Abstract
Multiparameter flow cytometry has matured tremendously since the 1990s, giving rise to a technology that allows us to study the immune system in unprecedented detail. In this article, we review the development of hardware, reagents, and data analysis tools for multiparameter flow cytometry and discuss future advances in the field. Finally, we highlight new applications that use this technology to reveal previously unappreciated aspects of cell biology and immunity.
Keywords: flow cytometry, T cells, tetramers, fluorochromes, data analysis
Introduction
The complexity of the immune system is remarkable: leucocytes are capable of expressing numerous proteins, combinations of which define functionally distinct cell subsets. To understand which subsets are important for a given immune response, researchers have relied on fluorescence-activated flow cytometry, the method of choice for enumerating and purifying cells. In this article, we review state-of-the-art technology in multiparameter flow cytometry and discuss recent research accomplishments stemming from it.
Beginning in the late 1960s, a series of studies defined unique subsets of B and T cells, setting the stage for multiparameter work. Among B cells, antibody-secreting plasma cells were first isolated in 1969 using single-laser flow cytometers1 and, in 1982, two-laser systems defined subpopulations expressing immunoglobulin M (IgM) and/or IgD.2,3 Subsequent studies defined several distinct peripheral B-cell populations; including B-1a, B-1b, B-2, and marginal zone B cells.4–8 Similarly, simple distinctions between CD4+ and CD8+ T cells were expanded to define naïve, memory, T helper type 1 (Th1), Th2, T cytotoxic type 1 (Tc1) and Tc2 subsets by expression of cell surface proteins.9–11 Beyond these phenotypic measurements, flow cytometry can also be used to measure cell function (e.g. cytokine production12 and cytotoxicity)13 or death14 on a cell-by-cell basis. In fact, a multitude of measurements can be performed now by flow cytometry; recent technical advances allow these measurements to be made simultaneously on individual cells.
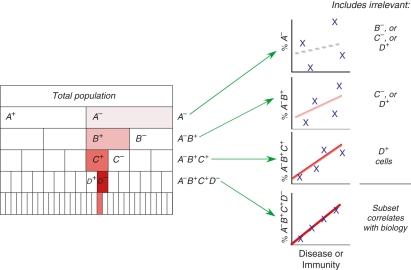
In theory, such multiparameter analysis is quite powerful. It provides more data from less sample, a key consideration when patient samples are limited. Multiparameter analysis also allows more accurate identification of populations, by excluding unwanted cells that bind some reagents.15,16 Most importantly, the technology can identify cells with complex phenotypes, such as those responsible for haematological malignancies17 or those that may be relevant to immunity in vaccine or disease settings.18 As Fig. 1 demonstrates, bulk measurements (using only a few parameters) may not have the power to detect the complex phenotypes relevant to immunity.
Figure 1.
Bulk measurements include irrelevant cell populations that introduce noise, and reduce the power to detect cells of interest. If a population of cells, defined by the phenotype A− B+ C+ D−, correlates strongly with disease or immunity, then assays with fewer than four markers include irrelevant populations that introduce noise into the assay. Note that the measurement of too many may also reduce power, unless the additional markers are excluded from analysis.
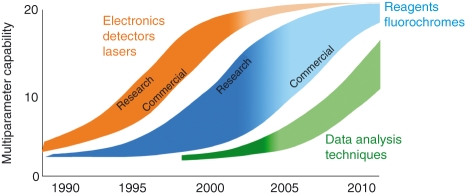
This rationale underlies the field of multiparameter flow cytometry and has driven decades of successive advances in instrumentation and reagents. In particular, an explosion of advances since the 1990s (Fig. 2) has driven the field from five19 to 13 colours (in 2001),20 and recently to 18 colours (in 2006).21 The earliest advances occurred for hardware; these have largely been incorporated into commercial cytometers. Advances in fluorochromes and reagents followed, but some new fluorochromes were used for years in research laboratories before they became commercially available; the repertoire of commercial fluorescent reagents is still expanding rapidly. Finally, the least developed of the flow cytometry technologies is data analysis, where there is great potential (and a strong need) for significant new advances.
Figure 2.
The development of multiparameter flow cytometry. The width of each curve represents the time lag between research discovery and commercialization, while the colour gradient indicates the degree to which each technology has been broadly adopted. For example, advances in hardware (electronics, detectors and lasers) largely occurred in the late 1990s, leading to 20-parameter instrumentation. Instrumentation first developed in 2001 is now widely available; therefore, the colour gradient of this curve is mostly dark. Advances in reagents lagged somewhat behind, and the period between research discovery and commercialization was much longer than for instrumentation. Notably, a number of monoclonal antibody–fluorochrome conjugates are not yet commercially available. Data analysis techniques in the larger flow community remain nascent and represent the area where the greatest need for development remains.
Hardware
At the heart of today’s multiparameter flow cytometers is complex hardware and electronics, including lasers, optical filters, detectors and data acquisition boards. Predecessors to today’s instruments were reported in 1997 (10-parameter flow cytometers)22 and 2001 (13-parameter flow cytometers);20 however, these relied on high-powered lasers, were extensively customized, and required highly skilled operators.23 Recent advances, and the graduation of technologies from research to commercial settings, have made multiparameter flow cytometry much more accessible.
This is particularly evident for laser technology. Early lasers were available in only limited wavelengths and required cooling systems. Today, lasers are small and provide stable excitation sources in a wide variety of wavelengths. Blue (488 nm) and red (633 nm) lasers are typically found in commercial flow cytometers, allowing excitation of a wide range of fluorochromes, from fluorescein isothiocyanate (FITC) to allophycocyanin (APC). Although blue lasers are typically used to excite phycoerythrin (PE) and related tandem dyes (such as Cy5PE), a recent report shows that green (532 nm) lasers provide more sensitive measurement of these dyes, allowing better resolution of dimly-staining populations from background.24 Furthermore, expensive high-powered green lasers may not be necessary to realize this benefit. Simple green laser pointers, priced at around $150, may be used in instruments modified to allow slower transit of cells through the laser beam.25 This has important implications for the development of portable, low-cost flow cytometers.26
Although the three lasers described above can excite a wide variety of fluorochromes, other lasers are available for specialized applications. Ultraviolet (UV; 350 nm) lasers are routinely employed to excite Hoechst dyes, which measure DNA content in cell cycle applications.27 These can also excite quantum dots (QDs), a new class of fluorochromes based on nanotechnology. However, cells exposed to UV lasers are highly autofluorescent, which reduces staining sensitivity. Moreover, UV lasers are large and require much maintenance. Consequently, for QDs, violet (408 nm) lasers are preferred; they induce strong signals with much less cellular autofluorescence.21,28 Additionally, with the advent of yellow (560 nm) and orange (610 nm) lasers,29 excitation sources are now available for the complete range of visible light. These lasers may facilitate the use of new fluorescent dyes (such as red fluorescent proteins)30 and offer additional flexibility in multiparameter hardware. A system that employs violet, blue, green and red lasers can excite a broad range of fluorochromes, providing significant potential for multiplexed analysis.21
The development of high-throughput systems represents the most recent commercial advance in flow cytometry. These systems allow serial analysis of hundreds, if not thousands, of samples automatically and are particularly valuable in drug discovery efforts. In research and clinical settings, high-throughput systems allow the staining of multiple samples at once, thereby reducing manual labour and the potential for experiment-to-experiment variability. However, because high-throughput systems consume so many samples at once, it is critical that the flow cytometer is under rigorous quality control, otherwise massive amounts of data are lost before an instrumentation problem is noticed. In 2006, we published a research protocol describing our three-part approach to quality assurance.31 This approach consists of: (1) a system optimization step (to ensure that laser, mirror and filter configuration is optimal), (2) a calibration step (to determine the sensitivity and dynamic range of the detector, and develop a target value for detector gain), and (3) a monitoring step (to track precision, accuracy and sensitivity of fluorescence measurements over time). Analogous commercial efforts were recently introduced.
In summary, recent advances in lasers (and previous advances in other hardware) have produced relatively mature instrument technology that may, in its current form, be nearing its peak. In the short term, improvements in high-throughput systems are likely to occur.32 In the long term, emerging advances include the use of silicone avalanche photodiodes,33 for enhanced detection of far-red and infrared light, and microfluidic/acoustical focusing34,35 for instrument miniaturization. Beyond these, advances to the next generation of flow cytometers will rely on non-fluorescent probes36–39 to extend the number of measurable parameters.
Reagents
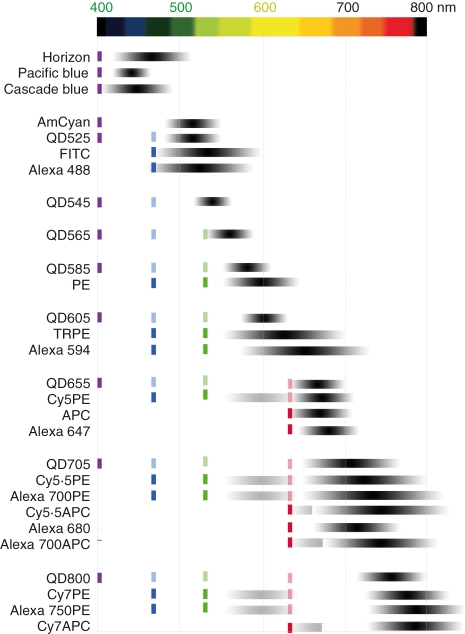
Our understanding of the complexity of the immune system drove the need for new fluorochromes beyond the classical organic dyes FITC, PE and APC. By coupling cyanines (Cy5, Cy5.5 and Cy7) and Alexa dyes with PE and APC to produce resonance energy tandem dyes, the repertoire of fluorochromes was broadened through the late 1980s and 1990s. For example, tandem dyes produced with Alexa fluors and PE or APC often perform as well as (or better than) similar organic dyes in terms of fluorescent signal and stability. Many of these dyes are also commonly used alone, directly conjugated to antibodies (e.g. Alexas 488, 647 and 680). More recently, the development of inorganic dyes has provided even more choices (Fig. 3), and allowed researchers to evaluate the best fluorochromes for their instrumentation.
Figure 3.
A selection of fluorochromes commonly used in flow cytometry. For each fluorochrome, the optimal laser source for excitation is indicated by coloured dark lines (violet = 405 nm, blue = 488 nm, green = 532 nm and red = 633 nm). Some fluorochromes, such as quantum dots, are suboptimally excited by a number of other laser lines, which are represented by the dimmer lines. Each fluorochrome’s range of emission is indicated by the grey/black bars, with the darkest region corresponding to the wavelengths of strongest emission. APC, allophycocyanin, FITC, fluorescein isothiocyanate; PE, phycoerythrin; QD, quantum dots.
In particular, the development of a new class of inorganic dyes, based on nanotechnology, has generated considerable interest. QDs are semiconductor nanocrystals with physical and fluorescence properties suitable for flow cytometry.21,28 Compared to organic fluorochromes, these properties seem tailor-made for multicolour flow cytometry. Most importantly, organic fluorochromes have narrow excitation spectra, such that multiplexed analysis of these fluorochromes usually requires multiple lasers for excitation.40 The QDs, however, have broad excitation spectra21,28 so that all (QD525–QD800) are excited by the same laser source with bright signals. Consequently, even a single-laser flow cytometer can be used for complex multicolour experiments.21
The emission properties of QDs also offer advantages over organic fluorochromes. Most organic fluorochromes have broader emission spectra than QDs,21,40 particularly on the red side of the emission band. Therefore, when organic fluorochromes are multiplexed, their signals overlap significantly with each other, often reducing the ability to detect dimly-staining populations.41 In contrast, a five-QD system can be constructed with minimal fluorochrome overlap and excellent sensitivity.21 Moreover, when used in staining panels with organic fluorochromes, very little light from the organic dyes is detected in QD channels.21 Bright QDs are therefore particularly useful in multicolour panels that attempt to detect low-level expression.23,28,42
As Fig. 3 shows, there are now over 30 different fluorochromes used as tags with monoclonal antibody conjugates; this significantly exceeds the number of detectors available in even the most technologically advanced systems.23 Most fluorochromes differ in fluorescent and/or chemical properties so the wide variety provides researchers with many options, both in terms of applications and antibody–fluorochrome combinations. The latter is a particularly important consideration because not all reagent combinations work together in multiparameter systems.42
In summary, the development of reagent technology has lagged somewhat behind instrumentation (Fig. 2), still, research efforts for the development of new fluorochromes have reached a point of diminishing returns. In contrast, better commercial availability of more and varied fluorochrome–antibody combinations is still increasing.
Data analysis
As multiparameter flow cytometry becomes more complex, the volume of information generated grows exponentially. A typical four-colour experiment, identifying only positive and negative populations, could generate 16 potential subsets for analysis, while eight- and 18-colour experiments could generate 256 and 262 144 possible subsets, respectively. This sometimes exceeds the capabilities of researchers, computer processors and software. Fortunately, strategies and tools for the presentation and analysis of complex, multiparameter datasets are slowly emerging; however, there remains a critical need for additional work in this area. Analysis systems that rely heavily on the researcher (i.e. ‘manually managed’) are the most developed, but in the future we may rely increasingly on computer-driven analysis.
Data visualization
Historically, flow cytometry data have been plotted on a simple log scale to accommodate the wide dynamic range of the data – and because many fluorescence distributions are log-normal. However, in multiparametric fluorescence analyses, the distributions of events at low fluorescence are no longer log-normal; furthermore, the measurements can include values that are approaching zero and even have negative values (this arises from relatively large errors in measurements propagated by fluorescence spillover compensation).41 The end result was distributions that were difficult to interpret, and that could include a tremendous number of events ‘piled up’ on the axis of the log scale.43 Recently, new methods to scale the data (e.g. Logicle scaling)43,44 have become widely available to address this problem. This scaling smoothly combines a logarithmic scaling for highly fluorescent distributions with a linear scaling for measurements near zero, resulting in data displays that are far more intuitive and understandable to researchers. While seemingly arcane, this change in the visualization of multiparameter data has revolutionized the analysis and presentation of these complex data.
Manually managed analysis
When data analysis is managed manually, the choice of which relationships are examined is typically hypothesis driven, and a limited number of parameters are compared. This can provide focused and biologically plausible results; however, such analyses can miss data relationships that researchers had not envisioned. Moreover, the analysis methods of the researchers may influence outcomes. In this regard, certain methods may be used to ensure that populations of interest are identified correctly, easily quantified, correctly compared and optimally presented.
Once cell populations are correctly identified, they can be quantified and their frequency compared across groups. Generating gates to define every distinct subset can be cumbersome; therefore, we define cells positive for each parameter individually, and then use Boolean gating algorithms to combine positive and negative subsets for each marker in every possible combination. The results, in a six-colour experiment, are 64 distinct subsets automatically generated from the single parameter gates. Clearly, this process is highly dependent on how the researcher determines the cut-off between positive and negative populations, a potential source of bias that could be avoided using computer-driven analysis techniques (e.g. clustering). In any case, the frequency of each subset can be displayed and compared against groups using the recently developed pestle and spice software suite (M. Roederer, Vaccine Research Center, NIAID, NIH), which can also mask specific variables or display the frequency of cells expressing a partial (e.g. three-colour) phenotype.
Optimal presentation of multiparameter data remains a challenge because the human mind is conditioned to view and understand data in two dimensions at a time. Over the years, options that overlay additional data on classical bivariate dot plots have emerged. Examples include the addition of rough quantitative information with contour, density and pseudocolour plots.45 Notably, however, these graphs share the disadvantage that they do not provide any information about data relationships beyond the two markers displayed. Recently, polychromatic plots were developed to overcome this limitation.46 In these plots, shades of colour are overlayed on bivariate graphs to communicate the values of other parameters. For example, for a bivariate dot plot of CD45RA versus CCR7, the expression of interleukin-2 (IL-2) and interferon-γ (IFN-γ) for each event in the histogram can be shown by assigning two colours (red and blue, respectively) to the cytokines; cells that coexpress IL-2 and IFN-γ are purple, while cells expressing IL-2 alone are red. By changing how these plots are scaled and which colour takes priority, certain populations with shared characteristics can be emphasized or de-emphasized. In this way, data can be explored manually in up to five dimensions.
In summary, there are a number of means by which to analyse multiparameter data manually; however, these tools depend on human input, so confusion, bias and artefacts may arise if samples are not analysed carefully and consistently. Also, these analyses can be labour intensive. Finally, manually managed analyses tend to ignore relationships between markers that had not been envisioned by the researcher. In this way, they constrain the discovery of new cell types relevant to disease and immunity.
Computer-driven analysis
Because of the high content of today’s flow cytometry data, the potential to find new, biologically important cell types is enormous; however, without increasingly sophisticated analysis tools, multiparameter data cannot be mined easily. In recent years, techniques developed for genomic and proteomic studies are now being applied to flow cytometry.
For example, once values (such as population frequencies) are generated in conventional flow cytometry software, the relationships among parameters can be explored using automated analysis tools, such as principal component analysis and hierarchical clustering. These techniques have been used to identify differences between in vitro cultured cell populations, age-related memory T-cell frequency, and the profiles of various inflammatory and malignant diseases.47–53 The disadvantage of this approach is that manual data-processing (to determine population frequencies) is still required.
In contrast, raw flow cytometry data (in the form of list mode data files) can be used to cluster data automatically (without any researcher effort or intervention).54–57 These methods are very similar to those used to analyse microarray data; however, list mode data have much higher information content than microarrays. For example, 100 microarrays contain around five million summarized measures, while just one 10-colour list mode file of 500 000 events contains more than six million data-points. This highlights one of the challenges facing this field: cluster analysis of high content, multicolour clinical data from large cohorts of patients will require enormous computing power.
The identification of the best clustering tools also remains an important challenge. A good clustering algorithm should describe not only visually evident populations within the data but also more intricate clusters distributed in multidimensional space. To describe populations and subsets based on phenotypic characteristics, a partitioning technique (such as k-means clustering) is frequently used. However, the asymmetric data distributions often found in flow cytometry data are not handled well by this algorithm. Moreover, with the simplest k-means tools the number of clusters to generate must be determined by the researcher.58,59 To eliminate this requirement, and allow completely unsupervised (computer-based) analysis, a number of recent studies have explored Gaussian Mixture modelling.55,60,61
In addition, to successfully employ automated/computer-driven analysis, it is very important to limit technical sources of variation in flow cytometry instrumentation, reagents and sample handling. In microarray studies, technical sources of variance are estimated simply by using control probes; however, in flow cytometry, multiple parameters (that are difficult to control for) can introduce variance. These include unique characteristics of the instrument, such as laser output and alignment, and staining-related issues such as pipetting, staining temperature and incubation time. To some extent, these sources of variance can be minimized with strict quality control and monitoring of instruments.31
As indicated in Fig. 2, data analysis remains a relatively nascent technology. There are few clear standards for data analysis,62–64 and the tools available are either extremely labour-intensive or require considerable expertise to apply. Moreover, basic questions remain. For example, what is a population, how can clustering tools identify populations, and how do we know whether the clusters discovered are ‘real’ and biologically relevant? Can clustering tools provide reproducible results when data are analysed on different instruments, on different days? These questions must be addressed before automated techniques for data analysis can be applied to clinical studies in a more extensive way.
Applications
In today’s era of systems biology,65,66 multiparameter flow cytometry can be a key tool for understanding protein networks. The remainder of this article highlights three applications where this technology is revealing previously unappreciated aspects of cell biology and immunity.
Recently, phosphoprotein-specific multicolour assays have been developed to explore the dynamics of the intracellular signalling pathways and map cell-signalling networks important in immunology and haematology.67 These assays combine intracellular and surface staining to determine the phosphorylation status of a number of intracellular signalling molecules, such as mitogen-activated protein kinases (MAPK), Jun N-terminal kinases (JNK), extracellular signal-regulated kinases (Erk), p38, Janus kinases/signal transducer and activator of transcription (JAK/Stat) and others. Studies using this technology have not only enabled the exploration of fundamental aspects of signalling, but have also paved the way for new clinical paradigms. By mapping aberrant signalling cascades in malignancies, and grouping diseases with shared networks, this technology can be used to reclassify cancers based on precise molecular (rather than nebulous pathological or clinical) traits, and may lead to novel disease therapies.68,69
Multiparameter flow cytometry has also reshaped our understanding of antigen-specific T-cell responses. In particular, recent studies show that cells with certain functional traits correlate with protective immunity to a wide variety of diseases or vaccines. These protective T cells cannot be identified solely by measurement of IFN-γ, but rather are defined by the combinations of cytokines they produce.
This was first described in the setting of human immunodeficiency virus type 1 (HIV-1) infection, where people with progressive disease have fewer cells expressing IL-2 alone or IL-2 + IFN-γ than people with non-progressing or treated disease.70,71 These studies were extended by developing nine-plus colour panels to measure tumour necrosis factor-α, macrophage inflammatory protein-1β, and the degranulation marker CD107a, which showed that cells expressing multiple functional molecules simultaneously correlated with reduced disease burden; cells producing IFN-γ alone were associated with progressive disease.72–74 Similarly, in settings where antigen load is low and transient (e.g. vaccinia immunization)75 more multifunctional cells are observed than when antigen is persistent (cytomegalovirus, HIV).72 Therefore, those immune responses that control viral infections were associated with multifunctional T cells, but it remained unclear whether these cells mediated control or whether their presence was a consequence of viral clearance.72 A mouse model for vaccine-induced protection provided the first evidence that this was not just a correlation.76 In this model, the protection afforded by different vaccine regimens could be predicted by the number of polyfunctional T cells induced before challenge. This study also revealed that cells making multiple cytokines express higher levels of IFN-γ on a per cell basis than cells that produce only one cytokine.76 The multifunctional measurement therefore revealed a strong correlate of vaccine-mediated protection as well as a mechanism underlying the protection. Because this important correlate of immunity is multidimensional, it could only be revealed through multiparameter flow cytometry.
Finally, in recent years, a wide variety of applications have emerged from QD technology. For example, in 2006, we analysed the maturity of various antigen-specific T-cell populations using a 17-colour staining panel.21 This panel consisted of seven QDs and 10 organic fluorochromes, which were measured simultaneously in the same sample. The QD reagents used were conjugates with conventional antibodies (against CD4, CD45RA and CD57), as well as peptide major histocompatibility complex class I (pMHCI) multimers designed to detect those antigen-specific T cells directed against HIV, Epstein–Barr virus and cytomegalovirus epitopes. Before this study, only FITC-, PE- and APC-tetramers were available, which limited panel design because many novel or dimly-staining antibodies are only found on these fluorochromes. The QDs also display higher valency than PE or APC, so more pMHCI molecules could be bound to a QD than to a PE or APC-streptavidin (SAV), allowing brighter signals and better staining resolution. This work demonstrated the power of a multiplexed approach: by identifying multiple phenotypically distinct subsets within each antigen-specific T-cell population, the remarkable intricacy of T-cell immunity can be appreciated. Moreover, QDs also allow us to measure many antigen-specific populations simultaneously, an important factor when sample availability is limited.
Conclusions
Since the development of a one-laser system for purification of plasma B cells, flow cytometry technology has come a long way. Early advances occurred in instrumentation, and over time these have produced relatively mature systems that can be customized extensively and are suitable for hundreds of applications. These applications would not be possible, however, without significant development of reagents and fluorochromes. Currently, in the research setting, enough fluorochromes are available to cover most instrumentation configurations; however, commercially this is not yet the case. Finally, data analysis technology lags significantly behind hardware and reagent technology, and is likely to provide the most dramatic advances in the future. The next few years will see this remarkable technology being applied to a wide variety of systems, undoubtedly spurring many advances in our knowledge of the immune system.
References
- 1.Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–9. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- 2.Hardy RR, Hayakawa K, Haaijman J, Herzenberg LA. B-cell subpopulations identified by two-colour fluorescence analysis. Nature. 1982;297:589–91. doi: 10.1038/297589a0. [DOI] [PubMed] [Google Scholar]
- 3.Hardy RR, Hayakawa K, Haaijman J, Herzenberg LA. B-cell subpopulations identifiable by two-color fluorescence analysis using a dual-laser FACS. Ann NY Acad Sci. 1982;399:112–21. doi: 10.1111/j.1749-6632.1982.tb25667.x. [DOI] [PubMed] [Google Scholar]
- 4.Hardy RR, Hayakawa K, Parks DR, Herzenberg LA. Demonstration of B-cell maturation in X-linked immunodeficient mice by simultaneous three-colour immunofluorescence. Nature. 1983;306:270–2. doi: 10.1038/306270a0. [DOI] [PubMed] [Google Scholar]
- 5.Kantor AB, Stall AM, Adams S, Herzenberg LA, Herzenberg LA. Differential development of progenitor activity for three B-cell lineages. Proc Natl Acad Sci USA. 1992;89:3320–4. doi: 10.1073/pnas.89.8.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kantor AB, Stall AM, Adams S, Herzenberg LA, Herzenberg LA. Adoptive transfer of murine B-cell lineages. Ann NY Acad Sci. 1992;651:168–9. doi: 10.1111/j.1749-6632.1992.tb24610.x. [DOI] [PubMed] [Google Scholar]
- 7.Martin F, Kearney JF. CD21high IgMhigh splenic B cells enriched in the marginal zone: distinct phenotypes and functions. Curr Top Microbiol Immunol. 1999;246:45–52. doi: 10.1007/978-3-642-60162-0_6. [DOI] [PubMed] [Google Scholar]
- 8.Martin F, Oliver AM, Kearney JF. Marginal zone and B1 B cells unite in the early response against T-independent blood-borne particulate antigens. Immunity. 2001;14:617–29. doi: 10.1016/s1074-7613(01)00129-7. [DOI] [PubMed] [Google Scholar]
- 9.Thomas ML, Lefran√ßois L. Differential expression of the leucocyte-common antigen family. Immunol Today. 1988;9:320–6. doi: 10.1016/0167-5699(88)91326-6. [DOI] [PubMed] [Google Scholar]
- 10.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145–73. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 11.Mosmann TR, Li L, Sad S. Functions of CD8 T-cell subsets secreting different cytokine patterns. Semin Immunol. 1997;9:87–92. doi: 10.1006/smim.1997.0065. [DOI] [PubMed] [Google Scholar]
- 12.Picker LJ, Singh MK, Zdraveski Z, Treer JR, Waldrop SL, Bergstresser PR, Maino VC. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood. 1995;86:1408–19. [PubMed] [Google Scholar]
- 13.Liu J, Roederer M. Differential susceptibility of leukocyte subsets to cytotoxic T cell killing: implications for HIV immunopathogenesis. Cytometry A. 2007;71:94–104. doi: 10.1002/cyto.a.20363. [DOI] [PubMed] [Google Scholar]
- 14.Darzynkiewicz Z, Bruno S, Del Bino G, Gorczyca W, Hotz MA, Lassota P, Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13:795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- 15.Perfetto SP, Chattopadhyay PK, Lamoreaux L, Nguyen R, Ambrozak D, Koup RA, Roederer M. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006;313:199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 16.Chattopadhyay PK, Melenhorst J, Ladell K, et al. Techniques to improve the direct ex vivo detection of low frequency antigen-specific CD8+ T cells with peptide-major histocompatibility complex class I tetramers. Cytometry A. 2008;21 doi: 10.1002/cyto.a.20642. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood BL. Myeloid malignancies: myelodysplastic syndromes, myeloproliferative disorders, and acute myeloid leukemia. Clin Lab Med. 2007;27:551–75. doi: 10.1016/j.cll.2007.05.006. vii. [DOI] [PubMed] [Google Scholar]
- 18.Makedonas G, Betts MR. Polyfunctional analysis of human T cell responses: importance in vaccine immunogenicity and natural infection. Springer Semin Immunopathol. 2006;28:209–19. doi: 10.1007/s00281-006-0025-4. [DOI] [PubMed] [Google Scholar]
- 19.Roederer M, Bigos M, Nozaki T, Stovel RT, Parks DR, Herzenberg LA. Heterogeneous calcium flux in peripheral T cell subsets revealed by five-color flow cytometry using log-ratio circuitry. Cytometry. 1995;21:187–96. doi: 10.1002/cyto.990210211. [DOI] [PubMed] [Google Scholar]
- 20.De Rosa SC, Roederer M. Eleven-color flow cytometry. A powerful tool for elucidation of the complex immune system. Clin Lab Med. 2001;21:697–712. Vii. [PubMed] [Google Scholar]
- 21.Chattopadhyay PK, Price DA, Harper TF, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006;12:972–7. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 22.Roederer M, De Rosa SC, Gerstein R, et al. Eight color, 10-parameter flow cytometry to elucidate complex leukocyte heterogeneity. Cytometry. 1997;29:328–39. doi: 10.1002/(sici)1097-0320(19971201)29:4<328::aid-cyto10>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 23.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004;4:648–55. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 24.Perfetto SP, Roederer M. Increased immunofluorescence sensitivity using 532 nm laser excitation. Cytometry A. 2007;71:73–9. doi: 10.1002/cyto.a.20358. [DOI] [PubMed] [Google Scholar]
- 25.Habbersett RC, Naivar MA, Woods TA, Goddard GR, Graves SW. Evaluation of a green laser pointer for flow cytometry. Cytometry A. 2007;71:809–17. doi: 10.1002/cyto.a.20454. [DOI] [PubMed] [Google Scholar]
- 26.Shapiro HM, Perlmutter NG. Personal cytometers: slow flow or no flow? Cytometry A. 2006;69:620–30. doi: 10.1002/cyto.a.20284. [DOI] [PubMed] [Google Scholar]
- 27.Telford WG. Analysis of UV-excited fluorochromes by flow cytometry using near-ultraviolet laser diodes. Cytometry A. 2004;61:9–17. doi: 10.1002/cyto.a.20032. [DOI] [PubMed] [Google Scholar]
- 28.Chattopadhyay PK, Yu J, Roederer M. Application of quantum dots to multicolor flow cytometry. Methods Mol Biol. 2007;374:175–84. doi: 10.1385/1-59745-369-2:175. [DOI] [PubMed] [Google Scholar]
- 29.Kapoor V, Karpov V, Linton C, Subach FV, Verkhusha VV, Telford WG. Solid state yellow and orange lasers for flow cytometry. Cytometry A. 2008;73:570–7. doi: 10.1002/cyto.a.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kapoor V, Subach FV, Kozlov VG, Grudinin A, Verkhusha VV, Telford WG. New lasers for flow cytometry: filling the gaps. Nat Methods. 2007;4:678–9. doi: 10.1038/nmeth0907-678. [DOI] [PubMed] [Google Scholar]
- 31.Perfetto SP, Ambrozak D, Nguyen R, Chattopadhyay P, Roederer M. Quality assurance for polychromatic flow cytometry. Nat Protoc. 2006;1:1522–30. doi: 10.1038/nprot.2006.250. [DOI] [PubMed] [Google Scholar]
- 32.Edwards B. Flow cytometry for high-throughput, high-content screening. Curr Opin Chem Biol. 2004;8:392–8. doi: 10.1016/j.cbpa.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 33.Lawrence WG, Varadi G, Entine G, Podniesinski E, Wallace PK. Enhanced red and near infrared detection in flow cytometry using avalanche photodiodes. Cytometry A. 2008;73:767–76. doi: 10.1002/cyto.a.20595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh D, Gu W, Kamotani Y, Grotberg JB, Takayama S. Microfluidics for flow cytometric analysis of cells and particles. Physiol Meas. 2005;26:R73–98. doi: 10.1088/0967-3334/26/3/R02. [DOI] [PubMed] [Google Scholar]
- 35.Leary JF. Ultra high-speed sorting. Cytometry A. 2005;67:76–85. doi: 10.1002/cyto.a.20160. [DOI] [PubMed] [Google Scholar]
- 36.Ornatsky O, Baranov VI, Bandura DR, Tanner SD, Dick J. Multiple cellular antigen detection by ICP-MS. J Immunol Methods. 2006;308:68–76. doi: 10.1016/j.jim.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 37.Jett JH. Raman spectroscopy comes to flow cytometry. Cytometry A. 2008;73:109–10. doi: 10.1002/cyto.a.20526. [DOI] [PubMed] [Google Scholar]
- 38.Razumienko E, Ornatsky O, Kinach R, Milyavsky M, Lechman E, Baranov V, Winnik MA, Tanner SD. Element-tagged immunoassay with ICP-MS detection: evaluation and comparison to conventional immunoassays. J Immunol Methods. 2008;336:56–63. doi: 10.1016/j.jim.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson DA, Brown LO, Gaskill DF, Naivar M, Graves SW, Doorn SK, Nolan JP. A flow cytometer for the measurement of Raman spectra. Cytometry A. 2008;73:119–28. doi: 10.1002/cyto.a.20520. [DOI] [PubMed] [Google Scholar]
- 40.Riegler J, Nann T. Application of luminescent nanocrystals as labels for biological molecules. Anal Bioanal Chem. 2004;379:913–9. doi: 10.1007/s00216-004-2706-y. [DOI] [PubMed] [Google Scholar]
- 41.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry A. 2001;45:194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 42.Mahnke YD, Roederer M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med. 2007;27:469–85. doi: 10.1016/j.cll.2007.05.002. v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006;7:681–5. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 44.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006;69:541–51. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]
- 45.Roederer M, Darzynkiewicz Z, Parks DR. Guidelines for the presentation of flow cytometric data. Methods Cell Biol. 2004;75:241–56. doi: 10.1016/s0091-679x(04)75010-4. [DOI] [PubMed] [Google Scholar]
- 46.Roederer M, Moody MA. Polychromatic plots: graphical display of multidimensional data. Cytometry A. 2008;73:868–74. doi: 10.1002/cyto.a.20610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brinkman RR, Gasparetto M, Lee SJ, Ribickas AJ, Perkins J, Janssen W, Smiley R, Smith C. High-content flow cytometry and temporal data analysis for defining a cellular signature of graft-versus-host disease. Biol Blood Marrow Transplant. 2007;13:691–700. doi: 10.1016/j.bbmt.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Finn WG, Carter KM, Raich R, Stoolman LM, Hero AO. Analysis of clinical flow cytometric immunophenotyping data by clustering on statistical manifolds: treating flow cytometry data as high-dimensional objects. Cytometry B Clin Cytom. 2008 doi: 10.1002/cyto.b.20435. in press. [DOI] [PubMed] [Google Scholar]
- 49.Lugli E, Pinti M, Nasi M, et al. Subject classification obtained by cluster analysis and principal component analysis applied to flow cytometric data. Cytometry A. 2007;71:334–44. doi: 10.1002/cyto.a.20387. [DOI] [PubMed] [Google Scholar]
- 50.Petrausch U, Haley D, Miller W, Floyd K, Urba WJ, Walker E. Polychromatic flow cytometry: a rapid method for the reduction and analysis of complex multiparameter data. Cytometry A. 2006;69:1162–73. doi: 10.1002/cyto.a.20342. [DOI] [PubMed] [Google Scholar]
- 51.Sachs K, Perez O, Pe’er D, Lauffenburger DA, Nolan GP. Causal protein-signaling networks derived from multiparameter single-cell data. Science. 2005;308:523–9. doi: 10.1126/science.1105809. [DOI] [PubMed] [Google Scholar]
- 52.Steinbrich-Zollner M, Gron JR, Kaiser T, et al. From transcriptome to cytome: integrating cytometric profiling, multivariate cluster, and prediction analyses for a phenotypical classification of inflammatory diseases. Cytometry A. 2008;73:333–40. doi: 10.1002/cyto.a.20505. [DOI] [PubMed] [Google Scholar]
- 53.Zamir E, Geiger B, Cohen N, Kam Z, Katz BZ. Resolving and classifying haematopoietic bone-marrow cell populations by multi-dimensional analysis of flow-cytometry data. Br J Haematol. 2005;129:420–31. doi: 10.1111/j.1365-2141.2005.05471.x. [DOI] [PubMed] [Google Scholar]
- 54.Jeffries D, Zaidi I, de Jong B, Holland MJ, Miles DJ. Analysis of flow cytometry data using an automatic processing tool. Cytometry A. 2008;73:857–67. doi: 10.1002/cyto.a.20611. [DOI] [PubMed] [Google Scholar]
- 55.Lo K, Brinkman RR, Gottardo R. Automated gating of flow cytometry data via robust model-based clustering. Cytometry A. 2008;73:321–32. doi: 10.1002/cyto.a.20531. [DOI] [PubMed] [Google Scholar]
- 56.Pedreira CE, Costa ES, Arroyo ME, Almeida J, Orfao A. A multidimensional classification approach for the automated analysis of flow cytometry data. IEEE Trans Biomed Eng. 2008;55:1155–62. doi: 10.1109/TBME.2008.915729. [DOI] [PubMed] [Google Scholar]
- 57.Toedling J, Rhein P, Ratei R, Karawajew L, Spang R. Automated in-silico detection of cell populations in flow cytometry readouts and its application to leukemia disease monitoring. BMC Bioinformatics. 2006;7:282. doi: 10.1186/1471-2105-7-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boddy L, Wilkins MF, Morris CW. Pattern recognition in flow cytometry. Cytometry. 2001;44:195–209. doi: 10.1002/1097-0320(20010701)44:3<195::aid-cyto1112>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 59.Wilkins MF, Hardy SA, Boddy L, Morris CW. Comparison of five clustering algorithms to classify phytoplankton from flow cytometry data. Cytometry. 2001;44:210–7. doi: 10.1002/1097-0320(20010701)44:3<210::aid-cyto1113>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 60.Boedigheimer MJ, Ferbas J. Mixture modeling approach to flow cytometry data. Cytometry A. 2008;73:421–9. doi: 10.1002/cyto.a.20553. [DOI] [PubMed] [Google Scholar]
- 61.Chan C, Feng F, Ottinger J, Foster D, West M, Kepler TB. Statistical mixture modeling for cell subtype identification in flow cytometry. Cytometry A. 2008;73:693–701. doi: 10.1002/cyto.a.20583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee JA, Spidlen J, Boyce K, et al. MIFlowCyt: the minimum information about a flow cytometry experiment. Cytometry A. 2008;73A:926–30. doi: 10.1002/cyto.a.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spidlen J, Gentleman RC, Haaland PD, et al. Data standards for flow cytometry. OMICS. 2006;10:209–14. doi: 10.1089/omi.2006.10.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spidlen J, Leif R, Moore W, Roederer M, Brinkman RR. International Society for the Advancement of Cytometry Data Standards Task F. Gating-ML: XML-based gating descriptions in flow cytometry. Cytometry A. 2008 doi: 10.1002/cyto.a.20637. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kitano H. Systems biology: a brief overview. Science. 2002;295:1662–4. doi: 10.1126/science.1069492. [DOI] [PubMed] [Google Scholar]
- 66.Nurse P. Systems biology: understanding cells. Nature. 2003;424:883. doi: 10.1038/424883a. [DOI] [PubMed] [Google Scholar]
- 67.Perez OD, Nolan GP. Simultaneous measurement of multiple active kinase states using polychromatic flow cytometry. Nat Biotechnol. 2002;20:155–62. doi: 10.1038/nbt0202-155. [DOI] [PubMed] [Google Scholar]
- 68.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6:146–55. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 69.Irish JM, Hovland R, Krutzik PO, Perez OD, Bruserud ò, Gjertsen BT, Nolan GP. Single cell profiling of potentiated phospho-protein networks in cancer cells. Cell. 2004;118:217–28. doi: 10.1016/j.cell.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 70.Younes SA, Yassine-Diab B, Dumont AR, Boulassel MR, Grossman Z, Routy JP, Sekaly RP. HIV-1 viremia prevents the establishment of interleukin 2-producing HIV-specific memory CD4+ T cells endowed with proliferative capacity. J Exp Med. 2003;198:1909–22. doi: 10.1084/jem.20031598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Harari A, Petitpierre S, Vallelian F, Pantaleo G. Skewed representation of functionally distinct populations of virus-specific CD4 T cells in HIV-1-infected subjects with progressive disease: changes after antiretroviral therapy. Blood. 2004;103:966–72. doi: 10.1182/blood-2003-04-1203. [DOI] [PubMed] [Google Scholar]
- 72.Seder RA, Darrah PA, Roederer M. T-cell quality in memory and protection: implications for vaccine design. Nat Rev Immunol. 2008;8:247–58. doi: 10.1038/nri2274. [DOI] [PubMed] [Google Scholar]
- 73.Duvall MG, Precopio ML, Ambrozak DA, et al. Polyfunctional T cell responses are a hallmark of HIV-2 infection. Eur J Immunol. 2008;38:350–63. doi: 10.1002/eji.200737768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Betts MR, Nason MC, West SM, et al. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood. 2006;107:4781–9. doi: 10.1182/blood-2005-12-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Precopio ML, Betts MR, Parrino J, et al. Immunization with vaccinia virus induces polyfunctional and phenotypically distinctive CD8(+) T cell responses. J Exp Med. 2007;204:1405–16. doi: 10.1084/jem.20062363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Darrah PA, Patel DT, De Luca PM, et al. Multifunctional TH1 cells define a correlate of vaccine-mediated protection against Leishmania major. Nat Med. 2007;13:843–50. doi: 10.1038/nm1592. [DOI] [PubMed] [Google Scholar]