Abstract
Helix-loop-helix (HLH) transcription factors are key regulators of neurogenesis, myogenesis and osteogenesis. Here the relative contributions of multiple classes of HLH factors to the expression of bone related genes during osteoblast maturation were compared. We examined the expression of a panel of HLH proteins (e.g., Twist1/2, USF1/2, c-Myc, Id1~4, E12/47, Stra13) and one Zn finger protein (Snail which recognizes a subset of E-boxes), during osteoblast differentiation and their functional contributions to bone phenotypic gene regulation. While expression of Twist1, Stra13, E12/47 and Snail transcripts remains relatively constant, expression of Twist2 as well as the inhibitory factors Id1, Id2, Id3 and Id4 decreases and USF1 is up-regulated during osteoblastic differentiation of MC3T3 cells. Forced expression of selected HLH transcription factors shows that Myc, Snail and USF factors increase expression of the bone markers osteocalcin (OC) and/or alkaline phosphatase (AP), while E12/47, Twist and Id factors decrease their expression. None of these factors affect Runx2 gene expression. Interestingly, Snail enhances expression of osteoblast markers, while Twist1 and Twist2 factors are cross-regulated and inhibit bone specific gene expression and other HLH proteins (e.g., Id) indirectly. Thus, our data suggest that the integrated activities of negative and positive E-box related regulatory factors control osteoblast differentiation.
Keywords: Osteocalcin, Runx2, E-box, Twist, Id, Snail, c-Myc
The basic helix-loop-helix (bHLH) transcription factors recognize E-box motifs and play key roles in a wide array of developmental processes such as cellular differentiation, lineage commitment and sex determination [Davis et al., 1990; Murre et al., 1989; Voronova and Baltimore, 1990]. The activity of bHLH factors is opposed by Id proteins (i.e., Id1, Id2, Id3, and Id4) that lack the basic DNA binding domain and act as dominant negative factors, because they can dimerize with bHLH proteins but fail to bind E boxes. Distinct HLH factors have been implicated in myogenesis [Olson, 1990; Hoshizaki et al., 1990], neurogenesis [Ben-Arie et al., 1997; Guillemot et al., 1993; Ma et al., 1996] and osteogenesis [El, V et al., 1997; Yoshida et al., 2005; Ogata et al., 1993; Peng et al., 2004].
Twist proteins are known to repress osteogenesis. Gene deletion experiments have shown that Twist-1 is required for formation of the mouse coronal suture [Yoshida et al., 2005]. Haploinsufficiency of the human Twist-1 gene contributes to the craniosynostosis disorder Saethre-Chotzen syndrome (SCS) [El, V et al., 1997]. In vitro, decreased Twist-1 synthesis downregulates Fgfr2 mRNA expression, which in turn inhibits Runx2 and downstream osteoblast-specific genes in human calvarial osteoblasts [Guenou et al., 2005]. Twist-1 inactivation reduces Runx2/CBFA1 expression and DNA binding to the osteocalcin promoter in osteoblasts [Yousfi et al., 2002]. Paradoxically, Twist-1 and Twist-2 (Dermo-1) overexpression inhibits osteoblast differentiation by suppressing the activity but not expression of Runx2 [Bialek et al., 2004; Kronenberg, 2004]. However, over-expression of Twist-1 increases periostin expression, a secreted protein that is highly expressed in early osteoblastic cells in vitro [Oshima et al., 2002]. Although it is evident that Twist proteins control osteogenic differentiation, how and when Twist-1 and Twist-2 regulate different stages of osteoblast phenotype commitment remain to be addressed.
Id proteins also represent key regulators of osteogenesis. BMP-2 enhances expression of Id-1 in early cultures of pre-osteoblastic cells, suggesting that enhancement of Id-1 expression may promote BMP2 induced differentiation of osteoblasts [Ogata et al., 1993]. The fact that BMP-induced bone formation in vivo is suppressed in Id-1/Id-3 heterozygous knockout mice provides further support for this hypothesis [Maeda et al., 2004]. However, constitutive overexpression of Id-1, Id-2 and Id-3 blocks osteoblast differentiation initiated by BMP-9 [Peng et al., 2004]. These findings suggest Id proteins may promote proliferation of early osteoblast progenitor cells and are down-regulated during terminal differentiation of committed osteoblasts.
Twist and Id proteins inhibit E2A proteins that normally upregulate expression of p21 WAF/CIP1 during differentiation of calvarial osteoblasts [Funato et al., 2001]. E2A and Twist proteins compete with the zinc-finger transcription factor Snail for binding to E-boxes to control p21WAF/CIP1 expression [Takahashi et al., 2004]. Other HLH proteins (e.g., c-Myc, USF1, USF2 and Stra13) are also expressed in osteoblasts or chondrocytes [Ebara et al., 1997; Wang et al., 2006; Nose et al., 1989; Shen et al., 2002]. Although accumulating evidence suggests that HLH proteins control osteoblast differentiation, there is limited insight into how the expression and function of distinct HLH proteins are integrated to regulate bone phenotype development.
In this study, we investigated the contributions of multiple classes of HLH proteins to the expression of bone related genes during osteoblast maturation. Our results corroborate the concept that Id and Twist proteins modulate bone-related gene expression [Tamura and Noda, 1994], but the data indicate that these effects may occur predominantly by indirect mechanisms.
MATERIALS AND METHODS
Cell Culture
MC3T3-E1 cells were maintained in α-minimal essential media supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA). NIH3T3 cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Grand island, NY) supplemented with 10% fetal bovine serum ROS17/2.8 cells were grown in F-12 media supplemented with 5% fetal bovine serum. For differentiation studies, MC3T3-E1 cells were fed every second day at confluence with the above medium containing 10 mM β–glycerol phosphate and 50 μg/ml ascorbic acid.
Transient Transfection
Cells in 6-well plates at 70% confluence were treated with 6μl of FuGENE 6 transfection reagent (Roche Diagnostics, Sommerville, NJ) and 2 μg of total DNA per well in accordance with the manufacturer’s protocol. The first 0.6-kb of the bone-related P1 promoter of the mouse and rat Runx2 gene (expressing the p57/MASNS isoform) [Drissi et al., 2000; van Wijnen et al., 2004] and the 1.1 kb rat osteocalcin promoter [Ryoo et al., 1997] were each fused to the firefly the luciferase reporter and used in transfection assays. Twelve HLH factors were co-transfected with luciferase reporters, including pIRES-hGFP-Twist1 (provided by Sachiko Iseki, Tokoyo Medical and Dental University) [Yoshida et al., 2005], pcDNA-flag-Twist2 (Li Li, Wayne State University) [Gong and Li, 2002], pSG5-USF1/2 and pSG5-c-myc (Michele Sawadogo, the University of Texas MD Anderson Cancer Center) [Choe et al., 2005], pcDNA-Id1, 2, 3 and 4, pcDNA-E12/47 (Barbara A. Christy, University of Texas Health Science Center) [Bounpheng et al., 1999], and pcDNA-flag Stra13 (Sean E. Egan, the Hospital for Sick Children, Toronto) [St-Pierre et al., 2002]. We also used an expression vector for the zinc-finger factor Snail (Masataka Nakamura, Tokoyo Medical and Dental University) in transfection assays [Takahashi et al., 2004].
Luciferase Reporter Assays
The firefly luciferase reporter plasmids, expression plasmids and Renilla luciferase reference gene (pRLh-null-Renilla) plasmid, as internal control, were co-transfected in each well. The total amount of DNA was maintained at a constant level (1 μg/well) by adding appropriate expression empty vector. After 24 h, the cells were harvested using 200 μl passive lysis buffer (Promega, Madison, WI) per well. Cell lysates (20 μl) were evaluated for luciferase activity using the Dual-Luciferase reporter assay kit (Promega, Madison, WI). The luciferase activity was measured according to the manufacturer’s instructions and normalized to values for Renilla luciferase. For all transcription studies, promoter activity (firefly/Renilla luciferase) is represented as fold induction compared to luciferase activity observed for promoterless pGL3 or pGL2 constructs.
RNA Isolation and Analysis
RNA was isolated from cultures of MC3T3-E1 cells using TRIzol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s protocol. After purification, 5 μg of total RNA was DNase treated using a DNA-free RNA column purification kit (Zymo Research, Orange, CA). RNA (1 μg) was then reverse transcribed using random hexamers as primers and the SuperScript 1st Strand Synthesis kit (Invitrogen) according to the manufacturer’s protocol. Gene expression was assessed by quantitative real-time PCR (including Runx2, osteocalcin, alkaline phosphatase, eleven distinct HLH factors and Snail). Primer Express software was used to predict optimum reverse transcription-PCR (RT-PCR) primer sets (Fig. 1A), except for GAPDH primers (Applied Biosystems, Foster City, CA). Quantitative PCR (QPCR) was performed using SYBR Green 2x master mixture (Applied Biosciences) and a two-step cycling protocol (anneal and elongate at 60°C, denature at 94°C). Specificity of primers was verified by dissociation of amplicons using SYBR Green as a detector. All transcript levels were normalized to that of GAPDH.
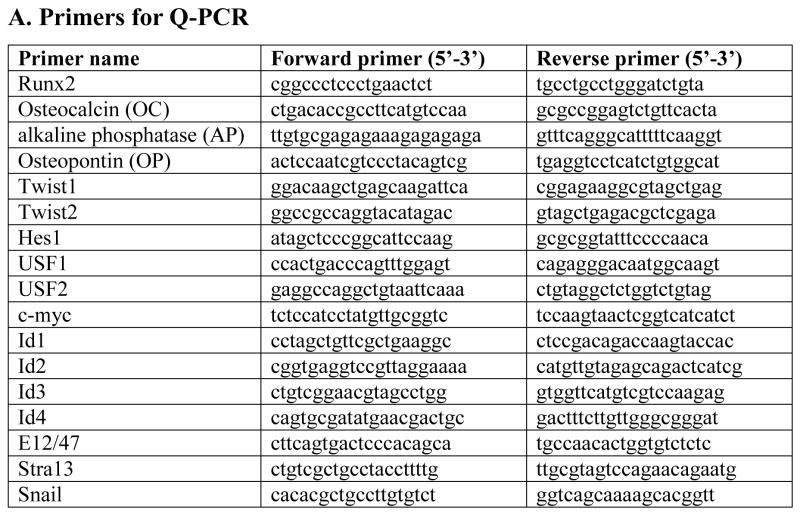
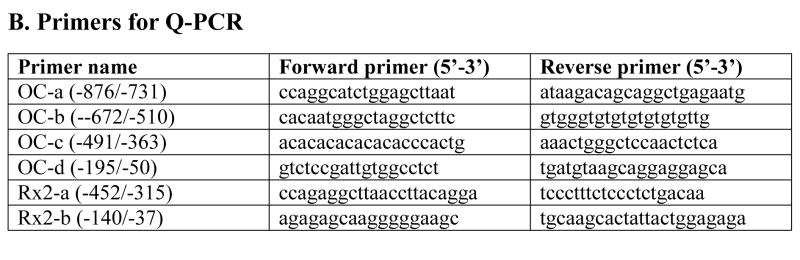
Fig. 1. Primers for quantitative PCR (QPCR) and chromatin immunoprecipitation assay (ChIP assay).
Part A lists primer sets that are used for detecting gene expression in MC3T3-E1 cells. These genes include four bone phenotypic genes (Runx2, osteocalcin, alkaline phosphatase, and osteopontin) and eleven HLH factors and Snail (as listed in the table). Part B lists all the primers which are used in ChIP assays to detect transcription factors binding to osteocalcin (OC-a, b, c, d) and Runx2 (Rx2-a, b) promoters.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) studies were performed as described previously [Young et al., 2007]. In brief, formaldehyde cross-linking was quenched by addition of glycine to a final concentration of 0.125 M at room temperature for 10 min, followed by rinsing with ice-cold 1×PBS before resuspension in lysis buffer. Sonication was performed 6 times at setting 3 (model 550 sonic dismembrator, Fisher Scientific, Pittsburgh, PA) for 10 s. The precleared cell lysate was incubated overnight at 4°C with 3 μg of anti-flag and anti-Twist1 antibodies and normal mouse IgG as a control. After reversal of cross-links at 65°C overnight, the DNA was recovered by phenol-chloroform extraction and ethanol precipitation using 5 μg of glycogen as carrier. An aliquot of each sample was assayed using quantitative PCR. To detect the presence of specific DNA fragments containing putative E-box motifs, several sets of primers in the distal Runx2 promoter (Rx2) and osteocalcin promoter (OC) were used, which are listed in Figure 1B.
RESULTS
The HLH proteins Twist-1, c-Myc and E2A, as well as Id-2, Id-3 and Id-4, are prominently expressed during differentiation of MC3T3-E1 cells
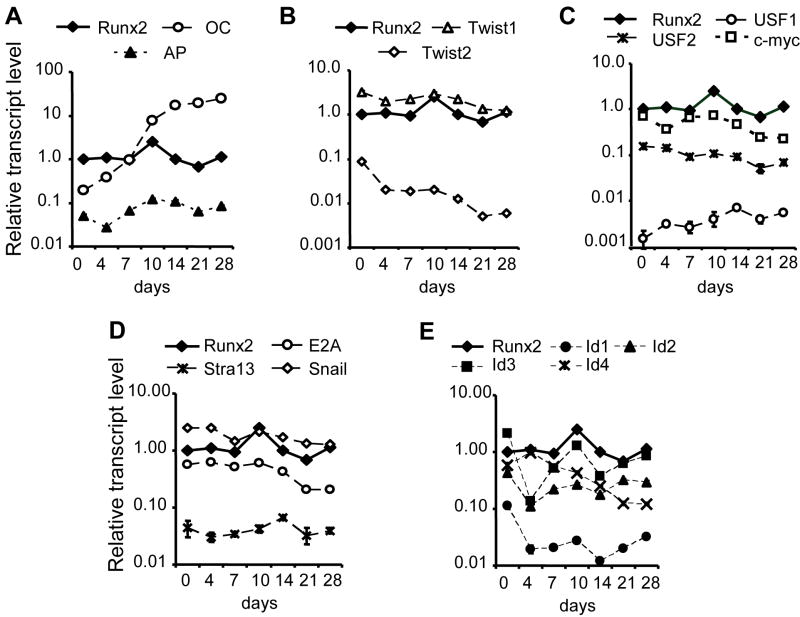
While the expression and biological function of selected subsets of HLH proteins have been studied, our understanding of the relative expression of HLH proteins and temporal aspects of their functions during osteoblast growth and differentiation is fragmented. Therefore, we examined a panel of eleven HLH factors during bone cell differentiation, all of which have been reported to be expressed in osteoblastic cells [Ogata et al., 1993; Bialek et al., 2004; Ebara et al., 1997; Shur et al., 2001; Shen et al., 2002]. In addition, we included a zinc finger protein, Snail, which is known to bind to HLH recognition sites to regulate transcription [Takahashi et al., 2004]. We determined the mRNA level of each HLH factor by Q-PCR in relation to established stages of osteoblast differentiation. For example, expression of the osteogenic regulator Runx2 and the bone-related marker Alkaline Phosphatase (AP) increases after day 4 when differentiation commences (Fig.2 A), and the late mineralization stage marker Osteocalcin (OC) is increased by ~100 fold at day 28 relative to levels at day 0.
Fig. 2. Differential regulation of multiple HLH transcription factors during osteoblast differentiation.
We analyzed the expression of thirteen selected transcription factors relative to standard bone marker genes during differentiation of MC3T3-E1 cells by quantitative real-time RT-PCR. RNA was isolated from differentiated MC3T3-E1 cells at different days up to day 28. The following genes were tested: the osteogenic transcription factor Runx2, the bone marker genes, osteocalcin (OC) and alkaline phosphatase (AP) (Panel A), Twist1 and Twist2 (Panel B), USF1/2 and c-myc (Panel C), E2A, Stra13 and Snail (Panel D), and the ID proteins ID1, ID2, ID3 and ID4 (Panel E). To permit comparison of the relative transcript levels of each of the genes, values were normalized to GAPDH and the level of Runx2 at day 0 was arbitrarily set as 1. The graphs shows a representative differentiation time course and the data points represent the means ± S.D. (n=3). The error bars in each data point are typically less than 10%.
Twist1 and Twist2 are reported to have similar functions in bone development through different mechanisms [Lee et al., 2000]. We observed that during differentiation of MC3T3, Twist1 mRNA is expressed at higher levels than Twist2. Twist1 levels decrease by 2.5-fold from day 0 to day 28. However, the level of Twist2 mRNA diminishes 10-fold during the same time span (Fig.2 B). The reciprocal modulations in Twist1 and Twist2 expression suggest that these proteins may have dynamic roles in regulating distinct phenotypic transitions during osteoblast differentiation (e.g., when cells reach confluency at day 4 or exhibit multilayering at day 10 when AP reaches peak expression). USF1, USF2 and c-myc belong to basic helix-loop-helix-leucine zipper (bHLH-zip) subset of HLH transcription factors. During differentiation of MC3T3-E1 cells, the USF2 and c-myc transcript levels remain almost constant (Fig.2 C). Although USF1 showed the lowest mRNA levels of these three factors, its transcript level on day 28 is increased 5-fold compared with day 0. The transcript levels of E2A and the zinc finger protein (Snail) at day 28 were ~2 fold lower than levels at day 0, while Stra13 expression remains relatively constant throughout differentiation (Fig.2 D).
Id proteins are negative regulators of basic helix-loop-helix proteins. Of the four Id genes identified in mammals, Id-1 and Id-3 are ubiquitously expressed, whereas Id-2 and Id-4 exhibit a more restricted pattern of expression [Kreider et al., 1992; Ruzinova and Benezra, 2003]. Our data show that all four Id genes are expressed in MC3T3-E1 cells (Fig.2 E). Id1 showed the lowest transcript level of the four Id genes. The expression of Id-1, 2 and 3 is dramatically reduced (>5 fold) at day 4 during the early stages of differentiation. Subsequently, the expression of Id-2 and Id-3, but not that of Id-1 or Id-4, increases to levels observed on day 0. Id4 expression is maximal of day 4 and steadily declines during differentiation. Taken together, these results indicate that multiple HLH genes are expressed at different levels and vary in their temporal expression during MC3T3-E1 differentiation. These differences in expression suggest that each of the HLH proteins may perform distinct biological functions during progression of osteoblast phenotype commitment.
The application of quantitative real time PCR to examine the mRNA levels of multiple HLH factors in parallel permits assessment of the relative abundance of each member in osteoblasts. The relatively high expression of the non-DNA binding HLH proteins Id-2, Id-3 and Id-4 and of the DNA binding HLH transcription factors Twist1, c-Myc and E2A in osteoblastic cells (compared to Id-1, Twist2, USF1, USF2 and Stra13), suggest that this set of six proteins may bind to or control binding to the majority of E-boxes in bone-specific promoters.
Differential regulation of bone marker gene expression by E-box binding transcription factors
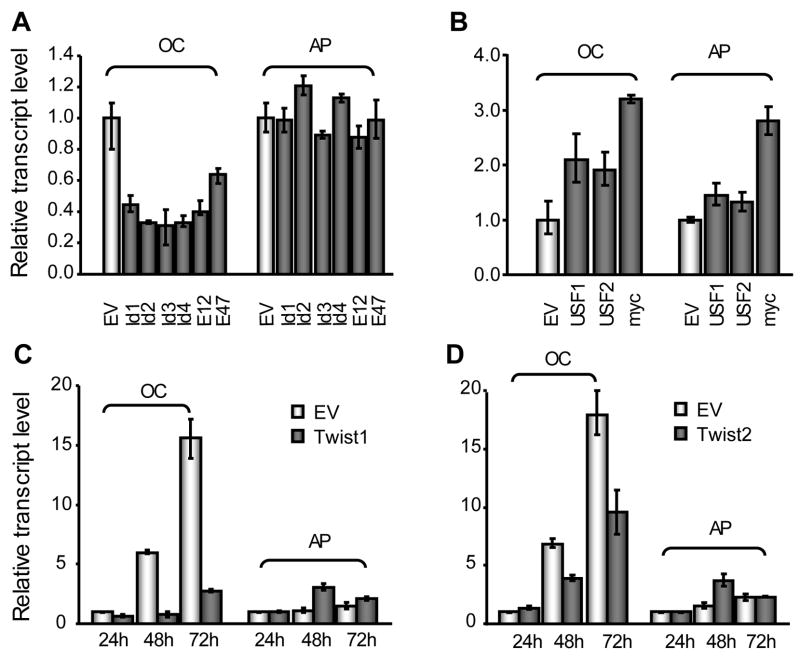
Because the levels of most HLH proteins we studied are naturally modulated during osteoblast differentiation, we experimentally modulated the levels of our panel of HLH proteins in MC3T3 cells to assess these functional effects on the expression of bone marker genes. Each HLH protein was transiently expressed and mRNA levels of bone-related genes examined at 24 h post-transfection. All four Id proteins, as well as the E2A derived proteins E12 and E47, inhibited OC transcript levels by 40 to 70% (Fig. 3A). However, Id proteins and E2A proteins have no effects on AP mRNA levels (Fig. 3A) or Runx2 gene expression (data not shown). Forced expression of USF1 and USF2 marginally increases AP transcript levels, while c-myc increases AP expression almost 3-fold (Fig. 3B). USF1, USF2 and c-Myc enhance OC gene expression by ~2 to 3 fold (Fig. 3B) but do not affect Runx2 mRNA levels (data not shown). These data suggest that the regulatory effects of these factors are restricted to specific osteoblast-related genes.
Fig. 3. Modulation of bone-related gene expression by forced expression of HLH factors in MC3T3-E1 cells.
The panels show the results of transiently expressing different HLH factors (0.5 μg) on the endogenous mRNA levels of OP and AP. The bar graphs in Panel A show the effect of exogenous expression of Id proteins (Id1-4), as well as E12 and E47; all six proteins suppress osteocalcin (OC) transcript level at 24 h post-transfection. Panel B show that USF1/2 and c-myc increase OC and AP mRNA levels after 24 h overexpression. Panels C and D show the inhibitory effects of Twist1 (Panel C) and Twist2 (Panel D) on OC transcript levels that are observed 48 h after transfection. In Panels C and D, cells were transfected with 0.5 μg expression constructor or empty vector (EV), and harvested at 24 h, 48 h, and 72 h post-transfection for RNA isolation and subsequent analysis by quantitative real time RT-PCR. The results were normalized to GADH transcript level. The means ± S.D. (n=3) are shown.
Twist1 and Twist2 affected bone phenotypic gene transcription through different mechanisms. Data from qRT-PCR assays indicated that neither factor affected OC or AP expression at 24h post-transfection (Fig. 3C, D). However, 48h after transfection, Twist1 and Twist2 inhibit OC transcript levels by 50% or more, with Twist1 suppressing OC transcript levels to a greater degree than Twist2. The inhibitory effects of Twist1 and Twist2 became more pronounced at 72 h after transfection. In contrast to suppression of the OC gene, both Twist1 and Twist2 transiently increased AP transcript levels after 48 h, but not at 72 h post-transfection. These results suggest that Twist1 and Twist2 indirectly regulate osteoblast differentiation and simultaneously may suppress or activate expression. However, because these regulatory effects of Twist proteins are only observed in later stages of transfection, it is likely that the effects of Twist1 and Twist2 are mediated by secondary factors that are directly controlled by these two Twist proteins.
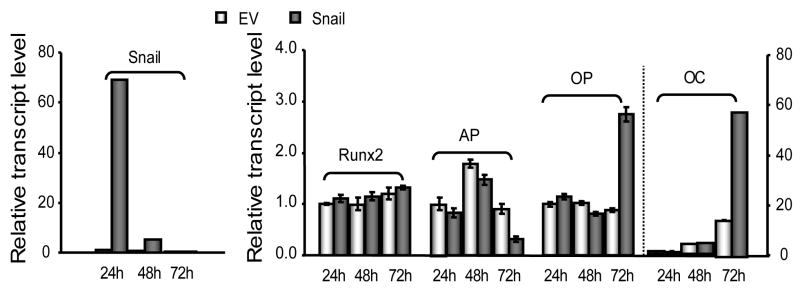
Similar to the observations with Twist1 and Twist2, Snail increases OC and osteopontin (OP) transcript levels, and inhibits AP gene expression only at 72 h after transfection (Fig. 4). However, Snail does not affect Runx2 gene expression. Snail mRNA levels are maximal at 24 h and decrease to the same level as that observed for empty vector by 72 h after transfection. These results clearly indicate that Snail, and presumably Twist1 and Twist2, are each only transiently required to modulate the expression of osteoblast related markers.
Fig. 4. Transient elevation of Snail expression indirectly regulates bone marker gene expression.
The graphs show qRT-PCR results obtained by transfecting MC3T3-E1 cells with Snail expression constructs (0.5 μg). Left panel shows the forced expression of Snail at 72 h post-transfection. Right panel shows that Snail increases OC and OP transcript levels, but inhibits AP gene expression at 72 h post-transfection.
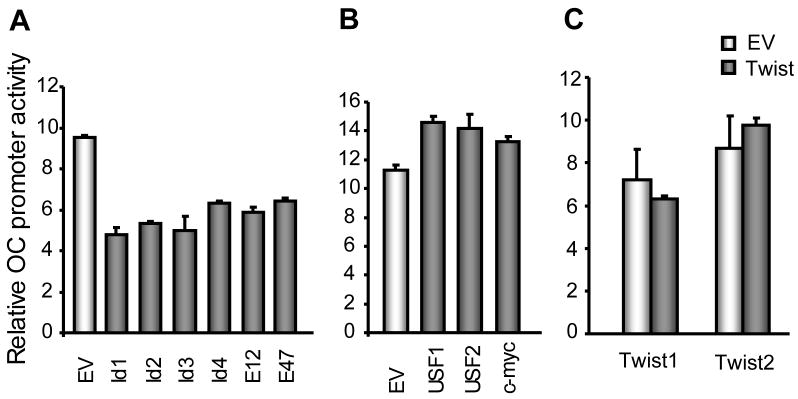
Transient reporter gene assays revealed that OC promoter activity is directly modulated by Id and E2A (E12/E47) proteins, but not by Twist1 or Twist2 (Fig. 5). For example, over-expression of Id proteins inhibited osteocalcin promoter activity up to 40% of control values observed in the presence of empty vector (Fig. 5A). Similar to Id proteins, E12 and E47 inhibit OC transcription based on results from luciferase assays (Fig. 5A). USF1, USF2 and c-myc activated OC-Luc reporter activity only modestly (Fig. 5B), while Twist1 and Twist2 do not affect OC or AP transcription at 24 h post-transfection (Fig. 5C). The latter results further suggest that effects of Twist1 and Twist2 on bone marker gene expression (see Fig. 3) are indirect. Twist proteins may exert their regulatory effects through activation of a proxy factor (e.g., another transcription factor) or perhaps mediate cross-talk with other gene regulators through protein/protein interactions.
Fig. 5. Suppression of OC promoter activity by selected HLH proteins.
The bar graphs in Panel A show that Id1, Id2, Id3 and Id4, as well as E12/47 decrease OC promoter activity by 40%–50%. In contrast, USF1/2 and c-myc have marginal positive effects on OC promoter activity (Panel B), while both Twist1 and Twist2 do not affects OC promoter activity (Panel C). MC3T3-E1 cells were co-transfected with 0.5 μg of 1.1-kb OC promoter reporter and 0.5 μg empty vectors (EV) or expression constructs, and harvested with lysis buffer after 24 h. Luciferase activity was measured and normalized to Renilla activity as described in methods.
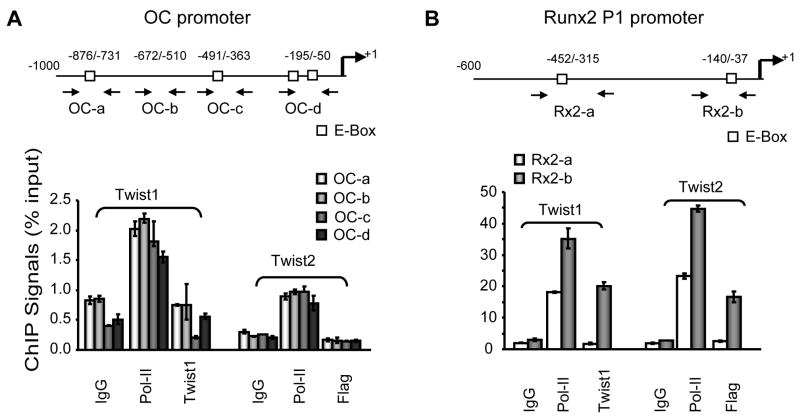
Chromatin immunoprecipitation analysis reveals Twist proteins regulate OC and Runx2 gene transcription through different mechanisms
To further investigate how Twist1 and Twist2 regulate OC gene transcription, we performed chromatin immunoprecipitation (ChIP) assays using exogenously expressed (and epitope-tagged) Twist proteins. We also examined the bone-specific Runx2 P1 promoter for comparison. We designed four pairs of primers to amplify the OC promoter regions containing putative E-boxes as previously reported [Tamura and Noda, 1994] (Fig. 6A), and two sets of primers to amplify Runx2 promoter fragments containing an E-box (Fig. 6B). Anti-Twist1 and anti-flag (for Twist2) antibodies, as well as antibodies against RNA polymerase II (positive control) and non-specific IgG (negative control) were used for immunoprecipitation. We examined four regions in the OC promoter that contain E-boxes (e.g., OC-a, OC-c and OC-d) or not (OC-b), and two regions in the Runx2 promoter one of which encompasses an E box. The results revealed that RNA polymerase II is specifically associated with the OC or Runx2 promoters, which is expected because both genes are actively transcribed (Fig. 6A, B). Twist1 and Twist2 associate only with the Runx2 gene but not the OC gene (Fig. 6A, B). The Twist1 and -2 interactions with the Runx2 promoter are specific to the region spanning the E-box in the proximal promoter (−140/−37), as there were no binding signals using primer Rx2-a that amplify the region between −452 and −315 containing the second E-box (Fig. 6B). These results indicate that while Twist1 and Twist2 may contribute to control of Runx2 expression through a promoter-dependent mechanism (i.e., protein/DNA or protein/protein), the inhibitory regulation of OC gene transcription by Twist1 and Twist2 appears to be indirect. Because Runx2 transcription is not influenced by forced expression of Twist1 and Twist2, it appears that these proteins are not rate-limiting for promoter activating under our experimental conditions. However, it is conceivable that control of Runx2 gene transcription by Twist1 and Twist2 may require the presence of additional co-factors.
Fig. 6. In vivo interaction of the osteocalcin (OC) and Runx2 P1 promoters by Twist1 and Twist2 in MC3T3-E1 cells.
Chromatin immunoprecipitation experiments (ChIP assays) were performed with MC3T3-E1 transfected expression constructs (0.5 μg) for Twist1 and Twist2 or the corresponding empty vector (EV). Panel A shows results obtained with primers sets (OC-a to OC-d) that amplify the proximal region of the mouse osteocalcin promoter. Each of the primer pairs with the exception of OC-b contains putative E-boxes. The horizontal arrows indicate the positions of the forward and reverse primers. Binding of both Twist1 and Twist2 to the OC promoter is below the level of detection. Panel B shows ChIP assay results obtained with two sets of primers that amplify the proximal regions of the Runx2 P1 promoter that contains putative E-boxes. Twist1 and Twist2 interact with the Runx2 P1 promoter. Input represents 1% of each chromatin fraction that was used for immunoprecipitation.
Regulation of other HLH transcription factors by Twist1 and Twist2 may contribute to OC gene transcription
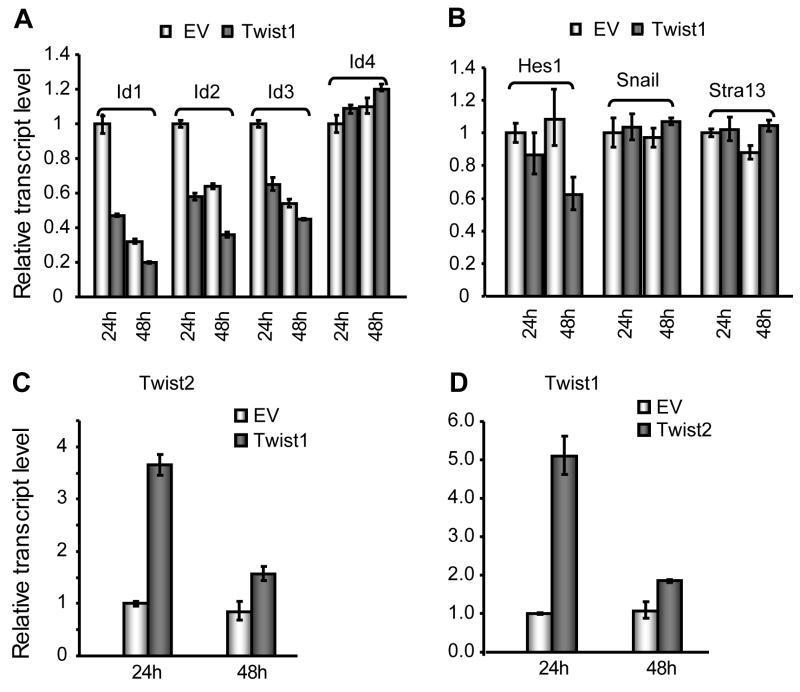
HLH transcription factors are capable of cross-regulation [Braun et al., 1992; O'Toole et al., 2003; Tamura and Noda, 1994], therefore we considered if molecular cross-talk between HLH proteins may contribute to the Twist-1 and -2 dependent modulations in bone marker gene expression. Results from qRT-PCR assays revealed that experimental elevation of Twist1 level suppresses expression of three Id genes (Id1, Id2 and Id3), but not Id4, by 40–50% at 24h post-transfection (Fig. 7A). Twist1 also has no effects on Snail or Stra13 transcript levels (Fig. 7B). Id4 and Snail are both downregulated during differentiation, while Stra13 mRNA levels are barely detectable throughout differentiation (see Fig. 2B), thus suggesting that Twist1 preferentially controls HLH genes that are more robustly expressed throughout osteoblast differentiation.
Fig. 7. Regulation of Id proteins and cross-regulation by Twist1 and Twist2.
We examined whether forced expression of Twist1 and Twist2 modulates expression of other HLH factors in MC3T3-E1 cells. The mRNA levels for Id1, Id2 and Id3 decrease upon transient elevation of Twist1 levels (Panel A). After 48 h, the mRNA levels of Hes1 are also suppressed (Panel B), but over- expression of Twist1 does not affect the transcript levels of Snail and Stra13. Panels C and D show reciprocal stimulation of expression of Twist2 by Twist1 (Panel C) and of Twist2 by Twist1 (Panel D) in transfected cells. Cells were transfected with 0.5 μg empty vectors (EV) or the indicated expression constructs, and harvested after 24 h and 48 h. RNA was isolated for analysis by quantitative real time RT-QPCR. Transcript levels were normalized to that of GAPDH.
Strikingly, our data show that exogenous Twist1 activates transcription of endogenous Twist2 gene expression, and exogenous Twist2 can reciprocally activate endogenous Twist1 expression (Fig. 7C and D). This cross-regulation is specific for these two bHLH proteins, because we did not find any effects of Twist1 and Twist2 on Snail or Stra13 transcript levels (Fig. 7B and data not shown). Interestingly, Twist1 and Twist2 do not exhibit cross-regulation in either NIH3T3 fibroblasts or ROS 17/2.8 osteosarcoma cells (data not shown), suggesting that regulatory interrelationships between Twist1 and Twist2 are particularly important for MC3T3 cells that represent osteoblastic progenitors that have not fully developed the phenotype of mature bone forming cells.
DISCUSSION
HLH transcription factors perform key functions in many developmental processes and tissues. It has been known for some time that there are HLH factors that can bind to a conserved E-box (5’ CACATG) in the proximal region of the osteocalcin promoter [Tamura and Noda, 1994; Quarles et al., 1997]. Whether HLH factors functionally control OC gene transcription has remained unresolved. Several studies have explored how osteogenesis is regulated by HLH factors, yet most of these studies have been restricted to examination of the functions of the Twist and Id class of proteins. While osteoblast-specific HLH factors have yet to be discovered, in this study we examined expression of multiple HLH genes during osteoblastic differentiation of MC3T3-E1 cells. The temporal expression of selected HLH factors is positively correlated with the expression of bone marker genes. We observed constitutive expression of Twist1 and E2A, as well as the E-box binding Zn finger transcription factor Snail throughout bone phenotype development, indicating that all three factors may contribute to multiple stages of osteoblast differentiation. Pronounced down regulation of Twist2 expression suggests that Twist2 is an inhibitor of osteoblast differentiation (see below). Relatively constant expression of USF2 and increased expression of USF1 are consistent with their ability to enhance OC and AP transcription. Based on the functional expression of a panel of HLH factors at distinct stages of osteogenic differentiation, we propose that multiple HLH factors may jointly regulate osteoblast differentiation.
Analysis of eleven distinct HLH factors and Snail revealed that these factors use distinct mechanisms to regulate bone marker gene expression and osteoblast differentiation. As previously reported, our data show that Id proteins and Twist are repressors for OC transcription [Yousfi et al., 2002], and our results indicate that Ids directly inhibit OC gene expression and promoter activity. While E2A is known as a transcriptional activator [Funato et al., 2001], we find that transient expression of E2A represses OC gene expression. Because E2A is expressed constantly during differentiation of MC3T3-E1 cells, it is possible that transcriptional activation or repression by E2A depends on the co-factors with which it may interact. While Id proteins and E2A act as repressors, our data clearly show that USF1, USF2 and c-myc directly activate OC and AP transcription, thus providing experimental evidence that these ubiquitous HLH proteins can functionally control expression of bone marker genes. USF1 has been shown to activate transforming growth factor β type II receptor (TbRII) promoter activity in primary cultures of fetal rat osteoblasts, consistent with a role of USF1 in osteoblast differentiation [Chang et al., 2002]. Taken together, our findings suggest that Id proteins, E2A, USF1, USF2 and c-myc are each capable of directly regulating MC3T3-E1 differentiation.
Our study reveals that forced expression of Twist1 and Twist2 inhibits OC transcription after 48h when exogenous Twist expression is maximal. These data complement previous data indicating that Twist proteins inhibit osteogenesis, although the mechanisms by which Twist proteins control osteoblast differentiation remain unclear. It is plausible that other (co-)factors that are directly controlled by these two Twist proteins may be essential for Twist dependent regulation. This concept may explain paradoxical results from different groups. For instance, over-expression of Twist-1 represses OC transcription by inhibiting Runx2 activity [Yousfi et al., 2002], but mutation of Twist-1 proteins decreases OC expression [Yoshida et al., 2005]. We find that forced expression of Twist-1 and Twist-2 proteins not only results in mutual activation of Twist mRNA levels, but also inhibit Id mRNA expression with the exception of Id4. It remains to be elucidated whether other HLH factors involved in osteoblast differentiation are controlled by Twist. However, our combined results indicate that indirect inhibition of OC transcription by Twist1 and Twist2 is controlled by an intricate set of HLH-related gene regulatory pathways.
Apart from OC, we also explored whether Twist proteins control Runx2 gene transcription. Forced expression of Twist1 and Twist2 does not modulate Runx2 transcript levels or promoter activity, consistent with previous studies [Guenou et al., 2005]. However, our data suggest that Twist proteins interact with the proximal part of the Runx2 P1 promoter which encompasses an E-box. Interestingly, we find that Twist proteins inhibit OC transcription but do not bind to the OC promoter, but these proteins interact with the Runx2 P1 promoter, but do not affect Runx2 transcription. These data indicate that Twist proteins regulate OC and Runx2 transcription through different mechanisms. The inhibition of the OC promoter may occur by protein/protein interactions between Twist and Runx2 proteins [Bialek et al., 2004], but Twist/Runx2 interactions are not sufficient to explain the mechanistic distinctions between the OC and Runx2 promoters.
In conclusion, our studies reveal that multiple HLH factors functionally support osteoblast differentiation and regulate transcription of bone marker genes through both direct and indirect mechanisms. We suggest that cross-talk between HLH factors may contribute to transcriptional control of osteogenesis.
Acknowledgments
We thank the following investigators for generously sharing expression constructs used in this study: Barbara Christy, University of Texas Health Science Center; Sean Egan, the Hospital for Sick Children, Toronto; Sachiko Iseki, Tokyo Medical and Dental University; Li Li, Wayne State University; Masataka Nakamura, Tokyo Medical and Dental University; and Michele Sawadogo, the University of Texas MD Anderson Cancer Center. We thank the members of our laboratories including Jacqueline Akech, Margaretha van der Deen, Ricardo Medina, Ronglin Xie and Yang Lou for stimulating discussions and technical advice. Special thanks to Carlotta Glackin for sharing reagents and unpublished data throughout the course of this study experiments. Also we thank Judy Rask for assistance with preparation of the manuscript.
Contract Grant Sponsor: NIH grant R01 AR039588. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
References
- Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Math1 is essential for genesis of cerebellar granule neurons. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- Bounpheng MA, Melnikova IN, Dimas JJ, Christy BA. Identification of a novel transcriptional activity of mammalian Id proteins. Nucleic Acids Res. 1999;27:1740–1746. doi: 10.1093/nar/27.7.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Bober E, Arnold HH. Inhibition of muscle differentiation by the adenovirus E1a protein: repression of the transcriptional activating function of the HLH protein Myf-5. Genes Dev. 1992;6:888–902. doi: 10.1101/gad.6.5.888. [DOI] [PubMed] [Google Scholar]
- Chang W, Parra M, Ji C, Liu Y, Eickelberg O, McCarthy TL, Centrella M. Transcriptional and post-transcriptional regulation of transforming growth factor beta type II receptor expression in osteoblasts. Gene. 2002;299:65–77. doi: 10.1016/s0378-1119(02)01013-2. [DOI] [PubMed] [Google Scholar]
- Choe C, Chen N, Sawadogo M. Decreased tumorigenicity of c-Myc-transformed fibroblasts expressing active USF2. Exp Cell Res. 2005;302:1–10. doi: 10.1016/j.yexcr.2004.08.013. [DOI] [PubMed] [Google Scholar]
- Davis RL, Cheng PF, Lassar AB, Weintraub H. The MyoD DNA binding domain contains a recognition code for muscle-specific gene activation. Cell. 1990;60:733–746. doi: 10.1016/0092-8674(90)90088-v. [DOI] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva de Sousa Lopes S, Choi J-Y, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Ebara S, Kawasaki S, Nakamura I, Tsutsumimoto T, Nakayama K, Nikaido T, Takaoka K. Transcriptional regulation of the mBMP-4 gene through an E-box in the 5'-flanking promoter region involving USF. Biochem Biophys Res Commun. 1997;240:136–141. doi: 10.1006/bbrc.1997.7618. [DOI] [PubMed] [Google Scholar]
- El GV, Le MM, Perrin-Schmitt F, Lajeunie E, Benit P, Renier D, Bourgeois P, Bolcato-Bellemin AL, Munnich A, Bonaventure J. Mutations of the TWIST gene in the Saethre-Chotzen syndrome. Nat Genet. 1997;15:42–46. doi: 10.1038/ng0197-42. [DOI] [PubMed] [Google Scholar]
- Funato N, Ohtani K, Ohyama K, Kuroda T, Nakamura M. Common regulation of growth arrest and differentiation of osteoblasts by helix-loop-helix factors. Mol Cell Biol. 2001;21:7416–7428. doi: 10.1128/MCB.21.21.7416-7428.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong XQ, Li L. Dermo-1, a multifunctional basic helix-loop-helix protein, represses MyoD transactivation via the HLH domain, MEF2 interaction, and chromatin deacetylation. J Biol Chem. 2002;277:12310–12317. doi: 10.1074/jbc.M110228200. [DOI] [PubMed] [Google Scholar]
- Guenou H, Kaabeche K, Mee SL, Marie PJ. A role for fibroblast growth factor receptor-2 in the altered osteoblast phenotype induced by Twist haploinsufficiency in the Saethre-Chotzen syndrome. Hum Mol Genet. 2005;14:1429–1439. doi: 10.1093/hmg/ddi152. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-y. [DOI] [PubMed] [Google Scholar]
- Hoshizaki DK, Hill JE, Henry SA. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- Kreider BL, Benezra R, Rovera G, Kadesch T. Inhibition of myeloid differentiation by the helix-loop-helix protein Id. Science. 1992;255:1700–1702. doi: 10.1126/science.1372755. [DOI] [PubMed] [Google Scholar]
- Kronenberg HM. Twist genes regulate Runx2 and bone formation. Dev Cell. 2004;6:317–318. doi: 10.1016/s1534-5807(04)00069-3. [DOI] [PubMed] [Google Scholar]
- Lee MS, Lowe G, Flanagan S, Kuchler K, Glackin CA. Human Dermo-1 has attributes similar to twist in early bone development. Bone. 2000;27:591–602. doi: 10.1016/s8756-3282(00)00380-x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- Maeda Y, Tsuji K, Nifuji A, Noda M. Inhibitory helix-loop-helix transcription factors Id1/Id3 promote bone formation in vivo. J Cell Biochem. 2004;93:337–344. doi: 10.1002/jcb.20154. [DOI] [PubMed] [Google Scholar]
- Murre C, McCaw PS, Vaessin H, Caudy M, Jan LY, Jan YN, Cabrera CV, Buskin JN, Hauschka SD, Lassar AB. Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell. 1989;58:537–544. doi: 10.1016/0092-8674(89)90434-0. [DOI] [PubMed] [Google Scholar]
- Nose K, Itami M, Satake M, Ito Y, Kuroki T. Abolishment of c-fos inducibility in ras-transformed mouse osteoblast cell lines. Mol Carcinog. 1989;2:208–216. doi: 10.1002/mc.2940020407. [DOI] [PubMed] [Google Scholar]
- O'Toole PJ, Inoue T, Emerson L, Morrison IE, Mackie AR, Cherry RJ, Norton JD. Id proteins negatively regulate basic helix-loop-helix transcription factor function by disrupting subnuclear compartmentalization. J Biol Chem. 2003;278:45770–45776. doi: 10.1074/jbc.M306056200. [DOI] [PubMed] [Google Scholar]
- Ogata T, Wozney JM, Benezra R, Noda M. Bone morphogenetic protein 2 transiently enhances expression of a gene, Id (inhibitor of differentiation), encoding a helix-loop-helix molecule in osteoblast-like cells. Proc Natl Acad Sci USA. 1993;90:9219–9222. doi: 10.1073/pnas.90.19.9219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN. MyoD family: a paradigm for development? Genes Dev. 1990;4:1454–1461. doi: 10.1101/gad.4.9.1454. [DOI] [PubMed] [Google Scholar]
- Oshima A, Tanabe H, Yan T, Lowe GN, Glackin CA, Kudo A. A novel mechanism for the regulation of osteoblast differentiation: Transcription of periostin, a member of the fasciclin I family, is regulated by the bHLH transcription factor, twist. J Cell Biochem. 2002;86:792–804. doi: 10.1002/jcb.10272. [DOI] [PubMed] [Google Scholar]
- Peng Y, Kang Q, Luo Q, Jiang W, Si W, Liu BA, Luu HH, Park JK, Li X, Luo J, Montag AG, Haydon RC, He TC. Inhibitor of DNA binding/differentiation helix-loop-helix proteins mediate bone morphogenetic protein-induced osteoblast differentiation of mesenchymal stem cells. J Biol Chem. 2004;279:32941–32949. doi: 10.1074/jbc.M403344200. [DOI] [PubMed] [Google Scholar]
- Quarles LD, Siddhanti SR, Medda S. Developmental regulation of osteocalcin expression in MC3T3-E1 osteoblasts: minimal role of the proximal E-box cis-acting promoter elements. J Cell Biochem. 1997;65:11–24. [PubMed] [Google Scholar]
- Ruzinova MB, Benezra R. Id proteins in development, cell cycle and cancer. Trends Cell Biol. 2003;13:410–418. doi: 10.1016/s0962-8924(03)00147-8. [DOI] [PubMed] [Google Scholar]
- Ryoo H-M, Hoffmann HM, Beumer TL, Frenkel B, Towler DA, Stein GS, Stein JL, van Wijnen AJ, Lian JB. Stage-specific expression of Dlx-5 during osteoblast differentiation: involvement in regulation of osteocalcin gene expression. Mol Endocrinol. 1997;11:1681–1694. doi: 10.1210/mend.11.11.0011. [DOI] [PubMed] [Google Scholar]
- Shen M, Yoshida E, Yan W, Kawamoto T, Suardita K, Koyano Y, Fujimoto K, Noshiro M, Kato Y. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J Biol Chem. 2002;277:50112–50120. doi: 10.1074/jbc.M206771200. [DOI] [PubMed] [Google Scholar]
- Shur I, Lokiec F, Bleiberg I, Benayahu D. Differential gene expression of cultured human osteoblasts. J Cell Biochem. 2001;83:547–553. doi: 10.1002/jcb.1249. [DOI] [PubMed] [Google Scholar]
- St-Pierre B, Flock G, Zacksenhaus E, Egan SE. Stra13 homodimers repress transcription through class B E-box elements. J Biol Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- Takahashi E, Funato N, Higashihori N, Hata Y, Gridley T, Nakamura M. Snail regulates p21(WAF/CIP1) expression in cooperation with E2A and Twist. Biochem Biophys Res Commun. 2004;325:1136–1144. doi: 10.1016/j.bbrc.2004.10.148. [DOI] [PubMed] [Google Scholar]
- Tamura M, Noda M. Identification of a DNA sequence involved in osteoblast-specific gene expression via interaction with helix-loop-helix (HLH)-type transcription factors. J Cell Biol. 1994;126:773–782. doi: 10.1083/jcb.126.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wijnen AJ, Stein GS, Gergen JP, Groner Y, Hiebert SW, Ito Y, Liu P, Neil JC, Ohki M, Speck N. Nomenclature for Runt-related (RUNX) proteins. Oncogene. 2004;23:4209–4210. doi: 10.1038/sj.onc.1207758. [DOI] [PubMed] [Google Scholar]
- Voronova A, Baltimore D. Mutations that disrupt DNA binding and dimer formation in the E47 helix-loop-helix protein map to distinct domains. Proc Natl Acad Sci U S A. 1990;87:4722–4726. doi: 10.1073/pnas.87.12.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee G, Hsu W, Yeh CH, Ho ML, Wang GJ. Identification of USF2 as a key regulator of Runx2 expression in mouse pluripotent mesenchymal D1 cells. Mol Cell Biochem. 2006;292:79–88. doi: 10.1007/s11010-006-9220-9. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Phylactou LA, Uney JB, Ishikawa I, Eto K, Iseki S. Twist is required for establishment of the mouse coronal suture. J Anat. 2005;206:437–444. doi: 10.1111/j.1469-7580.2005.00411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young DW, Hassan MQ, Yang X-Q, Galindo M, Javed A, Zaidi SK, Furcinitti P, Lapointe D, Montecino M, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Mitotic retention of gene expression patterns by the cell fate determining transcription factor Runx2. Proc Natl Acad Sci USA. 2007;104:3189–3194. doi: 10.1073/pnas.0611419104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousfi M, Lasmoles F, Kern B, Marie PJ. TWIST inactivation reduces CBFA1/RUNX2 expression and DNA binding to the osteocalcin promoter in osteoblasts. Biochem Biophys Res Commun. 2002;297:641–644. doi: 10.1016/s0006-291x(02)02260-x. [DOI] [PubMed] [Google Scholar]