Abstract
Expression of the Bacillus subtilis trpEDCFBA operon is regulated by the interaction of tryptophan-activated TRAP with 11 (G/U)AG trinucleotide repeats that lie in the leader region of the nascent trp transcript. Bound TRAP prevents folding of an antiterminator structure and favors formation of an overlapping intrinsic terminator hairpin upstream of the trp operon structural genes. A 5′-stem–loop (5′SL) structure that forms just upstream of the triplet repeat region increases the affinity of TRAP–trp RNA interaction, thereby increasing the efficiency of transcription termination. Single-stranded nucleotides in the internal loop and in the hairpin loop of the 5′SL are important for TRAP binding. We show here that altering the distance between these two loops suggests that G7, A8, and A9 from the internal loop and A19 and G20 from the hairpin loop constitute two structurally discrete TRAP-binding regions. Photochemical cross-linking experiments also show that the hairpin loop of the 5′SL is in close proximity to the flexible loop region of TRAP during TRAP–5′SL interaction. The dimensions of B. subtilis TRAP and of a three-dimensional model of the 5′SL generated using the MC-Sym and MC-Fold pipeline imply that the 5′SL binds the protein in an orientation where the helical axis of the 5′SL is perpendicular to the plane of TRAP. This interaction not only increases the affinity of TRAP–trp leader RNA interaction, but also orients the downstream triplet repeats for interaction with the 11 KKR motifs that lie on TRAP's perimeter, increasing the likelihood that TRAP will bind in time to promote termination.
Keywords: transcription attenuation, RNA binding protein, RNA structure, photochemical cross-linking, molecular modeling
INTRODUCTION
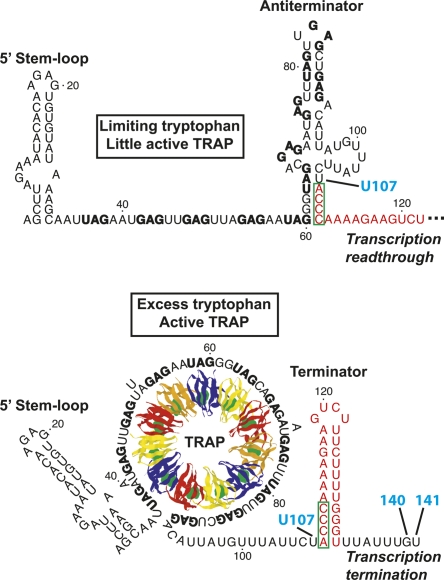
Protein–RNA interactions are important components of a large number of regulatory mechanisms. Expression of the Bacillus subtilis trpEDCFBA operon is regulated by interaction of the RNA binding protein TRAP with the nascent trp operon leader transcript in a transcription attenuation mechanism (Fig. 1; for review, see Gollnick et al. 2005). A hallmark of this attenuation mechanism is the presence of overlapping antiterminator and terminator structures that can form in the nascent trp leader transcript. When a sufficient level of intracellular tryptophan is present, TRAP is activated and binds to 11 equivalently spaced (G/U)AG triplet repeats, six of which lie within the antiterminator structure. Thus, bound TRAP prevents formation of the antiterminator, thereby favoring formation of the intrinsic terminator hairpin that halts transcription before RNA polymerase (RNAP) can reach the downstream structural genes. Under limiting tryptophan conditions, TRAP is not activated and does not bind to the nascent trp transcript. In this case, the antiterminator hairpin forms, which promotes transcription readthrough into the trp operon structural genes by preventing formation of the terminator hairpin. RNAP pausing at U107, the nucleotide just preceding the critical overlap between the antiterminator and terminator structures, participates in the attenuation mechanism, presumably by providing additional time for TRAP to bind to the nascent trp leader transcript (Yakhnin and Babitzke 2002; Yakhnin et al. 2006a).
FIGURE 1.
Model of the B. subtilis trpEDCFBA operon transcription attenuation mechanism. (Top) When tryptophan is limiting, TRAP is not activated and therefore does not bind to the nascent trp leader transcript. Thus, the default conformation of the trp leader RNA, which includes the 5′SL and antiterminator structures, allows transcriptional readthrough. (Bottom) When tryptophan is in excess, TRAP becomes activated by tryptophan and binds to the triplet repeats in the nascent transcript. Bound TRAP prevents formation of the antiterminator and favors formation of an intrinsic terminator hairpin, which stops RNA synthesis at G140 or U141, before the transcribing RNAP reaches the coding sequence of the downstream structural genes. Since the (boxed nucleotides) antiterminator and terminator structures overlap, the two structures are mutually exclusive. Pausing of RNAP at U107, the nucleotide just preceding the 4-nt overlap between the antiterminator and terminator structures, presumably allows more time for TRAP to bind to the nascent transcript and promote termination (Yakhnin and Babitzke 2002; Yakhnin et al. 2006a). (Red) Nucleotides that form the terminator hairpin; (bold type) the (G/U)AG triplet repeats.
TRAP also regulates translation of trpE, the first gene in the trp operon. TRAP binding to trp operon readthrough transcripts promotes formation of the trpE Shine–Dalgarno (SD) sequestering hairpin; formation of this structure inhibits TrpE synthesis by preventing ribosome binding (Merino et al. 1995; Du and Babitzke 1998). In the absence of bound TRAP, an alternative RNA secondary structure forms such that the trpE SD sequence is single-stranded and available for ribosome binding. Interestingly, RNAP pausing at U144, which is 3–4 nucleotides (nt) downstream from the sites of transcription termination, participates in this translation control mechanism, presumably by providing a second opportunity for TRAP to bind to the nascent transcript. As translation of trpE and trpD are coupled, formation of the trpE SD-sequestering hairpin regulates trpD expression as well. Formation of the trpE SD-sequestering hairpin also results in transcriptional polarity, leading to reduced expression of the downstream genes (Yakhnin et al. 2001). In addition, the anti-TRAP protein is expressed when the cellular level of charged tRNATrp is low (Valbuzzi et al. 2002; Chen and Yanofsky 2003). This protein appears to bind the KKR motifs of tryptophan-activated TRAP, thereby blocking the interaction between TRAP and the trinucleotide repeats (Chen and Gollnick 2008). Thus, anti-TRAP could influence both the attenuation and trpE translation control mechanisms.
TRAP consists of 11 identical subunits arranged in a ring structure, with the hydrophobic tryptophan binding pockets positioned between adjacent subunits (Fig. 1; Antson et al. 1995; Babitzke and Yanofsky 1995). NMR studies have suggested that the conformation of the loop region above the tryptophan binding pocket is dynamic in the absence of bound tryptophan (McElroy et al. 2002; Heddle et al. 2007). Binding of tryptophan induces a structural reordering of 11 KKR RNA-binding motifs in TRAP, allowing each motif to interact with a single trinucleotide repeat in the leader transcript (Yang et al. 1997; Antson et al. 1999; McElroy et al. 2006). The interaction between these triplets and TRAP has been well-characterized. The crystal structure of Bacillus stearothermophilus TRAP complexed with a synthetic RNA target indicates that Lys37, Lys56, and Arg58 (KKR) form hydrogen bonds with the A and the G residues in the second and third positions of the triplet repeat (Antson et al. 1999; Elliott et al. 1999). Gly18 and Asp39 form additional hydrogen bonds with the triplet repeats, and Phe32 likely participates in a stacking interaction (Hopcroft et al. 2004). Few contacts form between TRAP and the first nucleotide of the triplet, which explains the sequence variability at this position. No direct interactions occur between TRAP and the nucleotides separating the triplet repeats, although the sequence of these spacers can influence TRAP binding (Babitzke et al. 1995, 1996; Hopcroft et al. 2004). Since the KKR motifs encircle TRAP, the single-stranded triplet repeat region wraps around the perimeter of the protein upon binding (Fig. 1). The triplets at the 5′-end of the full-length TRAP binding site have been shown to associate first (Barbolina et al. 2005), and the crystal structure suggests that this interaction proceeds in a clockwise direction (Antson et al. 1999), with the flexible loop region of TRAP considered to be the top of the protein.
The intracellular concentration of TRAP in B. subtilis has been estimated to be ∼80 nM (200–400 molecules/cell) (McCabe and Gollnick 2004). In vitro, the binding affinity of TRAP for the triplet repeat region of trp RNA is ∼0.1 nM (Baumann et al. 1996). While these values suggest that the intracellular concentration of TRAP is sufficient to bind trp leader RNA, TRAP must bind rapidly to prevent formation of the antiterminator. A 5′ stem–loop (5′SL) structure that forms just upstream of the triplet repeat region (Fig. 1) also interacts with TRAP (Sudershana et al. 1999; Du et al. 2000). The 5′SL functions in the attenuation mechanism by increasing the affinity of TRAP for trp leader RNA when the transcript contains nine or fewer repeats, thus increasing the likelihood that TRAP will bind to the nascent transcript in time to promote termination (McGraw et al. 2007). Deletion or mutation of the 5′SL results in decreased termination efficiency and, therefore, higher expression of the trp operon. Nucleotides G7, A8, and A9 from the internal loop, as well as A19 and G20 from the hairpin loop, have been implicated in the mechanism of TRAP–5′SL interaction (McGraw et al. 2007). These nucleotides are strongly conserved in 5′SL structures among organisms closely related to B. subtilis, raising the possibility that the 5′SL participates in the trp operon attenuation mechanisms in these organisms as well. However, with the exception of Bacillus pumilus (Hoffman and Gollnick 1995) and Bacillus halodurans (Szigeti et al. 2004), the presumed transcription attenuation mechanisms of these other organisms have not been experimentally characterized.
Although 5′SL nucleotides important to TRAP binding have been identified in B. subtilis, little is known about the mechanism of TRAP–5′SL interaction. All available crystal structures of the TRAP–RNA complexes have been solved with synthetic RNA targets lacking the 5′SL. To develop a detailed mechanistic model of TRAP–5′SL interaction in B. subtilis, we characterized a set of 5′SL mutants. Footprinting and filter binding assays performed on these mutant transcripts suggest that the important nucleotides from the hairpin loop and the internal loop comprise two distinct TRAP-binding regions, and that the spacing between these regions is critical for TRAP–5′SL interaction. Results from photochemical cross-linking experiments indicate that during TRAP–5′SL interaction, the hairpin loop is near His34 and His51, which are in the flexible loop region of the protein. Additionally, the MC-Fold and MC-Sym structure prediction algorithms (Parisien and Major 2008) allowed us to develop a three-dimensional (3D) model of TRAP–5′SL interaction. This model has mechanistic implications for how the 5′SL increases the efficiency of TRAP-dependent transcription termination in the trp operon leader region.
RESULTS
The length of the 5′SL upper stem is critical for TRAP binding
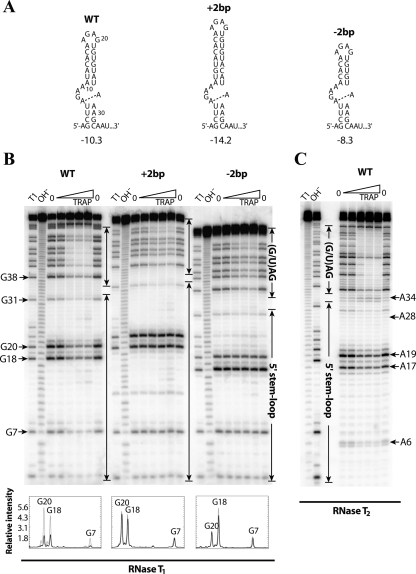
The B. subtilis 5′SL consists of a 4-base pairs (bp) lower stem, a 4 × 1 asymmetric internal loop, a 7-bp upper stem, and a 4-nt hairpin loop at the apex of the structure (Fig. 2A). Earlier structure mapping experiments that used chemical cleavage reagents (Du et al. 2000) suggested that the U5-A29 and A16-U21 base pairs, closing base pairs in the internal loop and in the hairpin loop, respectively, do not form. However, structure mapping experiments conducted with RNase T2 (Fig. 2C) and RNase A (data not shown), as well as the Mfold (Zuker 2003) and MC-Fold (Parisien and Major 2008) structure prediction algorithms, suggest that these base pairs do form. It is therefore likely that these closing base pairs breathe, which is reasonable given that they are A-U base pairs. The U5-A29 and A16-U21 base pairs are included in the 5′SL secondary structures shown here.
FIGURE 2.
In vitro characterization of 5′SL upper stem mutants. (A) Secondary structures of the wild-type (WT) and mutant 5′SL structures with an (+2bp) insertion or (−2bp) deletion of 2 bp in the upper stem. The predicted free energy value (in kilocalories per mole, kcal/mol) at 37°C (Zuker 2003) is shown below each structure. (Dashed line) The potential noncanonical A-A base pair within the internal loop. (B) RNase T1 footprinting of the WT 5′SL and of the upper-stem-length mutants at various TRAP concentrations. The transcripts contained five downstream triplet repeats. Compared to the WT transcript, G18 and G20 are 2 nt longer or shorter in the +2bp and −2bp mutant transcripts, respectively. A densitometry analysis of the bands corresponding to G7, G18, and G20 from the WT and mutant 5′SL structures is included below the gel. (Gray lines) The relative intensity of the annotated nucleotides in the absence of TRAP; (black lines) the relative intensity of these nucleotides at the highest TRAP concentration that was investigated (2500 nM). Band assignments are numbered relative to the WT 5′SL. (C) RNase T2 footprinting of the WT 5′SL with eight downstream triplet repeats. (B,C) Important residues are labeled to the side of the gels. (Enclosed arrows) Nucleotides corresponding to the (G/U)AG triplet repeat region and the 5′SL. Denaturing RNase T1 digestion (T1) and base hydrolysis (OH−) ladders are shown.
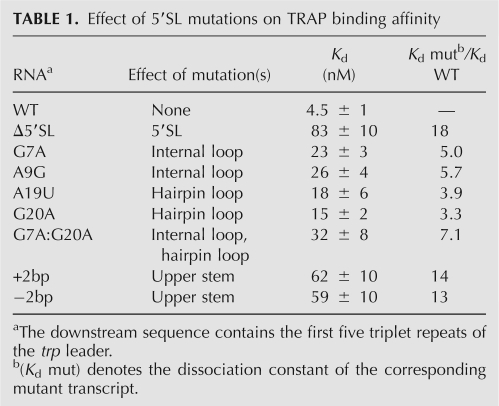
Previous mutagenesis studies showed that single-stranded nucleotides in the internal loop and the hairpin loop are important for TRAP binding (McGraw et al. 2007). To develop a structural model for TRAP–5′SL interaction, transcripts containing additional 5′SL mutations were characterized. Filter binding assays were used to measure apparent dissociation constants (K d) of TRAP–RNA interactions with mutant transcripts (Table 1). The downstream sequence contained five triplet repeats, as this was previously shown to be the number of repeats in which the 5′SL has its greatest influence on TRAP–trp RNA interaction while maintaining moderate affinity (McGraw et al. 2007). As previously observed, the G7A and A9G mutations in the internal loop and the A19U and G20A mutations in the hairpin loop decreased the affinity of TRAP–RNA interaction by three- to sixfold (Table 1). Moreover, the affinity of TRAP for the G7A:G20A double-mutant RNA, with changes in the internal and hairpin loops, was lower than either mutation alone, suggesting that these nucleotides are part of two discrete TRAP-binding regions.
TABLE 1.
Effect of 5′SL mutations on TRAP binding affinity
The length of the upper stem is strongly conserved among organisms containing a trp leader 5′SL, particularly those with B. subtilis-like 5′SL structures (McGraw et al. 2007). To examine the importance of the upper stem length in the B. subtilis 5′SL, RNAs containing the 5′SL with deletion or insertion of 2 bp in the upper stem (Fig. 2A, denoted as +2bp and −2bp, respectively) were generated and characterized in vitro. These mutant transcripts were designed such that the remainder of the 5′SL structure, including the identity of the closing base pairs of the internal and hairpin loops, was maintained, which was confirmed experimentally (see below). Strikingly, shortening or lengthening the upper stem by only 2 bp—corresponding to a helical rise of ∼6.6 Å in A-form dsRNA—reduced the affinity of TRAP–RNA interaction by 13-fold to 14-fold (Table 1). The effect of the +2bp and −2bp mutations was comparable to the 5′SL deletion, which resulted in an 18-fold decrease in affinity. Taken together, our filter binding results suggest that nucleotides in the hairpin loop and internal loop form two distinct TRAP-binding regions that need to be properly spaced and/or oriented.
To gain further insight into the importance of the 5′SL upper stem in TRAP interaction, footprinting assays using RNase T1, which cleaves RNA on the 3′ side of single-stranded G residues, were conducted on the upper stem mutants (Fig. 2B). This nuclease was chosen because G7 and G20 are unpaired and each lies within one of the two predicted TRAP-binding regions. In the absence of TRAP, only nucleotides that were predicted to be single-stranded were cleaved, consistent with the expected secondary structures for wild type (WT) and the mutants shown in Figure 2A. While G7, G18, and G20 were protected by bound TRAP in the WT 5′SL, no protection of the corresponding nucleotides was observed for the +2bp and −2bp mutant transcripts. Lack of RNA protection by TRAP is consistent with their greatly decreased affinity for TRAP (Table 1). Finally, TRAP-dependent protection of the downstream triplet repeat region was observed in all three cases, confirming that TRAP was bound to each of these transcripts, even when protection of the 5′SL was lost. However, compared to the WT transcript, protection of the triplet repeat region was not as strong for either mutant RNA, which reflects weaker binding that results from loss of TRAP–5′SL interaction in the background of five triplet repeats.
The hairpin loop is in close proximity to His34 and His51 of bound TRAP
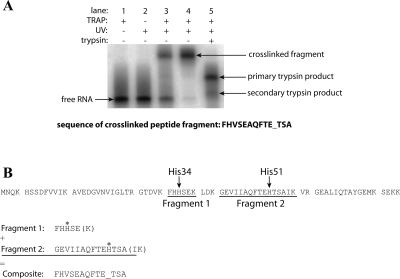
While the results described above indicate that two discrete regions of the 5′SL are important for TRAP interaction, it was unknown how they might interact with the protein. These two regions comprise G7, A8, and A9 from the internal loop and A19 and G20 from the hairpin loop. To determine where these nucleotides are positioned when the 5′SL is bound to TRAP, we conducted photochemical cross-linking experiments with trp leader RNAs in which U5 or U21, which are adjacent to the internal loop or the hairpin loop, respectively, was replaced with 5-iodouracil (5IU). This photoionizable uridine analog has been shown to cross-link to proteins with high specificity and typically has minimal effects on affinity (Willis et al. 1993; Stump and Hall 1995; Meisenheimer and Koch 1997). Moreover, the conservative U-to-5IU substitution ensured that the mutant 5′SL structures were as close to WT as possible.
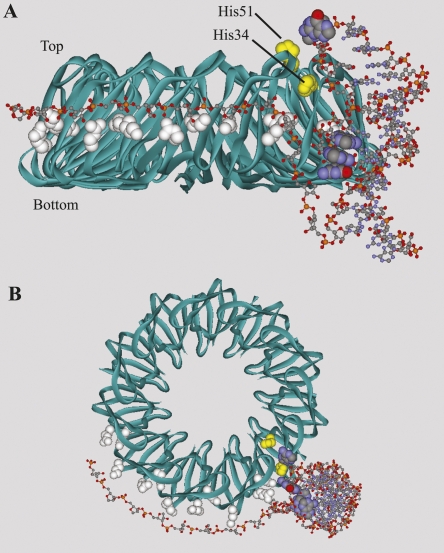
The modified RNAs were incubated with TRAP and then exposed to UV light (Fig. 3A). Free tryptophan was found to quench the cross-linking reaction (data not shown); thus, the T30V mutant of TRAP, which binds trp RNA in the absence of tryptophan (Yakhnin et al. 2000; Payal and Gollnick 2006), was used. Cross-linking was not observed with the RNA containing the 5IU substitution at U5. However, a cross-linking efficiency of 56% was achieved with the RNA containing the 5IU substitution at U21 (Fig. 3A), which was well above the nonspecific background cross-linking (∼5%) observed with the unmodified transcript. The cross-linked material was purified by denaturing PAGE, digested with trypsin, and re-purified by denaturing PAGE. N-terminal sequencing of the gel-purified material revealed that it was a mixture of two cross-linked peptide fragments; the modified U21 residue had cross-linked to His34 and His51 (Fig. 3B). Both of these histidine residues lie in the flexible loop region above the tryptophan binding pocket (Fig. 4, yellow space fill). These results suggest that neighboring A19 and G20, the two nucleotides in the hairpin loop that are known to be important for TRAP binding, are positioned near His34 and His51. It should be noted that our results only indicate that U21 is in close proximity to these two residues during TRAP–5′SL interaction, and do not necessarily implicate His34 or His51 in contacting the 5′SL. A second point of protein–RNA contact can be inferred between the first triplet repeat, which is separated from the base of the 5′SL by 3 nt, and a KKR motif on the perimeter of TRAP (Fig. 1).
FIGURE 3.
Photochemical cross-linking of U21 5IU-substituted RNA. (A) Denaturing PAGE separation of cross-linked macromolecules. (Lane 1) U21 5IU-substituted RNA incubated with T30V TRAP; (lane 2) U21 5IU-substituted RNA exposed to UV light; (lane 3) U21 5IU-substituted RNA incubated with T30V TRAP and exposed to UV light; (lane 4) purified cross-linked protein–RNA complex; (lane 5) partial trypsin digest of the purified cross-linked fragment. As TRAP is highly stable, trypsin digests of the purified cross-linked fragment under standard reaction conditions yielded incomplete cleavage of the peptide fragment (lane 5, primary trypsin product band). When the reaction was conducted under more denaturing conditions (see Materials and Methods; data not shown), the secondary trypsin product became the predominant species (lane 5, secondary trypsin product band). This secondary trypsin product fragment was used in the N-terminal sequencing reaction. The resulting sequence of the cross-linked peptide fragment is shown below the gel. Amino acids involved in chemical cross-links appear as blank cycles (underscored) during the sequencing reaction because the large RNA molecule covalently attached to the side chain interferes with the sequencing analysis. (B) Alignment of the cross-linked peptide fragment(s) with TRAP. The sequence of TRAP is shown broken into tryptic fragments. The results indicate that instead of cross-linking to a single residue, the photoactive nucleobase reacted with two separate amino acids (asterisks). These residues are His34 in Fragment 1 and His51 in Fragment 2. As the N-terminal sequencing reaction is indiscriminant, the instrument defaulted to amino acids in the more abundant fragment (Fragment 1) at a given position. When one of these amino acids was cross-linked (His34), the residue from the less abundant fragment (Fragment 2) was detected instead (Val43). When the end of the shorter fragment (Fragment 1) was reached, the instrument detected amino acids from the less abundant but longer fragment (Fragment 2). The cross-linked amino acid in this region (His51) appeared as a blank cycle. The C-terminal amino acids (shown in parentheses) of each fragment were not detected in the sequencing reaction.
FIGURE 4.
Three-dimensional model of TRAP–5′SL interaction. The MC-Sym-predicted tertiary structure of the 5′SL was merged with the crystal structure of B. subtilis TRAP (PDB ID 1WAP) and manually positioned in the orientation most consistent with all known experimental data. (Yellow spacefill) The two histidine residues that cross-linked to U5; (white spacefill) four KKR motifs; (standard spacefill) the two TRAP-binding domains of the 5′SL. While the histidine cross-linking site is highlighted in only one of the TRAP subunits, TRAP contains 11 equivalent 5′SL binding sites. The unstructured sugar-phosphate backbone downstream from the 5′SL was constructed manually and positioned to mimic binding of the triplet repeat region, as observed in the B. stearothermophilus TRAP-RNA cocrystal structure (PDB ID 1GTF). Specific modeling of KKR–triplet repeat interactions was not attempted. (A) Side view of the TRAP–5′SL complex. The 5′SL interacts with TRAP in an orientation where the helical axis of the RNA is perpendicular to the toroidal plane of TRAP. The three nucleotides within the internal loop that are known to be important for TRAP binding (G7, A8, and A9) are shown stacked inside the RNA duplex. These nucleotides are unpaired, however, and are free to flip outward and interact with the protein. (B) Top-down view of the TRAP–5′SL complex.
In an effort to identify specific amino acid–nucleotide (AA-NT) contacts formed during TRAP–5′SL interaction, we constructed a series of TRAP mutants and measured their affinity for transcripts containing a WT or mutant 5′SL. Based on phylogenetic conservation, 3D location, and chemical favorability for interaction, certain amino acids that seemed most likely to interact with 5′SL nucleotides were mutated to alanine. WT and mutant proteins were analyzed in a double-mutant cycle with WT and mutant RNAs by filter binding. The AA-NT pairs investigated were G7-Glu60, G7-Glu69, A19-Thr28, G20-Asp29, and G20-Glu50; however, the corresponding K d values did not support these specific interactions (data not shown). As other amino acids that lie in regions suspected of interacting with the 5′SL are important for binding of tryptophan or the downstream triplet repeats, many other potential TRAP–5′SL AA-NT pairs could not be experimentally characterized.
Molecular modeling of the 5′SL suggests a perpendicular interaction with TRAP
The secondary structure of the B. subtilis 5′SL has been extensively characterized using a combination of enzymatic and chemical probing techniques (Du et al. 2000; McGraw et al. 2007). The data afforded by these assays agree well; however, there is a discrepancy regarding the structure of the internal loop in the absence of bound TRAP. Even though A6, A8, A9, A28, and A29 were modified by dimethyl sulfate (DMS) at N1 (Du et al. 2000), which suggests that they are unpaired, none of these nucleotides were strongly cleaved by the single-stranded, A-specific RNase T2 (Fig. 2C). On the other hand, A17 and A19, the two unpaired A residues in the hairpin loop, were modified by DMS and strongly cleaved by RNase T2. In addition, A16 in the hairpin loop was only weakly cleaved by RNase T2 (Fig. 2C), despite being modified by DMS (Du et al. 2000). Thus, it appears that the A16-U21 closing base pair is only partially formed.
To gain further insight into the structure of the 5′SL and to generate a model for how the 5′SL docks with TRAP, we made use of the MC-Fold and MC-Sym structure prediction algorithms (Parisien and Major 2008). These differ from Mfold in that rather than relying on thermodynamic parameters, they assign and score small RNA building blocks observed in NMR and crystallographic data that best accommodate the sequence of interest. Matching motifs are compiled into two-dimensional (2D) and 3D structures.
Using our enzymatic T1 and T2 reactivity data as folding constraints, MC-Fold predicted a B. subtilis 5′SL secondary structure identical to that of the experimentally determined structure, except for the addition of a noncanonical base pair between A6 and A28 (Fig. 2A). Multiple-sequence predictions among four related 5′SLs were obtained using MC-Cons (Parisien and Major 2008) (see Materials and Methods) and reveal that the GAA bulge is conserved, although in one case (Bacillus licheniformis), it is a UGAA bulge. In addition to B. subtilis, these predictions suggest that the noncanonical A-A base pair is also present in Bacillus amyloliquefaciens. In one of the other two sequences (B. pumilus), a suboptimal structure was selected by MC-Cons that also contains the A-A base pair and an AGU bulge sequence, which preserves most of the exposed electronegative atoms of the GAA bulge (data not shown). Since there are no unpaired A residues within the 3′ side of the internal loop from the remaining organism (B. licheniformis), the 5′SL from B. licheniformis does not have the capacity to form the A-A base pair.
The A6-A28 base pair in the B. subtilis 5′SL involves the Hoogsteen faces of both bases, with hydrogen-bonding of A6N6 to A28N7 and A28N6 to A6N7. Interestingly, the unconventional geometry associated with this Hoogsteen trans base pair may explain the apparent discrepancies between the enzymatic and chemical cleavage data. As this base pair leaves the N1 of both A residues exposed to solvent, they are susceptible to DMS modification (Du et al. 2000); however, Hoogsteen base-pairing between these nucleotides would appear to prevent cleavage by RNase T2 (Fig. 2C). In addition, with the A6-A28 base pair, a compact internal loop could make A8 and A9 inaccessible to the larger ribonuclease, but accessible to the small DMS molecule. Taken together, the biochemical probing and computer modeling suggest that the internal loop region of the 5′SL is more structured than anticipated. Instead of a 4 × 1 asymmetric internal loop, it is possible that this region consists of a 3-nt GAA bulge, as also predicted in B. amyloliquefaciens and the best free-energy structure of the 5′SL from B. pumilus.
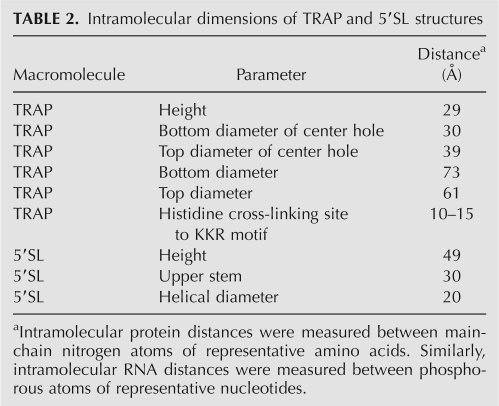
The MC-Fold-predicted secondary structure was imported into MC-Sym to build a 3D model of the 5′SL (Fig. 4). The dimensions of this computer-generated RNA tertiary structure were calculated and compared to those of the B. subtilis TRAP crystal structure (Table 2; Antson et al. 1995). This structural information, along with our biochemical data, was used to exclude or support possible modes of TRAP–5′SL interaction. As shown in Figure 4, our photochemical cross-linking data places U21 in close proximity to His34 and His51. In addition, the ∼180° helical rotation about the 7-bp upper stem of the 5′SL places important nucleotides in the hairpin and internal loops on the same face of the RNA structure, suggesting that these two regions interact with amino acids on the same general surface of TRAP. These constraints, combined with fixing the first triplet repeat to a KKR motif, leaves only a few possible modes of TRAP–5′SL interaction.
TABLE 2.
Intramolecular dimensions of TRAP and 5′SL structures
A first scenario wherein the 5′SL stretches across the top surface of the protein is unlikely because the upper diameter of the center hole (39 Å) is too large to maintain contacts with both TRAP-binding regions of the 5′SL simultaneously, which are separated by just 30 Å (Table 2). It is possible that the seven N-terminal amino acids of TRAP, which were not visible in the crystal structure, could stabilize this orientation; however, this model cannot explain the displacement of the 5′SL by the last few triplet repeats of the TRAP binding site (see Discussion). The lower diameter of the center hole of TRAP is small enough to allow simultaneous interaction with both TRAP-binding regions; however, this second possible orientation would place the hairpin loop on the lower edge of the protein opposite the cross-linking site, which is on the upper edge of TRAP. A third possibility wherein the 5′SL binds amino acids within or near the KKR motifs also seems unlikely. Mutagenic studies showed that potential triplet repeats within the hairpin loop (A16-G18) and internal loop (U5-G7 and A29-G31) do not function as such. In addition, spacer insertions after the 5′SL do not support this type of interaction (McGraw et al. 2007). These nucleotides are therefore not likely positioned near a KKR motif during TRAP–5′SL interaction. Furthermore, this third model would occlude a large number of KKR motifs from triplet repeats in the downstream RNA sequence. A fourth model wherein the 5′SL is positioned in the center hole of TRAP is also unlikely, as the high concentration of acidic residues in this region of the protein makes this interaction electrostatically unfavorable.
A final model of TRAP–5′SL interaction is consistent with the experiments presented here. This model orients the helical axis of the upper stem perpendicular to the plane of TRAP (Fig. 4) and is supported by the fact that the intramolecular distance between the two TRAP-binding regions in the 5′SL (30 Å) is roughly equivalent to the height of the TRAP molecule (29 Å) (Table 2). Moreover, this model positions the downstream triplet repeat region in an orientation that is favorable for interaction with the KKR motifs (Fig. 4) and is also supported by other experimental observations.
DISCUSSION
While the function of the 5′SL in the trp operon transcription attenuation mechanism had been established, little was known about the mechanism of TRAP–5′SL interaction. The experiments described here have allowed us to define a model for this key interaction. It is important to note that the proposed model is based on biochemical data and does not have the same resolution as a cocrystal structure. Nonetheless, this model agrees with all of the experimental data.
The 5′SL contains two discrete TRAP-binding regions that comprise G7, A8, and A9 from the internal loop and A19 and G20 from the hairpin loop. Altering the distance between these two regions by adding or subtracting 2 bp essentially abolished the contribution of the 5′SL to TRAP binding (Fig. 2; Table 1). Computer modeling showed that the nucleotides in these two regions that are important for TRAP binding are on the same face of the 5′SL (Fig. 4) and that the distance between them matches the height of the TRAP molecule (Table 2). The +2bp and −2bp mutant transcripts not only altered the distance between the two 5′SL regions, but also altered the phasing such that they were no longer on the same face of the 5′SL. Also, photochemical cross-linking experiments indicated that the hairpin loop is in close proximity to His34 and His51 (Figs. 3, 4). Additional points of protein-RNA contact are provided by interaction between a KKR motif and the first triplet repeat (Figs. 1, 4), as well as between an unidentified protein surface and the 5′ side of the internal loop. These data suggest that the helical axis of the 5′SL is oriented perpendicular to the plane of the protein (Fig. 4), allowing the two TRAP-binding regions of the 5′SL to interact with amino acid residues that are distinct from the KKR motifs. This model is also supported by a number of previously unexplained experimental observations.
The G7A:G20A double mutant disrupts important nucleotides in both TRAP-binding regions of the 5′SL (Table 1), since the effects of the two individual mutations are cumulative. As these are not the only two nucleotides that interact with TRAP, it is not surprising that this double mutation does not completely negate the effect of the 5′SL on TRAP binding. Remarkably, altering the spacing, and hence the phasing, between the two TRAP-binding regions by deleting or inserting 2 bp reduced the affinity of TRAP–RNA interaction by 13- or 14-fold, respectively (Table 1). These mutations were several times more destabilizing than any of the single point mutants investigated and were similar to the effect of deleting the entire 5′SL (an 18-fold reduction in affinity). Furthermore, RNase T1 footprinting assays showed that these mutations lead to a complete loss of protection of nucleotides in the internal and hairpin loops (Fig. 2B). Thus, altering the spacing and phasing between the two discrete TRAP-binding regions essentially eliminates TRAP–5′SL interaction and, therefore, 5′SL function. Weaker protection of the triplet repeat region in these two mutant transcripts also underscores the importance of the 5′SL to TRAP binding (Fig. 2B).
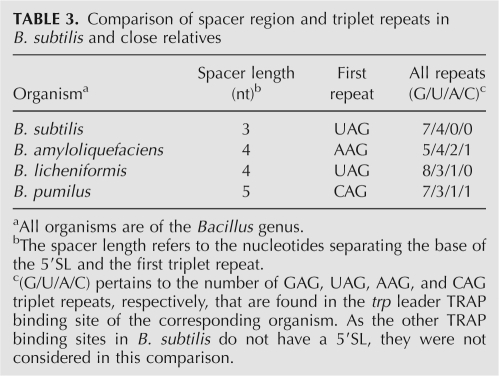
It is interesting to note that among 5′SL structures from organisms closely related to B. subtilis (McGraw et al. 2007), the number of unpaired nucleotides in the internal loop is inversely related to the length of the upper stem, suggesting that a shorter stem can be accommodated if the internal loop region is more flexible. For example, while the B. subtilis 5′SL contains a 7-bp upper stem and a 4 × 1 asymmetric internal loop, the 5′SL from B. pumilus contains an 8-bp upper stem and a 3-nt bulge. This correlation suggests that the distance between the two independent TRAP-binding regions in the 5′SL is strongly conserved, despite variations in upper stem length. As suggested by the prediction of a noncanonical A-A base pair within the internal loop of the B. subtilis 5′SL, it is possible that the 5′SL internal loops from these related organisms are also structured.
Photochemical cross-linking experiments indicate that U21, which is adjacent to the functionally important nucleotides A19 and G20, is in close proximity to His34 and His51 during TRAP–5′SL interaction (Fig. 3). These histidines are 10 to 15 Å from the KKR motifs, excluding the possibility of G18, A19, and G20 acting as a triplet repeat, consistent with earlier biochemical data (McGraw et al. 2007). Instead, it is likely that A19 and G20 interact with amino acids from an RNA binding surface near the two cross-linked histidine residues (Fig. 4). Unfortunately, strategic attempts to identify these amino acids using site-directed mutagenesis were unsuccessful.
All attempts to cross-link the U5 5IU-substituted RNA to TRAP were unsuccessful. This is most likely caused by the lack of a suitable amino acid in the vicinity of U5, rather than lack of interaction with TRAP. Consistent with this interpretation, none of the amino acids that commonly cross-link with 5IU (histidine, tyrosine, methionine, and phenylalanine) are found near U5 in the proposed model of TRAP–5′SL interaction shown in Figure 4 (data not shown). Also, U5 is pointed away from the protein in this model (Fig. 4A). Another point of protein–RNA contact occurs between U36, A37, and G38, which make up the first triplet repeat just downstream from the 5′SL, and Lys37, Lys56, and Arg58, which are the three amino acids that form the KKR motifs (Fig. 1; Antson et al. 1999). As the triplet repeat–KKR interaction is responsible for preventing formation of the antiterminator, it is likely that the lower stem, which is separated from the first triplet repeat by only 3 nt, is in the vicinity of the KKR region of TRAP.
On the basis of the intramolecular dimensions of TRAP and the MC-Sym-derived 5′SL tertiary structure, a model of TRAP–5′SL interaction was generated. In this model, the helical axis of the 5′SL is perpendicular to the toroidal plane of TRAP (Fig. 4). As the two TRAP-binding regions of the 5′SL lie on the same face of the RNA structure, this orientation allows both regions to bind the protein simultaneously. Moreover, the distance between the two TRAP-binding regions is sufficient to span the side of the protein (Table 2). Although not probed experimentally, nonspecific interactions between phosphate groups in the upper stem of the 5′SL and the abundant lysine and arginine residues on the side of TRAP could further stabilize this perpendicular orientation (Fig. 4A).
Downstream, the 3-nt spacer between the 5′SL and the first triplet repeat (UAG) may loop upward to allow the triplet to dock easily with a KKR motif. Although GAG repeats are preferred, UAG repeats, such as in the first triplet repeat, occur when additional flexibility of the RNA is beneficial (Hopcroft et al. 2004). This correlation is also observed with the fifth (UAG) and sixth (UAG) triplet repeats, which flank a suboptimal GG spacer. Flexibility for the first triplet repeat is supported experimentally in that G38 in the first triplet repeat is not as strongly protected by TRAP as the other triplet repeat nucleotides (Fig. 2B, WT panel). In addition, the trp leaders from organisms closely related to B. subtilis contain a slightly longer spacer (4 to 5 nt) and a non-GAG triplet repeat following the 5′SL (Table 3), further suggesting the flexibility of this region. These RNA features are not found in the leaders of some of the other 5′SL-containing organisms that are more distantly related to B. subtilis, such as Bacillus stearothermophilus, Bacillus caldotenax, and Geobacillus kaustophilus. Furthermore, the 5′SL in these three organisms lack the conserved residues in the internal and hairpin loops (McGraw et al. 2007), suggesting that these structures may not contact TRAP. The potential lack of TRAP–5′SL interaction in these organisms might be compensated for by the presence of more than 11 triplet repeats in their respective trp leaders.
TABLE 3.
Comparison of spacer region and triplet repeats in B. subtilis and close relatives
A perpendicular interaction between TRAP and the 5′SL is also supported by experimental observations that previously could not be explained mechanistically (McGraw et al. 2007). Since the proposed 5′SL binding site overlaps one or two KKR motifs (Fig. 4A), the 5′SL would have to dissociate from the protein to accommodate the last few triplet repeats. In agreement with this interpretation, when the downstream sequence extends to 10 or 11 triplet repeats, the 5′SL no longer affects TRAP affinity, and a complete loss of TRAP-mediated protection of the 5′SL is observed (Du et al. 2000; McGraw et al. 2007). These observations suggest that the 5′SL is displaced by the 3′-end of the triplet repeat region, which support the model in Figure 4. Importantly, the models in which the 5′SL binds across the top or bottom surface of TRAP or binds in the center hole of TRAP cannot explain these displacement data. In addition, footprinting of the trp leader with RNase T2 showed that TRAP protects most of the unpaired A residues in the 5′SL and triplet repeat region. However, A33 and A34, which lie in the 3-nt spacer between the 5′SL and the first triplet, exhibit enhanced cleavage by this nuclease (Fig. 2C). Enhanced cleavage in the presence of TRAP is consistent with looping of these nucleotides away from the 5′SL and the protein to allow the first triplet repeat to bind a nearby KKR motif (Fig. 4). Similarly, it was shown that increasing the length of this linker region up to 13 nt had no effect on trp operon expression (McGraw et al. 2007), which is consistent with a simple tethering role.
Surface plasmon resonance (SPR) studies were also conducted to determine whether the 5′SL directly influenced the kinetics of TRAP binding to trp leader RNA. The rate of association between TRAP and RNA was the same with and without the 5′SL for a given number of downstream repeats (data not shown). These studies thus suggest that the 5′SL does not directly affect the on-rate of TRAP; however, the 5′SL increases the affinity of TRAP for trp leader RNA, thereby increasing the efficiency of termination (McGraw et al. 2007). This observation is consistent with the finding that the rate of TRAP binding is critical for the attenuation mechanism (Barbolina et al. 2007). Since the on-rate is unaffected by the 5′SL, the 5′SL appears to function by allowing TRAP to bind to shorter transcripts, and therefore earlier during transcription.
The anti-TRAP protein, which is expressed when the cellular level of charged tRNATrp is low, appears to block TRAP–RNA interaction by binding to the KKR motifs of tryptophan-activated TRAP (Valbuzzi et al. 2002; Chen and Yanofsky 2003; Chen and Gollnick 2008). Since the 5′SL-binding site on TRAP also overlaps one or two KKR motifs, the 5′SL may not be able to bind TRAP if the protein molecule was completely bound by anti-TRAP. Thus, it appears that binding of anti-TRAP would prevent binding of the 5′SL, as well as of the downstream triplet repeats. Conversely, since the 5′SL only blocks access to one or two KKR motifs, binding of the 5′SL to TRAP might not prevent interaction of TRAP with anti-TRAP.
In B. subtilis, the trpG, trpP, and ycbK transcripts each contain a TRAP-binding site that overlaps the cognate SD sequence and/or translation initiation region, such that bound TRAP inhibits translation of the associated gene (Du et al. 1997; Yakhnin et al. 2004, 2006b). These TRAP-binding sites do not contain a 5′SL structure. While TRAP only leads to modest regulation of ycbK and trpG, TRAP-mediated regulation of trpP is similar in magnitude to that of the trp operon (Yakhnin et al. 2007), demonstrating that a 5′SL is not required for efficient TRAP-mediated control of gene expression. However, since the timeframe for TRAP binding in the attenuation mechanism is limited (unlike the translational control mechanisms of trpG, trpP, and ycbK), the contribution of the 5′SL to TRAP binding is critical for proper regulation of the trp operon.
What mechanistic details of TRAP–5′SL interaction are responsible for the increased termination efficiency afforded by the 5′SL? One advantage is that TRAP contains 11 equivalent 5′SL-binding sites, which allows the 5′SL to more readily bind the protein. In addition, the helical twist about the lower stem orients the downstream single-stranded region tangentially to TRAP (Fig. 4B), placing the first few triplet repeats near KKR motifs. Binding of the 5′SL also positions the triplet repeats in the required clockwise orientation and favors association with the repeats in the requisite 5′-to-3′ direction (Barbolina et al. 2005). Association of TRAP with the full-length triplet repeat region appears to be mechanistically complex due to self-structure and looping of the RNA (Murtola et al. 2008; data not shown). By ensuring that the first few triplet repeats more easily find the KKR motifs through increased affinity and proper positioning of the RNA, the 5′SL presumably allows the triplet repeats to interact with TRAP as they emerge from RNAP during transcription. The 5′SL therefore increases the likelihood that TRAP will bind the nascent trp transcript in time to promote termination.
MATERIALS AND METHODS
TRAP purification and RNA synthesis
TRAP was purified as previously described (Yakhnin et al. 2000). No modifications to the purification protocol were required for the production of the TRAP mutants. RNA transcripts for in vitro analyses were generated using the RNAmaxx kit (Stratagene). BamHI-linearized plasmid pPB1105 was used as the DNA template for synthesizing the transcript containing the WT 5′SL with five downstream repeats. Plasmids pAM2, pAM20, pAM4, and pAM17 were used to generate transcripts containing the G7A, A9G, A19U, and G20A mutants, respectively. Since these transcripts were produced from BamHI-linearized templates, they contain three additional nucleotides at the 3′ end. RNAs with modifications in the upper stem of the 5′SL and five downstream repeats were generated by Milligan transcription (Milligan and Uhlenbeck 1989). This technique was also used to generate the G7A:G20A mutant transcript. After transcription, reaction mixtures were treated with 1 U of RNase-free DNase (Ambion), followed by phenol/chloroform extraction and ethanol precipitation. RNA was resuspended, treated with alkaline phosphatase (New England Biolabs), and 5′-end-labeled with [γ-32P]ATP (Perkin-Elmer). Labeled RNAs were purified on 12% denaturing gels and quantified by scintillation counting. Prior to analysis, all RNAs were renatured in 50 mM NaCl by heating for 1 min to 90°C, followed by slow cooling to room temperature.
Filter binding, RNA structure mapping, and RNA footprinting assays
Filter binding and footprinting assays were performed as described previously (McGraw et al. 2007). Briefly, 0.5–0.01 nM 5′-end-labeled RNA was renatured by heating for 1 min to 90°C followed by slow cooling to room temperature. Renatured transcripts were incubated for 25 min at 37°C with various concentrations of TRAP in reaction buffer containing 1 mM L-tryptophan, 40 mM Tris-HCl at pH 8.0, 250 mM KCl, 4 mM MgCl2, 7.5%–10% glycerol, and 0.2 mg/mL Escherichia coli tRNA. For RNA structure mapping and footprinting, the reaction buffer contained 6.5 ng/μL yeast RNA instead of E. coli tRNA and 20 μg/μL bovine serum albumin. The TRAP concentrations used in the footprinting assay shown in Figure 2B were 5, 50, 500, and 2500 nM, while the concentrations used in the footprinting assay in Figure 2C were 3, 30, 300, and 1000 nM. Then, 0.04 U of RNase T1 (Roche) or 0.1 U of RNase T2 was added to the reaction mixtures, followed by incubation for 15 min at 37°C. The previously reported structure mapping experiments using chemical cleavage reagents (Du et al. 2000) were also conducted at 37°C. Reactions were halted with the addition of 2× loading buffer (95% formamide, 0.2% SDS, 20 mM EDTA, 0.025% bromophenol blue, and 0.025% xylene cyanol). Samples of each reaction were electrophoresed on 12% denaturing sequencing gels and imaged using a PhosphorImager (Typhoon 8600 Variable Mode Imager; Molecular Dynamics). After incubation, filter-binding reactions were blotted onto 0.2-μm nitrocellulose (Whatman) and Hybond N+ (Amersham) membranes and imaged using a PhosphorImager. Spots corresponding to each protein concentration were integrated using Imagequant 5.2 (Molecular Dynamics) and fit to the binding equation f bound = f max[TRAP]/([TRAP] + K d); where f max is the maximum fraction bound and K d is the dissociation binding constant. The addition of a Hill coefficient did not significantly alter the K d values or improve the quality of the fit and was excluded.
Photochemical cross-linking
RNAs containing five downstream triplet repeats and the 5′SL with a 5-iodouracil (5IU) substitution at U5 or U21 were purchased from Dharmacon. All manipulations prior to UV exposure were performed in reduced light. A total of 10 nmol of RNA was renatured by heating for 1 min to 90°C in 50 μL of 1× TEN50 buffer (1 mM Tris-HCl at pH 7.5, 0.1 mM EDTA, 50 mM NaCl), followed by slow cooling to room temperature. An additional 5 μL of 0.1 μM 5′-end-labeled RNA was included in the reaction as a tracer. The reaction volume was increased to 800 μL with the addition of 25 mM sodium cacodylate (pH 7.5) and a 1.2 molar excess of T30V TRAP. The T30V mutant, which binds trp leader RNA in the absence of tryptophan (Yakhnin et al. 2000; Payal and Gollnick 2006), was used in the cross-linking procedure because free tryptophan quenched the cross-linking reaction. The final concentrations of RNA (12.5 μM) and T30V TRAP (15 μM) were well above the K d of the T30V-trp leader RNA complex (∼15 nM). The reaction mixture was incubated for 25 min at 37°C, then placed on ice and exposed to UV light. Irradiation was conducted with a handheld UV lamp equipped with an 8-W Spectroline UV-B bulb (λmax = 312 nm) at a distance of ∼2 cm for 30 min. A polystyrene Petri dish was placed over the samples to filter out stray radiation, since wavelengths of <300 nm can lead to nonspecific cross-linking. The reaction mixture was concentrated using a 10 kDa molecular weight cutoff (MWCO) Nanosep spin column (Pall Life Sciences) and electrophoresed through a 15% denaturing polyacrylamide gel. The cross-linked fragment was isolated and dialyzed into 100 mM Tris-HCl at pH 8.0, containing 4 M urea using a 3 kDa MWCO Nanosep spin column. This reaction mixture was denatured by heating for 5 min to 94°C and plunging on ice, then brought to a final concentration of 2 M urea with the addition of 100 mM Tris-HCl (pH 8.0). A total of 20 μg of proteomics grade trypsin (Sigma) was added, and the reaction was incubated for 4 h at 37°C. After digestion, the reaction mixture was electrophoresed through a 15% denaturing polyacrylamide gel to isolate the digested peptide–RNA fragment. The extracted sample was concentrated and dialyzed into DEPC-treated water using a 3 kDa MWCO Nanosep column, then submitted for N-terminal sequencing at the Pennsylvania State University Macromolecular Core Facility. Since the amino acids that were cross-linked to the photoionizable nucleotide were covalently linked to a large (∼20 kDa) RNA fragment, they appeared as blank cycles during the sequencing reaction.
Molecular modeling
Intramolecular protein distances in B. subtilis TRAP (PDB ID 1WAP) were determined by measuring the distance between backbone nitrogen atoms of various amino acids using DeepView 3.7 (Guex and Peitsch 1997). RNA secondary structures were modeled with MC-Fold, using data from RNase T1 and RNase T2 structure mapping assays (McGraw et al. 2007) as experimentally determined structural constraints. The most favorable (best free-energy in the context of enzymatic data) B. subtilis 5′SL structure predicted among 1000 suboptimals by MC-Fold is very similar to the experimentally determined structure (Du et al. 2000; McGraw et al. 2007), although the MC-Fold structure includes a noncanonical A-A base pair in the internal loop region. We performed this procedure for each of the four related sequences: B. subtilis, B. pumilis, B. amyloliquefaciens, and B. licheniformis. For each sequence, the best 100 suboptimal predictions were input into MC-Cons (Parisien and Major 2008), a program that identifies the closest secondary structure in each set that minimizes the overall structural distance. The secondary structure topology of all four MC-Cons selected structures is alike, but the B. licheniformis structure differs from the others by the absence of the A-A noncanonical base pair and the presence of a longer lower stem (+2 bp) and a longer bulge loop (+1 nt).
The MC-Cons selected B. subtilis secondary structure is also the best free-energy structure in the context of the enzymatic data. It was input into MC-Sym, resulting in a computer-predicted 3D model of the 5′SL. The dimensions of the RNA structure were measured between phosphorus atoms of the desired nucleotides. The model of the 5′SL was then merged with the crystal structure of B. subtilis TRAP in DeepView 3.7 and DSviewer Pro 5.0 (Discovery Studio Modeling Environment, Release 5.0, Accelerys Software, Inc.) and manually positioned to show the mode of TRAP–5′SL interaction that is most consistent with all known experimental data. The unstructured sugar-phosphate backbone downstream of the 5′SL was added manually and positioned near the KKR motifs to mimic binding of the triplet repeats to TRAP.
ACKNOWLEDGMENTS
We thank Alexander Yakhnin for technical assistance and Anne Stanley for help with N-terminal sequencing. We also thank Paul Gollnick for plasmids used to overexpress several TRAP mutants. This work was supported by grant GM52840 to P.B. and grant GM58709 to P.C.B. from the National Institutes of Health. A.M. holds a Canadian Institutes of Health Strategic Training in Bioinformatics fellowship.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1314409.
REFERENCES
- Antson A.A., Otridge J., Brzozowski A.M., Dodson E.J., Dodson G.G., Wilson K.S., Smith T.M., Yang M., Kurecki T., Gollnick P. The structure of trp RNA-binding attenuation protein. Nature. 1995;374:693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- Antson A.A., Dodson E.J., Dodson G., Greaves R.B., Chen X., Gollnick P. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401:235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Yanofsky C. Structural features of L-tryptophan required for activation of TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis . J. Biol. Chem. 1995;270:12452–12456. doi: 10.1074/jbc.270.21.12452. [DOI] [PubMed] [Google Scholar]
- Babitzke P., Bear D.G., Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a toroid-shaped molecule that binds transcripts containing GAG or UAG repeats separated by two nucleotides. Proc. Natl. Acad. Sci. 1995;92:7916–7920. doi: 10.1073/pnas.92.17.7916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitzke P., Yealy J., Campanelli D. Interaction of the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis with RNA: Effects of the number of GAG repeats, the nucleotides separating adjacent repeats, and RNA secondary structure. J. Bacteriol. 1996;178:5159–5163. doi: 10.1128/jb.178.17.5159-5163.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbolina M.V., Li X., Gollnick P. Bacillus subtilis TRAP binds to its RNA target by a 5′ to 3′ directional mechanism. J. Mol. Biol. 2005;345:667–679. doi: 10.1016/j.jmb.2004.10.071. [DOI] [PubMed] [Google Scholar]
- Barbolina M.V., Kristoforov R., Manfredo A., Chen Y., Gollnick P. The rate of TRAP binding to RNA is crucial for the transcription attenuation control of the B. subtilis trp operon. J. Mol. Biol. 2007;370:925–938. doi: 10.1016/j.jmb.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann C., Otridge J., Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J. Biol. Chem. 1996;271:12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- Chen Y., Gollnick P. Alanine scanning mutagenesis of anti-TRAP (AT) reveals residues involved in binding to TRAP. J. Mol. Biol. 2008;377:1529–1543. doi: 10.1016/j.jmb.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Yanofsky C. Tandem transcription and translation regulatory sensing of uncharged tryptophan tRNA. Science. 2003;301:211–213. doi: 10.1126/science.1084902. [DOI] [PubMed] [Google Scholar]
- Du H., Babitzke P. trp RNA-binding attenuation protein-mediated long distance RNA refolding regulates translation of trpE in Bacillus subtilis . J. Biol. Chem. 1998;273:20494–20503. doi: 10.1074/jbc.273.32.20494. [DOI] [PubMed] [Google Scholar]
- Du H., Tarpey R., Babitzke P. The trp RNA-binding attenuation protein regulates TrpG synthesis by binding to the trpG ribosome binding site of Bacillus subtilis . J. Bacteriol. 1997;179:2582–2586. doi: 10.1128/jb.179.8.2582-2586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Yakhnin A.V., Dharmaraj S., Babitzke P. trp RNA-binding attenuation protein-5′ stem–loop RNA interaction is required for proper transcription attenuation control of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol. 2000;182:1819–1827. doi: 10.1128/jb.182.7.1819-1827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott M.B., Gottlieb P.A., Gollnick P. Probing the TRAP–RNA interaction with nucleoside analogs. RNA. 1999;5:1277–1289. doi: 10.1017/s1355838299991057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollnick P., Babitzke P., Antson A., Yanofsky C. Complexity in regulation of tryptophan biosynthesis in Bacillus subtilis . Annu. Rev. Genet. 2005;39:47–68. doi: 10.1146/annurev.genet.39.073003.093745. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Heddle J.G., Okajima T., Scott D.J., Akashi S., Park S.Y., Tame J.R. Dynamic allostery in the ring protein TRAP. J. Mol. Biol. 2007;371:154–167. doi: 10.1016/j.jmb.2007.05.013. [DOI] [PubMed] [Google Scholar]
- Hoffman R.J., Gollnick P. The mtrB gene of Bacillus pumilus encodes a protein with sequence and functional homology to the trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis . J. Bacteriol. 1995;177:839–842. doi: 10.1128/jb.177.3.839-842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopcroft N.H., Manfredo A., Wendt A.L., Brzozowski A.M., Gollnick P., Antson A.A. The interaction of RNA with TRAP: The role of triplet repeats and separating spacer nucleotides. J. Mol. Biol. 2004;338:43–53. doi: 10.1016/j.jmb.2004.02.038. [DOI] [PubMed] [Google Scholar]
- McCabe B.C., Gollnick P. Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis . J. Bacteriol. 2004;186:5157–5159. doi: 10.1128/JB.186.15.5157-5159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy C., Manfredo A., Wendt A., Gollnick P., Foster M. TROSY-NMR studies of the 91-kDa TRAP protein reveal allosteric control of a gene regulatory protein by ligand-altered flexibility. J. Mol. Biol. 2002;323:463–473. doi: 10.1016/s0022-2836(02)00940-3. [DOI] [PubMed] [Google Scholar]
- McElroy C., Manfredo A., Gollnick P., Foster M.P. Thermodynamics of tryptophan-mediated activation of the trp RNA-binding attenuation protein. Biochemistry. 2006;45:7844–7853. doi: 10.1021/bi0526074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGraw A.P., Bevilacqua P.C., Babitzke P. TRAP-5′ stem–loop interaction increases the efficiency of transcription termination in the Bacillus subtilis trpEDCFBA operon leader region. RNA. 2007;13:2020–2033. doi: 10.1261/rna.719507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenheimer K.M., Koch T.H. Photocross-linking of nucleic acids to associated proteins. Crit. Rev. Biochem. Mol. Biol. 1997;32:101–140. doi: 10.3109/10409239709108550. [DOI] [PubMed] [Google Scholar]
- Merino E., Babitzke P., Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translation as well as transcriptional regulation of the Bacillus subtilis trp operon. J. Bacteriol. 1995;177:6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J.F., Uhlenbeck O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Murtola T., Vattulainen I., Falck E. Insights into activation and RNA binding of trp RNA-binding attenuation protein (TRAP) through all-atom simulations. Proteins. 2008;71:1995–2011. doi: 10.1002/prot.21878. [DOI] [PubMed] [Google Scholar]
- Parisien M., Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- Payal V., Gollnick P. Substitutions of Thr30 provide mechanistic insight into tryptophan-mediated activation of TRAP binding to RNA. Nucleic Acids Res. 2006;34:2933–2942. doi: 10.1093/nar/gkl383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump W.T., Hall K.B. Crosslinking of an iodo-uridine-RNA hairpin to a single site on the human U1A N-terminal RNA binding domain. RNA. 1995;1:55–63. [PMC free article] [PubMed] [Google Scholar]
- Sudershana S., Du H., Mahalanabis M., Babitzke P. A 5′ RNA stem–loop participates in the transcription attenuation mechanism that controls expression of the Bacillus subtilis trpEDCFBA operon. J. Bacteriol. 1999;181:5742–5749. doi: 10.1128/jb.181.18.5742-5749.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szigeti R., Milescu M., Gollnick P. Regulation of the tryptophan biosynthetic genes in Bacillus halodurans: Common elements but different strategies than those used by Bacillus subtilis . J. Bacteriol. 2004;186:818–828. doi: 10.1128/JB.186.3.818-828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valbuzzi A., Gollnick P., Babitzke P., Yanofsky C. The anti-trp RNA-binding attenuation protein (Anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J. Biol. Chem. 2002;277:10608–10613. doi: 10.1074/jbc.M111813200. [DOI] [PubMed] [Google Scholar]
- Willis M.C., Hicke B.J., Uhlenbeck O.C., Cech T.R., Koch T.H. Photocrosslinking of 5-iodouracil-substituted RNA and DNA to proteins. Science. 1993;262:1255–1257. doi: 10.1126/science.7694369. [DOI] [PubMed] [Google Scholar]
- Yakhnin A.V., Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism in vitro. Proc. Natl. Acad. Sci. 2002;99:11067–11072. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin A.V., Trimble J.J., Chiaro C.R., Babitzke P. Effects of mutations in the L-tryptophan binding pocket of the trp RNA-binding attenuation protein of Bacillus subtilis . J. Biol. Chem. 2000;275:4519–4524. doi: 10.1074/jbc.275.6.4519. [DOI] [PubMed] [Google Scholar]
- Yakhnin H., Babiarz J.E., Yakhnin A.V., Babitzke P. Expression of the Bacillus subtilis trpEDCFBA operon is influenced by translational coupling and ρ termination factor. J. Bacteriol. 2001;183:5918–5926. doi: 10.1128/JB.183.20.5918-5926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin H., Zhang H., Yakhnin A.V., Babitzke P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation of the tryptophan transport gene, trpP (yhaG), by blocking ribosome binding. J. Bacteriol. 2004;186:278–286. doi: 10.1128/JB.186.2.278-286.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnin A.V., Yakhnin H., Babitzke P. RNA polymerase pausing regulates translation initiation by providing additional time for TRAP–RNA interaction. Mol. Cell. 2006a;24:547–557. doi: 10.1016/j.molcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Yakhnin H., Yakhnin A.V., Babitzke P. The trp RNA-binding attenuation protein (TRAP) of Bacillus subtilis regulates translation initiation of ycbK, a gene encoding a putative efflux protein, by blocking ribosome binding. Mol. Microbiol. 2006b;61:1252–1266. doi: 10.1111/j.1365-2958.2006.05278.x. [DOI] [PubMed] [Google Scholar]
- Yakhnin H., Yakhnin A.V., Babitzke P. Translation control of trpG from transcripts originating from the folate operon promoter of Bacillus subtilis is influenced by translation-mediated displacement of bound TRAP, while translation control of transcripts originating from a newly identified trpG promoter is not. J. Bacteriol. 2007;189:872–879. doi: 10.1128/JB.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Chen X., Militello K., Hoffman R., Fernandez B., Baumann C., Gollnick P. Alanine-scanning mutagenesis of Bacillus subtilis trp RNA-binding attenuation protein (TRAP) reveals residues involved in tryptophan binding and RNA binding. J. Mol. Biol. 1997;270:696–710. doi: 10.1006/jmbi.1997.1149. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]