71644425 |
| 229943_at | Ret finger protein 2 | RFP2 | 0·71658355 |
| 218671_s_at | ATPase inhibitory factor 1 | ATPIF1 | 0·71663755 |
| 232909_s_at | Fetal Alzheimer antigen | FALZ | 0·7174373 |
| 225816_at | PHD finger protein 17 | PHF17 | 0·7179169 |
| 211713_x_at | KIAA0101 | KIAA0101 | 0·71805906 |
| 229394_s_at | Glucocorticoid receptor DNA binding factor 1 | GRLF1 | 0·71806455 |
| 212572_at | Serine/threonine kinase 38 like | STK38L | 0·71829355 |
| 209382_at | Polymerase (RNA) III | POLR3C | 0·7190526 |
| 225913_at | KIAA2002 protein | KIAA2002 | 0·7191407 |
| 210776_x_at | Transcription factor 3 | TCF3 | 0·7192611 |
| 223053_x_at | Ssu72 RNA polymerase II CTD phosphatase homolog | HSPC182 | 0·71963733 |
| 226297_at | Homeodomain interacting protein kinase 3 | HIPK3 | 0·7197105 |
| 1558801_at | Nicotinamide nucleotide transhydrogenase | NNT | 0·71974134 |
| 222369_at | Hypothetical protein FLJ13848 | FLJ13848 | 0·71995664 |
| 230352_at | Phosphoribosyl pyrophosphate synthetase 2 | PRPS2 | 0·7200596 |
| 202365_at | Hypothetical protein MGC5139 | MGC5139 | 0·7206042 |
| 205902_at | Potassium intermediate/small conductance calcium-activated channel | KCNN3 | 0·72095835 |
| 233936_s_at | Zinc finger protein 403 | DIF3 | 0·7217099 |
| 201319_at | Myosin regulatory light chain MRCL3 | MRCL3 | 0·72199404 |
| 229582_at | Chromosome 18 open reading frame 37 | C18orf37 | 0·7221231 |
| 204115_at | Guanine nucleotide binding protein (G protein), gamma 11 | GNG11 | 0·7221238 |
| 212010_s_at | Hypothetical protein H41 | H41 | 0·7224393 |
| 218421_at | Ceramide kinase | CERK | 0·722603 |
| 221500_s_at | Syntaxin 16 | STX16 | 0·7226911 |
| 222433_at | Enabled homolog | ENAH | 0·72285813 |
| 211936_at | Heat shock 70 kDa protein 5 | HSPA5 | 0·7232242 |
| 203136_at | Rab acceptor 1 | RABAC1 | 0·7233282 |
| 209902_at | Ataxia telangiectasia and Rad3 related | ATR | 0·7234323 |
| 212780_at | Son of sevenless homolog 1 | SOS1 | 0·723696 |
| 211503_s_at | RAB14, member RAS oncogene family | RAB14 | 0·72393346 |
| 204174_at | Arachidonate 5-lipoxygenase-activating protein | ALOX5AP | 0·7239532 |
| 223090_x_at | Transmembrane protein vezatin | VEZATIN | 0·72426665 |
| 226392_at | RAS p21 protein activator 2 | RASA2 | 0·7243885 |
| 227990_at | Step II splicing factor SLU7 | SLU7 | 0·72443694 |
| 218823_s_at | Potassium channel tetramerisation domain containing 9 | KCTD9 | 0·7246428 |
| 202804_at | ATP-binding cassette, sub-family C (CFTR/MRP), member 1 | ABCC1 | 0·7248081 |
| 227808_at | DnaJ (Hsp40) homolog, subfamily C, member 15 | DNAJD1 | 0·7252901 |
| 208798_x_at | Golgi autoantigen, golgin subfamily a, 8A | GOLGIN-67 | 0·72628635 |
| 213292_s_at | Sorting nexin 13 | SNX13 | 0·72653073 |
| 222996_s_at | CXXC finger 5 | CXXC5 | 0·72690076 |
| 201237_at | Capping protein (actin filament) muscle Z-line, alpha 2 | CAPZA2 | 0·7269243 |
| 224990_at | Hypothetical protein LOC201895 | LOC201895 | 0·72730917 |
| 232008_s_at | Bobby sox homolog | BBX | 0·7273624 |
| 225951_s_at | LOC440309 | CHD2 | 0·727875 |
| 213005_s_at | Ankyrin repeat domain 15 | ANKRD15 | 0·72830474 |
| 213385_at | Chimerin (chimaerin) 2 | CHN2 | 0·72833586 |
| 212952_at | Calreticulin | CALR | 0·72959316 |
| 219356_s_at | Chromatin modifying protein 5 | SNF7DC2 | 0·7297071 |
| 215785_s_at | Cytoplasmic FMR1 interacting protein 2 | CYFIP2 | 0·7301472 |
| 227167_s_at | Mesenchymal stem cell protein DSC96 | 0·73024756 | |
| 204951_at | Ras homolog gene family, member H | RHOH | 0·7306572 |
| 229050_s_at | Hypothetical protein MGC16037 | MGC16037 | 0·73081815 |
| 224778_s_at | TAO kinase 1 | TAOK1 | 0·7310451 |
| 218191_s_at | LMBR1 domain containing 1 | C6orf209 | 0·7311856 |
| 200728_at | ARP2 actin-related protein 2 homolog | ACTR2 | 0·7312589 |
| 228959_at | CDNA | 0·73128814 | |
| 224827_at | Dendritic cell-derived ubiquitin-like protein | DC-UbP | 0·73133826 |
| 218478_s_at | Zinc finger, CCHC domain containing 8 | ZCCHC8 | 0·732107 |
| 201864_at | GDP dissociation inhibitor 1 | GDI1 | 0·7322083 |
| 212069_s_at | KIAA0515 | KIAA0515 | 0·73250145 |
| 202769_at | Cyclin G2 | CCNG2 | 0·7329358 |
| 221751_at | Solute carrier family 2, member 3 pseudogene 1 | PANK3 | 0·7336308 |
| 223054_at | DnaJ (Hsp40) homolog, subfamily B, member 11 | DNAJB11 | 0·73389965 |
| 208765_s_at | Heterogeneous nuclear ribonucleoprotein R | HNRPR | 0·7345736 |
| 212080_at | Similar to CDNA sequence BC021608 | LOC143941 | 0·73511535 |
| 212838_at | Dynamin binding protein | DNMBP | 0·73558724 |
| 243910_x_at | Cullin-associated and neddylation-dissociated 1 | TIP120A | 0·73592746 |
| 205034_at | Cyclin E2 | CCNE2 | 0·7360657 |
| 205267_at | POU domain, class 2, associating factor 1 | POU2AF1 | 0·7366251 |
| 202108_at | Peptidase D | PEPD | 0·7367287 |
| 205367_at | Adaptor protein with pleckstrin homology and src homology 2 domains | APS | 0·7371733 |
| 1562836_at | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | DDX6 | 0·73732203 |
| 217823_s_at | Ubiquitin-conjugating enzyme E2, J1 | UBE2J1 | 0·7376396 |
| 204658_at | Transformer-2 alpha | TRA2A | 0·7381278 |
| 225893_at | MRNA; cDNA DKFZp686D04119 | 0·73877865 | |
| 226874_at | Kelch-like 8 (Drosophila) | KLHL8 | 0·73884803 |
| 217118_s_at | Chromosome 22 open reading frame 9 | C22orf9 | 0·7391449 |
| 209463_s_at | TAF12 RNA polymerase II | TAF12 | 0·73976564 |
| 226134_s_at | Musashi homolog 2 | MSI2 | 0·7397834 |
| 1553906_s_at | FYVE, RhoGEF and PH domain containing 2 | FGD2 | 0·739792 |
| 209748_at | Spastin | SPG4 | 0·7398436 |
| 212995_x_at | Hypothetical protein FLJ14346 | FLJ14346 | 0·7402725 |
| 200967_at | Peptidylprolyl isomerase B | PPIB | 0·74033284 |
| 204391_x_at | Tripartite motif-containing 24 | TIF1 | 0·74084216 |
| 221520_s_at | Cell division cycle associated 8 | CDCA8 | 0·7408515 |
| 223059_s_at | Chromosome 10 open reading frame 45 | C10orf45 | 0·7411011 |
| 224890_s_at | Similar to CG14977-PA | LOC389541 | 0·7412293 |
| 206061_s_at | Dicer1, Dcr-1 homolog | DICER1 | 0·74136984 |
| 201885_s_at | Cytochrome b5 reductase 3 | DIA1 | 0·74161553 |
| 212665_at | TCDD-inducible poly(ADP-ribose) polymerase | TIPARP | 0·74172264 |
| 201242_s_at | ATPase, Na+/K+ transporting, beta 1 polypeptide | ATP1B1 | 0·74187577 |
| 218167_at | Archaemetzincins-2 | LOC51321 | 0·7419247 |
| 214715_x_at | Zinc finger protein 160 | ZNF160 | 0·7425452 |
| 200654_at | Procollagen-proline, 2-oxoglutarate 4-dioxygenase, beta polypeptide | P4HB | 0·74279094 |
| 202171_at | Zinc finger protein 161 | ZNF161 | 0·7429878 |
| 224250_s_at | SECIS binding protein 2 | SECISBP2 | 0·7430157 |
| 222673_x_at | Similar to hypothetical protein MGC17347 | LOC159090 | 0·74343455 |
| 1557961_s_at | 602536302F1 NIH_MGC_59 | 0·7434646 | |
| 202660_at | Family with sequence similarity 20, member C | ITPR2 | 0·7436526 |
| 217901_at | Desmoglein 2 | DSG2 | 0·7440809 |
| 222691_at | solute carrier family 35, member B3 | CGI-19 | 0·7441896 |
| 205739_x_at | Zinc finger protein 588 | ZNF588 | 0·74437064 |
| 224415_s_at | Histidine triad nucleotide binding protein 2 | HINT2 | 0·7444139 |
| 209898_x_at | Intersectin 2 | ITSN2 | 0·74446803 |
| 203660_s_at | Pericentrin (kendrin) | PCNT2 | 0·74484545 |
| 203947_at | Hypothetical protein LOC283267 | CSTF3 | 0·74533767 |
| 212382_at | Transcription factor 4 | TCF4 | 0·7455324 |
| 212560_at | Chromosome 11 open reading frame 32 | SORL1 | 0·7456149 |
| 218095_s_at | TPA regulated locus | TPARL | 0·74569106 |
| 216187_x_at | Kinesin 2 | KNS2 | 0·74619925 |
| 224599_at | CGG triplet repeat binding protein 1 | CGGBP1 | 0·74662995 |
| 230795_at | Histone H4/o | HIST2H4 | 0·74673605 |
| 201266_at | Thioredoxin reductase 1 | TXNRD1 | 0·7467521 |
| 201195_s_at | Solute carrier family 7, member 5 | SLC7A5 | 0·74680513 |
| 221760_at | Mannosidase, alpha, class 1A, member 1 | MAN1A1 | 0·7468957 |
| 225285_at | Branched chain aminotransferase 1, cytosolic | BCAT1 | 0·747043 |
| 217776_at | Retinol dehydrogenase 11 | RDH11 | 0·7471061 |
| 219259_at | semaphorin 4A | SEMA4A | 0·74722433 |
| 223165_s_at | Inositol hexaphosphate kinase 2 | IHPK2 | 0·74733293 |
| 201791_s_at | 7-dehydrocholesterol reductase | DHCR7 | 0·74754477 |
| 208993_s_at | Peptidyl-prolyl isomerase G | PPIG | 0·7477576 |
| 202951_at | Serine/threonine kinase 38 | STK38 | 0·7478889 |
| 203648_at | TatD DNase domain containing 2 | TATDN2 | 0·74821633 |
| 202716_at | Protein tyrosine phosphatase, non-receptor type 1 | PTPN1 | 0·74856865 |
| 227601_at | KIAA1627 protein | KIAA1627 | 0·74859655 |
| 204028_s_at | RAB GTPase activating protein 1 | RABGAP1 | 0·74878204 |
| 209539_at | Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 | ARHGEF6 | 0·7491753 |
| 202027_at | Chromosome 22 open reading frame 5 | C22orf5 | 0·74919754 |
| 1555762_s_at | Megakaryoblastic leukemia 1 | MKL1 | 0·7492855 |
| 212205_at | H2A histone family, member V | H2AFV | 0·7496612 |
| 219553_at | Non-metastatic cells 7, protein expressed in | NME7 | 0·7496923 |
| 208723_at | Ubiquitin specific peptidase 11 | USP11 | 0·749806 |
| 222499_at | SIL1 homolog, endoplasmic reticulum chaperone | SIL1 | 0·74986565 |
Next we examined the effect of CD40 stimulation on apoptosis induction in MLMA cells. As shown in Fig. 6(b), when CD40 was stimulated by the addition of CD40-ligand (CD40L) in the presence of interleukin-4, induction of apoptosis in MLMA cells was mediated by both BCR- and CD20-cross-linking. Furthermore, when both CD40 and BAFF were simultaneously stimulated, better inhibition of BCR-induced and CD20-induced apoptosis was observed than those mediated by each of them alone.
Discussion
In the present study, we have clearly shown that BAFF can inhibit apoptosis mediated by BCR or CD20 in MLMA cells that exhibit a mature B-cell phenotype. The BCR is thought to play a crucial role in clonal selection and clonal expansion in the process of B-cell development to expand high-affinity clones against exogenous antigens and eliminate self-acting or low-affinity clones.31 Although its precise function is yet to be clarified, CD20 is thought to play a role in B-cell development by mediating lipid raft-related signalling.32 Our findings indicate that BAFF contributes to the regulation of B-cell development by modulating apoptotic elimination of B cells mediated by BCR and CD20. Furthermore, our results also indicate that BAFF can enhance cell proliferation in MLMA cells.
A number of studies have attempted to elucidate the molecular basis of the function of BAFF.24,33–39 A major focus of recent investigations has been the pro-survival signalling of BAFF-R. Activation of the alternative NF-κB pathway (processing of NF-κB2 and the nuclear translocation of p52/RelB heterodimers) is a major outcome of BAFF-R-stimulation,2,24,36 whereas BAFF-R also weakly activates the classical NF-κB pathway mediated by NF-κB1 and low-level nuclear translocation of p50/RelA DNA-binding activity is induced. Coincident with previous reports, we also observed that BAFF induced cleavage of both NF-κB2 and NF-κB1 in MLMA cells (Fig. 4). APRIL only activates NF-κB1 via either BCMA or TACI and did not inhibit CD20- and BCR-mediated apoptosis in MLMA cells, so the anti-apoptotic effect of BAFF is thought to be mediated mainly by NF-κB2 activation.
Recent studies have shown that NF-κB directly binds to the promoter region of the Bcl-2 gene and induces transcriptional activation.39 Since Bcl-2 has an anti-apoptotic function, the elevated level of Bcl-2 protein is thought to be important for BAFF-mediated B-cell survival. In addition, BAFF is reported to temporarily inactivate GSK-3β via AKT-mediated phosphorylation.34 Since GSK-3β has been found to cause apoptosis by inducing the degradation of Mcl-1 (an anti-apoptotic Bcl-2 family member) and compromising mitochondrial membrane integrity,40 the BAFF-mediated phosphorylation of GSK-3β is thought to also participate in the anti-apoptotic effect of BAFF. Consistent with previous reports, we observed that BAFF treatment also induced both an increase in Bcl-2 expression and the transient phosphorylation of GSK-3β in MLMA cells. Therefore, it is likely that the inhibitory effect of BAFF against apoptosis mediated by CD20 or BCR is also mainly the result of NF-κB2-mediated Bcl-2 expression and the transient inactivation of GSK-3β.
The pro-apoptotic BH3-only Bcl-2 family member Bim was shown to sequestrate Bcl-2 and play an essential role for BCR cross-linking-induced apoptosis.41 Moreover, Mcl-1 was shown to inhibit Bim selectively and to be essential both early in lymphoid development and later on in the maintenance of mature B lymphocytes.42 Therefore, future investigation of the involvement of Bim and MCL-1 in the BAFF-induced inhibition of CD20-mediated and BCR-mediated apoptosis in MLMA cells should be interesting.
Interestingly, our findings indicated that BAFF treatment also induces the expression of a series of genes related to cell survival. For example, both the microarray analysis and the flow cytometric analysis revealed increased expression of CD40 after the treatment. Since CD40 is known to mediate pro-survival signalling upon interaction with CD40L expressed on activated T cells,43 it is suggested that BAFF-mediated up-regulation of CD40 expression also contributes to B-cell survival in vivo. Indeed, we observed in this study that simultaneous stimulation with CD40 and BAFF resulted in better inhibition of BCR-induced and CD20-induced apoptosis than stimulation with by each of them alone. Therefore, it may be possible that BAFF-mediated up-regulation of CD40 inhibits apoptosis induction synergistically with the effect of BAFF in vivo.
The microarray analysis also revealed up-regulation of several genes involved in either the inhibition of apoptosis or the proliferation of B cells (Table 1). For example, Myb has been demonstrated to directly up-regulate Bcl-2 and suppresses apoptosis.44,45 EBI3 is a subunit of interleukin-27 that increases proliferation of B cells.46 CFLAR is known to inhibit the activation of caspase 8.47 Therefore, our data may indicate that mechanisms other than NF-κB2-mediated Bcl-2 up-regulation are involved in the anti-apoptotic effect of BAFF in MLMA cells. Furthermore, because direct cross-talk between the BCR-signalling and BAFF-signalling systems has been reported,48 BAFF-mediated signals may also be able to directly influence the BCR-mediated apoptotic signalling system.
It is well documented that BAFF-mediated signalling is involved in the survival of malignant B cells.2,49 For example, BAFF is reported to be an autocrine pro-survival and proliferation factor for B-cell chronic lymphocytic leukaemia and multiple myeloma.2,49–51 BAFF is also thought to promote cell survival and proliferation in Hodgkin and non-Hodgkin lymphoma. Therefore, BAFF and BAFF-R might be potential molecular targets in the treatment of B-cell malignancies.52–54 Using a combination of blocking of BAFF-signalling and activation of apoptosis induction, such as CD20 cross-linking, a novel therapeutic approach would be developed.
In conclusion, BAFF-mediated signalling inhibited CD20-mediated or BCR-mediated apoptosis in MLMA cells. Although more detailed experiments are clearly needed, MLMA cells should provide a model for investigating the molecular basis of BAFF’s effect on B cells in vitro and will help to elucidate how B cells survive in an immune system in which BAFF-mediated signalling is involved.
Acknowledgments
We thank S. Yamauchi for excellent secretarial work. This work was supported by a grant from the Japan Health Sciences Foundation for Research on Publicly Essential Drugs and Medical Devices (KHA1004), Health and Labour Sciences Research Grants (the third term comprehensive 10-year-strategy for cancer control H19-010, Research on Children and Families H18-005 and H19-003, Research on Human Genome Tailor made and Research on Publicly Essential Drugs and Medical Devices H18-005), and a Grant for Child Health and Development from the Ministry of Health, Labour and Welfare of Japan. It was also supported by CREST, JST, and the Budget for Nuclear Research of the Ministry of Education, Culture, Sports, Science and Technology, based on screening and counselling by the Atomic Energy Commission.
References
- 1.Plas DR, Rathmell JC, Thompson CB. Homeostatic control of lymphocyte survival: potential origins and implications. Nat Immunol. 2002;3:515–21. doi: 10.1038/ni0602-515. [DOI] [PubMed] [Google Scholar]
- 2.Mackay F, Silveira PA, Brink R. B cells and the BAFF/APRIL axis: fast-forward on autoimmunity and signaling. Curr Opin Immunol. 2007;19:327–36. doi: 10.1016/j.coi.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Schneider P, MacKay F, Steiner V, et al. BAFF, a novel ligand of the tumor necrosis factor family, stimulates B cell growth. J Exp Med. 1999;189:1747–56. doi: 10.1084/jem.189.11.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu HB, Hu WH, Johnson H. TALL-1 is a novel member of the TNFfamily that is down-regulated by mitogens. J Leukoc Biol. 1999;65:680–3. [PubMed] [Google Scholar]
- 5.Mukhopadhyay A, Ni J, Zhai Y, Yu GL, Aggarwal BB. Identification and characterization of a novel cytokine, THANK, a TNF homologue that activates apoptosis, nuclear factor-κB, and c-Jun NH2-terminal kinase. J Biol Chem. 1999;274:15978–81. doi: 10.1074/jbc.274.23.15978. [DOI] [PubMed] [Google Scholar]
- 6.Moore PA, Belvedere O, Orr A, et al. BLyS: member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–3. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 7.Gross JA, Johnston J, Mudri S, et al. TACI and BCMA are receptors for a TNF homologue implicated in B-cell autoimmune disease. Nature. 2000;404:995–9. doi: 10.1038/35010115. [DOI] [PubMed] [Google Scholar]
- 8.Mackay F, Woodcock SA, Lawton P, Ambrose C, Baetscher M, Schneider P, Tschopp J, Browning JL. Mice transgenic for BAFF develop lymphocytic disorders along with autoimmune manifestations. J Exp Med. 1999;190:1697–710. doi: 10.1084/jem.190.11.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khare SD, Sarosi I, Xia XZ, et al. Severe B cell hyperplasia and autoimmune disease in TALL-1 transgenic mice. Proc Natl Acad Sci USA. 2000;97:3370–5. doi: 10.1073/pnas.050580697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schiemann B, Gommerman JL, Vora K, Cachero TG, Shulga-Morskaya S, Dobles M, Frew E, Scott ML. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–4. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- 11.Gross JA, Dillon SR, Mudri S, et al. TACI-Ig neutralizes molecules critical for B cell development and autoimmune disease. Impaired B cell maturation in mice lacking BLyS. Immunity. 2001;15:289–302. doi: 10.1016/s1074-7613(01)00183-2. [DOI] [PubMed] [Google Scholar]
- 12.Vora KA, Wang LC, Rao SP, et al. Cutting edge: germinal centers formed in the absence of B cell-activating factor belonging to the TNF family exhibit impaired maturation and function. J Immunol. 2003;171:547–51. doi: 10.4049/jimmunol.171.2.547. [DOI] [PubMed] [Google Scholar]
- 13.Schneider P. The role of APRIL and BAFF in lymphocyte activation. Curr Opin Immunol. 2005;17:282–9. doi: 10.1016/j.coi.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 14.Smith SH, Cancro MP. Cutting edge: B cell receptor signals regulate BLyS receptor levels in mature B cells and their immediate progenitors. J Immunol. 2003;170:5820–3. doi: 10.4049/jimmunol.170.12.5820. [DOI] [PubMed] [Google Scholar]
- 15.Ng LG, Sutherland AP, Newton R, et al. B cell-activating factor belonging to the TNF family (BAFF)-R is the principal BAFF receptor facilitating BAFF costimulation of circulating T and B cells. J Immunol. 2004;173:807–17. doi: 10.4049/jimmunol.173.2.807. [DOI] [PubMed] [Google Scholar]
- 16.Mackay F, Leung H. The role of the BAFF/APRIL system on T cell function. Semin Immunol. 2006;18:284–9. doi: 10.1016/j.smim.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Thompson JS, Schneider P, Kalled SL, et al. BAFF binds to the tumor necrosis factor receptor-like molecule B cell maturation antigen and is important for maintaining the peripheral B cell population. J Exp Med. 2000;192:129–35. doi: 10.1084/jem.192.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xia XZ, Treanor J, Senaldi G, et al. TACI is a TRAF-interacting receptor for TALL-1, a tumor necrosis factor family member involved in B cell regulation. J Exp Med. 2000;192:137–43. doi: 10.1084/jem.192.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsters SA, Yan M, Pitti RM, Haas PE, Dixit VM, Ashkenazi A. Interaction of the TNF homologues BLyS and APRIL with the TNFreceptor homologues BCMA and TACI. Curr Biol. 2000;10:785–8. doi: 10.1016/s0960-9822(00)00566-2. [DOI] [PubMed] [Google Scholar]
- 20.Shu HB, Johnson H. B cell maturation protein is a receptor for the tumor necrosis factor family member TALL-1. Proc Natl Acad Sci USA. 2000;97:9156–61. doi: 10.1073/pnas.160213497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu Y, Bressette D, Carrell JA, et al. Tumor necrosis factor (TNF) receptor superfamily member TACI is a high affinity receptor for TNF family members APRIL and BLyS. J Biol Chem. 2000;275:35478–85. doi: 10.1074/jbc.M005224200. [DOI] [PubMed] [Google Scholar]
- 22.Thompson JS, Bixler SA, Qian F, et al. BAFF-R, a newly identified TNF receptor that specifically interacts with BAFF. Science. 2001;293:2108–11. doi: 10.1126/science.1061965. [DOI] [PubMed] [Google Scholar]
- 23.Gras MP, Laabi Y, Linares-Cruz G, et al. BCMAp: an integral membrane protein in the Golgi apparatus of human mature B lymphocytes. Int Immunol. 1995;7:1093–106. doi: 10.1093/intimm/7.7.1093. [DOI] [PubMed] [Google Scholar]
- 24.Kayagaki N, Yan M, Seshasayee D, et al. BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-κB2. Immunity. 2002;10:515–24. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- 25.Chaouchi N, Vazquez A, Galanaud P, Leprince C. B cell antigen receptor-mediated apoptosis. Importance of accessory molecules CD19 and CD22, and of surface IgM crosslinking. J Immunol. 1995;154:3096–104. [PubMed] [Google Scholar]
- 26.Shan D, Ledbetter JA, Press OW. Apoptosis of malignant human B cells by ligation of CD20 with monoclonal antibodies. Blood. 1998;91:1644–52. [PubMed] [Google Scholar]
- 27.Kiyokawa N, Kokai Y, Ishimoto K, Fujita H, Fujimoto J, Hata JI. Characterization of the common acute lymphoblastic leukaemia antigen (CD10) as an activation molecule on mature human B cells. Clin Exp Immunol. 1990;79:322–7. doi: 10.1111/j.1365-2249.1990.tb08090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takenouchi H, Kiyokawa N, Taguchi T, Matsui J, Katagiri YU, Okita H, Okuda K, Fujimoto J. Shiga toxin binding to globotriaosyl ceramide induces intracellular signals that mediate cytoskeleton remodeling in human renal carcinoma-derived cells. J Cell Sci. 2004;117:3911–22. doi: 10.1242/jcs.01246. [DOI] [PubMed] [Google Scholar]
- 29.Kiyokawa N, Lee EK, Karunagaran D, Lin SY, Hung MC. Mitosis-specific negative regulation of epidermal growth factor receptor, triggered by a decrease in ligand binding and dimerization, can be overcome by overexpression of receptor. J Biol Chem. 1997;272:18656–65. doi: 10.1074/jbc.272.30.18656. [DOI] [PubMed] [Google Scholar]
- 30.Mimori K, Kiyokawa N, Taguchi T, et al. Costimulatory signals distinctively affect CD20- and B-cell-antigen-receptor-mediated apoptosis in Burkitt’s lymphoma/leukemia cells. Leukemia. 2003;17:1164–74. doi: 10.1038/sj.leu.2402936. [DOI] [PubMed] [Google Scholar]
- 31.Levine MH, Haberman AM, Sant’Angelo DB, Hannum LG, Cancro MP, Janeway CA Jr, Shlomchik MJ. A B-cell receptor-specific selection step governs immature to mature B cell differentiation. Proc Natl Acad Sci USA. 2000;97:2743–8. doi: 10.1073/pnas.050552997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deans JP, Li H, Polyak MJ. CD20-mediated apoptosis: signalling through lipid rafts. Immunology. 2002;107:176–82. doi: 10.1046/j.1365-2567.2002.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enzler T, Bonizzi G, Silverman GJ, Otero DC, Widhopf GF, Anzelon-Mills A, Rickert RC, Karin M. Alternative and classical NF-κB signaling retain autoreactive B cells in the splenic marginal zone and result in lupus-like disease. Immunity. 2006;25:403–15. doi: 10.1016/j.immuni.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 34.Patke A, Mecklenbrauker I, Erdjument-Bromage H, Tempst P, Tarakhovsky A. BAFF controls B cell metabolic fitness through a PKCb- and Akt-dependent mechanism. J Exp Med. 2006;203:2551–62. doi: 10.1084/jem.20060990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qian Y, Qin J, Cui G, et al. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Curr Opin Immunol. 2007;19:327–36. doi: 10.1016/j.immuni.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 36.Claudio E, Brown K, Park S, Wang H, Siebenlist U. BAFF-induced NEMO-independent processing of NF-κB2 in maturing B cells. Nat Immunol. 2002;3:958–65. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 37.Craxton A, Draves KE, Gruppi A, Clark EA. BAFF regulates B cell survival by downregulating the BH3-only family member Bim via the ERK pathway. J Exp Med. 2005;202:1363–74. doi: 10.1084/jem.20051283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mecklenbräuker I, Kalled SL, Leitges M, Mackay F, Tarakhovsky A. Regulation of B-cell survival by BAFF-dependent PKCdelta-mediated nuclear signaling. Nature. 2004;431:456–61. doi: 10.1038/nature02955. [DOI] [PubMed] [Google Scholar]
- 39.Viatour P, Beitires AM, Chariot A, Deregowski V, Leval L, Merville MP, Bours V. NF-κB2/p100 induces Bcl-2 expression. Leukemia. 2003;17:1349–56. doi: 10.1038/sj.leu.2402982. [DOI] [PubMed] [Google Scholar]
- 40.Maurer U, Charvet C, Wagman AS, Dejardin E, Green DR. Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Mol Cell. 2006;21:749–60. doi: 10.1016/j.molcel.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A. Loss of the pro-apoptotic BH3-only Bcl-2 family member Bim inhibits BCR stimulation-induced apoptosis and deletion of autoreactive B cells. J Exp Med. 2003;198:1119–26. doi: 10.1084/jem.20030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Opferman JT, Letai A, Beard Csorcinelli MD, Ong CC, Korsmeyer SJ. Development and maintenance of B and T lymphocytes requires antiapoptotic MCL-1. Nature. 2003;426:671–6. doi: 10.1038/nature02067. [DOI] [PubMed] [Google Scholar]
- 43.Kehry MR. CD40-mediated signaling in B cells. Balancing cell survival, growth, and death. J Immunol. 1996;156:2345–8. [PubMed] [Google Scholar]
- 44.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–44. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 45.Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 1996;10:2720–31. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- 46.Larousserie F, Charlot P, Bardel E, Froger J, Kastelein RA, Devergne O. Differential effects of IL-27 on human B cell subsets. J Immunol. 2006;176:5890–7. doi: 10.4049/jimmunol.176.10.5890. [DOI] [PubMed] [Google Scholar]
- 47.Chandrasekaran Y, McKee CM, Ye Y, Richburg JH. Influence of TRP53 status on FAS membrane localization, CFLAR (c-FLIP) ubiquitinylation, and sensitivity of GC-2spd (ts) cells to undergo FAS-mediated apoptosis. Biol Reprod. 2006;74:560–8. doi: 10.1095/biolreprod.105.045146. [DOI] [PubMed] [Google Scholar]
- 48.Patke A, Mecklenbrauker I, Tarakhovsky A. Survival signaling in resting B cells. Curr Opin Immunol. 2004;16:251–5. doi: 10.1016/j.coi.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 49.Haiat S, Billard C, Quiney C, Ajchenbaum-Cymbalista F, Kolb JP. Role of BAFF and APRIL in human B-cell chronic lymphocytic leukaemia. Immunology. 2006;118:281–92. doi: 10.1111/j.1365-2567.2006.02377.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novak AJ, Darce JR, Arendt BK, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–94. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 51.Moreaux J, Legouffe E, Jourdan E, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–57. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiu A, Xu W, He B, et al. Hodgkin lymphoma cells express TACI and BCMA receptors and generate survival and proliferation signals in response to BAFF and APRIL. Blood. 2007;109:729–39. doi: 10.1182/blood-2006-04-015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He B, Chadburn A, Jou E, Schattner EJ, Knowles DM, Cerutti A. Lymphoma B cells evade apoptosis through the TNF family members BAFF/BLyS and APRIL. J Immunol. 2004;172:3268–79. doi: 10.4049/jimmunol.172.5.3268. [DOI] [PubMed] [Google Scholar]
- 54.Novak AJ, Grote DM, Stenson M, et al. Expression of BLyS and its receptors in B-cell non-Hodgkin lymphoma: correlation with disease activity and patient outcome. Blood. 2004;104:2247–53. doi: 10.1182/blood-2004-02-0762. [DOI] [PubMed] [Google Scholar]