Abstract
Summary: DNAPlotter is an interactive Java application for generating circular and linear representations of genomes. Making use of the Artemis libraries to provide a user-friendly method of loading in sequence files (EMBL, GenBank, GFF) as well as data from relational databases, it filters features of interest to display on separate user-definable tracks. It can be used to produce publication quality images for papers or web pages.
Availability: DNAPlotter is freely available (under a GPL licence) for download (for MacOSX, UNIX and Windows) at the Wellcome Trust Sanger Institute web sites: http://www.sanger.ac.uk/Software/Artemis/circular/
Contact: artemis@sanger.ac.uk
1 INTRODUCTION
Circular and linear DNA diagrams provide a powerful tool for illustrating the features of a genome in their full genomic context. CGView (Stothard and Wishart, 2005) is a circular genome viewer that produces PNG, JPEG or scalable vector graphics (SVG) format. It is primarily designed to be part of a pipeline and not an interactive and editable viewer. GenomeDiagram (Pritchard et al., 2006) is a Python module, which can also be used to generate diagrams. GenomePlot (Gibson and Smith, 2003), GenoMap (Sato and Ehira, 2003) and Microbial Genome Viewer (MGV; Kerkhoven et al., 2004), are GUI based but do not provide the ability to directly read in files in the common sequence formats or a filtering tool to define tracks.
Circos (http://mkweb.bcgsc.ca/circos/) is web-based genome comparison visualizer that is configured from flat files. It can be configured for comparisons within a genome or between multiple genomes. There are also commercial packages available that draw circular and linear plots.
Below we present DNAPlotter, a collaborative project combining graphics from the Jemboss (http://emboss.sf.net/Jemboss/) DNA viewer and Artemis (http://www.sanger.ac.uk/Software/Artemis/) libraries to read files and filter features, which has been designed to be a graphical means of customizing circular and linear diagrams.
2 IMPLEMENTATION
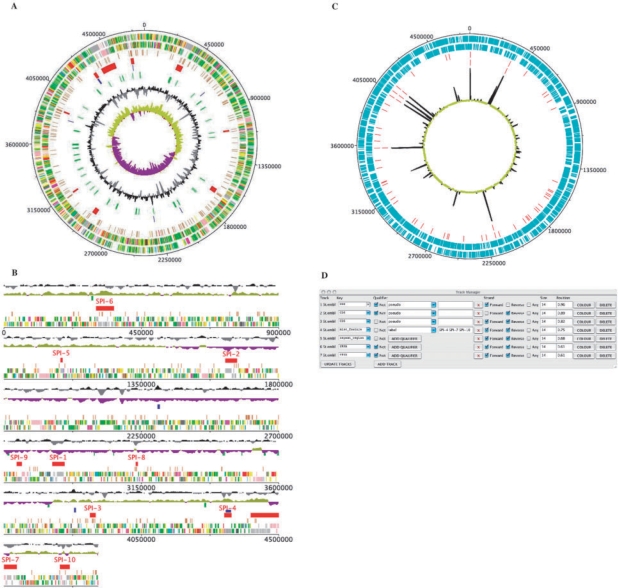
DNAPlotter can be run as a standalone application or from within Artemis (Berriman and Rutherford, 2003; Rutherford et al., 2000). Written in Java, DNAPlotter is platform independent. The main advantage of this program is that it is a fully interactive interface in which the track features can be customized and configuration changes are applied immediately to the figure. Under the Options menu there is a checkbox for switching between the circular (Fig. 1A) and linear (Fig. 1B) representation of the DNA and associated features.
Fig. 1.
(A, B) showing Salmonella typhi genome as a circular and linear plot, respectively. The tracks from the outside represent: (1) Forward CDS; (2) Reverse CDS; (3) Pseudogenes 4. Salmonella Pathogenicity Islands (red); (5) repeat regions (blue); (6) rRNA and tRNA (green); (7) %GC plot 8. GC skew [(GC)/(G+C)]. (C) A generated example showing a transcriptome graph (black and yellow) on a circular plot for a prokaryotic genome. The tracks from the outside represent: (1) Forward CDS; (2) Reverse CDS; (3) tRNA; (4) rRNA. (D) Snapshot of the track manager showing filtering criteria.
DNAPlotter reads the common sequence formats (EMBL, Genbank, GFF) using the Artemis file reading library. The program reads in a sequence file, which may or may not contain features, such as coding sequences. DNAPlotter displays the sequence as a circular or linear plot. Additional feature files can be read in and overlaid on the sequence. As Artemis code is used for DNAPlotter, features can also be read from the Generic Model Organism Database (GMOD, http://www.gmod.org) Chado relational database schema (Carver et al., 2008).
DNAPlotter uses Artemis libraries to filter features to be displayed on tracks for both circular and linear plots. The built-in track manager (Fig. 1C) enables the user to define what gets displayed. Each row in the track manager defines the filter for selecting what is shown on that track. It also allows a list of labels or gene names to be pasted in. So, for example, a list of regions or genes can be specified. The size of each track and its position is specified in the track manager.
The colour for the features can be taken from the colour qualifier defined for the feature or, if not specified, from the Artemis predefined colour code. Alternatively, the colour can be set for all features belonging to a track.
Tracks can also be added by reading in additional files. This means that it is straightforward, for example, to mark the position of genes for which orthologues have been calculated.
Details for a feature are displayed in mouse over tool-tips. By double clicking on a feature the display properties for that feature can be edited, e.g. coordinates, line width and colour. Labels and arrows can be added to a feature. Alternatively, these properties can be adjusted using the DNA Wizard utility. Features can also be dragged in the circular plot and this will move the position of all features in the track.
The GC content and GC skew [(G − C)/(G + C)] graphs can also be displayed. There are options for the graphs to set the window and step size for the calculation. The graph positions and colour scheme can also be changed. Users can also load in their own graph data in the form of a single value for each base position.
The resulting images can be saved as JPEG, PNG, BMP, WBMP or PostScript. PostScript is the best format for high resolution and publication quality images. The circular and linear plots can also be sent directly to a PostScript printer.
ACKNOWLEDGEMENTS
We would like to thank Maria Fookes and Tim Perkins (Wellcome Trust Sanger Institute) for the data Figure 1C is based on.
Funding: The Wellcome trust.
Conflict of Interest: none declared.
REFERENCES
- Berriman M, Rutherford K. Viewing and annotating sequence data with Artemis. Brief. Bioinform. 2003;4:124–132. doi: 10.1093/bib/4.2.124. [DOI] [PubMed] [Google Scholar]
- Carver T, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008 doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R, Smith DR. Genome visualization made fast and simple. Bioinformatics. 2003;19:1449–1450. doi: 10.1093/bioinformatics/btg152. [DOI] [PubMed] [Google Scholar]
- Kerkhoven R, et al. Visualization for genomics: the microbial genome viewer. Bioinformatics. 2004;20:1812–1814. doi: 10.1093/bioinformatics/bth159. [DOI] [PubMed] [Google Scholar]
- Pritchard L, et al. GenomeDiagram: a python package for the visualization of large-scale genomic data. Bioinformatics. 2006;22:616–617. doi: 10.1093/bioinformatics/btk021. [DOI] [PubMed] [Google Scholar]
- Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- Sato N, Ehira S. GenoMap, a circular genome data viewer. Bioinformatics. 2003;19:1583–1584. doi: 10.1093/bioinformatics/btg195. [DOI] [PubMed] [Google Scholar]
- Stothard P, Wishart DS. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]