Abstract
Antigenic variation by the malaria parasite Plasmodium falciparum results from switches in expression between members of the multicopy var gene family. These genes encode the variant surface protein PfEMP-1, the primary determinant of the antigenic and cytoadherent properties of infected erythrocytes. Only a single var gene is expressed at a time while the remaining members of the family remain transcriptionally silent. How mutually exclusive var gene expression is regulated is poorly understood; however, it is generally thought to involve alterations in chromatin assembly and modification, resulting in a type of cellular memory. Recently, several aspects of the chromatin structure surrounding var genes have been described, in particular the histone modifications associated with the active and silent states of the genes as well as their subnuclear localization. Here, we demonstrate that this chromatin structure also includes the incorporation of long sense and antisense noncoding RNAs. These sterile transcripts initiate from a bidirectional promoter located within a conserved intron found in all var genes that was previously implicated in var gene silencing. Mapping of the 5′ and 3′ ends of the sterile transcripts indicates that they are nonpolyadenylated. RNA fluorescent in situ hybridization (RNA-FISH) analysis detects both the sense and antisense noncoding RNAs in distinct spots within the nucleus similar to the pattern described for the var genes themselves. Further, analysis by RNA chromatin immunoprecipitation (ChIP) indicates that the noncoding RNAs are physically associated with chromatin. These sterile transcripts therefore might act in a manner analogous to noncoding RNAs associated with silent, condensed chromatin found in other eukaryotic systems.
Keywords: chromatin, noncoding RNAs, silencing, transcription
INTRODUCTION
Malaria is a vector-borne infectious disease endemic to tropical and subtropical regions, placing over 40% of the world's population at risk (Snow et al. 2005). Protozoan parasites of the genus Plasmodium, the etiological cause of malaria, are transmitted by Anopheles mosquitoes and undergo a complex multistage life cycle. The clinical symptoms of Plasmodium infection are caused almost exclusively by the asexual erythrocytic stages of the parasite, which multiply inside the host's circulating red blood cells (RBCs). The severe pathogenic effects of Plasmodium falciparum, the most virulent Plasmodium species, are linked to cytoadhesion of infected RBCs to the vascular endothelium, allowing them to avoid clearance by the spleen. In addition, parasites undergo a process of antigenic variation by which they continuously alter the molecules displayed on the surface of infected RBCs, thus enabling them to avoid the antibody response of the host. Antigenic variation by P. falciparum results from switches in expression between the approximately 60 members of the var gene family (Baruch et al. 1995; Su et al. 1995; Smith et al. 1995). These genes encode the variant surface protein Plasmodium falciparum erythrocyte membrane protein 1 (PfEMP-1), the primary determinant of both the antigenic and cytoadherent properties of infected erythrocytes. Only a single var gene is expressed at any given time, while the remaining members of the family remain transcriptionally silent (Scherf et al. 1998).
Several studies provide evidence for an involvement of chromatin remodeling in mediating var gene repression: (1) silencing of var genes is S-Phase dependent, suggesting alterations of chromatin structure (Deitsch et al. 2001); (2) the histone deacetylase Sir2 is involved in repression of subtelomeric var genes (Duraisingh et al. 2005; Freitas-Junior et al. 2005); (3) trimethylation of histones (H3K9) is decreased in active var promoters (Chookajorn et al. 2007; Lopez-Rubio et al. 2007); and (4) acetylation of histones H3 and H4 is associated with active var genes (Freitas-Junior et al. 2005). Furthermore, changes in subnuclear localization have been observed to be associated with var gene activation (Duraisingh et al. 2005; Ralph et al. 2005b; Dzikowski et al. 2007), which has led to the hypothesis of a defined var subnuclear transcription site.
The details of the mechanism maintaining var gene silencing are largely unknown; however, recent work indicates that each var gene is regulated by two separate promoters, one upstream of the coding region and a second within the relatively conserved intron (Calderwood et al. 2003; Dzikowski et al. 2007). The upstream promoter from the single expressed gene produces var mRNA and is active in the ring stages of the cell cycle (8–20 h after invasion), while the intron promoter produces a noncoding RNA and is active in pigmented trophozoite and schizont stages (24–48 h after invasion) (Kyes et al. 2003). In experiments using transgenic parasite lines, silencing of a var upstream promoter driving luciferase as reporter gene requires the presence of a var intron (Frank et al. 2006). A more-detailed analysis with intron derivatives revealed a direct correlation of the promoter activity of the intron and its silencing capacity (Calderwood et al. 2003; Gannoun-Zaki et al. 2005; Dzikowski et al. 2007).
It is not clear how the intron and the var upstream promoter interact to achieve silencing and potentially trigger structural changes in the parasite nucleus. One possibility is the production of noncoding, nuclear RNAs. Noncoding RNAs are involved in the regulation of monoallelic gene expression in several organisms, where they have been shown to be essential for the establishment of epigenetic marks and are associated with structural changes such as chromatin modification and/or spatial reorganization within the nucleus (Yang and Kuroda 2007). Studies reporting the discovery of the var gene family described truncated RNA transcripts of var exon 2 and, due to a lack of an open reading frame, they were named “sterile” RNAs (Su et al. 1995). These sterile RNAs appear late in the replicative cycle in contrast to var mRNA and thus are unlikely to represent alternative processing products of var preRNA; they rather seem to originate from the transcriptionally active promoter within the var intron. In transient transfection studies, the intron promoter was shown to be functional when inserted into a plasmid in either orientation, suggesting a bidirectional transcriptional activity (Calderwood et al. 2003). However, it had not previously been shown if the intron could drive transcription in both directions simultaneously or if this bidirectional promoter produces the corresponding transcripts in the parasite. Further, neither the fate nor the structure of these noncoding RNAs was defined.
In this study, we characterize in detail the bidirectional promoter activity of var introns and identify antisense RNA transcripts of var exon1 produced by the intron promoters. Further, investigations into the subcellular localization of these noncoding RNAs suggest that they associate with chromatin at var loci and might thereby participate in the structural organization and epigenetic regulation of this gene family. In order to better understand the nuclear retention of the exon2 noncoding RNAs, we map the 5′ and 3′ ends of these, as well as the var mRNAs. Thus, in addition to the epigenetic marks previously described for var genes, noncoding RNAs may also represent a component of the chromatin structure regulating antigenic variation in P. falciparum.
RESULTS
var introns contain simultaneous, bidirectional promoter activities
While var genes can be grouped according to their upstream sequences (ups) and their genomic location (Kraemer and Smith 2003; Lavstsen et al. 2003), var introns all share a conserved sequence structure, displaying three regions of distinct base composition (Calderwood et al. 2003). When placed upstream of a reporter gene, var intron promoters can be active in either orientation (Calderwood et al. 2003); however, it was not known whether this reflects active transcription in both directions at the same time or if the direction of transcription is unidirectional and influenced by the surrounding DNA. In order to determine if var introns are transcriptionally active in both directions simultaneously, the plasmid pHLIRH was constructed, in which an intron is flanked by the reporter genes firefly- and Renilla luciferase in opposite orientations (Fig. 1A). Several var introns were cloned into this reporter construct and used in transient transfection assays. Six different var introns were chosen representing the different upstream promoter types (ups A, B, and C) and different chromosomal positions of the corresponding var genes (Fig. 1C), including an atypical intron of a “var-like” gene. The latter intron is unusually short (∼200 base pairs [bp]) and does not show the conserved asymmetry in nucleotide composition observed in introns from the rest of the var gene family.
FIGURE 1.
Bidirectional promoter activity of var introns. Promoter activity of var introns was measured in transient transfection assays using the reporter construct pHLIRH (A), containing the reporter genes firefly and Renilla luciferase. Parasites were transfected and luciferase activity (firefly, light gray; Renilla dark gray) was measured from 2.5 mL cultures after ∼68 h (ring stage, light-colored columns) and ∼92 h (schizonts, dark columns), respectively (B). Luminescence values were standardized to 1% parasitemia. Genomic loci and upstream promoter types of var genes corresponding to the chosen var introns (grouping of var5′ promoter types according to Lavstsen et al. [2003]) (C).
All five introns with a conserved structure showed significant promoter activity in both directions (Fig. 1B). The values measured from schizont stages were generally higher than for the ring stages, consistent with previous studies and the occurrence of the sterile intron-derived exon 2 transcripts in trophozoite and schizont stages (Calderwood et al. 2003; Kyes et al. 2003). In addition, a related study directly compared intron promoter activity to other stage-specific promoters and found that it is indeed specific to late trophozoites and schizonts (Epp et al. 2008). In contrast, the atypical var intron PFF0020c did not show any transcriptional activity, indicating the importance of the conserved structure of the intron sequences for their promoter activity. It is unlikely that the bidirectional transcriptional activity is simply due to the AT richness of the intron sequence. We have previously determined that the promoter activity of the intron is dependent on a specific, conserved region near its center (Calderwood et al. 2003) and that the intron from the hsp86 gene, with similar length and AT content, cannot substitute for the var intron in terms of regulating var gene expression (Deitsch et al. 2001) and does not possess promoter activity.
Detection of noncoding antisense RNA of exon 1
Studies reporting the discovery of the var gene family described truncated RNA transcripts of var exon 2, and, due to a lack of an open reading frame or a detectable protein product, they were named sterile RNAs (Su et al. 1995). As a consequence of the bidirectional promoter activity detected in the plasmid constructs, we hypothesized that var introns might additionally produce noncoding RNAs extending into exon 1. For direct identification of RNA transcripts, three specific probes for exon 1 of var gene PFF0845c were used in Northern blots (Fig. 2). To distinguish between the “on” and “off” state of a var gene, two P. falciparum clones were used: A3, in which PFF0845c is the predominantly expressed var gene, and G6, in which no PFF0845c transcription can be detected (Frank et al. 2007).
FIGURE 2.
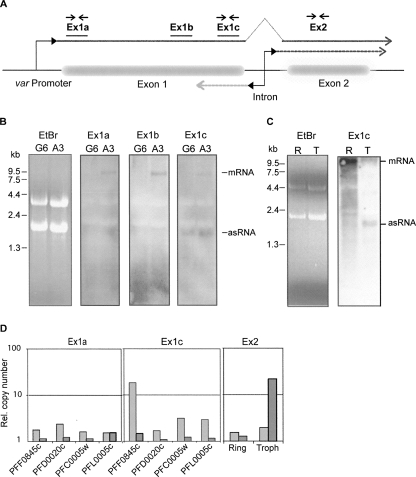
Identification of exon 1 antisense RNA by Northern blot and quantitative RT-PCR. (A) The structure of a typical var gene is depicted schematically, and the relative positions of probes for Northern blot (solid lines) and PCR primer pairs (arrows) are indicated. Dashed lines represent the putative RNA transcripts originating in the intron in the sense (dark gray) and antisense (light gray) orientation. (B) Northern blot detection of RNA with specific probes for var gene PFF0845c. RNA was prepared from asynchronous cultures of NF54 clones A3 (PFF0845c “on”) and G6 (PFF0845c “off”). Major bands corresponding to var mRNA and antisense (as) transcripts are indicated. (C) Northern blot analysis using a PFF0845c exon 1 specific probe. RNA samples from synchronized A3 cultures were prepared at two different time points: in ring stage (R), ∼16 h after invasion and trophozoite stage (T), ∼36 h after invasion. For both B and C, ethidium bromide (EtBr) stained samples are included to show that approximately equal amounts of RNA were loaded in each lane. (D) var RNA transcripts in the transgenic DCJ line (Dzikowski et al. 2006) were detected by quantitative PCR using strand-specific cDNA as template created using either sense or antisense primers. Two primer pairs within exon 1 were specific for each of four var genes, while the primer pair within exon 2 recognized conserved sequences present in several var genes. The relative amounts of antisense and sense RNA, as determined by real-time PCR, are shown as light and dark gray columns, respectively. RNA was prepared from trophozoite stages (∼36 h post-invasion) of synchronous cultures, and for analysis with exon 2 primers additionally from ring stages (∼16 h).
All three probes along exon 1 specifically recognized the ∼9.5-kb var mRNA in the A3 clone, but not in G6. The probe Ex1c, which recognizes a sequence at the 3′ end of exon 1 close to the intron, identified a second transcript of ∼1.7 kb. This RNA is not recognized by the two other probes, located further 5′ within exon 1, suggesting that it is an antisense RNA produced from the intron promoter. In contrast to the mRNA, the 1.7-kb transcript is present in both parasite clones, A3 and G6. A Northern blot using a synchronized parasite culture shows that the mRNA is synthesized in ring stage parasites, while the synthesis of the antisense 1.7-kb transcript is restricted to the trophozoite stage (Fig. 2C), which corresponds to the stage-specific activities of var5′ and intron promoters, respectively (Kyes et al. 2003).
In an alternative approach, RNA transcripts were identified by strand-specific reverse transcription followed by real-time PCR (Fig. 2D). This technique allowed us to distinguish between “sense” and “antisense” transcripts and to measure the relative abundance of the different RNA species. Using RNA extracted from synchronized, late stage parasites, the exon 1 transcripts of four different var genes were analyzed with two different primer pairs specific for each gene along exon 1 (Fig. 2A). In addition, a pair of conserved primers was used to detect RNA transcripts of exon 2. To obtain a homogenous RNA sample, the transgenic parasite line DCJ was used, in which upon selection with blasticidin only the transgene is active, and all the remaining var genes are transcriptionally silent (Dzikowski et al. 2006). As a consequence, all var genes analyzed in this experiment are in the transcriptionally silent state. In all four var genes, significantly more antisense than sense transcripts were detected within exon 1. This indicates that the intron promoter is active in these var genes and produces antisense RNA extending into exon1. There are generally more antisense transcripts detected by the primers close to the intron, suggesting that the majority of transcripts terminate within exon 1. Accordingly, conserved primers recognizing exon 2 detected more sense than antisense transcripts from trophozoite RNA, corresponding to the previously identified sterile transcripts.
Subcellular localization of intron-derived var transcripts
To determine the cellular localization of the novel antisense transcripts, we applied RNA fluorescent in situ hybridization (RNA-FISH) analysis. Single-stranded RNA probes were created that were specific for var gene PFF0845c, recognizing the 3′ region of exon 1. Strand-specific probes were used to distinguish between var mRNA and antisense transcripts of exon 1 (Fig. 1A, Ex1c). In all cases, multiple nuclei from a single slide were counted and scored. The numbers reported here are representative of multiple experiments. In the A3 line, which predominantly expresses PFF0845c, the mRNA is recognized in ring stages (14/14 nuclei) (Fig. 3). It localizes in part to the nucleus and in part to the cytoplasm, as expected for an exported mRNA and as previously observed for an mRNA transcript (Li et al. 2008). The antisense transcript was identified in late stages as a distinct spot within the nucleus. It is detected in both parasite lines, A3 (19/22 nuclei; PFF0845c active) and DCJ (18/18 nuclei; PFF0845c off), indicating that antisense transcripts are synthesized in the active as well as the silent var genes. The antisense transcript was first detected in late trophozoites and remained evident through segmented schizonts, which were almost completely segregated into merozoites. Each of the new progeny parasites contained a single spot, as expected due to equal distribution of the haploid genome. As there was no signal detected in ring stages of either A3 (0/11 nuclei) or DCJ (0/13 nuclei), degradation of antisense RNA, therefore, must occur in free merozoites or shortly after reinvasion of new RBCs. This might coincide with “resetting” of a genetic mark to allow switching between different var genes. The fact that FISH signals were only observed with the appropriate single-stranded probe and at the appropriate stage of the cell cycle indicates that they are hybridizing to RNA and not to genomic DNA.
FIGURE 3.
Detection of var mRNA and antisense transcript by RNA-FISH. Single-stranded RNA probes were used to specifically detect mRNA (− strand) and exon 1 antisense transcript (+ strand) of var gene PFF0845c. A3 (PFF0845c “on”) and DCJ (PFF0845c “off”) parasites were analyzed 16–20 h post-invasion (ring stage), ∼30 hpi (trophozoite), and ∼40 hpi (schizont). Parasite nuclei were visualized by DAPI (blue). Overlays were generated of fluorescence pictures obtained by labeled RNA probes and DAPI stain, and one representative picture is shown for each sample.
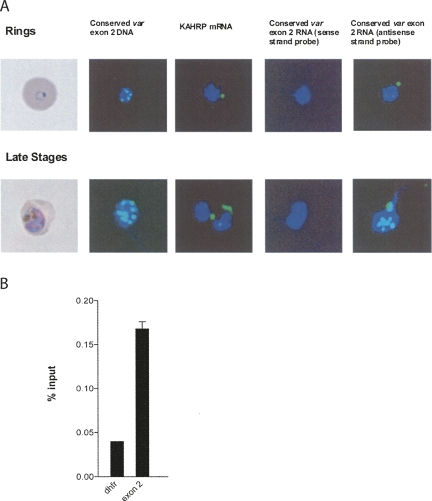
To determine the localization of the sterile transcripts of exon 2, we similarly utilized RNA-FISH (Fig. 4). Parasites were examined in ring and late trophozoite stages using an RNA antisense probe corresponding to exon 2 of a representative var gene. Because exon 2 is largely conserved, this probe hybridizes to both the expressed var mRNA as well as a large percentage of the sterile transcripts. In ring stage parasites where the full-length message is most abundant, the hybridization signal was almost always observed as a single spot either at the edge of the nucleus or within the cytoplasm (11/12 nuclei), as expected for a translated message. This spot is thought to represent the point where the nuclear pore exports the mRNA to the endoplasmic reticulum. A similar pattern was observed for an unrelated RNA probe for the mRNA of the KAHRP (knob-associated histidine rich protein) gene (28/30 nuclei). This hybridization pattern is consistent with previous Northern blot data that indicates that at this stage in the cell cycle only the full-length transcript is detected (Kyes et al. 2003), which is presumably exported to the cytoplasm and translated into PfEMP1. The sterile transcripts are not detected at this stage. In contrast, in late stage parasites the exon 2 sterile transcripts were concentrated at discrete points within the nucleus in a punctate pattern in virtually all cells examined (32/33 cells), suggesting that exon 2 sterile transcripts localize exclusively to the nucleus in late trophozoites. To see if var genes display a similar pattern, we used an exon 2 specific DNA probe to localize the genes themselves. As expected, the chromosomal genes organize into a pattern that closely resembles the RNA localization (11/11 nuclei), raising the possibility that the var noncoding RNAs remain closely associated with the var gene clusters.
FIGURE 4.
Subcellular localization of the var sterile transcripts. (A) The clusters of var genes previously described can be seen in both rings and late trophozoites using DNA probes against exon 2. A larger number of clusters is seen in late trophozoites, indicating DNA replication has begun. RNA-FISH shows the localization of var transcripts in both rings (cytoplasmic, corresponding to the full-length, translated message) and the late trophozoites (nuclear, corresponding to the sterile transcripts). Sense strand RNA probes do not hybridize to either stage of parasites. The KAHRP RNA probe demonstrates cytoplasmic labeling as expected for a translated mRNA. (B) Chromatin immunprecipitation (ChIP) of var sterile transcripts with antibodies against histone H3 indicates that the sterile transcripts are associated with chromatin. Synchronized, late stage parasites were isolated and chromatin immunoprecipitated as described (Chookajorn et al. 2007). RNA was then isolated and cDNA analyzed using quantitative RT-PCR. The var sterile transcripts were more than fourfold more highly represented in the chromatin fraction than a representative mRNA dhfr. Precipitation with nonhistone antibodies (against PFI1780w) or experiments completed without reverse transcriptase did not give any signal.
var noncoding RNAs are associated with chromatin
The hybridization pattern of both the sense and antisense noncoding RNAs indicated that they might remain localized within the nucleus, perhaps associated with the var genes from which they were transcribed. Various noncoding RNAs associated with gene silencing and dosage compensation in other organisms have been shown to be necessary for proper chromatin assembly and regulation of gene expression. This is perhaps best described for Xist in mammals or rox1 and 2 in fruit flies, where these RNA molecules localize to their specific chromosomal targets within the nucleus, becoming incorporated into the chromatin structure, effectively “painting” their target chromosome and leading to a condensed RNA/chromatin body within the nucleus (Meller et al. 1997; Wutz et al. 2002). These “bodies,” called “Barr” bodies, and the RNA molecules they contain can be visualized microscopically within the nuclei of these cells (Lee et al. 1999; Rasmussen et al. 2001). If the var noncoding transcripts behave in a similar fashion, they would be predicted to be associated with chromatin.
To determine if the var noncoding RNAs are associated with chromatin within the parasite nucleus, chromatin immunoprecipitation (ChIP) was performed as previously described (Chookajorn et al. 2007). After immunoprecipitation, RNA was isolated and cDNA prepared, and the association of the sterile transcripts with chromatin compared to that of an mRNA. The control mRNA was the human dhfr gene in the transgenic line B15C2 (Dzikowski et al. 2006). In this line, dhfr is transcribed from an intron promoter, but it is terminated by the polyadenylation signal from the hrp2 gene, and thus is efficiently exported and translated into protein, making the parasites resistant to selection with WR99210. Therefore, both the mRNA and the sterile RNA assayed in this experiment are transcribed from intron promoters and thus are expressed at relatively similar levels and at the same point in the cell cycle. As shown in Figure 4B, the var sterile transcripts were fourfold more efficiently precipitated with antibodies specific to core histones than the dhfr mRNA, providing further evidence that they are in fact nuclear and chromatin associated. Precipitation with nonhistone antibodies (against PFI1780w) or experiments completed without reverse transcriptase did not give any signal. The primary difference between the dhfr mRNA and the sterile transcripts assayed in this experiment is the presence of a heterologous polyadenylation signal in the dhfr transcript. This raised the possibility that the sterile transcripts might be terminated differently than mRNAs.
var noncoding transcripts are not polyadenylated
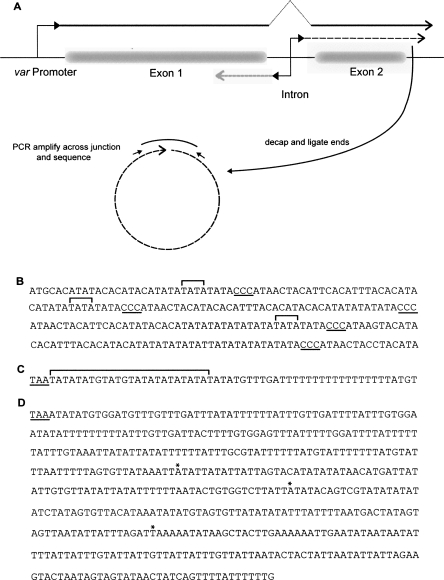
Efficient nuclear export of mRNAs has been shown to involve the addition of a poly(A) tail. For example, Dower et al. (2004) showed that removal of the poly(A) tail from yeast transcripts, via a self-cleaving ribozyme element inserted after the stop codon, abolished their nuclear export. The var mRNAs and sense ncRNAs share the same 3′ region, and yet the mRNAs are exported from the nucleus and translated while the ncRNAs are retained. We, therefore, were interested to see if the 3′ ends of both var noncoding transcripts and mRNAs are differentially processed. To determine if the var noncoding transcripts are polyadenylated, an RNA circularization technique originally developed by Mandl et al. (1991) was used. The technique involves circularizing RNA molecules with RNA ligase and sequencing across the junction, simultaneously mapping the 5′ and 3′ ends (Fig. 5A). The advantage of this technique is that it not only allows simultaneous sequencing of the two ends, but also that is does not require the presence of a poly(A) tail. Briefly, tobacco acid pyrophospahatase (TAP) is used to decap the transcript. It is then treated with RNA ligase to circularize the ends, and cDNA is synthesized using a primer specific to the 5′ region of the gene of interest. In this case, a primer to the conserved region of exon 2 was used, allowing sense ncRNAs from more than one gene to be examined. For the antisense transcripts, primers specific to var genes PFF0845c and PFD0005W were designed. PCR was then performed with primers flanking the junction regions, and subsequent products were subcloned and sequenced.
FIGURE 5.
Identification of the 5′ and 3′ ends of var transcripts. (A) Schematic diagram showing the RNA circularization method for simultaneous mapping of 5′ and 3′ ends. (B) The DNA sequence of a portion of the intron from var gene PFD0005w. The 5′ ends of the sterile RNAs transcribed from this gene are marked with brackets. The ccc repeat motif is underlined. (C) The DNA sequence downstream from the coding region of PFD0005w. The stop codon of the open reading frame is underlined. The sterile RNAs transcribed from this gene ended within 20 bp of the stop codon as marked by the bracket. (D) The DNA sequence downstream from the coding region of var gene PFD1005c. The polyadenylation sites of the mRNA transcribed from this gene are marked with asterisks. The stop codon is underlined.
var sterile RNA was harvested from late trophozoite stage DCJ parasites. As noted above, DCJ parasites only produce var sterile RNAs, allowing us to map the ends without contamination by mRNA (Dzikowski et al. 2006). The 5′ transcriptional start site of the var sense ncRNAs was mapped to a TATA repeat region just upstream of a 20-bp sequence beginning with CCC. var introns contain several (2–5) of these CCC repeat regions, and the transcription start site was found just prior to any of these. Therefore, the length of intron transcribed before the start of exon 2 is variable (Fig. 5B). This held true for four different genes. The 3′ end of these transcripts was found to be nonpolyadenylated. The var sterile transcript generally ended within 20 bp of the exon 2 stop codon, within an ATAT repeat region (Fig. 5C). Multiple transcripts were examined from the four genes in question. Both the 5′ and the 3′ ends varied between different transcripts of the same gene; however, when the total length of the transcripts was determined based on their 5′ and 3′ ends, they always coincided with the ∼2.0 kb determined by Northern hybridization (Fig. 2). Additionally, circularized sequences were only produced in the presence of TAP, suggesting that the sterile RNAs are indeed capped. Similar results were obtained for the antisense ncRNAs from genes PFF0845c and PFD0005W, with both the 5′ and 3′ ends being somewhat variable, but always coinciding with a total length of ∼1.7 kb. Unlike the 5′ ends of the sense ncRNAs, the ends of the antisense transcripts were not found at a specific motif, but rather varied within a ∼50-bp region. Once again these transcripts were determined to be capped but not polyadenylated.
The transcription start site for var mRNAs had been previously mapped (Deitsch et al. 1999). In order to determine the 3′ end of the mRNAs, 3′ rapid amplification of cDNA ends (RACE) was performed. The C3 parasite line, which predominantly expresses var gene PFD1005c, was used as a source of mRNA from a known var gene, allowing specific amplification of the 3′ end. Polyadenylation sites were mapped to ∼200 or 400 bp downstream from the exon2 stop codon (Fig. 5D). This is consistent with the observation that poly(A) sites of other Plasmodium genes are also variable (Ruvolo et al. 1993; Oguariri et al. 2006). Regardless, it is apparent that although the var mRNAs and sense ncRNAs terminate within the same region of the gene, they are differentially processed. The lack of poly(A) tail on the sterile RNA may contribute to its retention in the nucleus.
DISCUSSION
var introns contain several features, including an unusual base composition and the presence of putative initiator sequences, that are conserved throughout the var gene family (Calderwood et al. 2003). However, unlike var upstream sequences, var introns cannot be grouped into different sequence types (Kraemer and Smith 2003; Lavstsen et al. 2003), and all five introns with a conserved sequence tested in this study displayed bidirectional promoter activity. This was expected due to the apparent requirement of intron promoter activity for proper var gene regulation (Dzikowski et al. 2007). Although the detailed functions of the different elements remain elusive, the conserved sequence features of var introns are likely to be important for their promoter activity and function. In contrast, a short intron of an atypical var gene did not show transcriptional activity in our promoter assays. The corresponding var gene PFF0020c belongs to a distinct subgroup of so-called Type 3 var genes (PFI1820w, PFA0015c) (Lavstsen et al. 2003). Although transcripts of one of these genes have been detected in a previous study (Tham et al. 2007), little is known about their function or if they are expressed in the mutually exclusive manner typical of the rest of the var gene family. It is possible that they are regulated by a somewhat different mechanism.
In this study, we identified antisense transcripts of var exon 1 by three different methods: Northern blot, strand-specific RT-PCR, and RNA-FISH. While this is in contrast to the result of a micro-array-based study that did not detect antisense transcripts (Ralph et al. 2005a), it is consistent with a recent study by Kyes et al. (2007) in which exon 1 transcripts close to the intron are detected by nuclear run-on analysis. Depending on the detection method and due to the presence of conserved domains, it can be difficult to specifically detect transcripts from individual var genes. In particular, detection of antisense RNA by RT-PCR is prone to false positive amplification due to presence of sense strand mRNA molecules. In our study, we avoided this problem by utilizing the parasite line DCJ (Dzikowski et al. 2006), which upon selection with blasticidin exclusively expresses the blasticidin resistance gene and does not produce any var mRNA. Therefore, we can exclude the possibility that the detected RNA transcripts in this parasite line are derived from mRNA. Northern blot identified a well-defined band corresponding to an antisense RNA species of ∼1.7 kb for the gene PFF0845c. The size may vary for each var gene; however, the nuclear run-on assays of Kyes et al. (2007) also detected transcripts in this region of exon 1; thus it seems that, in general, antisense transcripts can only be detected in the 3′ region of exon 1, relatively close to the intron. This finding implies that antisense transcription does not typically extend through the var upstream promoter, and therefore this is unlikely to be a mechanism of promoter interaction.
On first sight, these results contradict a study by Ralph et al. (2005a), in which genome-wide analysis of var RNA was applied to investigate a possible correlation of antisense RNA with var gene activity. In comparing transcripts from parasites selected for binding to chondroitin sulfate A versus CD36, no significant levels of exon 1 antisense RNAs were detected, and minor amounts of antisense RNA were attributed to transcriptional noise of actively transcribed genes. In contrast, we detected transcription of antisense RNA in both transcriptionally silent and active var genes and propose the var intron promoter as the origin of the noncoding RNAs independent of the var upstream promoter. However, in our study we also find that not all var genes produce equal amounts of antisense RNA. In particular, the intron promoters vary regarding their promoter strength and the direction of the main transcriptional activity. As a result var genes with intron promoters with bias for one direction might not always show detectable levels of both transcripts, but instead produce one major type of transcript, either the exon 1 antisense or exon 2 sense transcript. Depending on the detailed mechanism it might be possible that the two noncoding transcripts have redundant functions, requiring both or either one of the RNAs to be produced by a single var intron. Ralph et al. (2005a) did not detect changes in antisense RNA levels between active and inactive var genes. We do not find correlations of the intron promoter between active and inactive var genes either, but rather observe antisense RNA produced in on as well as off var genes. Therefore, we would not expect to find a change in the ratio between active and silent var genes.
Several studies have shown that the presence of the intron promoter is required for silencing and proper recognition of a var promoter (Calderwood et al. 2003; Gannoun-Zaki et al. 2005; Frank et al. 2006; Dzikowski et al. 2007). In this study, we used Northern blot and RNA-FISH to compare the expression pattern of gene-specific intron transcripts in parasite lines in which the corresponding var gene was either active or silent. We specifically detected intron-derived transcripts produced from an on var gene, and we found no significant difference in antisense transcripts between on and off var genes. This implies that intron promoters are active in all var genes, regardless of the transcriptional status of the upstream promoter. Thus, it seems unlikely that the intron promoter and/or the intron-derived transcripts are a direct signal for var gene silencing. Instead, the intron, together with the upstream promoter, might be required for var gene recognition and inclusion in the pathway that limits expression to a single var gene. Alternatively, it might be important for the proper timing of the var upstream promoter and restrict its transcriptional activity to ring stage parasites. Interestingly, the var1csa gene (PFE1640w) is conserved in numerous isolates similar to var2csa; however, in the FCR3 line in which it was first identified, the intron of this gene has a substantial deletion that eliminates its promoter activity. Its expression violates the general var gene transcription pattern in both ways: first, it is not recognized by the monoallelic expression pathway for the var gene family and is expressed in addition to an active var gene, and, second, its expression is not restricted to ring stage parasites, and mRNA is synthesized later in the cell cycle (Kyes et al. 2003; Winter et al. 2003).
Noncoding RNAs are implicated in several examples of epigenetic regulation in different organisms. While the canonical RNA interference machinery has not been identified in Plasmodium, noncoding RNAs might carry out important regulatory functions independent of RNAi. In this study we show by RNA-FISH that the noncoding RNAs produced from the var intron remain localized to the nucleus, displaying a pattern of hybridization resembling closely the bouquet-like pattern of 7–10 spots observed for var genes by DNA-FISH analysis (Freitas-Junior et al. 2000). Although the co-localization of DNA and RNA cannot be shown directly, this result along with the RNA-ChIP analysis lead us to speculate that the noncoding RNAs might be associating directly with var genes. It is possible that the lack of a poly(A) tail is sufficient to retain the sterile RNAs in the nucleus (Dower et al. 2004). Since the exon 2 sterile transcript appears to be capped, it is likely that the differential processing of the 3′ end is the main mechanism of retention. The process by which the sterile RNAs are identified for alternative processing and associated with chromatin has yet to be to be investigated.
The role of the noncoding RNAs described here in var gene regulation might share many attributes with that of noncoding RNAs in other epigenetically regulated systems. For example, in X-chromosome inactivation in mammals, the noncoding Xist RNA binds in cis and spreads along the chromosome to mark it for silencing, creating chromatin/RNA bodies not unlike the noncoding RNA-containing var gene clusters observed here (Akhtar 2003). Dosage compensation in fruit flies also relies on the production of cis-acting, noncoding RNAs that associate with the chromosomes (Stuckenholz et al. 1999). Similarly, episomal var promoters cannot be silenced in trans by introns contained on separate cotransfected episomes (data not shown), and presumably chromosomal var genes are flanked by boundary elements to prevent silencing from spreading to adjacent var genes.
This form of imprinting or epigenetic gene silencing may be common in P. falciparum. An analysis of the sequenced genome has suggested a predisposition for epigenetic regulation (Aravind et al. 2003). In the prokaryotic world, pathogens routinely possess a more limited transcriptional regulatory apparatus compared to free-living organisms (Andersson et al. 1998; Stephens et al. 1998). While a large family of putative transcription factors was recently identified using a bioinformatic approach (Balaji et al. 2005), P. falciparum is missing a number of gene regulatory proteins that are present in virtually all other sequenced eukaryotes (Aravind et al. 2003). For example, the genome contains all of the classic histone proteins but lacks the H1 linker (H1) or the forkhead proteins (FKH) (Gardner et al. 2002). Several other recognizable eukaryotic transcription factor domains are also missing, such as bHLH and GATA finger domains. What P. falciparum does retain are all of the core basal transcription factors and much of chromatin structural and remodeling components of the average eukaryotic genome. This may include a number of chromatin-associated factors capable of binding noncoding RNA. The mammalian HP1 protein has been shown to have RNA binding activity that is essential for its targeting to specific areas of heterochromatin (Muchardt et al. 2002). In much the same way, sterile transcripts may direct chromatin-associated factors to target specific var genes for silencing. Given the data presented here, the recent identification in silico of more than 30 novel ncRNAs of unknown function (Chakrabarti et al. 2007; Mourier et al. 2008) and evidence that 17% of annotated genes express detectable antisense RNA transcripts, it is likely that RNA-mediated gene regulation is an integral part of P. falciparum biology (Patankar et al. 2001).
MATERIALS AND METHODS
Plasmodium falciparum culture
All experiments utilized the P. falciparum NF54 line (or derivatives thereof) cultivated at 5% hematocrit in RPMI 1640 medium, 0.5% Albumax II (Invitrogen), 0.25% sodium bicarbonate, and 0.1 mg/mL gentamicin. Parasites were incubated at 37°C in an atmosphere of 5% oxygen, 5% carbon dioxide, and 90% nitrogen. The transgenic line DCJ has been previously described (Dzikowski et al. 2006). It was generated in the NF54 genetic background and carries the bsd gene encoding blasticidin S deaminase integrated into a subtelomeric var locus. Blasticidin pressure selects for activation of the var transgene and results in a reversible knock out of the entire var gene family. Parasite lines A3 and G6 have been described previously (Frank et al. 2007). They were obtained by limiting dilution of P. falciparum NF54, and the predominantly expressed var genes were identified by real-time PCR as PFF0845c (A3) and PFD1005c (G6).
Bidirectional luciferase reporter constructs
Plasmids pVLH, pIRH, and pVLH/int had been described previously (Calderwood et al. 2003). The luciferase coding region and hrp2 3′ UTR of pVLH were amplified by PCR introducing restriction sites for PstI (5′) and KpnI (3′) using the primers FflucPstI: 5′-AACTGCAGGCATGGAAGACGCCAAAAAC-3′ and hrp3′downKpnI: 5′-GGGGTACCCGCCTCTCCCCGCGCGTTGG-3′. Subsequently, PstI and KpnI sites were used to insert the PCR products into pIRH to yield plasmid pHLIRH, containing the PFB1055c intron as promoter for both reporter genes. Additional intron sequences were first amplified from genomic DNA with the following primers, thereby adding the restriction sites BamHI and SalI to the 5′ and 3′ ends, respectively, and inserted into the HLIRH construct folllowing digestion with restriction enzymes BamHI and SalI. Gene-specific primers for var introns are the following: PFC0005w: 5′-CGGGATCCGAAGGTAAAAGAGAATATATATGTG-3′/5′-GCGTCGACTTCTAAAATAATAAAAGAGG-3′; PFF0845c: 5′-CGGGATCCCTAAAGGTATTATATATG-3′/5′-GCGTCGACGATCAATAGTAGATTTGG-3′; PFD0020c: 5′-CGGGATCCGCGCTTTTATTTTTGAAGG-3′/5′-GCGZTCGACGGTCCACAGGAGATTTAGG-3′; PFF0020c (var-like): 5′-CGGGATCCCTCTAGGATTGTTATTATTTAAGG-3′/5′-GCGTCGACGTTTTGGTTTTGTTCTCATC-3′; and PFL0005w: 5′-CGGGATCCCTAAAGGTATTATATATA-3′/5′-GCGTCGACCGAACAGATGATTTGG-3′.
Northern blots
RNA preparations from synchronized parasites were harvested ∼16 h (rings) or ∼40 h after invasion. RNA purification and Northern blots were carried out as described earlier (Kyes et al. 2000). We loaded 6.5 μg RNA per lane. Incubation with probes was done overnight at 42°C. Stringency washes with 2× SSC, 0.1% SDS (2 × 5 min) and 0.5× SSC, 0.1% SDS (2 × 15 min) were carried out at 50°C. Probes were prepared using the “DIG-High Prime Kit” (Roche Diagnostics) according to the manufacturer's instructions. Probe templates were generated by PCR and gel purified before labeling. The following primer pairs specific for PFF0845c were used: Probe Ex1a: 5′-CAGCAACCACTACATACAG-3′/5′-CGTAGTGAGAGTGTTTTGC-3′; Probe Ex1b: 5′-CCTGCAATCGATTGCG-3′/5′-GAAGTTGTTATGCGTGTTAG-3′; Probe Ex1c: 5′-GGCGGTTGTGAACCAGC-3′/5′-CGAAACAAAAGGCAACCG-3′; and Probe Ex2: 5′-GAATCGGATGACTACAC-3′/5′-GAGATACGTTTAATACAC-3′.
Strand-specific RT-PCR and quantitative PCR
RNA was extracted from synchronized ring stage parasites 16–18 h or late trophozoites 40–42 h post-invasion. RNA extraction was performed with the TRIZOL LS Reagent (Invitrogen) as previously described (Kyes et al. 2000). RNA to be used for cDNA synthesis was purified on a PureLink column (Invitrogen) according to the manufacturer's protocol. Isolated RNA was then treated with Deoxyribonuclease I (Invitrogen) to degrade contaminating gDNA. cDNA synthesis was performed with Superscript II Rnase H reverse transcriptase (Invitrogen) as described by the manufacturer. For strand-specific reverse transcription, 3 pmol of specific primers per 20 μL reaction were used instead of random primers. cDNA was synthesized from 800 ng total RNA in a reaction volume of 20 μL. For each cDNA synthesis reaction, a control reaction without reverse transcriptase was performed with identical amounts of template and primers.
Forward and reverse primers of primer pairs Ex1a, Ex1c, and Ex2 were used for strand-specific RT-PCR detection of PFF0845c transcripts. For detection of exon 1 transcripts of four additional var genes the following gene-specific primers were used: PFC0005w: Exon1 5′: 5′-GGAGAAACTAGGAGGTTTG-3′/5′-CACAACTGGAAATTGTGTC-3′; and Exon1 3′: 5′-GCGTTCGAGAGGTCAATTC-3′/5′-GGCAACCCACAACAACCG-3′. PFL0005w: Exon1 5′: 5′-CGGAGGAGGAAAAACAAGAG-3′/5′-ATGTGGTCTCAAATACGGCA-3′; and Exon1 3′: 5′-CGAGAACGTTACGTCAAAC-3′/5′-CTGATTGTACTGTCGCAG-3′. PFB1055c: Exon1 5′: 5′-CAATTTTGGGTGTGGAATCA-3′/5′-GATACACTTGGTGGCCAGTG-3′; and Exon1 3′: 5′-GTGGCAAAGTATGTGGTG-3′/5′- CACATTGTACGAAAAATGG-3′. PFD0020c: Exon1 5′: 5′-ATATGGGAAGGGATGCTCTG-3′/5′-TCAATTCCTTCGATGGTTCA-3′; and Exon1 3′: 5′-GATGTGGTGCCACAAGAACC-3′/5′-CCTAGTGAAGGCACAGAGG-3′.
To quantify strand-specific cDNAs, the corresponding forward and reverse primers were used as primer pairs for real-time PCR. Real-time PCR was carried out as described previously (Dzikowski et al. 2006). All reactions included primers for the control genes seryl-tRNA synthetase (PF07_0073), fructose biphsphate aldolase (PF14_0425), and actin (PFL2215w), as described by Salanti et al. (2003), as well as arginyl-tRNA synthetase (PFL0900c) and glutaminyl-tRNA synthetase (PF13_0170), as described by Dzikowski et al. (2006).
Fluorescent in situ hybridization
RNA FISH was performed with slight modifications as described by Thompson (2002) on ring and late stage P. falciparum parasites. Conserved var exon 2, exon 1 of var gene PFF0845c, and KAHRP antisense RNA probes were labeled by in vitro transcription in the presence of fluorescein. RNA FISH was also performed using sense RNA probes as controls. DNA FISH was carried out on ring and late stage parasites using a conserved var exon 2 DNA probe labeled with fluorescein as previously described (Dzikowski et al. 2007). After washing, the slides were mounted in antifade medium and visualized using an Olympus M081 fluorescent microscope. In all cases, multiple nuclei from a single slide were counted and scored. The numbers reported in the text are from a single experiment, but are representative of multiple experiments.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as previously described (Chookajorn et al. 2007). Briefly, cells were lysed in 0.1% saponin, followed by the addition of formamide to 0.5% and incubated at room temperature (RT) for 10 min. Cross-linking was arrested by addition of glycine to 125 mM and reaction incubated for 5 min at RT. The solution was centrifuged and the pellet washed twice with ice-cold TBS (20 mM Tris.HCl at pH 7.6, 150 mM NaCl) then resuspended in 2 mL ice-cold IP buffer (50 mM HEPES at pH 7.5, 140 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% Na-deoxycholate, 5 mM DTT, 1× Roach protease inhibitors/PMSF; MW = 174.2). The mixture was homogenized 200 times on ice then sonicated six times by Branson Digital Sonifer Model 450 at setting 50% for 20 sec on, 2 min off. We added 60 μL of Protein A (50%) to a 250-μL aliquot (a 25 μL aliquot was kept for input control), and the solution was incubated for 1 h at RT and spun down at maximum speed. Antibodies specific to histone H3 (Ab1791; Abcam) or control antibodies against PFI1780w (Chookajorn et al. 2007) were added to the supernatant and incubated overnight at RT. We added 60 μL of Protein A (50%) beads with protease inhibitor and the suspension was incubated for 3 h at RT. Beads were pelleted and washed two times for 5 min on nutator at RT with 1 mL of IP buffer, once for 5 min on nutator at RT with 1 mL of IP buffer/500 mM NaCl, once for 5 min on nutator at RT with 1 mL of 10 mM Tris/HCl (pH 8.0), 0.25 M LiCl, 0.5% NP-40, 0.5% Na-Deoxycholate, and 1 mM EDTA, and once for 5 min on nutator at RT with 1 mL of TE. The precipitate was eluted from antibody beads by adding 100 μL of 50 mM Tris/HCl at pH 8.0, 10 mM EDTA, 1% SDS, 5 mM DTT and incubating at 65°C for 15 min. Beads were pelleted at full speed and the eluate transferred to a fresh tube. Beads were washed with 150 μL TE, 0.67% SDS, 5 mM DTT, mixed, and pelleted again. Eluates were pooled and incubated overnight at 65°C. After the cross-links were reversed, 200 μL TE (plus 5 mM DTT), 20 μg glycogen, and 100 μg Proteinase K were added and incubated for at least 1 h at 65°C. Phenol/chloroform extraction and ethanol precipitation were performed with resuspension in 100 μL ddH2O. The isolated nucleic acids were then analyzed by quantitative RT-PCR as described above.
Mapping of transcipt 5′ and 3′ ends
RNA circularization was performed with modifications as described by Mandl et al.(1991). Briefly, RNA was harvested from late stage DCJ parasites. RNA extraction was performed with the TRIZOL LS Reagent (Invitrogen) as previously described (Kyes et al. 2000). We treated 1 μg RNA with 20 U TAP (Ambion) at 37°C for 1 h to remove the 5′ cap. It was then purified over an illustra ProbeQuant G-50 Micro Column (GE Healthcare) according to the manufacturer's instructions. Purified RNA was then treated with 80 U RNaseOut (Invitrogen), 100 U RNA ligase (Ambion), and 10 μL DMSO at 4°C overnight. The RNA was again purified over a ProbeQuant column and subjected to cDNA synthesis using Superscript II Rnase H reverse transcriptase (Invitrogen) as described by the manufacturer. For the sense ncRNAs, an exon 2 specific primer was used in cDNA synthesis (5′-TCATTAATTAAATCTATACCA-3′) and the cDNA was then subjected to PCR across the ligation junction using primer pairs (5′-GTTGTGTTGTTACCACTAGGTTC-3′) and (5′-GGGATTATTAGATAAATTAAAAG-3′). These primers were selected from an alignment of exon 2s from four var genes (PFE0005w, PFA0005w, PF10_0001, MAL7P1.212). For the antisense ncRNAs, combinations of the following primers were used for both cDNA synthesis and PCR amplification: PFF0845c, forward primers: 5′-GTGTCATGACCAACATAGCGAC-3′; 5′-GAGGACTTACTCCTTGAAGAAG-3′; 5′-CCACCAAAACGAAACAAAAGGC-3′; 5′-GCGACAATCCACAAGAAAAATG-3′; and 5′-GAATGGGAAGAAATAAAAAAAC-3′. PFF0845c, reverse primers: 5′-CCATGTTCTGAATTTAAAGTT-3′; 5′-GTGGGGCGACGGCTTCTGTCG-3′; 5′-CACACGCTTCCCCATAACCGC-3′; and 5′-GCATCACTCCCGCAATTACAC-3′. PFD0005w, forward primers: 5′-GGCGAACAAACAGAAACAGAG-3′; 5′-GAAGGCAAGTGTGGCGAAGAC-3′; and 5′-CCCCCGCCCTTGTGACCTCC-3′. PFD005w, reverse primers: 5′-GTATCGTTATCACTAACACGC-3′; 5′-CGTTATTCCATCGCACTTCCC-3′; and 5′-TTGTGCAGCTGTATCGCACG-3′. Total PCR products were then subcloned into pGEMT Easy (Promega) and transformed into XL10-gold Ultracompetent cells (Stratagene) as per the manufacturer's instructions. Resulting colonies were prepared and sequenced using M13F and M13R universal primers. To observe the polyadenylation sites of var mRNA, RNA was harvested from the ring stage C3 parasite line. 3′ RACE was performed with the 3′ RACE Kit (Invitrogen) according to the manufacturer's instructions using gene-specific primers to var gene PFD1005c, which is clonally expressed by the C3 parasite line.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health Grant AI 52390 and by a grant from the Deutsche Forschungsgemeinschaft (DFG) to C.E.. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of the William Randolph Hearst Foundation. K.W.D. is a Stavros S. Niarchos Scholar.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1080109.
REFERENCES
- Akhtar A. Dosage compensation: An intertwined world of RNA and chromatin remodelling. Curr. Opin. Genet. Dev. 2003;13:161–169. doi: 10.1016/s0959-437x(03)00016-9. [DOI] [PubMed] [Google Scholar]
- Andersson S.G.E., Zomorodipour A., Andersson J.O., Sicheritz-Ponten T., Alsmark U.C.M., Podowski R.M., Naslund A.K., Eriksson A.S., Winkler H.H., Kurland C.G. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Aravind L., Iyer L.M., Wellems T.E., Miller L.H. Plasmodium biology: Genomic gleanings. Cell. 2003;115:771–785. doi: 10.1016/s0092-8674(03)01023-7. [DOI] [PubMed] [Google Scholar]
- Balaji S., Babu M.M., Iyer L.M., Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch D.I., Pasloske B.L., Singh H.B., Bi X., Ma X.C., Feldman M., Taraschi T.F., Howard R.J. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Calderwood M.S., Gannoun-Zaki L., Wellems T.E., Deitsch K.W. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- Chakrabarti K., Pearson M., Grate L., Sterne-Weiler T., Deans J., Donohue J.P., Ares M., Jr Structural RNAs of known and unknown function identified in malaria parasites by comparative genomics and RNA analysis. RNA. 2007;13:1923–1939. doi: 10.1261/rna.751807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chookajorn T., Dzikowski R., Frank M., Li F., Jiwani A.Z., Hartl D.L., Deitsch K.W. Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch K.W., del Pinal A., Wellems T.E. Intracluster recombination and var transcription switches in the antigenic variation of Plasmodium falciparum . Mol. Biochem. Parasitol. 1999;101:107–116. doi: 10.1016/s0166-6851(99)00062-6. [DOI] [PubMed] [Google Scholar]
- Deitsch K.W., Calderwood M.S., Wellems T.E. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- Dower K., Kuperwasser N., Merrikh H., Rosbash M. A synthetic A tail rescues yeast nuclear accumulation of a ribozyme-terminated transcript. RNA. 2004;10:1888–1899. doi: 10.1261/rna.7166704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh M.T., Voss T.S., Marty A.J., Duffy M.F., Good R.T., Thompson J.K., Freitas-Junior L.H., Scherf A., Crabb B.S., Cowman A.F. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum . Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Dzikowski R., Frank M., Deitsch K. Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzikowski R., Li F., Amulic B., Eisberg A., Frank M., Patel S., Wellems T.E., Deitsch K.W. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp C., Raskolnikov D., Deitsch K.W. A regulatable transgene expression system for cultured Plasmodium falciparum parasites. Malar. J. 2008;7:86. doi: 10.1186/1475-2875-7-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M., Dzikowski R., Constantini D., Amulic B., Burdougo E., Deitsch K. Strict pairing of var promoters and introns is required for var gene silencing in the malaria parasite Plasmodium falciparum . J. Biol. Chem. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank M., Dzikowski R., Amulic B., Deitsch K. Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 2007;64:1486–1498. doi: 10.1111/j.1365-2958.2007.05736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Bottius E., Pirrit L.A., Deitsch K.W., Scheidig C., Guinet F., Nehrbass U., Wellems T.E., Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum . Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior L.H., Hernandez-Rivas R., Ralph S.A., Montiel-Condado D., Ruvalcaba-Salazar O.K., Rojas-Meza A.P., Mancio-Silva L., Leal-Silvestre R.J., Gontijo A.M., Shorte S., et al. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Gannoun-Zaki L., Jost A., Mu J.B., Deitsch K.W., Wellems T.E. A silenced Plasmodium falciparum var promoter can be activated in vivo through spontaneous deletion of a silencing element in the intron. Eukaryot. Cell. 2005;4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.J., Hall N., Fung E., White O., Berriman M., Hyman R.W., Carlton J.M., Pain A., Nelson K.E., Bowman S., et al. Genome sequence of the human malaria parasite Plasmodium falciparum . Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer S.M., Smith J.D. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol. Microbiol. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- Kyes S., Pinches R., Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum . Mol. Biochem. Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Kyes S.A., Christodoulou Z., Raza A., Horrocks P., Pinches R., Rowe J.A., Newbold C.I. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol. Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes S., Christodoulou Z., Pinches R., Kriek N., Horrocks P., Newbold C. Plasmodium falciparum var gene expression is developmentally controlled at the level of RNA polymerase II-mediated transcription initiation. Mol. Microbiol. 2007;63:1237–1247. doi: 10.1111/j.1365-2958.2007.05587.x. [DOI] [PubMed] [Google Scholar]
- Lavstsen T., Salanti A., Jensen A.T.R., Arnot D.E., Theander T.G. Sub-grouping of Plasmodium falciparum 3D7 var genes based on sequence analysis of coding and noncoding regions. Malar. J. 2003;2:27. doi: 10.1186/1475-2875-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.T., Davidow L.S., Warshawsky D. Tsix, a gene antisense to Xist at the X-inactivation center. Nat. Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- Li F., Sonbuchner L., Kyes S.A., Epp C., Deitsch K.W. Nuclear noncoding RNAs are transcribed from the centromeres of Plasmodium falciparum and are associated with centromeric chromatin. J. Biol. Chem. 2008;283:5692–5698. doi: 10.1074/jbc.M707344200. [DOI] [PubMed] [Google Scholar]
- Lopez-Rubio J.J., Gontijo A.M., Nunes M.C., Issar N., Hernandez R.R., Scherf A. 5′ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandl C.W., Heinz F.X., Puchhammer-Stockl E., Kunz C. Sequencing the termini of capped viral RNA by 5′-3′ ligation and PCR. Biotechniques. 1991;10:484–486. [PubMed] [Google Scholar]
- Meller V.H., Wu K.H., Roman G., Kuroda M.I., Davis R.L. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- Mourier T., Carret C., Kyes S., Christodoulou Z., Gardner P.P., Jeffares D.C., Pinches R., Barrell B., Berriman M., Griffiths-Jones S., et al. Genome-wide discovery and verification of novel structured RNAs in Plasmodium falciparum . Genome Res. 2008;18:281–292. doi: 10.1101/gr.6836108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C., Guilleme M., Seeler J.S., Trouche D., Dejean A., Yaniv M. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1 α. EMBO Rep. 2002;3:975–981. doi: 10.1093/embo-reports/kvf194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguariri R.M., Dunn J.M., Golightly L.M. 3′ gene regulatory elements required for expression of the Plasmodium falciparum developmental protein, Pfs25. Mol. Biochem. Parasitol. 2006;146:163–172. doi: 10.1016/j.molbiopara.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Patankar S., Munasinghe A., Shoaibi A., Cummings L.M., Wirth D.F. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of antisense transcripts in the malarial parasite. Mol. Biol. Cell. 2001;12:3114–3125. doi: 10.1091/mbc.12.10.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S.A., Bischoff E., Mattei D., Sismeiro O., Dillies M.A., Guigon G., Coppee J.Y., David P.H., Scherf A. Transcriptome analysis of antigenic variation in Plasmodium falciparum–var silencing is not dependent on antisense RNA. Genome Biol. 2005a;6:R93. doi: 10.1186/gb-2005-6-11-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph S.A., Scheidig-Benatar C., Scherf A. Antigenic variation in Plasmodium falciparum is associated with movement of var loci between subnuclear locations. Proc. Natl. Acad. Sci. 2005b;102:5414–5419. doi: 10.1073/pnas.0408883102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.P., Wutz A., Pehrson J.R., Jaenisch R. Expression of Xist RNA is sufficient to initiate macrochromatin body formation. Chromosoma. 2001;110:411–420. doi: 10.1007/s004120100158. [DOI] [PubMed] [Google Scholar]
- Ruvolo V., Altszuler R., Levitt A. The transcript encoding the circumsporozoite antigen of Plasmodium berghei utilizes heterogeneous polyadenylation sites. Mol. Biochem. Parasitol. 1993;57:137–150. doi: 10.1016/0166-6851(93)90251-r. [DOI] [PubMed] [Google Scholar]
- Salanti A., Staalsoe T., Lavstsen T., Jensen A.T.R., Sowa M.P.K., Arnot D.E., Hviid L., Theander T.G. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Scherf A., Hernandez-Rivas R., Buffet P., Bottius E., Benatar C., Pouvelle B., Gysin J., Lanzer M. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum . EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.D., Chitnis C.E., Craig A.G., Roberts D.J., Hudson-Taylor D.E., Peterson D.S., Pinches R., Newbold C.I., Miller L.H. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow R.W., Guerra C.A., Noor A.M., Myint H.Y., Hay S.I. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens R.S., Kalman S., Lammel C., Fan J., Marathe R., Aravind L., Mitchell W., Olinger L., Tatusov R.L., Zhao Q.X., et al. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis . Science. 1998;282:754–759. doi: 10.1126/science.282.5389.754. [DOI] [PubMed] [Google Scholar]
- Stuckenholz C., Kageyama Y., Kuroda M.I. Guilt by association: Noncoding RNAs, chromosome-specific proteins and dosage compensation in Drosophila . Trends Genet. 1999;15:454–458. doi: 10.1016/s0168-9525(99)01855-7. [DOI] [PubMed] [Google Scholar]
- Su X., Heatwole V.M., Wertheimer S.P., Guinet F., Herrfeldt J.V., Peterson D.S., Ravetch J.V., Wellems T.E. A large and diverse gene family (var) encodes 200–350 kDa proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythrocytes. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Tham W.H., Payne P.D., Brown G.V., Rogerson S.J. Identification of basic transcriptional elements required for rif gene transcription. Int. J. Parasitol. 2007;37:605–615. doi: 10.1016/j.ijpara.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Thompson J. In situ detection of RNA in blood- and mosquito-stage malaria parasites. Methods Mol. Med. 2002;72:225–233. doi: 10.1385/1-59259-271-6:225. [DOI] [PubMed] [Google Scholar]
- Winter G., Chen Q.J., Flick K., Kremsner P., Fernandez V., Wahlgren M. The 3D7var5.2 (var COMMON) type var gene family is commonly expressed in nonplacental Plasmodium falciparum malaria. Mol. Biochem. Parasitol. 2003;127:179–191. doi: 10.1016/s0166-6851(03)00004-5. [DOI] [PubMed] [Google Scholar]
- Wutz A., Rasmussen T.P., Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 2002;30:167–174. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- Yang P.K., Kuroda M.I. Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell. 2007;128:777–786. doi: 10.1016/j.cell.2007.01.032. [DOI] [PubMed] [Google Scholar]