Abstract
The regulatory machinery that governs genetic and epigenetic control of gene expression is compartmentalized in nuclear microenvironments. Temporal and spatial parameters of regulatory complex organization and assembly are functionally linked to biological control and are compromised with the onset and progression of tumorigenesis providing a novel platform for cancer diagnosis and treatment.
Keywords: nuclear microenvironments, epigenetic control, transcription, nuclear matrix, chromatin
For the past century control of gene expression has been a dominant theme in fundamental biological and clinically relevant research. A platform for discovery has been provided by advances in nucleic acid, protein and carbohydrate biochemistry, structural biology and microscopy. The capabilities to synthesize and sequence nucleic acids and proteins have been instrumental. The pathway to understanding gene expression and the molecular basis of pathologies was initiated by the seminal discovery that DNA is the genetic material (Avery et al., 1944). Subsequent breakthroughs that have been pivotal in elucidating gene regulatory mechanisms include resolving the double helical structure of DNA (Watson and Crick, 1953), defining parameters of transcription, replication, recombination and repair, mapping signaling pathways that transduce and integrate regulatory cues within and between cells and linking genes with cancer and a broad spectrum of diseases. The accrual of insight into biological control from encyclopedic initiatives in genomics, proteomics and bioinformatics has been spectacular. Yet, there is additionally a requirement for resolution of mechanisms that support the organization, assembly, and integration of gene regulatory information into physiologically responsive networks. And, the mechanisms that epigenetically support cell fate and lineage commitment must be further defined.
Both genetic and epigenetic control mediate the regulation of proliferation, cell growth and phenotype, as well as establish and sustain the properties of normal and tumor cells. Recognizing the scope of mechanisms that are associated with genetic and epigenetic control we will focus on the Runx family of transcription factors to illustrate involvement of regulation at both levels. Temporal and spatial configuration of regulatory machinery for compartmentalization of genetic and epigenetic control in nuclear microenvironments will be considered within the context of gene activation and suppression.
I: Gene Expression Within the Three Dimensional Context of Nuclear Architecture
A. Multiple levels of nuclear organization contribute to combinatorial control of gene expression
Fidelity of gene expression necessitates integrating a broad spectrum of regulatory signals that govern proliferation, differentiation and maintenance of cell and tissue phenotypes. To accommodate the requirements for short term and sustained expression of cell growth and tissue-specific genes, it is necessary to identify and functionally characterize the promoter regulatory elements as well as cohorts of protein/DNA and protein/protein interactions that determine the extent to which genes are transcribed. However, it is becoming increasingly evident that the catalogue of regulatory elements and proteins is insufficient to support transcriptional control in the nucleus of intact cells in an in vivo environment. Rather, gene regulatory mechanisms must be understood in relation to the subnuclear organization of nucleic acids and regulatory proteins.
There is growing appreciation that transcriptional control requires multiple levels of nuclear organization. It is essential to package 2.5 yards of DNA as chromatin within the limited confines of the nucleus. Gene promoter elements must be rendered competent for protein/DNA and protein/protein interactions in a manner that permits binding and functional activities of primary transcription factors as well as co-activators and co-repressors. Less understood but pivotally relevant to physiologic control is the localization of the regulatory machinery for gene expression, replication, and repair at subnuclear sites where the macromolecular complexes that support DNA and RNA synthesis are localized (Reviewed in Taatjes DJ et al., 2004; Isogai and Tjian, 2003; Zaidi et al., 2005; Zaidi et al., 2007; Misteli, 2004; Wei et al., 1998; Htun et al., 1996; DeFranco, 2002; Handwerger and Gall, 2006; Kosak and Groudine, 2004; Cremer et al., 2006; Shav-Tal et al., 2006; Branco and Pombo, 2007; Cai et al., 2003; Bissell et al., 1999).
While the mechanisms that govern gene expression remain to be formally defined, there is growing awareness that the fidelity of gene expression necessitates the coordination of transcription factor metabolism and the spatial organization of genes and regulatory proteins within the three dimensional context of nuclear architecture. The components of nuclear organization include the sequence of gene regulatory elements, chromatin structure and higher order organization of the transcriptional regulatory machinery in subnuclear domains. All of these parameters involve mechanisms that include transcription factor synthesis, nuclear import and retention (reviewed in Henderson, 2003; Yashiroda and Yoshida, 2003; Kau et al., 2004), post translational modification of factors, and directing factors to subnuclear sites that support the organization (Zeng et al., 1997; Zeng et al., 1998) and assembly of regulatory machinery for gene expression. Remodeling of chromatin and nucleosome organization to accommodate requirements for protein/DNA and protein/protein interactions, at promoter elements are essential modifications for both activation and suppression of genes and physiological control of transcription (Reviewed in Drobic et al., 2006; Singh et al., 2000; Carorozza et al., 2003; de la Serna et al., 2006; Nakemura et al., 2002). This is a key component of epigenetic control that mediates competency for gene activation or suppression and conveys phenotype and lineage commitment to progeny cells during mitotic division. The reconfiguration of gene promoters and assembly of specialized subnuclear domains reflect the orchestration of both regulated and regulatory mechanisms. There are analogous and complex regulatory requirements for processing of gene transcripts. Here it has been similarly demonstrated that the regulatory components of splicing and export of messenger RNA to the cytoplasm are dependent on the architectural organization of nucleic acids and regulatory proteins (Spector, 2001; Shopland et al., 2002; Smith et al., 1999). There is growing evidence that the focal localization of regulatory machinery in nuclear microenvironments supports the integration of regulatory signals in a manner that facilitates competency for physiological responsiveness (Reviewed in Taatjes et al., 2004; Zaidi et al., 2005; Zaidi et al., 2007; Misteli, 2004; Kosak and Groudine, 2004; Shav-Tal et al., 2006). The biological relevance of nuclear microenvironments is reflected by the punctate subnuclear localization of factors that mediate transcription, processing of gene transcripts, DNA replication and DNA repair at discreet domains and retention of regulatory factors with target gene promoters during mitosis to epigenetically maintain phenotype in progeny cells (Figure 1).
Figure 1. Nucleic acids and regulatory proteins are compartmentalized in nuclear microenvironments.
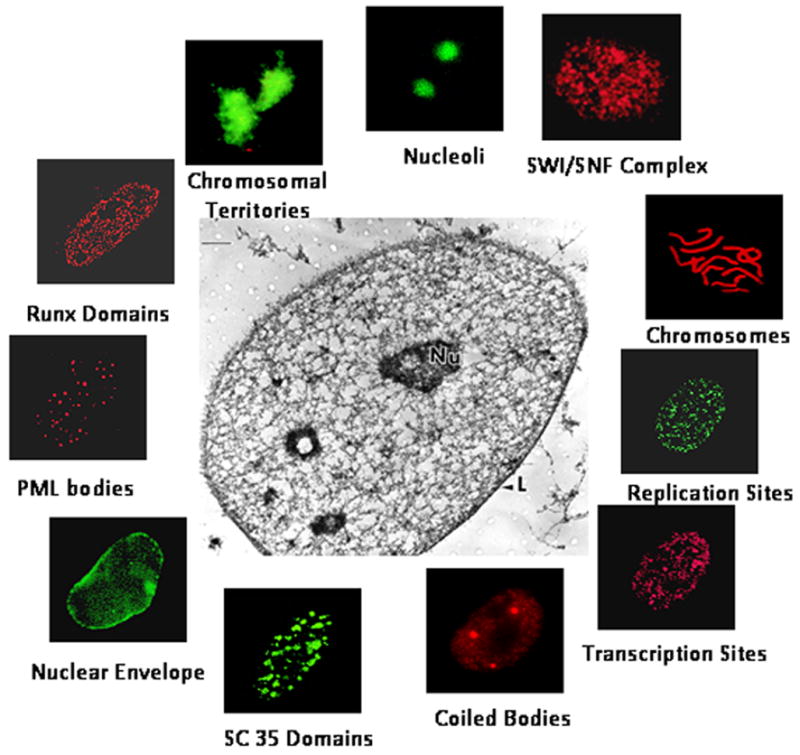
Nuclear functions are organized into distinct, non-overlapping subnuclear domains. Nuclear matrix, the underlying network of anastomising network of filaments and fibers provides structural basis for the functional compartmentalization of nuclear functions (Center). Immunofluorescence microscopy of the nucleus in situ has revealed the distinct subnuclear distribution of vital nuclear processes, including (but not limited to) DNA replication sites, chromatin remodeling, e.g., mediated by the SWI/SNF complex and Runx factors, structural parameters of the nucleus, such as the nuclear envelope, chromosomes, and chromosomal territories, Runx domains for transcriptional control of tissue-specific genes; and RNA synthesis and processing involving, for example, transcription sites, SC35 domains, coiled bodies and nucleoli. Subnuclear PML bodies of unknown function have been examined in numerous cell types. All these domains are associated with the nuclear matrix.
From a biological perspective, each parameter of factor metabolism requires stringent control and must be linked to structure-function interrelationships that mediate transcription and processing of gene transcripts. However, rather than representing regulatory obstacles, the complexities of nuclear biochemistry and morphology provide the required specificity for physiological responsiveness to a broad spectrum of signaling pathways to modulate transcription under diverse circumstances. Equally important, evidence is accruing that modifications in nuclear architecture and nuclear structure-function interrelationships accompany and appear to be causally related to compromised gene expression under pathological conditions, particularly in cancer, providing a platform for novel dimensions to diagnosis and treatment.
B. Organization and assembly of regulatory machinery in nuclear microenvironments
Runx transcription factors provide a paradigm for the focal organization and assembly of transcriptional regulatory machinery in nuclear microenvironments. These lineage-specific master regulatory proteins (Speck and Gilliland, 2002; Galindo et al., 2005; Barseguian et al., 2002; McNeil et al., 1999; Ito et al., 2005; Durst and Hiebert, 2004; Huang et al., 2008; Lian et al., 2004; Westendorf and Hiebert, 1999) control hematopoietic (Runx1), osteogenic (Runx2), and gastrointestinal/neural (Runx3) differentiation at two levels of nuclear organization. Activity is mediated by interactions with multiple sites of target gene promoters where they strategically provide scaffolds for the recruitment and integration of regulatory signals (e.g., TGFβ, SRC), as well as the recruitment of histone modifying enzymes and chromatin remodeling factors (e.g., HATs, HDAC, SWI/SNF) to influence promoter accessibility and placement of a broad spectrum of coregulatory proteins that contribute to transcriptional activation and suppression. Relevance for promoter localization of Runx transcription factors has been provided by loss or decline of biological activity when promoter binding sites of target genes are mutated or when functional domains of the Runx transcription factors are selectively mutated (Gutierrez et al., 2004). Gene expression within the three dimensional context of nuclear architecture is additionally supported by the organization of Runx regulatory machinery in punctate intranuclear domains (Zeng et al., 1997; Zeng et al., 1998; Zaidi et al., 2001). Here the necessity for fidelity of location within the nucleus is supported by the identification of a Runx-specific intranuclear targeting signal that is required for the execution of regulatory signals, Runx-dependent histone modifications and chromatin remodeling and differentiation both in vitro and in vivo (Figure 2) (Gutierrez et al., 2004; Choi et al., 2001; Gutierrez et al., 2007; Javed et al., 1999).
Figure 2. Positive and negative control of Runx responsive genes: Rat osteocalcin as a paradigm for tissue specific transcriptional control.
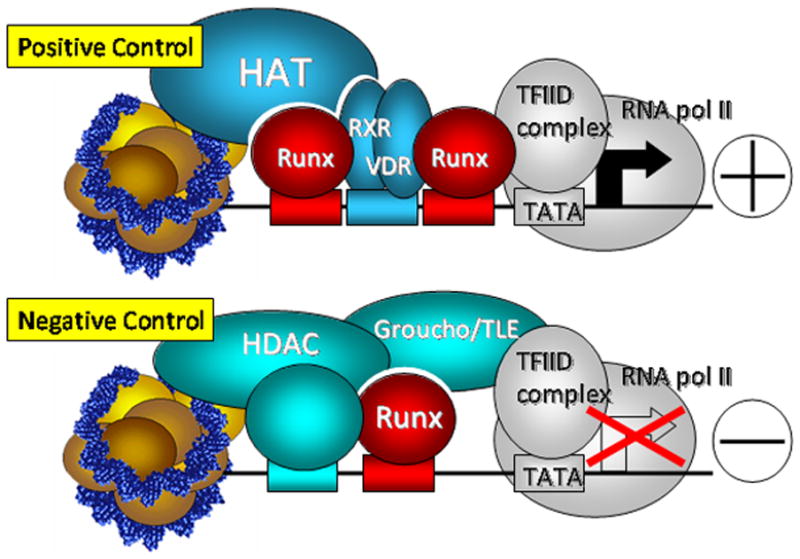
Runx transcription factors are multifunctional proteins that can synergize with different sequences-specific DNA-binding proteins (e.g., AP1, C/EBP) and associate with positive (e.g., Smads, p300) or negative (e.g., groucho/TLE, HDACs) gene regulatory cofactors depending on the promoter context of bone-tissue specific genes. Possible interactions accommodating the positive (Top) and negative (Bottom) regulation of transcription are shown. Depending on the cell type, coregulatory proteins may modulate transcriptional levels of the gene rather than completely repress the gene as illustrated.
Beyond the pivotal role for intranuclear organization of Runx regulatory complexes to support differentiation and development (e.g., osteogenesis and myeloid differentiation), there is a requirement for subnuclear localization of Runx proteins to initiate and sustain transformation and tumor progression. Localization of Runx2 within the nucleus is required for metastatic breast cancer and prostate cancer cells to form osteolytic lesions in bone (Javed et al., 2005) and competency for Runx1 intranuclear trafficking is necessary for myeloid differentiation and mutations that prevent intranuclear localization of Runx1 in myeloid progenitor cells results in a leukemic phenotype (Vradii et al., 2005).
Despite the compelling evidence for a focal organization of regulatory machinery within the nucleus to support biological activity, as illustrated by Runx regulatory complexes, there are key parameters of control that are essential to be clarified. The model for focal organization of factors to establish threshold concentrations for interactions with coregulatory proteins and target genes remains to be formally demonstrated. Rate limiting constituents of regulatory complex formation must be determined. It is essential to discriminate between colocalization and functional interactions. Determinants for the turnover and modifications of components in regulatory complexes should be identified and characterized. The extent to which targeting and retention are the definitive determinants for focal formation and stability of regulatory domains is open ended. The involvement of intranuclear trafficking and dynamic self assembly in the organization and turnover of regulatory sites for gene expression should be further explored.
Checkpoints that monitor the subnuclear distribution of regulatory factors and the sorting steps that ensure structural and functional fidelity of nuclear domains must be defined biochemically and mechanistically. However, there is growing support for informational content to organization of nuclear domains that is illustrated by the subnuclear organization of Runx regulatory machinery.
Recently, mathematical algorithms designated intranuclear informatics, have been developed to identify and assign unique quantitative signatures that define regulatory protein localization within the nucleus (Young et al., 2004). Quantitative parameters that can be assessed include nuclear size and variability in domain number, size, spatial randomness, and radial positioning.
The significance and implications of intranuclear informatics can be shown by three distinct biological examples. Regulatory proteins with different activities can be subjected to intranuclear informatics analysis, which assigns each protein a unique architectural signature. The overlap between the architectural signatures of different proteins is often correlated to their functional overlap. Alternatively, the subnuclear organization of the protein domain can be linked with subnuclear targeting, biological function and disease. For example, Runx2, and its subnuclear targeting defective mutant (mSTD) show distinct architectural signatures, indicating that the biological activity of a protein can be defined and quantified as subnuclear organization. Finally, the data can be used to define functional conservation. For example, this technique can be used to show that the post-mitotic restoration of the spatially ordered Runx subnuclear organization is functionally conserved. From the signatures that reflect regulatory protein localization within the nucleus and modifications that are associated with physiological responsiveness, transformation and tumorigenesis, a quantitative basis is provided for defining phenotype and detection/diagnosis of disease. It is also realistic to incorporate such signatures in strategies for novel dimensions to therapy.
C. Focal organization of transcription factors within the nucleus support regulatory networks for physiological responsiveness
The biological significance of focally organized regulatory complexes in nuclear microenvironments may reflect defined nuclear domains where threshold concentrations of regulatory factors for optimal formation of macromolecular complexes reside. The complexity of nuclear organization and nuclear structure-gene expression relationships ensures biological responsiveness. Each architecture-linked regulatory parameter is vulnerable to perturbations that can compromise control of cell growth, proliferation, and differentiation. However, each of these parameters is a potential target for therapy. An adjuvant therapeutic approach might be based on changes in radio- and chemo-sensitivity as a consequence of hypothermia-induced changes in the composition, assembly, and architectural organization of regulatory machinery within the cancer cell nucleus (Coffee et al., 2006). Challenges include: (i) methods of quantitative analysis that reproducibly capture subtle differences in subnuclear protein localization between normal and cancer cells; and (ii) development of small molecule inhibitors that specifically and selectively target components of nuclear organization that are perturbed during tumorigenesis.
These challenges can in part be overcome by an integrated biological approach. The heterogeneity of interactions that are supported by Runx transcription factors as scaffolding proteins serves as a basis for mechanisms that can accommodate diverse parameters of biological control. Architectural signatures that are derived from mathematical algorithms such as intranuclear informatics have the potential to discriminate between intranuclear localization of proteins in normal and cancer cells. Intranuclear informatics can be combined with proteomics (changes in protein-DNA and protein-protein interactions) and genomics (altered gene expression profiles) to develop a novel platform for identification and targeting of perturbed regulatory pathways in cancer cells (Figure 3). The convergence and integration of signaling networks in nuclear microenvironments provides an architecturally-based option for selectively targeting cancer-related changes in control of transcription, replication, and repair.
Figure 3. An integrated approach for mechanistic insights into nuclear structure-gene expression interrelationships.

Knowledge of co-regulatory factors that interact with scaffolding proteins obtained by the combined application of cellular, biochemical, and molecular approaches provides basis for developing an interactome for each scaffolding protein (top left panel). Components of the interactome can be visualized, albeit to a limited extent, by in situ immunofluorescence microscopy. Such “proteomics” approach provides mechanistic insight into combinatorial control of gene expression in normal and cancer cells. In parallel, genome-wide profiles can be generated that reflect changes in gene expression during tumorigenesis (“genomics”; bottom left panel). Importantly, newly developed mathematical algorithms can identify unique quantitative signatures for regulatory proteins (Right panel, “intranuclear informatics”) and apply subtle changes in these ‘architectural signatures’ to distinguish normal proteins (e.g. WT Runx2) from subnuclear targeting deficient variants (e.g. mSTD Runx2). Such an integrated approach is required to gain mechanistic insights into nuclear structure-gene expression interrelationships for biological control as well as for cancer diagnosis and treatment.
II. Architectural Parameters of Epigenetic Control
A. Runx transcription factors contribute to epigenetic regulation
Runx proteins illustrate a key parameter of epigenetic control that supports physiological responsiveness. The location of Runx transcription factors at proximal and upstream sites of targeted gene promoters supports the placement of histone-modifying and chromatin remodeling factors at regulatory domains which control basal and enhancer-mediated activity (Gutierrez et al., 2007; Javed et al., 1999; Gutierrez et al., 2004). Serving as scaffolds for assembling cohorts of regulatory factors that reconfigure chromatin organization and selectively modulate accessibility of promoter sequences to regulatory signals and proteins, an important component of biological control is provided that is based on a signature which does not depend on DNA sequences (Figure 2). This is an example of epigenetic regulatory information that establishes promoter landscape as architecturally assembled regulatory cues that can be conveyed to progeny cells during cell division. From a biological perspective such “epigenetic signatures” can sustain gene expression that establishes and ensures the persistence of phenotypes during development and tissue remodeling. A basis is also provided to support transformation and tumor progression in a manner where the tumor phenotype is retained as the cell population expands and the disease progresses.
There has been an evolution in our appreciation for the informational content of epigenetic control. Initial approaches focused on the chromatin organization of candidate genes and the localization of enzymology for histone modifications in the proximity of sequences where chromatin structure supports a phenotype. Runx transcription factor interactions with basal, tissue defining and upstream enhancer sequences of the bone specific osteocalcin gene provide scaffolds for the placement of HATs and HDACs (Westendorf et al., 2002; Yang et al, 2007). This mechanism supports epigenetic control by a master regulatory factor that is required for skeletogenesis and bone remodeling. Similarly, it is a requirement for Runx-mediated epigenetic control of skeletal genes in metastatic breast cancer and prostate cancer cells that are functionally linked to formation of osteolytic or osteoblastic lesions in bone (Barnes et al., 2004; Barnes et al., 2003; Pratap et al., 2005).
Recently, genome-wide profiling strategies have been developed that permit a global assessment of parameters for chromatin organization (Liu et al., 2005; Janjova et al., 2008). These global approaches provide complex but instructive signatures for epigenetic parameters of genome structure and organization. At the level of individual genes, the architectural context in which specific genes are embedded is revealed. Epigenetic control is not restricted to histone and chromatin signatures. DNA methylation is an additional, well documented, component of epigenetic regulatory mechanisms (Yoo and Jones, 2006). As with histone modifications, genome-wide profiling has enhanced understanding of epigenetic control that is functionally linked to biological regulation as well as to a broad spectrum of diseases that include cancer. Beyond the insight into regulatory mechanisms that are supported by histone modifications and DNA methylation, these components of epigenetic control serve as a basis for tumor diagnosis. Equally as relevant, HDAC inhibitors and DNA methylation inhibitors are being effectively used for cancer chemotherapy (Marks et al., 2004; Yoo and Jones, 2006).
B. Mitotic retention and segregation of transcriptional regulatory machinery
Post-mitotic gene expression necessitates restoration of nuclear organization. Regulatory complexes must be assembled in progeny cells as they emerge from cell division. There is an immediate and stringent requirement for expression of cell cycle, cell growth, and phenotypic genes. Using the focal nuclear organization of Runx transcription factors as a paradigm, immunofluorescence microscopy has directly shown that Runx transcription factors are focally retained on mitotic chromosomes and partitioned to progeny cells (Zaidi et al., 2003; Young et al., 2007a; Young et al., 2007b; Ali et al., 2008). The symmetrical localization of Runx transcription factors on mitotic chromosomes (Figure 4) and confirmation by chromatin immunoprecipitation analysis, indicate that Runx transcription factors remain associated with target genes as cells progress to mitosis (Young et al., 2007a; Young et al., 2007b).
Figure 4. Phenotypic transcription factors associate with mitotic chromosomes to epigenetically convey necessary regulatory information to progeny cells for lineage maintenance and commitment.
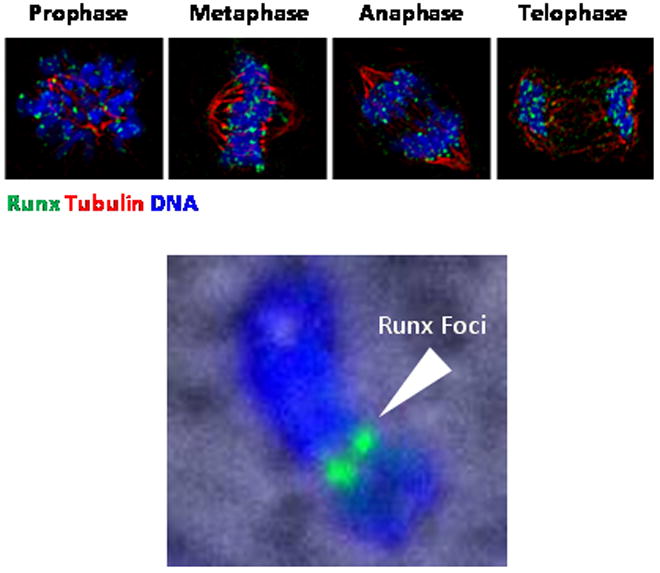
Phenotypic transcription factors that include Runx transcription factors are associated with mitotic chromosomes during cell division. This sequence-specific association of regulatory proteins with their target genes that regulate cell growth, proliferation, and differentiation bookmarks these genes for expression in the G1 phase of the cell cycle. Shown here are actively dividing human osteosarcoma SAOS-2 cells, immunostained with Tubulin (red) to identify mitotic spindle and Runx2 (green), which is a master regulator of bone formation. Cells are counterstained with DAPI (blue) to visualize mitotic chromosomes. Top panels show all phases of mitosis, while the bottom panel shows an enlarged human acrocentric chromosome exhibiting two large Runx foci (indicated by arrowhead).
Consequently the regulatory machinery for Runx control of gene expression remains in place during cell division rendering genes competent to reinitiate a program of transcription post-mitotically. The key question is the extent to which mitotic retention and segregation of regulatory proteins is a general regulatory mechanism. Several lines of evidence from gene expression profiling studies indicate mitotic retention of Runx transcription factors with more than 30 target gene promoters that are components of mechanisms which support multiple parameters of biological control (Young et al., 2007b). Association of regulatory factors that include SP1 (He and Davie, 2006), C/EBP, TBP, and TTF2 (Jiang et al., 2004; Segil et al., 1996; Tang et al., 2003) with chromosomes and/or genes during mitosis establishes the generality of this mechanism as a component of epigenetic control beyond histone modifications and DNA methylation.
Despite the compelling evidence for mitotic retention of transcription factors as a parameter of epigenetic control, there are numerous fundamental questions that must be resolved. How is association of transcription factors with target genes compatible with the global repression of genes during mitosis? Are transcription factors alone or transcription factors that are complexed with cohorts of co-regulatory proteins retained at target genes and conveyed to progeny cells? Are unique mechanisms in place to support association of transcription factors with target genes that are compatible with conformational properties of genes that are associated with chromatin condensation and decondensation during the entry and exit from mitosis? Are gene-associated regulatory proteins determinants for formation of interphase chromosomal territories? Resolution of these questions should reveal additional dimensions to nuclear structure – gene expression relationships that relate to epigenetic control.
C. Epigenetic control coordinates regulation of proliferation, cell growth, and phenotype
Several lines of evidence support association of transcription factors and co-regulatory proteins with RNA polymerase I and RNA polymerase II target genes during mitosis (Zaidi et al., 2003; Young et al., 2007a; Young et al., 2007b). Involvement in epigenetic control of gene expression for cell fate and lineage commitment is suggested by mitotic retention of tissue-specific regulatory proteins with promoters that are functionally linked to the establishment and maintenance of cell phenotype (Young et al., 2007b; Ali et al., 2008). In addition to mitotic retention of phenotypic genes, regulatory factors remain associated with genes that encode key components of signaling pathways, cell cycle control, and growth control (Young et al., 2007b). Occupancy of ribosomal gene promoters with key regulatory factors indicates that a major component of the regulatory machinery for protein synthesis is poised for resumption of expression when cells emerge from mitosis.
Recent results implicate phenotypic transcription factors in epigenetically mediating coordinate regulation of proliferation, cell cycle, and growth control. The Runx2 skeletal transcription factor associates with promoters of genes that support tissue-specific gene expression and expression of cell cycle regulatory genes that are transcribed by RNA polymerase II (Zaidi et al., 2003). In addition, Runx2 controls DNA polymerase I-mediated ribosomal gene transcription (Young et al., 2007a). During mitosis Runx2 resides at large discrete foci at nucleolar organizing regions where the ribosomal genes are located. The Runx2-UBF foci transition to nucleoli at sites of ribosomal RNA synthesis during interphase (Figure4). Functional studies directly establish Runx control of ribosomal gene transcription and protein synthesis (Young et al., 2007a). Similarly, the hematopoietic Runx1 and gastrointestinal/neural Runx3 transcription factors co-localize with ribosomal genes during mitosis and interphase to regulate protein synthesis. A similar mechanism is operative for control of ribosomal genes by MyoD during myogenesis and by C/EBP during adipogenesis (Ali et al., 2008).
Interrelationships between epigenetic control of tissue-specific genes, cell cycle and growth control appear to be operative in sustaining the transformed phenotype. The translocation of fusion protein AML/ETO associates with ribosomal genes during interphase and mitosis and contributes ribosomal gene expression and regulation of protein synthesis. Taken together, these findings are consistent with a critical molecular link between cell fate, proliferation and growth control.
III. Nuclear Structure – Gene Expression Relationships
The compartmentalization of regulatory machinery for gene expression is becoming increasingly evident. The focal organization of nucleic acids and regulatory proteins that support RNA polymerase I and RNA polymerase II-mediated transcription during interphase and mitosis are consistent with an architectural basis for contributions of genetic and epigenetic control to lineage-specific coordination of cell cycle and, cell growth, and phenotype regulation in nuclear microenvironments. Temporal and spatial organization of regulatory machinery in nuclear microenvironments conveys insight into mechanisms that support biological control. Equally relevant, a basis is provided for novel dimensions to understanding parameters of nuclear organization that are compromised in tumors can serve as a platform for diagnosis and therapy.
Acknowledgments
This work was supported by grants from the National Institutes of Health (CA82834, AR48818). The authors thank Patricia Jamieson for editorial assistance with the preparation of the manuscript.
Grants: This work was supported by grants from the National Institutes of Health (CA82834, AR48818).
References
- Ali SA, Zaidi SK, Dacwag CS, Salma N, Young DW, Shakoori AR, Montecino MA, Lian JB, van Wijnen AJ, Imbalzano AN, Stein GS, Stein JL. Phenotypic transcription factors epigenetically mediate cell growth control. Proc Natl Acad Sci USA. 2008 doi: 10.1073/pnas.0800970105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery OT, MacLeod CM, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus Type III. Journal of Experimental Medicine. 1944;79(2):137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes GL, Hebert KE, Kamal M, Javed A, Einhorn TA, Lian JB, Stein GS, Gerstenfeld LC. Fidelity of Runx2 activity in breast cancer cells is required for the generation of metastases-associated osteolytic disease. Cancer Res. 2004;64:4506–4513. doi: 10.1158/0008-5472.CAN-03-3851. [DOI] [PubMed] [Google Scholar]
- Barnes GL, Javed A, Waller SM, Kamal MH, Hebert KE, Hassan MQ, Bellahcene A, Van Wijnen AJ, Young MF, Lian JB, Stein GS, Gerstenfeld LC. Osteoblast-related transcription factors Runx2 (Cbfa1/AML3) and MSX2 mediate the expression of bone sialoprotein in human metastatic breast cancer cells. Cancer Res. 2003;63:2631–2637. [PubMed] [Google Scholar]
- Barseguian K, Lutterbach B, Hiebert SW, Nickerson J, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proc Natl Acad Sci USA. 2002;99:15434–15439. doi: 10.1073/pnas.242588499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissell MJ, Weaver VM, Lelièvre SA, Wang F, Petersen OW, Schmeichel KL. Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res. 1999;59:1757–1763s. [PubMed] [Google Scholar]
- Branco MR, Pombo A. Chromosome organization: new facts, new models. Trends Cell Biol. 2007;17:127–134. doi: 10.1016/j.tcb.2006.12.006. [DOI] [PubMed] [Google Scholar]
- Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nature Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- Carorozza MJ, Utley RT, Workman JL, Cote J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. doi: 10.1016/S0168-9525(03)00115-X. [DOI] [PubMed] [Google Scholar]
- Choi JY, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS. Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA. 2001;98:8650–8655. doi: 10.1073/pnas.151236498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffee DS, Getzenberg RH, DeWeese TL. Hyperthermic biology and cancer therapies: a hypothesis for the “Lance Armstrong effect”. JAMA. 2006;296:445–448. doi: 10.1001/jama.296.4.445. [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer M, Dietzel S, Müller S, Solovei I, Fakan S. Chromosome territories – a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- de la Serna I, Ohkawa Y, Imbalzano AN. Chromatin remodeling in mammalian differentiation: lessons from ATP-dependent remodellers. Nature Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- DeFranco DB. Navigating steroid hormone receptors through the nuclear compartment. Mol Endocrinol. 2002;16:1449–1455. doi: 10.1210/mend.16.7.0880. [DOI] [PubMed] [Google Scholar]
- Drobic B, Dunn KL, Espino PS, Davie JR. Abnormalities of chromatin in tumor cells. EXS. 2006;96:25–47. doi: 10.1007/3-7643-7378-4_2. [DOI] [PubMed] [Google Scholar]
- Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- Galindo M, Pratap J, Young DW, Hovhannisyan H, Im HJ, Choi JY, Lian JB, Stein JL, Stein GS, van Wijnen AJ. The bone-specific expression of RUNX2 oscillates during the cell cycle to support a G1 related anti-proliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez J, Paredes R, Cruzat F, Hill DA, van Wijnen AJ, Lian JB, Stein GS, Stein JL, Imbalzano AN, Montecino M. Chromatin remodeling by SWI/SNF results in nucleosome mobilization to preferential positions in the rat osteocalcin gene promoter. J Biol Chem. 2007;282:9445–9457. doi: 10.1074/jbc.M609847200. [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Liu J, Javed A, Montecino M, Stein GS, Lian JB, Stein JL. The vitamin D response element in the distal osteocalcin promoter contributes to chromatin organization of the proximal regulatory domain. J Biol Chem. 2004;279:43581–43588. doi: 10.1074/jbc.M408335200. [DOI] [PubMed] [Google Scholar]
- Hajkova P, Ancelin K, Waldmann T, Lacoste N, Lange UC, Cesari F, Lee C, Almouzni G, Schneider R, Surani MA. Chromatin dynamics during epigenetic reprogramming in the mouse germ line. Nature. 2008 Mar 19; doi: 10.1038/nature06714. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- He S, Davie JR. Sp1 and Sp3 foci distribution throughout mitosis. J Cell Sci. 2006;119:1063–1070. doi: 10.1242/jcs.02829. [DOI] [PubMed] [Google Scholar]
- Henderson B. Nuclear transport as a target for cancer therapies. Drug Disc Today. 2003;8:249. doi: 10.1016/s1359-6446(03)02628-x. [DOI] [PubMed] [Google Scholar]
- Htun H, Barsony j, Renyi I, Gould DL, Hager GL. Visualization of glucocorticoid receptor translocation and intranuclear organization in living cells with a green fluorescent protein chimera. Proc Natl Acad Sci USA. 1996;93:4845–4850. doi: 10.1073/pnas.93.10.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Zhang P, Hirai H, Elf S, Yan X, Chen Z, Koschmieder S, Okuno Y, Dayaram T, Growney JD, Shivdasani RA, Gilliland DG, Speck NA, Nimer SD, Tenen DG. PU.1 is a major downstream target of AML1(RUNX1) in adult mouse hematopoiesis. Nat Genet. 2008;40:51–60. doi: 10.1038/ng.2007.7. [Erratum in Nat Genet 2008 40:255.] [DOI] [PubMed] [Google Scholar]
- Isogai Y, Tjian R. Targeting genes and transcription factors to segregated nuclear compartments. Curr Opin Cell Biol. 2003;15:296–303. doi: 10.1016/s0955-0674(03)00052-8. [DOI] [PubMed] [Google Scholar]
- Ito K, Liu Q, Salto-Tellez M, Yano T, Tada K, Ida H, Huang C, Shah N, Inoue M, Rajnakova A, Hiong KC, Peh BK, Han HC, Ito T, Teh M, Yeoh KG, Ito Y. RUNX3, a novel tumor suppressor, is frequently inactivated in gastric cancer by protein mislocalization. Cancer Res. 2005;65:7743–7750. doi: 10.1158/0008-5472.CAN-05-0743. [DOI] [PubMed] [Google Scholar]
- Javed A, Barnes GL, Pratap J, Antkowiak T, Gerstenfeld LC, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Impaired intranuclear trafficking of Runx2 (AML3/CBFA1) transcription factors in breast cancer cells inhibits osteolysis in vivo. Proc Natl Acad Sci USA. 2005;102:1454–1459. doi: 10.1073/pnas.0409121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liu M, Spencer CA, Price DH. Involvement of transcription termination factor 2 in mitotic repression of transcription elongation. Mol Cell. 2004;14:375–385. doi: 10.1016/s1097-2765(04)00234-5. [DOI] [PubMed] [Google Scholar]
- Kau TR, Way JC, Silver PA. Nuclear transport and cancer: from mechanism to intervention. Nature Rev Cancer. 2004;4:106–117. doi: 10.1038/nrc1274. [DOI] [PubMed] [Google Scholar]
- Kosak ST, Groudine M. Gene order and dynamic domains. Science. 2004;306:644–647. doi: 10.1126/science.1103864. [DOI] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, Stein JL, Stein GS. Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit Rev Eukaryot Gene Expr. 2004;14:1–41. [PubMed] [Google Scholar]
- Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS Biol. 2005;3(10):e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Miller T, Kelly WK. Histone deacetylase inhibitors. Adv Cancer Res. 2004;91:137–168. doi: 10.1016/S0065-230X(04)91004-4. [DOI] [PubMed] [Google Scholar]
- McNeil S, Zeng C, Harrington KS, Hiebert S, Lian JB, Stein JL, van Wijnen AJ, Stein GS. The t(8;21) chromosomal translocation in acute myelogenous leukemia modifies intranuclear targeting of the AML1/CBFalpha2 transcription factor. Proc Natl Acad Sci USA. 1999;96:14882–14887. doi: 10.1073/pnas.96.26.14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Spatial positioning; a new dimension in genome function. Cell. 2004;119:153–156. doi: 10.1016/j.cell.2004.09.035. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Mori T, Tada S, Krajewski W, Rozovskaia T, Wassell R, Dubois G, Mazo A, Croce CM, Canaani E. ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell. 2002;10:1119–1128. doi: 10.1016/s1097-2765(02)00740-2. [DOI] [PubMed] [Google Scholar]
- Pratap J, Javed A, Languino LR, van Wijnen AJ, Stein JL, Stein GS, Lian JB. The Runx2 osteogenic transcription factor regulates matrix metalloproteinase 9 in bone metastatic cancer cells and controls cell invasion. Mol Cell Biol. 2005;25:8581–8591. doi: 10.1128/MCB.25.19.8581-8591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segil N, Guermah M, Hoffmann A, Roeder RG, Heintz N. Mitotic regulation of TFIID: inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996;10:2389–2400. doi: 10.1101/gad.10.19.2389. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Darzacq X, Singer RH. Gene expression within a dynamic nuclear landscape. EMBO J. 2006;25:3469–3479. doi: 10.1038/sj.emboj.7601226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shopland LS, Johnson CV, Lawrence JB. Evidence that all SC-35 domains contain mRNAs and that transcripts can be structurally constrained within these domains. J Struct Biol. 2002;140:131–139. doi: 10.1016/s1047-8477(02)00507-5. [DOI] [PubMed] [Google Scholar]
- Singh H, Sekinger EA, Gross DS. Chromatin and cancer: causes and consequences. J Cell Biochem (Suppl) 2000;35:61–68. [PubMed] [Google Scholar]
- Smith KP, Moen PT, Wydner KL, Coleman JR, Lawrence JB. Processing of endogenous pre-mRNAs in association with SC-35 domains is gene specific. J Cell Biol. 1999;144:617–629. doi: 10.1083/jcb.144.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nature Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Taatjes DJ, Marr MT, Tjian R. Regulatory diversity among metazoan co-activator complex. Nature Rev Mol Cell Biol. 2004;5:403–410. doi: 10.1038/nrm1369. [DOI] [PubMed] [Google Scholar]
- Tang QQ, Otto TC, Lane MD. CCAAT/enhancer-binding protein beta is required for mitotic clonal expansion during adipogenesis. Proc Natl Acad Sci USA 2003. 2003;100:850–855. doi: 10.1073/pnas.0337434100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vradii D, Zaidi SK, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Point mutation in AML1 disrupts subnuclear targeting, prevents myeloid differentiation, and effects a transformation-like phenotype. Proc Natl Acad Sci USA. 2005;102:7174–7179. doi: 10.1073/pnas.0502130102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R. Segregation of transcription and replication sites into higher order domains. Science. 1998;281:1502–1506. doi: 10.1126/science.281.5382.1502. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Hiebert SW. Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J Cell Biochem. 1999;32–33(Suppl):51–58. doi: 10.1002/(sici)1097-4644(1999)75:32+<51::aid-jcb7>3.3.co;2-j. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007;26:91–101. doi: 10.1038/sj.onc.1209760. [DOI] [PubMed] [Google Scholar]
- Yashiroda Y, Yoshida M. Nucleo-cytoplasmic transport of proteins as a target for therapeutic drugs. Curr Med Chem. 2003;10:741–748. doi: 10.2174/0929867033457791. [DOI] [PubMed] [Google Scholar]
- Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nature Rev Drug Disc. 2006;5:37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Pratap J, Galindo M, Zaidi SK, Lee SH, Yang X, Xie R, Javed A, Underwood JM, Furcinitti P, Imbalzano AN, Penman S, Nickerson JA, Montecino MA, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature. 2007a;445:442–446. doi: 10.1038/nature05473. [DOI] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang XQ, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007b;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Zaidi SK, Furcinitti PS, Javed A, van Wijnen AJ, Stein JL, Lian JB, Stein GS. Quantitative signature for architectural organization of regulatory factors using intranuclear informatics. J Cell Sci. 2004;117:4889–4896. doi: 10.1242/jcs.01229. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi JY, van Wijnen AJ, Stein JL, Lian JB, Stein GS. A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci. 2001;114:3093–3102. doi: 10.1242/jcs.114.17.3093. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS. The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep. 2005;6:128–133. doi: 10.1038/sj.embor.7400337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS. Nuclear microenvironments in biological control and cancer. Nature Rev Cancer. 2007;7(6):454–463. doi: 10.1038/nrc2149. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Pockwinse SM, Javed A, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic partitioning and selective reorganization of tissue-specific transcription factors in progeny cells. Proc Natl Acad Sci U S A. 2003;100:14852–14857. doi: 10.1073/pnas.2533076100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence JB, Penman S, Hiebert S, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Intranuclear targeting of AML/CBFalpha regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA. 1998;95:1585–1589. doi: 10.1073/pnas.95.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]


