Abstract
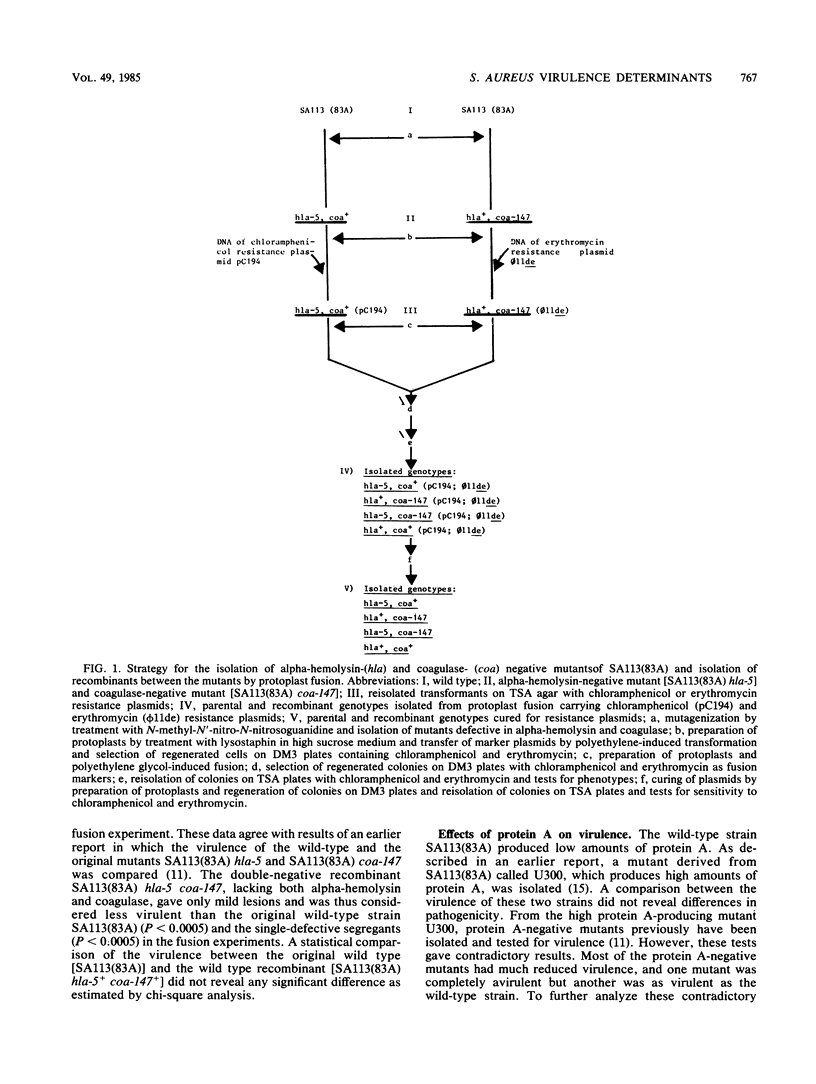
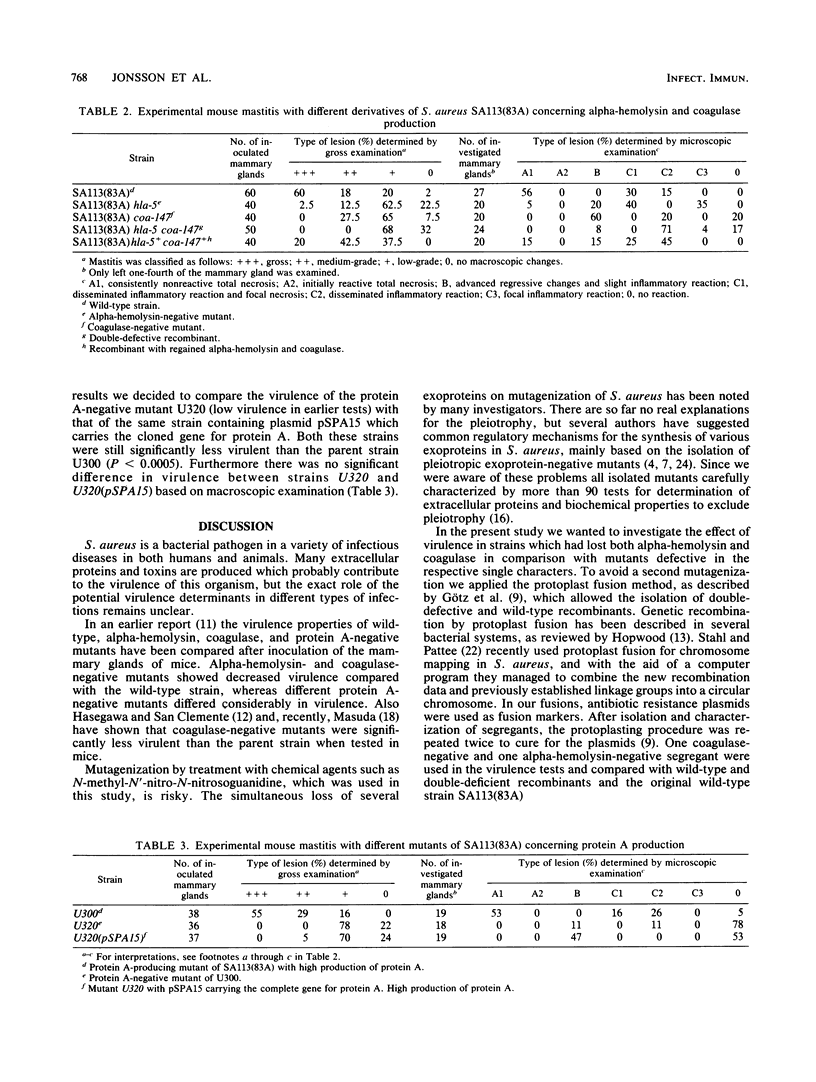
Mutants of a genetically well-characterized strain of Staphylococcus aureus [SA113(83A)] were isolated after mutagenization. Alpha-hemolysin- (hla), coagulase- (coa), and protein A- (spa) negative mutants were characterized by more than 90 biochemical tests for production of extracellular proteins and biochemical profile to exclude pleiotropy. Protoplast fusion was then used to isolate double-defective (hla and coa) recombinants and recombinants with regained properties, i.e., production of alpha-hemolysin and coagulase. Studies of such mutants and recombinants in the mouse mastitis model showed that one alpha-hemolysin [SA113(83A) hla-5] and one coagulase-negative [SA113(83A) coa-147] mutant were lower in virulence compared with the wild-type strain SA113(83A). The double-negative mutant SA113(83A) hla-5 coa-147 showed a drastic decline in virulence and only induced very mild changes, as determined by microscopic examinations of infected mammary gland tissue. The recombinant with regained properties, however, was as virulent as the wild-type strain. This suggests that alpha-hemolysin and coagulase are virulence determinants of S. aureus. A high-level protein A-producing mutant (U300) showed the same virulence as the parent strain SA113(83A) in this model. One low virulence protein A-negative mutant (U320) did not markedly increase in virulence when a plasmid containing the cloned gene for protein A (pSPA15) was introduced into this mutant. By these and earlier observations, it seems likely that protein A is not an important virulence determinant in mastitis of mice. The reduced virulence of the protein A-negative mutant U320 compared with the wild-type SA113(83A) may be due to pleiotropic loss of some other unknown virulence determinant(s). Our data confirm earlier findings that pleiotropic changes are common in protein A-negative mutants.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chandler R. L. Experimental bacterial mastitis in the mouse. J Med Microbiol. 1970 May;3(2):273–282. doi: 10.1099/00222615-3-2-273. [DOI] [PubMed] [Google Scholar]
- Forsgren A., Nordström K., Philipson L., Sjöquist J. Protein A mutants of Staphylococcus aureus. J Bacteriol. 1971 Jul;107(1):245–250. doi: 10.1128/jb.107.1.245-250.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GORRILL R. H., MCNEIL E. M. STAPHYLOCOCCAL INFECTION IN THE MOUSE. I. THE EFFECT OF ROUTE OF INJECTION. Br J Exp Pathol. 1963 Aug;44:404–415. [PMC free article] [PubMed] [Google Scholar]
- Gray G. S., Kehoe M. Primary sequence of the alpha-toxin gene from Staphylococcus aureus wood 46. Infect Immun. 1984 Nov;46(2):615–618. doi: 10.1128/iai.46.2.615-618.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz F., Ahrné S., Lindberg M. Plasmid transfer and genetic recombination by protoplast fusion in staphylococci. J Bacteriol. 1981 Jan;145(1):74–81. doi: 10.1128/jb.145.1.74-81.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraldsson I., Jonsson P. Histopathology and pathogenesis of mouse mastitis induced with Staphylococcus aureus mutants. J Comp Pathol. 1984 Apr;94(2):183–196. doi: 10.1016/0021-9975(84)90039-2. [DOI] [PubMed] [Google Scholar]
- Hasegawa N., San Clemente C. L. Virulence and immunity of Staphylococcus aureus BB and certain deficient mutants. Infect Immun. 1978 Nov;22(2):473–479. doi: 10.1128/iai.22.2.473-479.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A. Genetic studies with bacterial protoplasts. Annu Rev Microbiol. 1981;35:237–272. doi: 10.1146/annurev.mi.35.100181.001321. [DOI] [PubMed] [Google Scholar]
- Iordanescu S., Surdeanu M. Two restriction and modification systems in Staphylococcus aureus NCTC8325. J Gen Microbiol. 1976 Oct;96(2):277–281. doi: 10.1099/00221287-96-2-277. [DOI] [PubMed] [Google Scholar]
- Löfdahl S., Guss B., Uhlén M., Philipson L., Lindberg M. Gene for staphylococcal protein A. Proc Natl Acad Sci U S A. 1983 Feb;80(3):697–701. doi: 10.1073/pnas.80.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S. An efficient method for the isolation of a mutant with an extremely low producibility of coagulase from a Staphylococcus aureus strain. Microbiol Immunol. 1983;27(9):801–805. doi: 10.1111/j.1348-0421.1983.tb00645.x. [DOI] [PubMed] [Google Scholar]
- Novick R., Sanchez-Rivas C., Gruss A., Edelman I. Involvement of the cell envelope in plasmid maintenance: plasmid curing during the regeneration of protoplasts. Plasmid. 1980 May;3(3):348–358. doi: 10.1016/0147-619x(80)90048-7. [DOI] [PubMed] [Google Scholar]
- Sako T., Sawaki S., Sakurai T., Ito S., Yoshizawa Y., Kondo I. Cloning and expression of the staphylokinase gene of Staphylococcus aureus in Escherichia coli. Mol Gen Genet. 1983;190(2):271–277. doi: 10.1007/BF00330650. [DOI] [PubMed] [Google Scholar]
- Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983 May-Jun;22(2-3):181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- Stahl M. L., Pattee P. A. Confirmation of protoplast fusion-derived linkages in Staphylococcus aureus by transformation with protoplast DNA. J Bacteriol. 1983 Apr;154(1):406–412. doi: 10.1128/jb.154.1.406-412.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Guss B., Nilsson B., Götz F., Lindberg M. Expression of the gene encoding protein A in Staphylococcus aureus and coagulase-negative staphylococci. J Bacteriol. 1984 Aug;159(2):713–719. doi: 10.1128/jb.159.2.713-719.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]


