Abstract
RH1 (2,5-Diaziridinyl-3-(hydroxymethyl)-6-methyl-1,4-benzoquinone) is a novel antitumor diaziridinyl benzoquinone derivative designed to be bioactivated by the two-electron reductase, NAD(P)H:quinone oxidoreductase (NQO1) and is currently in clinical trials. NQO1 is expressed at high levels in many solid tumors. RH1 cytotoxicity has previously been shown to be NQO1-dependent. The purpose of this study was to investigate whether other reducing enzymes such as cytochrome b5 reductase (b5R), cytochrome P450 reductase (P450R), NRH:quinone oxidoreductase 2 (NQO2), and xanthine oxidase/xanthine dehydrogenase (XO/XDH) also contribute to the bioactivation and cytotoxicity of RH1 in human tumor cells. For these studies, we established a series of stable MDA468 breast cancer cell lines over-expressing various levels of NQO1, b5R, P450R and NQO2 and compared RH1-induced growth inhibition (MTT and SRB analysis) and interstrand DNA crosslinking (COMET analysis) in both parental MDA468 cells and transfected clones. RH1 toxicity correlated with NQO1 and NQO2 but not with either b5R or P450R activity levels in the respective series of transfected MDA468 cell clones. Enzymatic assays showed that RH1 was an in vitro substrate for xanthine oxidase. However, XO/XDH protein and activity could not be detected in a variety of human tumor cell lines. These studies suggest that NQO1 and NQO2 are the principal enzymatic determinants of RH1 bioactivation in MDA468 tumor cells and that b5R, P450R and XDH/XO are unlikely to play major roles. Our studies also suggest that NQO2 may be particularly relevant as a bioactivation system for RH1 in NQO1-deficient tumors such as leukemias and lymphomas.
Introduction
RH1 (2,5-Diaziridinyl-3-(hydroxymethyl)-6-methyl-1,4-benzoquinone) is a novel antitumor diaziridinyl benzoquinone derivative which is currently in phase I clinical trials. This drug was designed to be bioreductively activated by the two-electron reductase NAD(P)H:quinone oxidoreductase (NQO1, DT-diaphorase, EC 1.6.99.2) (Winski et al., 1998). NQO1 plays a pivotal role in the bioactivation of antitumor quinone drugs such as mitomycin C, EO9 (apaziquone), AZQ (diaziquinone) and MeDZQ (2,5-diaziridinyl-3,6-dimethyl-1,. 4-benzoquinone), to cytotoxic species capable of alkylating and crosslinking DNA (Fisher et al., 1992; Cummings et al., 1998; Walton et al., 1991). RH1 is a very efficient substrate for NQO1 and reduction by NQO1 to the hydroquinone form results in the activation of the aziridine rings and subsequent DNA alkylation and interstrand cross-linking (Ward et al., 2000; Dehn et al., 2005). NQO1 has been shown to be expressed at high levels in some normal tissue but also in many human solid tumors including lung, colon, pancreas and breast (Cresteil and Jaiswal, 1991; Malkinson et al., 1992; Mikami et al., 1998). Since RH1 is a very efficient substrate for NQO1, it was considered an ideal agent to be used in an NQO1-directed tumor targeting strategy. RH1 has demonstrated significant anti-tumor activity both in vitro (Winski et al., 1998, 2001) and in vivo (Cummings et al. 2003; Dehn et al., 2004).
Although RH1 was discovered to be effectively activated by NQO1, it has long been suspected that other reductases such as the two-electron reductases NRH:quinone oxidoreductase 2 (NQO2, EC 1.10.99.2), xanthine dehydrogenase (XDH, EC 1.1.1.204) or the one-electron reductases NADH:cytochrome b5 reductase (b5R, EC 1.6.2.2), NADPH:cytochrome P450 reductase (P450R, EC 1.6.2.4), and xanthine oxidase (XO, EC 1.1.3.22) may also contribute to the bioactivation and cytotoxicity of RH1 in tumor cells. Many anti-tumor quinones, which exert their toxicity via NQO1, are also substrates for the above reductases. Quinones can be reduced to hydroquinones via two-electron reduction. They can also be reduced by one-electron reductases to semiquinones, which can then undergo redox cycling in the presence of oxygen, generating superoxide anion and hydrogen peroxide (Powis, 1989). Mitomycin C and E09 have been shown to be substrates for one-electron reductases such as P450R, b5R and XO and the generation of oxidative stress following reduction by one-electron reductases has been shown to contribute to the overall cytotoxicity of these drugs (Bligh et al., 1990; Hodnick and Satorelli, 1993; Saunders et al., 2000; Pan et al., 1984). Whether RH1 is bioactivated by one-electron reductases is still an open question. Recent studies have shown that RH1 could serve as a substrate for P450R (Hasinoff et al., 2006; Begleiter et al., 2007). However, over-expression of P450R in two human breast cancer cell lines T47D (Begleiter et al., 2007) and MDA-MB231 (Kim et al., 2004) did not render cells more sensitive to RH1 cytotoxicity. A study of RH1 toxicity in the NCI 60 tumor cell line panel found a high sensitivity to RH1 in leukemia and lymphoma cell lines which have very low or absent NQO1 activity (Tudor et al., 2005). These findings suggest that, in addition to activation by NQO1, RH1-induced cytotoxicity might also involve alternative pathways. In this study, we investigated the contribution of multiple one and two-electron reductases to RH1 induced cytotoxicity.
Materials and methods
Materials
Cytochrome c, 2,6-dichlorophenol-indophenol (DCPIP), dicoumarol, menadione, 3-(4,5-dimethylthiazol-2,5-diphenyl)tetrazolium (MTT), NADH, NADPH, propidium iodide, resveratrol, sulforhodamine B (SRB), trichloroacetic acid, xanthine, xanthine oxidase and all other chemicals were purchased from Sigma Chemical Co. (St. Louis, MO). Dihydronicotinamide riboside (NRH) was prepared by phosphodiesterase treatment of NADH to generate dihydronicotinamide mononucleotide (NMNH), which was then treated with alkaline phosphatase to remove the phosphate group using a reported method (Friedlos et al., 1992). NRH was then purified from the reaction mixture by reverse phase HPLC. RH1 was supplied by NCI (NSC697726) and stock solution prepared in DMSO.
Cell Culture and Transfection
The human breast cancer cell line MDA-MB-468 (MDA468) was obtained from ATCC and is deficient in NQO1 activity as a result of a homozygous expression of the NQO1*2 polymorphism. Cells were grown in RPMI 1640 supplemented with 10% (v/v) fetal bovine serum, 2 mM L-glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were maintained in a humidified incubator containing 5% carbon dioxide at 37°C. MDA468 cells were stably transfected by electroporation with either the CMV-driven expression vector, pcDNA 3.0 (Invitrogen, San Diego, CA), containing human wtNQO1 cDNA, or the EF-1a driven IRES expression vector containing human P450R cDNA (a kind gift from Dr. A.V. Patterson, University of Auckland, New Zealand), human b5R cDNA or human NQO2 cDNA. All the selected clones (NQO1, b5R, P450R, and NQO2 transfected) were analyzed by measuring both protein levels (immunoblotting) and by measuring enzymatic activities.
Enzyme Activity Assays
Exponentially growing cells (approximately 5 × 106 total cells) were harvested into 100 μl 25 mM Tris-HCl (pH 7.4) containing 250 mM sucrose and 5 μM FAD, followed by probe sonication on ice. Supernatant from the sonicate was recovered after centrifugation and protein concentration determined using the method of Lowry (Lowry et al., 1951).
NQO1 activity was measured by following the NADH-dependent reduction of 2,6-dichlorophenol-indophenol (DCPIP) at 600 nm in a Beckman spectrophotometer (Ernster, 1967). Reactions (1 ml) contained 25 mM Tris-HCl (pH 7.4), 0.7 mg/mL bovine serum albumin, 0.2 mM NADH, and 40 μM DCPIP. Reactions were started by the addition of a small volume (5-10 μl) of cell supernatant and the linear decrease in absorbance was monitored at 600 nm for 1 min at 30°C. Reactions were performed in the absence and presence of 20 μM dicumarol. NQO1 activity is defined as the dicumarol-inhibitable reduction of 2,6-dichlorophenol-indophenol, calculated based on an extinction coefficient of 21 mM−1cm−1 and expressed as nmol DCPIP reduced/min/mg protein.
NQO2 activity was measured as NRH-dependent menadione reductase activity using MTT as the final electron acceptor (Wu et al., 1997). The assay mixture contained 50 mM phosphate buffer (pH 7.4), 200 μM NRH, 10 μM menadione, and 0.25 mg/ml MTT. Reactions were initiated with the addition of a small volume of cell supernatant and the linear increase in absorbance with time was recorded at 578 nm for 5 min. Reaction were performed in the absence and presence of 10 μM quercetin, a confirmed inhibitor of NQO2 (Wu et al., 1997). NQO2 activity was defined as the quercetin-inhibitable reduction of MTT, calculated based on an extinction coefficient of 13 mM−1cm−1 and expressed as nmol MTT formazan/min/mg protein.
P450R activity was measured as NADPH-dependent reduction of cytochrome c at 550-540 nm (Vermillion and Coon, 1978). Reactions (1 ml) contained 300 mM phosphate buffer (pH 7.6), 0.1 mM EDTA, 100 μM NADPH, and 40 μM cytochrome c. Reactions were started by the addition of cytochrome c and the linear increase in absorbance with time was recorded at 550-540 nm for 2 min at 30°C. P450R activity was calculated based on an extinction coefficient of 21 mM−1cm−1 and expressed as nmol cytochrome c reduced/min/mg protein.
b5R activity was measured as NADH:ferricyanide reductase at 420 nm (Mihara and Sato, 1998). Reactions (1 ml) contained 110 mM MOPS buffer (pH 7.0), 0.1 mM EDTA, 100 μM NADH, and 200 μM potassium ferricyanide. Reactions were started by the addition of NADH and the linear decrease in absorbance with time was recorded at 420 nm for 3 min at 30°C. b5R activity was calculated based on an extinction coefficient of 1.02 mM−1cm−1 and expressed as nmol potassium ferricyanide reduced/min/mg protein.
XO and XDH activities were determined spectrophotometrically (Stripe and Della, 1969) by measuring the rate of uric acid formation from xanthine at 295 nm in the absence of NAD+ (XO activity), and the rate of NADH formation at 340 nm in the presence of NAD+ (XDH activity). XO/XDH activities were also determined fluorimetrically by measuring the production of the fluorescent isoxanthiopterin from the non-fluorescent petrine, as previously described (Beckman et al., 1989).
Measurement of Oxygen Consumption
Oxygen consumption was measured using a Clark electrode in a 3 ml reaction at 37°C. Cells were collected, counted, and a total of 2 × 106 cells were resuspended in 3 ml complete media. Dissolved oxygen content in the 3 ml reaction system was monitored for 10 min to determine basal oxygen consumption rate and an additional 10 min following the injection of 25 μM menadione into the system. Rates of oxygen consumption were determined from linear regions of the oxygraph over 5 min. Dissolved oxygen concentrations were adjusted for temperature (37°C) and altitude (5280ft).
Growth Inhibition Assay
Growth inhibition was measured using both the MTT colorimetric assay (Mosmann, 1983) and the sulforhodamine B (SRB) colorimetric assay (Vichai and Kirtikara, 2006) to ensure similar results using two different endpoints (MTT: mitochondrial activity; SRB: total cellular protein). For the MTT assay, cells were seeded into triplicate 96-well plates at 2000 cells per well and allowed to attach for 16 h. Cells were then treated with RH1 in complete medium for 2 h, followed by incubation with drug free medium for additional 72 h at 37°C. Medium was removed by aspiration, and MTT (50 μg) in medium (50 μl) was added to each well and incubated for a further 4 h. Cell viability was determined by measuring the cellular reduction of MTT to the crystalline formazan product which was dissolved by the addition of 100 μl DMSO. Optical density was determined at 550 nm using a Molecular Devices Thermomax microplate reader. The IC50 values were defined as the concentration of RH1 that resulted in 50% reduction in cell number compared to the DMSO treated control.
In the SRB assay, drug treatment procedures was identical to the MTT assay, except that cells were seeded at 5000 cells per well. At the end of 72 h incubation with drug free medium, medium was removed, and cells were fixed with 10% (w/v) trichloroacetic acid, stained with 0.4% (w/v) sulforhodamine B (dissolved in 1% (v/v) acetic acid), and the protein bound dyes were then dissolved in 10 mM Tris base solution and absorption at 565 nm was determined using a Molecular Devices Thermomax microplate reader.
Clonogenic Assay
The inhibition of the colony-forming ability of MDA468 and NQO1/NQO2 transfected clones was assayed using the clonogenic assay as previously described (Winski et al., 2001). Briefly, 800 cells in 100mm plate were treated with RH1 for 2 h followed by incubation in fresh medium for 10 d. Colonies were stained with crystal violet and counted manually.
Comet Assay
DNA damage was evaluated using the single-cell gel electrophoresis method, commonly known as the alkaline comet assay, as described previously (Tice et al., 2000), including a modified version (Ward et al., 1997) to detect DNA cross-linking. Briefly, cells were seeded in 6-well plates at 1 × 105 cells/well and let attach for 16 h. After drug treatment, cells were harvested and 1 × 104 cells were then subjected to the comet assay. Comet slides were stained with PI and viewed using fluorescence microscopy under a Nikon invert microscope (Nikon Eclipse TE300) at 20× magnification. Images were captured using an attached CoolSNAPES CCD camera. One hundred cells, 50 each on duplicate slides were captured and scored using a software package (Komet Version 5, Kinetic Imaging, Belfast, UK). The tail moment was recorded for each comet as an indication of the extent of DNA single strand damage. The percent of DNA in comet tail was also recorded and presented as a supplementary table (Table S2).
For measurement of DNA cross-linking, a fixed amount of single strand breaks was induced post-treatment into control and RH1-treated cells at each concentration point by incubating with 200 μM H2O2 for 20 min on ice. Cross-linked DNA is unable to migrate from the head of the comet, and the extent of DNA cross-linking can be indirectly measured by analyzing the relative reduction of DNA migration induced by H2O2 compared with untreated H2O2 controls.
Metabolism of RH1 by XO
Metabolism of RH1 by XO under hypoxic conditions (RH1 competes with oxygen as electron acceptor) was performed in a sealed cuvette. The enzyme reaction mixture (1 ml) contained 100 mM phosphate buffer (pH 7.8), 0.2 mM xanthine, 25 mU xanthine oxidase (20 μl, diluted in phosphate buffer). RH1 was first diluted in 100mM phosphate buffer and then added to the reaction mixture. Hypoxic conditions were established by flushing the phosphate buffer and xanthine mixture with high purity argon for 5 min in a sealed cuvette. Reactions were carried out with a stream of argon gas gently bubbling the surface of the mixture throughout the experiment. The reaction was initiated by injection of xanthine oxidase and RH1. Uric acid formation from xanthine was monitored by measuring the increase in absorbance at 294 nm for 10 min at 30°C using a HP8452 spectrophotometer; RH1 consumption was measured as the decrease in absorbance at 326nm simultaneously. For allopurinol inhibition of XO, the enzyme was pre-incubated with 50 μM allopurinol for 10 minutes before injection into the reaction system.
Metabolism of RH1 by NQO2
Metabolism of RH1 by NQO2 was analyzed by HPLC as previously described (Beall et al., 1998). The reaction mixture contained 50 mM potassium phosphate buffer (pH 7.4), 1 mg/ml BSA, 200 μM NRH, 50 μM RH1 and 0.25 μg rhNQO2. Reactions (500 μl) were performed at 30°C in the absence and presence of 10 μM resveratrol for 30 min and then stopped by the addition of ice-cold methanol (500 μl) and samples were immediately analyzed by HPLC at 340 nm.
Statistical analysis
Statistical analysis was performed using one-way analysis of variance followed by appropriate post hoc tests: Dunnett test for comparison of multiple observations to a single control; Student's t test for pair-wise comparisons. Data are represented as mean ± SD of at least three replicate experiments.
Results
Role of NQO1 in RH1 toxicity
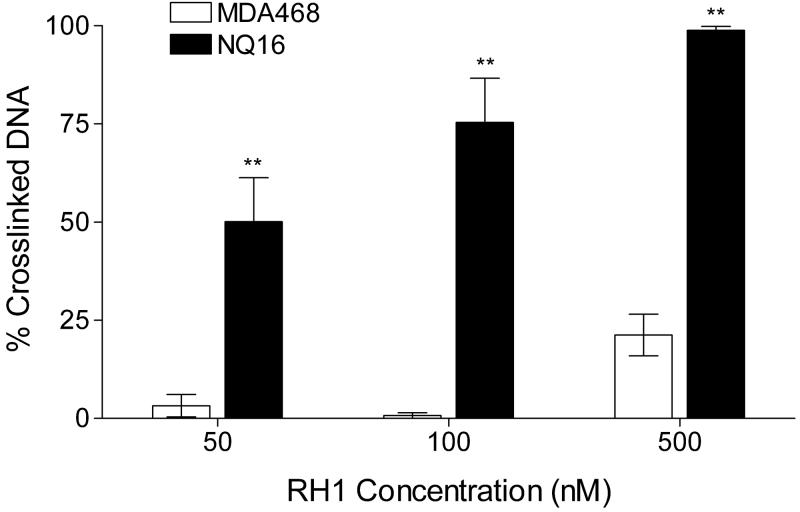
We have previously reported the establishment of a pair of isogenic human breast cancer cell lines, the MDA468 and MDA468/NQ16 cells, which differ only in NQO1 activity (Dehn et al., 2004). MDA468 parental cells have a homozygous *2 polymorphism in NQO1 and hence non-detectable NQO1 activity, due to rapid proteasomal degradation of NQO1*2 protein. The NQ16 cell line was generated by stable transfection of human wildtype NQO1 (Dehn et al., 2004). In this study we confirmed previous reports of high levels of NQO1 expression in the NQ16 cell line by immunoblot analysis (data not shown) and NQO1 activity assay (Table 1). As expected, RH1 was significantly more toxic to NQ16 cells than to parental MDA468 cells as measured by both MTT and SRB growth inhibition assay. The IC50 value for RH1 (Table 1) was about 15-fold lower in NQ16 cells compared with the IC50 for the parental MDA468 cells. The difference in RH1 toxicity between MDA468 and NQ16 cells was also confirmed by clonogenic assays (The ratio of sensitivity to RH1 was 44 for the MDA468/NQ16 cell pair. See Fig. S1 for clonogenic survival curve). RH1-induced DNA cross-linking was compared in MDA468 and NQ16 cells using the modified comet assay. Treatment of NQ16 cells with 50, 100, and 500 nM of RH1 for 2 h resulted in a significant dose-dependent increase in DNA cross-linking (Fig. 1), whereas in MDA468 cells, a small amount of DNA cross-linking could only be observed at the highest RH1 dose (500 nM).
Table 1.
NQO1 activity and IC50 values for RH1 in MDA468 parental cells and cells stably transfected with NQO1 (Clone NQ16).
| Cell lines |
||
|---|---|---|
| MDA468 | NQ16 | |
| NQO1 activity (nmole DCPIP/min/mg protein) | NDa | 1061.4 ± 50.5 |
| IC50 (nM) by MTT assay | 110.5 ± 13 | 6.1 ± 0.1b |
| IC50 (nM) by SRB assay | 99.2 ± 6.7 | 7.4 ± 1.9b |
nd, non-detectable, < 5 nmole DCPIP/min/mg protein
A significant difference (P<0.01, Student's t-test) between IC50s for MDA468 and NQ16 cells.
Figure 1. Formation of RH1-induced DNA cross-links in MDA468 and NQ16 cells.
Cells were treated with RH1 for 2 h. DNA cross-linking was measured by the modified comet assay and expressed as percentage decrease in tail moment compared with untreated controls. The percent of DNA in comet tail was also recorded and listed in Table S2. Results are expressed as the mean ± standard deviation of three separate determinations. * Significantly different from parental MDA468 cells treated with same RH1 concentration, p < 0.05, ** p < 0.01.
Role of P450R in RH1 toxicity
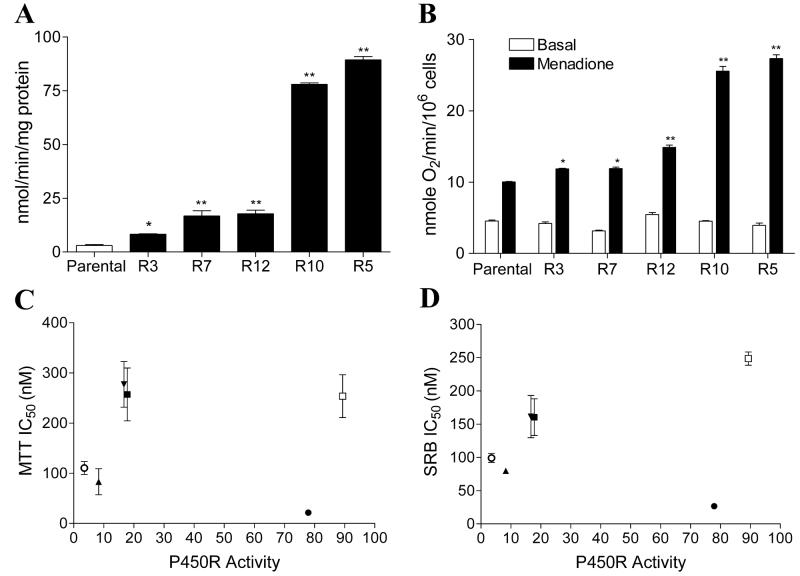
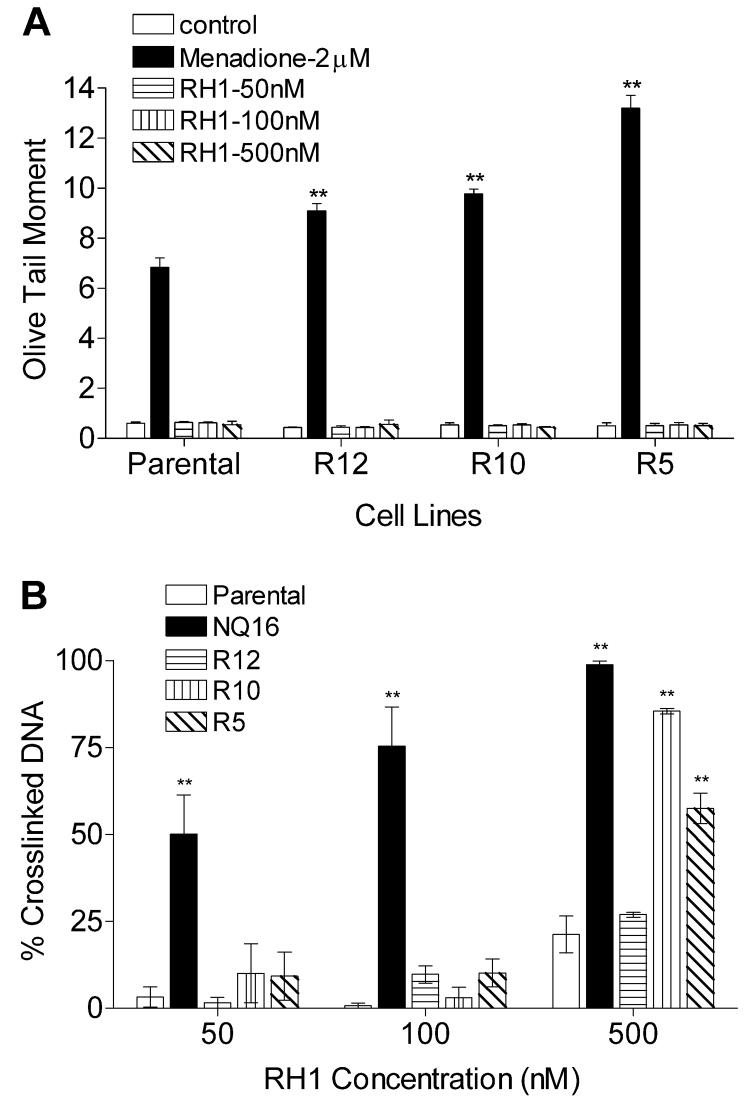
A wide range of P450R activity was detected in transfected clones generated as described in Materials and Methods. Activity was continuously measured in transfected clones to ensure maintenance of stable P450R expression. Immunoblot analysis was also performed to confirm the over-expression of P450R protein in the transfected clones (data not shown). P450R activity levels of parental MDA468 and transfected clones R3, R7, R12, R10, and R5 (in the order of increasing activity) are shown in Fig. 2A. To ensure that the transfected P450R was functioning as a bioreductive enzyme in MDA468 cells, oxygen consumption was measured in parental and P450R transfected MDA468 cells before and after treatment with 25 μM menadione. An increase in oxygen consumption following menadione treatment indicates the bioreduction of the redox cycling quinone menadione by P450R. All five transfected clones exhibited a significant increase in oxygen consumption following menadione treatment and the level of increase in oxygen uptake was proportional to P450R activity in all five clones (Fig. 2B). The IC50s of RH1 for all five P450R clones were determined by both MTT and SRB growth inhibition assay. No correlation was observed between P450R activity and IC50 values for RH1 in all P450R clones when the IC50 values were plotted against P450R activity (Fig. 2C-D). Three clones with the highest P450R activity, namely R12, R10 and R5, were selected for RH1 genotoxicity analysis. DNA single strand breaks and cross-linking were measured using the alkaline comet assay and the modified comet assay, respectively. No DNA single strand breaks could be detected in either parental MDA468 cells or P450R transfected clones following 2 h RH1 treatment at all doses tested (Fig. 3A). To exclude the possibility of DNA single strand breaks masked by quick DNA damage repair, formation of DNA single stand breaks were also monitored following shorter RH1 exposure. In these experiments DNA single strand breaks were not observed even at time points as early as 10 min (data not shown). The level of DNA cross-linking was not different between parental MDA468 and P450R transfected clones at low doses of RH1 exposure (50 and 100 nM). However, when cells were treated with 500 nM RH1 for 2 h, a significant increase in DNA cross-linking in the two clones with the highest P450R activity (clone R10 and R5) relative to parental MDA468 cells was observed (Fig. 3B).
Figure 2. Characterization of MDA468 cells stably transfected with human P450R.
(A) P450 reductase activity in parental MDA468 cells and P450R transfected clones R3, R7, R12, R10, R5. P450R activity was measured using the reduction of cytochrome c. (B) Transfected P450R can function as a bioreductive enzyme in MDA468 cells. Oxygen consumption was measured in parental and P450R transfected MDA468 cells using a Clark electrode before and after treatment with 25 μM menadione. Results are expressed as the mean ± standard deviation of three separate determinations. * Significantly different from parental cells, p < 0.05, ** p < 0.01. (C-D) Correlation analysis between P450R activity and IC50 values for RH1 in various P450R transfected MDA468 clones. IC50 values were determined by the MTT assay (C) or the SRB assay (D). IC50 values are expressed as the mean ± standard deviation of at least four separate determinations. P450R activity represents the mean of at least four separate determinations. ○, parental MDA468; ▲, R3; ▼, R7; ■, R12; ●, R10; □, R5.
Figure 3. Formation of DNA cross-linking and single strand breaks in parental and P450R transfected MDA468 cells following RH1 treatment.
DNA cross-linking and single strand breaks were measured by comet assay following 2 h treatment with RH1. (A) DNA single strand breaks were expressed as the olive tail moment. Menadione treatment (2 μM) was included as a positive control. (B) DNA cross-linking was expressed as the percentage decrease in comet tail moment in RH1 treated cells comparing to untreated H2O2 (200 μM for 20 min) control. NQ16 cells were included as a positive control. The percent of DNA in comet tail was also recorded and listed in Table S2. Results are expressed as the mean ± standard deviation of three separate determinations. ** Significantly different from parental cells with same drug treatment, p < 0.01.
Role of b5R in RH1 cytotoxicity
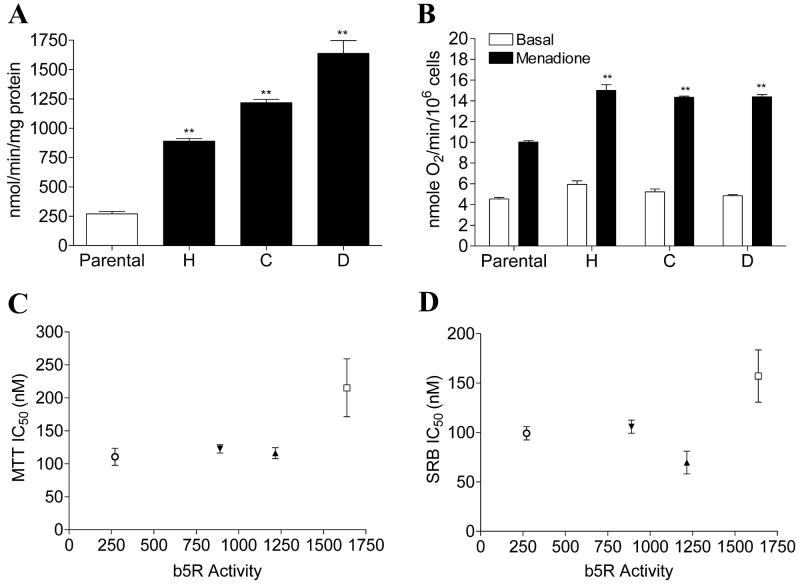
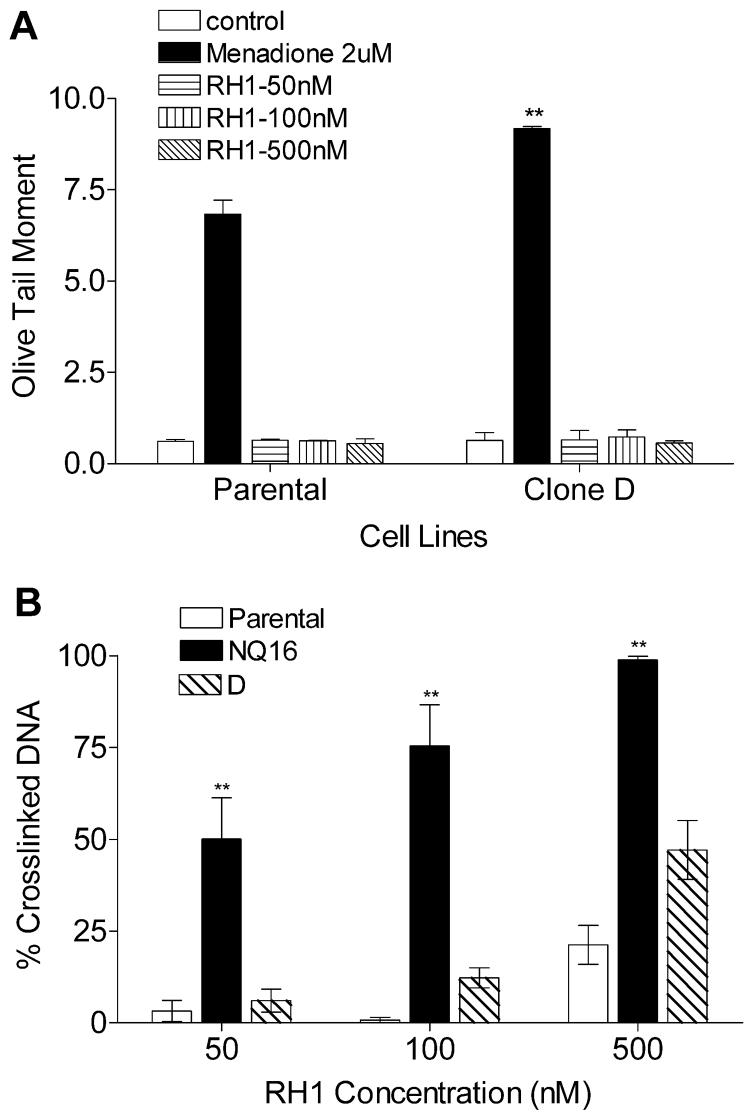
Three b5R transfected MDA468 clones were initially selected based on immnunoblot analysis (data not shown) and b5R catalytic activity was then continuously measured in transfected clones to ensure maintenance of stable b5R expression. b5R activity levels of parental MDA468 and b5R transfected clones H, C, and D (in the order of increasing activity) were shown in Fig. 4A. All three b5R clones exhibited a significant increase in oxygen consumption following menadione treatment, indicating that the transfected b5R was catalytically active as a bioreductive enzyme in MDA468 cells (Fig. 4B). The IC50s of RH1 for all three b5R clones were determined by both MTT and SRB growth inhibition assay. No correlation was observed between b5R activity and IC50 values for RH1 in all b5R clones when the IC50 values were plotted against b5R activity (Fig. 4C-D). The clone with the highest b5R activity, namely clone D, was selected for RH1 genotoxicity analysis. DNA single strand breaks and cross-linking were measured using the alkaline comet assay and the modified comet assay, respectively. No single strand breaks could be detected in either parental MDA468 cells or b5R tranfected clone D following 2 h RH1 treatment (Fig. 5A). The level of DNA cross-linking was not different between parental MDA468 and clone D at low doses of RH1 exposure (50 and 100 nM); a small increase in DNA cross-linking in clone D compared to parental MDA468 cells was observed following treatment with 500 nM RH1 for 2 h (Fig. 5B).
Figure 4. Characterization of MDA468 cells stably transfected with human b5R.
(A) b5R activity in parental MDA468 cells and b5R transfected clones H, C, D. b5R activity was measured using the reduction of potassium ferricyanide. (B) Transfected b5R can function as a bioreductive enzyme in MDA468 cells. Oxygen consumption was measured in parental and b5R transfected MDA468 cells using a Clark electrode before and after treatment with 25 μM menadione. Results are expressed as the mean ± standard deviation of three separate determinations. ** Significantly different from parental cells, p < 0.01. (C-D) Correlation analysis between b5R activity and IC50 values for RH1 in various b5R transfected MDA468 clones. IC50 values were determined by the MTT assay (C) or the SRB assay (D). IC50 values are expressed as the mean ± standard deviation of at least four separate determinations. b5R activity represents the mean of at least four separate determinations. ○, parental MDA468; ▲, clone H; ▼, clone C; □, clone D.
Figure 5. Formation of DNA cross-linking and single strand breaks in parental and b5R transfected MDA468 clones following RH1 treatment.
DNA cross-linking and single strand breaks were measured by comet assay following 2 h treatment with RH1. (A) DNA single strand breaks were expressed as olive tail moment. Menadione treatment (2 μM) was included as a positive control. (B) DNA cross-linking was expressed as the percent of decrease in comet tail moment in RH1 treated cells comparing to untreated H2O2 (200 μM for 20 min) control. NQ16 cells were included as a positive control. The percent of DNA in comet tail was also recorded and listed in Table S2. Results are expressed as the mean ± standard deviation of three separate determinations. ** Significantly different from parental cells with same drug treatment, p < 0.01.
Role of XO in RH1 bioactivation and cytotoxicity
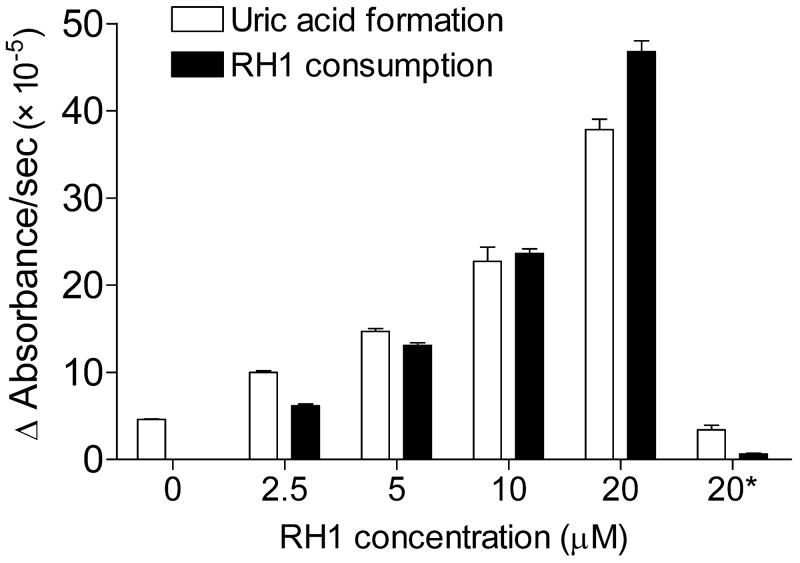
To test whether RH1 could serve as a substrate for XO, the effect of RH1 on the XO-mediated conversion of xanthine to uric acid and the concomitant reduction of RH1 was assessed. The addition of RH1 led to a dose-dependent increase in the rate of both uric acid formation and RH1 consumption (Fig. 6), which was completely inhibited in the presence of 50 μM allopurinol, a potent inhibitor of XO/XDH. However, pretreatment with allopurinol did not decrease RH1 toxicity in MDA468 cells (Fig. S2). XO/XDH levels in MDA468 cells were below detection levels of both colorimetric and fluorescence assays used for detection of XO/XDH activity.
Figure 6. Effect of RH1 on XO-mediated uric acid formation from xanthine and the concomitant consumption of RH1 under hypoxic conditions.
Uric acid formation was measured at 293 nm, RH1 consumption at 326 nm. * Reaction was carried out in the presence of 50 μM allopurinol. Results are expressed as the mean ± standard deviation of three separate determinations.
Role of NQO2 in RH1 bioactivation and cytotoxicity
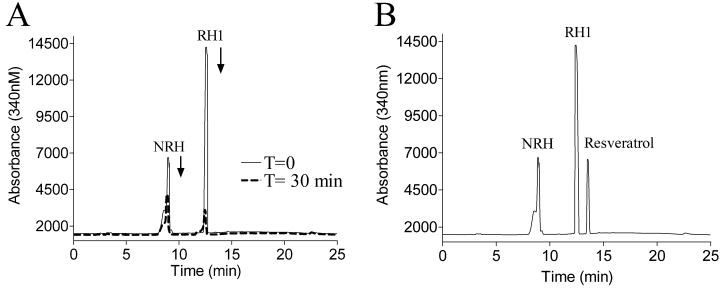
The ability of purified recombinant human NQO2 to metabolize RH1 was analyzed by HPLC in the presence of the cofactor NRH. As shown in Fig. 7A, RH1 was quickly metabolized within 30 minutes, accompanied by a concomitant consumption of NRH. The addition of 10 μM resveratrol, a confirmed NQO2 inhibitor, totally abolished the reduction of RH1 by rhNQO2 (Fig. 7B).
Figure 7. Reduction of RH1 by recombinant human NQO2.
RH1 metabolism by NQO2 was analyzed by HPLC after 30 min reaction in the absence (A) and presence (B) of 10 μM resveratrol. (A) RH1 and NRH absorbance at 340 nm at time zero (solid line) and after 30 min of reaction (dotted line). (B) RH1 and NRH peaks remained unchanged over 30 min in the presence of 10 μM resveratrol. Results shown are representative of three independent experiments.
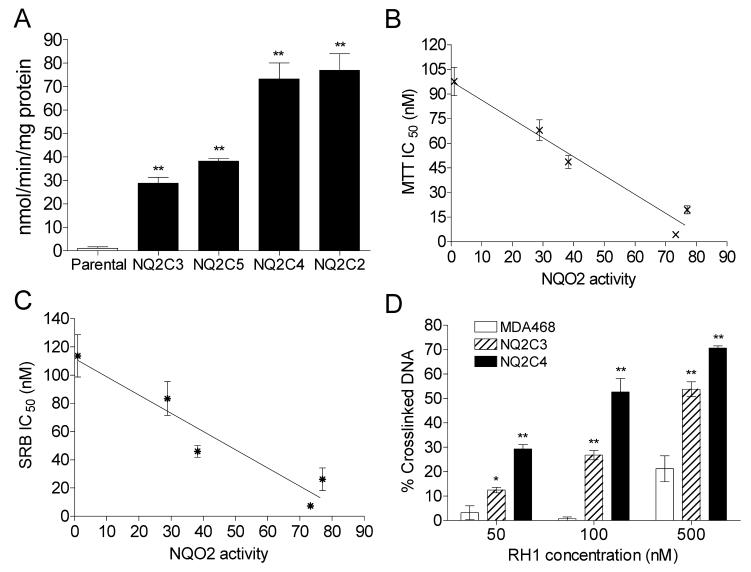
A series of MDA468 cell clones over-expressing NQO2 were then generated by stable transfection. Elevated NQO2 levels were confirmed by both immunoblot analysis (data not shown) and activity assay. NQO2 activity levels of parental MDA468 and transfected clones NQ2C3, NQ2C5, NQ2C4, NQ2C2 (in the order of increasing activity) are shown in Fig. 8A. The IC50 values for RH1 for all four NQO2 clones were determined by both MTT and SRB growth inhibition assay. A significant correlation [r2 = 0.94(MTT); 0.88(SRB)] was observed between NQO2 activity levels and IC50 values for RH1 in all NQO2 clones when the IC50 values were plotted against NQO2 activity (Fig. 8B-C). In MTT assays, the IC50 value for RH1 was about 20-fold lower in NQ2C4 cells compared with the IC50 for the parental MDA468 cells. The difference in RH1 toxicity between MDA468 and NQO2 transfected cells was also confirmed by the clonogenic assay (The ratio of clonogenic sensitivity to RH1 was 11 for the MDA468/NQ2C4 cell pair. See fig. S1 for clonogenic survival curve). Two clones with medium (NQ2C3) and high (NQ2C4) NQO2 activity were selected for RH1 genotoxicity analysis. A significant dose-dependent increase in DNA cross-linking was observed in both clones following 2 h RH1 treatment (Fig. 8D). The percentage of cross-linked DNA was much higher in clone NQ2C4 (high NQO2 activity level) than in clone NQ2C3 (medium NQO2 activity level).
Figure 8. Characterization of MDA468 cells stably transfected with human NQO2.
(A) NQO2 activity in parental MDA468 cells and NQO2 transfected clones NQ2C3, NQ2C5, NQ2C4, and NQ2C2. NQO2 activity was measured using the NRH-dependent reduction of menadione. (B-C) Correlation analysis between NQO2 activity and IC50 values for RH1 in various NQO2 transfected MDA468 clones. IC50 values were determined by the MTT assay (B) or the SRB assay (C). IC50 values are expressed as the mean ± standard deviation of at least four separate determinations. NQO2 activity represents the mean of four separate determinations. (D) Formation of DNA cross-linking in parental and NQO2 transfected MDA468 cells following RH1 treatment. DNA cross-linking was measured by modified comet assay following 2 h treatment with RH1. DNA cross-linking was expressed as the percentage decrease in comet tail moment in RH1 treated cells comparing to untreated H2O2 (200 μM for 20 min) control. The percent of DNA in comet tail was also recorded and listed in Table S2. Results are expressed as the mean ± standard deviation of three separate determinations. * Significantly different from parental cells with same drug treatment, p < 0.05, ** p < 0.01.
Discussion
NQO1 has emerged as a promising target for the design of bioreductive anti-tumor quinone drugs and we have reported a relationship between NQO1 levels and susceptibility to RH1 toxicity previously (Winski et al., 2001, Dehn et al., 2004). Using a panel of BE cells (human colon adenocarcinoma) over-expressing a range of NQO1 activity, Winski et al. found that a threshold of NQO1 activity was needed for RH1-induced toxicity (Winski et al., 2001). Using an isogenic cell line pair MDA468 and MDA468/NQ16 (MDA468 cells stably transfected with NQO1), Dehn et al. showed that RH1 exhibited antitumor actvity in NQ16 cells both in vitro and in vivo (Dehn et al., 2004). Since we were using the MDA468 cell line as the parental cell line in the current study, we first confirmed the significantly higher toxicity of RH1 in NQ16 cells than in MDA468 cells (Table 1, Fig. 1). The results were consistent with our previous data (Dehn et al., 2004, 2005) and confirmed a role for NQO1 in RH1 toxicity.
Although RH1 was discovered to be effectively activated by NQO1, the specificity of RH1 activation by NQO1 has been questioned. A recent study (Hasinoff and Begleiter, 2006) has shown that RH1 can serve as a substrate for P450R; however, Begleiter et al. did not observe any increase in RH1 toxicity when they overexpressed P450R in the human breast cancer cell line T47D (Begleiter et al., 2007). However, since the T47D cell line has considerable NQO1 activity (Begleiter et al., 2007), the more efficient reduction of RH1 by NQO1 may camouflage any effect of P450R. Kim et al. used another human breast cancer cell line MDA231, which does not have NQO1 activity, and found that overexpression of P450R had little impact on RH1 toxicity (Kim et al., 2004). In our current study, we used a human breast cancer cell line MDA468, which also has non-detectable NQO1 activity because it carries the homozygous NQO1 *2 polymorphism (Dehn et al., 2004; Siegel et al., 2001). We then stably overexpressed the one-electron reductases P450R and b5R, and the two-electron reductase NQO2 in the same MDA468 background. The establishment of a series of P450R, b5R, and NQO2 over-expressing clones in the same background allowed us to investigate the possible role of P450R, b5R, and NQO2 in RH1-induced DNA damage and growth inhibition. In our study, we selected five stable P450R transfected clones, three stable b5R transfected clones and four stable NQO2 transfected clones to compare with the parental MDA468. To ensure that transfection of the MDA468 cell line with one particular reductase did not alter the level of other reducing enzymes, the enzymatic activity of NQO1, NQO2, P450R, b5R, and XO/XDH were measured in the parental MDA468 and all the transfected clones. In all the clones tested, the transfection only elevated the activity of the reductase being transfected, without affecting the activity of any other reductases measured (Table S1).
The IC50 values of RH1 in both P450R and b5R transfected clones measured using both MTT and SRB assays were randomly scattered and no apparent correlation was observed when IC50 values were plotted against P450R or b5R activity. The only P450R-transfected clone that had a lower IC50 than parental MDA468 was R10 and the only b5R-transfected clone with a decreased IC50 value was clone C (using the SRB assay). One criticism of isogenic cell pairs is that the expression plasmid containing the cDNA of interest may insert anywhere into the genome and may affect the expression of other proteins. Therefore, by comparing a series of transfected clones rather than using only one clone, we were able to minimize the influence of random insertion during transfection.
To examine the ability of RH1 to induce DNA damage in P450R/b5R transfected MDA468 cells, we selected three clones with the highest P450R activity and one clone with the highest b5R activity and performed the alkaline comet assay for DNA single strand breaks and the modified comet assay for DNA-crosslinking. The lack of formation of DNA strand breaks by RH1 in the transfected clones was somewhat unexpected, since the RH1 semiquinone free radical generated by one electron reduction would be oxidized back to the quinone form in aerobic conditions, generating reactive oxygen species (ROS) (Hasinoff et al., 2006, Lusthof et al., 1992), which can cause DNA strand breaks. We found no difference in DNA cross-linking between parental and either P450R or b5R transfected MDA468 cells at low RH1 dose (50 nM and 100 nM). However, a significant increase in DNA cross-linking was observed in clone R10 and R5 (P450R) following 500 nM RH1 treatment for 2 h. However, a concentration of 500nM RH1 is almost 25 times greater than the IC50 value for clone R10 and twice the IC50 value for clone R5. Similarly, a non-significant increase in DNA cross-linking was observed in clone D (b5R) only at the highest RH1 dose (500 nM, three times of IC50 for clone D). These results indicate that at high drug concentrations, high levels of P450R or b5R activity may contribute to RH1-induced DNA cross-linking; but given the respective IC50 values, it seems unlikely that this has any influence on cell death. While the concentrations of RH1 have not been reported in human or animal tumors following RH1 administration, pharmacokinetic studies in mice have shown that RH1 has a short plasma half-life (<5min, Loadman et al., 2000); in addition, experiments that examined DNA crosslinking in human peripheral blood mononuclear cells following RH1 treatment indicated that 100% crosslinking could be achieved in these cells at RH1 concentrations near 50 nM (Danson et al., 2007). These data suggest that high levels of RH1 in tumors are unlikely due to rapid clearance and potential toxicities. We therefore reasoned that neither P450R nor b5R plays a significant role in activation of RH1 in cancer cells with normal levels of P450R and b5R activity, especially when considerable levels of NQO1 activity is also present. According to a reductase enzyme activity screen across the NCI tumor cell line panel (Fitzsimmons et al, 1996), the mean P450R activity is 14.8 nmol/min/mg protein and the mean b5R activity is 38.0 nmol/min/mg protein, both of which are similar to MDA468 parental cells and far less than the transfected clones. In contrast, NQO1 activities in the NCI tumor cell panel range from 0–5,000 nmol/min/mg protein with a mean of 199.5 nmol/min/mg protein. Such high levels of NQO1 would be expected to efficiently metabolize RH1.
Although our enzymatic studies showed that RH1 could serve as a substrate for xanthine oxidase in vitro, pretreatment of MDA468 cells with the XO/XDH inhibitor allopurinol failed to protect the cells from RH1 toxicity. We could not detect any XO/XDH activity in MDA468 cells, in agreement with previous data from our lab which showed that XO activity was non-detectable in HT-29 and BE human colon carcinoma cells (Winski et al., 1998). XO activity screening for the NCI tumor cell line panel is not available at present. Therefore, the over-expression of XO in MDA468 cells was not pursued.
An important finding of the current study is that we have shown, for the first time, that NQO2 can play an important role in RH1 bioactivation and cytotoxiciy in tumor cells. NQO2 differs from NQO1 in that it uses dihydronicotinamide riboside (NRH) rather than NAD(P)H as an electron donor (Wu et al., 1997). As expected, RH1 was a substrate for purified recombinant human NQO2 using NRH as a cofactor. MDA468 cells stably transfected with NQO2 were more susceptible to RH1-induced growth inhibition and DNA cross-linking, with a strong correlation between NQO2 activity levels and IC50 values for RH1 in all NQO2 transfected clones. These results strongly suggest that NQO2 is capable of mediating RH1 bioactivation and cytotoxicity in tumor cells. The possibility of limited NRH availability in cells has been recognized (Knox et al., 2000). However, in our studies transfected NQO2 did not need exogenous NRH to bioactivate RH1 in the MDA468 cell system. This suggest that either the MDA468 cells have sufficient NRH to catalyze turnover of NQO2 or alternatively, that other pyridine nucleotides in the cell can serve as a co-factor for NQO2. Interestingly, recent work showed that NQO2 can catalyze the reduction of mitomycin C using NADH as co-factor (Jamieson et al., 2006). The respective efficiencies of NQO1 and NQO2 at bioactivation of RH1 require further study. However, since NQO2 protein and activity levels are normally very low in solid tumors (Knox et al., 2000); it is possible that the contribution of NQO2 to RH1 toxicity could be overridden by NQO1. An important implication of our work is that NQO2 may be an important determinant of RH1 bioactivation and antitumor effect particularly in tumors which have low NQO1 levels. Leukemia and lymphoma cell lines are deficient in NQO1 levels (Tudor et al., 2005, Fitzsimmons et al, 1996) but are very sensitive to RH1 treatment (Tudor et al., 2005). NQO2 levels have previously been found to be high in the K562 leukemia cell line (Buryanovskyy et al., 2004) and this observation has been recently confirmed in two independent studies (Bantscheff et al., 2007; Rix et al., 2007). We have also performed immunoblot analysis of a number of leukemia lines (KG1A, K562, Jurkat, MOLT4, HL60) and have found NQO2 protein levels to be high across the panel (unpublished data). The elevated NQO2 levels in leukemia cell lines may explain the observed sensitivity of these tumors to RH1.
In conclusion, our study confirmed that NQO1 is an important reductase catalyzing the bioactivation of RH1 in tumor cells. P450R and b5R may contribute to the toxicity of RH1 in tumor cells at very high enzyme activity levels and high drug doses. However, the importance of P450R and b5R in the bioactivation and cytotoxicity of RH1 would be expected to be minor in cancer cells with normal levels of b5R and P450R activity. In addition, RH1 can serve as a substrate for XO, but XO was not a major contributor to RH1 toxicity in our studies. Importantly, NQO2 was found to be strongly associated with RH1 bioactivation and cytotoxicity in tumor cells. These results suggest that in addition to NQO1, levels of NQO2 have to be considered as a potential biomarker of RH1 efficacy in human tumors.
Supplementary Material
Acknowledgments
Grant support: This research was supported by NIH Grant RO1 CA-51210 (DR), and by a MRC grant (UK) GO500366 to I.J.S.
Abbreviations
- RH1
2,5-Diaziridinyl-3-(hydroxymethyl)-6-methyl-1,4-benzoquinone
- NQO1
NAD(P)H:quinone oxidoreductase 1
- NQO2
NRH:quinone oxidoreductase 2
- P450R
NADPH:cytochrome P450 reductase
- b5R
NADH:cytochrome b5 reductase
- XO
xanthine oxidase
- XDH
xanthine dehydrogenase
- SRB
sulforhodamine B
- NRH
dihydronicotinamide riboside
- EO9
apaziquone
- AZQ
diaziquinone
- MeDZQ
2,5-diaziridinyl-3,6-dimethyl-1,4-benzoquinone
References
- Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035–44. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- Beall HD, Winski S, Swann E, Hudnott AR, Cotterill AS, O'Sullivan N, Green SJ, Bien R, Siegel D, Ross D, Moody CJ. Indolequinone antitumor agents: correlation between quinone structure, rate of metabolism by recombinant human NAD(P)H:quinone oxidoreductase, and in vitro cytotoxicity. J Med Chem. 1998;41(24):4755–66. doi: 10.1021/jm980328r. [DOI] [PubMed] [Google Scholar]
- Beckman JS, Parks DA, Pearson JD, Marshall PA, Freeman BA. A sensitive fluorometric assay for measuring xanthine dehydrogenase and oxidase in tissues. Free Radic Biol Med. 1998;6:607–15. doi: 10.1016/0891-5849(89)90068-3. [DOI] [PubMed] [Google Scholar]
- Begleiter A, Leith MK, Patel D, Hasinoff BB. Role of NADPH cytochrome P450 reductase in activation of RH1. Cancer Chemother Pharmacol. 2007;60:713–23. doi: 10.1007/s00280-007-0417-8. [DOI] [PubMed] [Google Scholar]
- Bligh HF, Bartoszek A, Robson CN, Hickson ID, Kasper CB, Beggs JD, Wolf CR. Activation of mitomycin C by NADPH: cytochrome P-450 reductase. Cancer Res. 1990;50:7789–7792. [PubMed] [Google Scholar]
- Buryanovskyy L, Fu Y, Boyd M, Ma Y, Hsieh TC, Wu JM, Zhang Z. Crystal structure of quinone reductase 2 in complex with resveratrol. Biochemistry. 2004;43(36):11417–26. doi: 10.1021/bi049162o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresteil T, Jaiswal AK. High levels of expression of the NAD(P)H:quinone oxidoreductase (NQO1) gene in tumor cells compared to normal cells of the same origin. Biochem Pharmacol. 1991;42:1021–1027. doi: 10.1016/0006-2952(91)90284-c. [DOI] [PubMed] [Google Scholar]
- Cummings J, Ritchie A, Butler J, Ward TH, Langdon S. Activity profile of the novel aziridinylbenzoquinones MeDZQ and RH1 in human tumour xenografts. Anticancer Res. 2003;23:3979–83. [PubMed] [Google Scholar]
- Cummings J, Spanswick VJ, Tomasz M, Smyth JF. Enzymology of mitomycin C metabolic activation in tumour tissue. Biochem Pharmacol. 1998;56:405–414. doi: 10.1016/s0006-2952(98)00073-2. [DOI] [PubMed] [Google Scholar]
- Danson S, Ranson M, Denneny O, Cummings J, Ward TH. Validation of the comet-X assay as a pharmacodynamic assay for measuring DNA cross-linking produced by the novel anticancer agent RH1 during a phase I clinical trial. Cancer Chemother Pharmacol. 2007;60(6):851–61. doi: 10.1007/s00280-007-0432-9. [DOI] [PubMed] [Google Scholar]
- Dehn DL, Inayat-Hussain SH, Ross D. RH1 induces cellular damage in an NAD(P)H:quinone oxidoreductase 1-dependent manner: relationship between DNA cross-linking, cell cycle perturbations, and apoptosis. J Pharmacol Exp Ther. 2005;313:771–779. doi: 10.1124/jpet.104.081380. [DOI] [PubMed] [Google Scholar]
- Dehn DL, Winski SL, Ross D. Development of a new isogenic cellxenograft system for evaluation of NAD(H):quinone oxidoreductase-directed antitumor quinones: evaluation of the activity of RH1. Clin Cancer Res. 2004;10:3147–3155. doi: 10.1158/1078-0432.ccr-03-0411. [DOI] [PubMed] [Google Scholar]
- Ernster L. DT-diaphorase. Methods Enzymol. 1967;10:309–317. [Google Scholar]
- Fisher GR, Donis J, Gutierrez PL. Reductive metabolism of diaziquone (AZQ) in the S9 fraction of MCF-7 cells. II. Enhancement of the alkylating activity of AZQ by NAD(P)H: quinone-acceptor oxidoreductase (DT-diaphorase) Biochem Pharmacol. 1992;44:1625–1635. doi: 10.1016/0006-2952(92)90481-w. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons SA, Workman P, Grever M, Paull K, Camalier R, Lewis AD. Reductase enzyme expression across the National Cancer Institute Tumor cell line panel: correlation with sensitivity to mitomycin C and EO9. J Natl Cancer Inst. 1996;88:259–269. doi: 10.1093/jnci/88.5.259. [DOI] [PubMed] [Google Scholar]
- Friedlos F, Jarman M, Davies LC, Boland MP, Knox RJ. Identification of novel reduced pyridinium derivatives as synthetic co-factors for the enzyme DT diaphorase (NAD(P)H dehydrogenase (quinone), EC 1.6.99.2) Biochem Pharmacol. 1992;44(1):25–31. doi: 10.1016/0006-2952(92)90033-f. [DOI] [PubMed] [Google Scholar]
- Hasinoff BB, Begleiter A. The reductive activation of the antitumor drug RH1 to its semiquinone free radical by NADPH cytochrome P450 reductase and by HCT116 human colon cancer cells. Free Radic Res. 2006;40:974–978. doi: 10.1080/10715760600833077. [DOI] [PubMed] [Google Scholar]
- Hodnick WF, Sartorelli AC. Reductive activation of mitomycin C by NADH:cytochrome b5 reductase. Cancer Res. 1993;53:4907–4912. [PubMed] [Google Scholar]
- Jamieson D, Tung AT, Knox RJ, Boddy AV. Reduction of mitomycin C is catalysed by human recombinant NRH:quinone oxidoreductase 2 using reduced nicotinamide adenine dinucleotide as an electron donating co-factor. Br J Cancer. 2006;95(9):1229–33. doi: 10.1038/sj.bjc.6603414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Patterson AV, Stratford IJ, Hendry JH. The importance of DT-diaphorase and hypoxia in the cytotoxicity of RH1 in human breast and non-small cell lung cancer cell lines. Anticancer Drugs. 2004;15(1):71–7. doi: 10.1097/00001813-200401000-00011. [DOI] [PubMed] [Google Scholar]
- Knox RJ, Jenkins TC, Hobbs SM, Chen S, Melton RG, Burke PJ. Bioactivation of 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) by human NAD(P)H quinone oxidoreductase 2: a novel co-substrate-mediated antitumor prodrug therapy. Cancer Res. 2000;60(15):4179–86. [PubMed] [Google Scholar]
- Loadman PM, Phillips RM, Lim LE, Bibby MC. Pharmacological properties of a new aziridinylbenzoquinone, RH1 (2,5-diaziridinyl-3-(hydroxymethyl)-6-methyl-1,4-benzoquinone), in mice. Biochem Pharmacol. 2000;59(7):831–7. doi: 10.1016/s0006-2952(99)00391-3. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Lusthof KJ, de Mol NJ, Richter W, Janssen LH, Butler J, Hoey BM, Verboom W, Reinhoudt DN. Redox cycling of potential antitumor aziridinyl quinones. Free Radic Biol Med. 1992;13(6):599–608. doi: 10.1016/0891-5849(92)90034-e. [DOI] [PubMed] [Google Scholar]
- Malkinson AM, Siegel D, Forrest GL, Gazdar AF, Oie HK, Chan DC, Bunn PA, Mabry M, Dykes DJ, Harrison SD, Jr, Ross D. Elevated DT-diaphorase activity and messenger RNA content in human non-small cell lung carcinoma: relationship to the response of lung tumor xenografts to mitomycin C. Cancer Res. 1992;52:4752–4757. [PubMed] [Google Scholar]
- Mihara K, Sato R. Detergent-solubilized NADH-cytochrome b5 reductase. Meth Enzymol. 1978;52:102–8. doi: 10.1016/s0076-6879(78)52011-9. [DOI] [PubMed] [Google Scholar]
- Mikami K, Naito M, Ishiguro T, Yano H, Tomida A, Yamada T, Tanaka N, Shirakusa T, Tsuruo T. Immunological quantitation of DT-diaphorase in carcinoma cell lines and clinical colon cancers: advanced tumors express greater levels of DT-diaphorase. Jpn J Cancer Res. 1998;89:910–915. doi: 10.1111/j.1349-7006.1998.tb00648.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Pan SS, Andrews PA, Glover CJ. Reductive activation of mitomycin C metabolites catalyzed by NADPH-cytochrome P450 reductase and xanthine oxidase. J Biol Chem. 1984;259:959–966. [PubMed] [Google Scholar]
- Powis G. Free radical formation by antitumor quinones. Free Radic Biol Med. 1989;6:63–101. doi: 10.1016/0891-5849(89)90162-7. [DOI] [PubMed] [Google Scholar]
- Rix U, Hantschel O, Dürnberger G, Remsing Rix LL, Planyavsky M, Fernbach NV, Kaupe I, Bennett KL, Valent P, Colinge J, Köcher T, Superti-Furga G. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood. 2007;110(12):4055–63. doi: 10.1182/blood-2007-07-102061. [DOI] [PubMed] [Google Scholar]
- Saunders MP, Jaffar M, Patterson AV, Nolan J, Naylor MA, Phillips RM, Harris AL, Stratford IJ. The relative importance of NADPH: cytochrome c (P450) reductase for determining the sensitivity of human tumour cells to the indolequinone EO9 and related analogues lacking functionality at the C-2 and C-3 positions. Biochem Pharmacol. 2000;59:993–996. doi: 10.1016/s0006-2952(99)00405-0. [DOI] [PubMed] [Google Scholar]
- Siegel D, Anwar A, Winski SL, Kepa JK, Zolman KL, Ross D. Rapid polyubiquitination and proteasomal degradation of a mutant form of NAD(P)H: Quinone oxidoreductase 1. Mol Pharmcol. 2001;59:263–268. doi: 10.1124/mol.59.2.263. [DOI] [PubMed] [Google Scholar]
- Stirpe F, Della Corte E. The regulation of rat liver xanthine oxidase. Conversion in vitro of the enzyme activity from dehydrogenase (type D) to oxidase (type O) J Biol Chem. 1969;244:3855–63. [PubMed] [Google Scholar]
- Tice RR, Agurell E, Anderson D, Burlinson B, Hartmann A, Kobayaski H, Miyamae Y, Rojas E, Ryu JC, Sasaki YF. Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ Mol Mutagen. 2000;35:206–221. doi: 10.1002/(sici)1098-2280(2000)35:3<206::aid-em8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- Tudor G, Alley M, Nelson CM, Huang R, Covell DG, Gutierrez P, Sausville EA. Cytotoxicity of RH1: NAD(P)H:quinone acceptor oxidoreductase (NQO1)-independent oxidative stress and apoptosis induction. Anti-Cancer Drugs. 2005;16:381–391. doi: 10.1097/00001813-200504000-00004. [DOI] [PubMed] [Google Scholar]
- Vermilion JL, Coon MJ. Purified liver microsomal NADPH-cytochrome P-450 reductase. Spectral characterization of oxidation-reduction states. J Biol Chem. 1978;253:2694–2704. [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1(3):1112–6. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Walton MI, Smith PJ, Workman P. The role of NAD-(P)H:quinone reductase (EC 1.6.99.2, DT-diaphorase) in the reductive bioactivation of the novel indoloquinone antitumour agent EO9. Cancer Commun. 1991;3:199–206. doi: 10.3727/095535491820873164. [DOI] [PubMed] [Google Scholar]
- Ward TH, Butler J, Shahbakhti H, Richards JT. Comet assay studies on the activation of two diaziridinylbenzoquinones in K562 cells. Biochem Pharmacol. 1997;53:1115–1121. doi: 10.1016/s0006-2952(96)00818-0. [DOI] [PubMed] [Google Scholar]
- Ward TH, Coe N, Hargreaves R, Bulter J, McGown AT. Crosslinking studies on the novel bioreductive anti-cancer drug RH1. Clin Cancer Res. 2000;6(Suppl):4527s. [Google Scholar]
- Winski SL, Hargreaves RH, Butler J, Ross D. A new screening system for NAD(P)H:quinone oxidoreductase (NQO1)-directed antitumor quinones: identification of a new aziridinylbenzoquinone, RH1, as a NQO1-directed antitumor agent. Clin Cancer Res. 1998;4:3083–3088. [PubMed] [Google Scholar]
- Winski SL, Swann E, Hargreaves RH, Dehn DL, Butler J, Moody CJ, Ross D. Relationship between NAD(P)H:quinone oxidoreductase 1 (NQO1) levels in a series of stably transfected cell lines and susceptibility to antitumor quinones. Biochem Pharmacol. 2001;61:1509–1516. doi: 10.1016/s0006-2952(01)00631-1. [DOI] [PubMed] [Google Scholar]
- Wu K, Knox R, Sun XZ, Joseph P, Jaiswal AK, Zhang D, Deng PS, Chen S. Catalytic properties of NAD(P)H:quinone oxidoreductase-2 (NQO2), a dihydronicotinamide riboside dependent oxidoreductase. Arch Biochem Biophys. 1997;347(2):221–8. doi: 10.1006/abbi.1997.0344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.