Abstract
The 5′ external transcribed spacer (5′ETS) is critical for 18S rRNA formation and is the longest noncoding region in a ribosomal RNA transcript. Here we show that processing in mouse 5′ETS involves two cleavage events. Processing at site A′ corresponds to the previously described “primary cleavage,” which precedes other processing steps. Processing at the novel site A0 occurs 1 kb downstream from A′ yielding two new rRNA precursors: 43S and 29S. The excised 5′-A′ and A′-A0 fragments are rapidly degraded under normal conditions. Depletion of the exosome component EXOSC10/PM-Scl100 (ortholog of yeast Rrp6p) results in a strong accumulation of the A′-A0 spacer fragment in mouse cells. We discuss the finding of a second processing site in mammalian 5′ETS in relation to the involvement of the U3 snoRNA in pre-rRNA processing and present a revised map of the mouse 18S rRNA processing pathway.
Keywords: RNA processing, ribosome biogenesis, mammalian cells, U3 snoRNA, exosome
INTRODUCTION
Mammalian ribosomal RNA (rRNA) is transcribed by RNA polymerase I as a large precursor molecule of ∼14 kb in size. The primary transcript (47S pre-rRNA) is processed into mature 18S, 5.8S, and 28S rRNAs by endonucleolytic cleavages, exonucleolytic trimming, and covalent nucleotide modifications. About half of the primary rRNA transcript is comprised of the regions termed external transcribed spacers (5′ETS and 3′ETS) and internal transcribed spacers (ITS1 and ITS2) that are removed during rRNA maturation (Eichler and Craig 1994).
The 5′ETS, the first part of pre-rRNA transcribed by RNA polymerase I, is the longest noncoding region in the transcript, ∼4 kb in mouse and ∼3.7 kb in human. Although the length and nucleotide sequence of 5′ETS can vary significantly between species, phylogenetically conserved regions of complementarity exist between 5′ETS and the essential box C/D small nucleolar (sno)RNA U3 (Borovjagin and Gerbi 2000). U3 snoRNA is an integral component of the early 90S preribosomal particles that contain a large number of protein factors involved in 40S subunit assembly (Dragon et al. 2002; Grandi et al. 2002). Mutational analysis suggests that U3 is required for processing within 5′ETS, at the 5′ETS/18S boundary and within ITS1 (Hughes and Ares 1991; Beltrame and Tollervey 1992). In Saccharomyces cerevisiae, cleavage at site A0 located in 5′ETS 90 nucleotides (nt) upstream of the 5′ end of 18S rRNA requires U3 snoRNA (Hughes 1996). In Xenopus laevis and Trypanosoma brucei, two distinct cleavages in 5′ETS, A′ and A0 are dependent on U3 snoRNA (Hartshorne and Toyofuku 1999; Borovjagin and Gerbi 2001). Only one 5′ETS cleavage was identified in the early studies of mammalian rRNA processing. This cleavage, termed the “primary processing event,” rapidly occurs in the pre-rRNA transcript around nucleotide 650 in mouse and 414 in human (Kass et al. 1987), and is U3-dependent (Kass et al. 1990). A recent study, however, suggested that a second cleavage in 5′ETS may also occur in human cells (Rouquette et al. 2005).
Processing of pre-rRNA requires a large number of auxiliary protein factors (reviewed in Henras et al. 2008). A conserved complex of 3′ to 5′ exonucleases termed the exosome is involved in multiple steps in pre-rRNA maturation (Mitchell et al. 1997; Raijmakers et al. 2004; Houseley et al. 2006). The mammalian RNase D component of the exosome EXOSC10 (also known as PM-Scl100) is an ortholog of the S. cerevisiae Rrp6p (Allmang et al. 1999). In our studies of EXOSC10 we observed accumulation of a novel 5′ETS-derived fragment after the depletion of this protein from mouse cells. This finding led us to reexamine the pre-rRNA processing steps occurring in mouse cells. Here we show that mouse 5′ETS contains a previously undescribed processing site that corresponds to the A0 site observed in other organisms including Xenopus and yeast. Our results also demonstrate a conserved role for the Rrp6-like enzymes in pre-rRNA spacer degradation, and suggest that 5′ETS processing, dependent on U3 snoRNA binding, is similar for all eukaryotes.
RESULTS
Accumulation of a 5′ETS fragment in the absence of EXOSC10
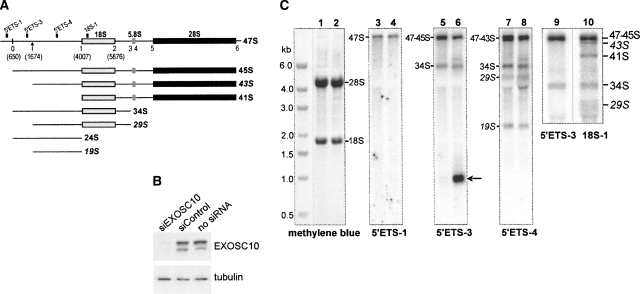
To explore the role of EXOSC10 in rRNA processing, we used siRNA to deplete this protein from NIH 3T3 cells, followed by Northern analysis of cellular RNA with probes complementary to different regions of the pre-rRNA transcript (Fig. 1A). Efficient down-regulation of the EXOSC10 protein was verified by immunoblotting (Fig. 1B). After EXOSC10 depletion, changes in the levels of several pre-rRNAs were observed, the most striking of which was accumulation of a 1-kb pre-rRNA fragment detected with probe 5′ETS-3 (Fig. 1C, lane 6). This fragment was not apparent in cells with normal EXOSC10 levels (Fig. 1C, lane 5). Other changes included the appearance of aberrant products of cleavage in ITS1 (a band visible above 34S in Fig. 1C, lanes 6,8) and accumulation of 12S to 5.8S intermediates (data not shown).
FIGURE 1.
Depletion of EXOSC10 unmasks a second processing site in the 5′ETS of mouse pre-rRNA. (A) Structure of the mouse 47S pre-rRNA transcript showing major cleavage sites and relative position of hybridization probes. Novel pre-rRNAs generated by cleavage at the second site described here (indicated by an arrow) are shown in italics. (B) EXOSC10 protein level is reduced after transfection with specific siRNA, but not after transfections with a heterologous siRNA as compared with mock-transfected cells. Immunoblotting of cell lysates was perfomed with antibodies against PM-Scl100; beta-tubulin was used as a loading control. (C) Hybridization analysis of pre-rRNA. Total RNA was isolated from siControl-transfected (lanes 1,3,5,7,9,10) or siEXOSC10-transfected cells (lanes 2,4,6,8), separated by formaldehyde-agarose gel electrophoresis, blotted on a membrane, stained with methylene blue to reveal 28S, 18S rRNAs, and marker bands, and hybridized with the indicated oligonucleotide probes. Arrow points to the spacer fragment accumulating in EXOSC10-depleted cells. Lanes 9 and 10 show a close-up view of hybridizations in which the gel was run for a longer time to improve separation between 43S and 47/45S pre-rRNAs.
Hybridization analysis of processing within 5′ETS
To understand the origin of the novel 1-kb fragment observed after EXOSC10 depletion, we examined pre-rRNAs that contained 5′ETS-derived regions in a series of hybridizations. The 1-kb fragment was not detected with probe 5′ETS-1 hybridizing upstream of the primary cleavage site (Fig. 1C, lanes 3–4). We therefore asked whether a second cleavage may exist in 5′ETS downstream from the primary site (Fig. 1A, shown by the arrow). This cleavage would also be expected to generate shorter forms of the known 45S, 34S, and 24S pre-rRNAs (Fig. 1A). Indeed, close analysis of hybridizations revealed three precursors that were not detected in previous studies. A shorter form of 34S pre-rRNA, which we designated as 29S, was found to migrate in the wake of 28S rRNA in hybridizations with probe 5′ETS-4 (Fig. 1C, lanes 7–8). 29S pre-rRNA did not hybridize with probe 5′ETS-3 (Fig. 1C, lanes 5–6), while both 5′ETS-3 and 5′ETS-4 detected 34S pre-rRNA on the same blots (Fig. 1C, lanes 5–8). This hybridization pattern suggests that 29S differs from 34S pre-rRNA by the lack of a 5′-proximal segment. The size of 29S pre-rRNA (∼5 kb) makes it partially obscured on blots by the highly abundant 28S rRNA (4.7 kb). However, 29S pre-rRNA was reproducibly detected with several other probes including 18S-1 that hybridizes with all 18S precursors (Fig. 1C, lane 10; data not shown).
The second predicted pre-rRNA, 43S (see Fig. 1A), was detected in hybridizations with probe 18S-1 as a distinct band migrating slightly ahead of the abundant 45S pre-rRNA (Fig. 1C, lane 10). The 43S pre-rRNA was detectable with a number of other ITS1- and ITS2-specific probes (data not shown), but not with probe 5′ETS-3 (Fig. 1C, lane 9). The lack of a region hybridizing with 5′ETS-3 is consistent with the notion that 43S is a product of 45S pre-rRNA derived by a second cleavage in 5′ETS.
Finally, hybridization with probe 5′ETS-4 revealed a prominent band migrating above 18S rRNA that we refer to as 19S (Fig. 1C, lanes 7–8). A large 5′ETS fragment formerly described as 24S rRNA was proposed to extend from the primary cleavage site to the 5′ end of 18S rRNA (Eichler and Craig 1994). However, the size of the 19S RNA detected by 5′ETS-4 hybridizations is 2.4 kb, which is 1 kb shorter than the distance between the primary cleavage site (nucleotide 650) and site 1 (nucleotide 4007). We conclude that the spacer fragment detected by hybridizations with probe 5′ETS-4 is generated by a second cleavage in 5′ETS located ∼1 kb downstream from the primary cleavage site.
Collectively, these data indicate that two cleavages, separated by ∼1 kb, take place within the mouse 5′ETS. Processing of 45S and 34S pre-rRNAs at the novel site generates 43S and 29S pre-rRNAs, respectively. The latter pre-rRNAs are subsequently cleaved at site 1, releasing the 19S spacer fragment (Fig. 1A). The 43S and 29S pre-rRNAs migrate close to abundant rRNA species during gel electrophoresis, which may have hindered their identification in the past.
Mapping of the 5′ETS cleavage sites
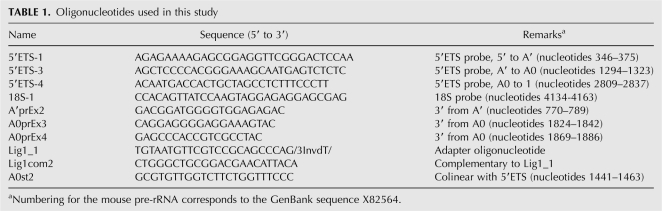
To map the second cleavage site within 5′ETS more precisely, we performed primer extensions on pre-rRNA using oligonucleotide primer A0prEx4 (see Table 1 for primer description). This assay revealed two strong reverse transcriptase stops at nucleotide 1673/1674 (Fig. 2A, lane 1). The identical stop positions were detected with primer A0prEx3 (data not shown). To reexamine the position of the primary cleavage site, we performed primer extensions with primer A′prEx2. Several stops were detected, the strongest of which corresponded to a cleavage at nucleotide 650 (Fig. 2A, lane 2), consistent with previous studies (Kass et al. 1987). Based on these results, the distance between the newly identified site and the primary processing site is ∼1025 nt, in good agreement with the hybridization results shown above.
TABLE 1.
Oligonucleotides used in this study
FIGURE 2.
Analysis of the 5′ and 3′ ends formed by the novel cleavage in mouse 5′ETS. (A) Primer extension analysis with primer A0prEx4 to map the novel cleavage site (lane 1) and A′prEx2 to map the primary cleavage site (lane 2) are shown next to the matching sequencing reactions. The nucleotide sequence shown with the 5′ end at the top is colinear with pre-rRNA; major and minor polymerase stops are indicated by the large and small arrowheads, respectively. (B) 3′ ends of RNA fragments derived by processing at nucleotide 1673/1674 in EXOSC10-depleted cells. See Materials and Methods for details. Nontemplated bases are shown in italics. Numbering in pre-rRNA corresponds to the GenBank sequence X82564.
To corroborate the primer extension data, we examined the 3′ ends of the RNA fragments accumulated in EXOSC10-depleted cells. We ligated an anchor oligonucleotide to the 3′ end of RNA isolated from these cells using T4 RNA ligase, followed by RT-PCR with primers designed to amplify sequences between nucleotide 1441 in 5′ETS and the attached anchor. Sequencing of several cloned PCR products showed that their 3′ ends terminated 980–1025 base pairs downstream from the primary cleavage site (Fig. 2B). The 3′ end of the longest clone (E23) was found to abut the second major primer extension stop detected at nucleotide 1673 (Fig. 2A, lane 1). The observed variability in the 3′ ends may result from cleavage at nucleotide 1673/1674 followed by partial 3′ to 5′ exonucleolytic degradation. Although none of our sequenced clones contained oligo(A) tails which are characteristic for many yeast exosome substrates (Houseley et al. 2006), some sequences contained nontemplated nucleotides at their 3′ ends. The precise mechanism of degradation of the mammalian 5′ETS spacer in the absence of EXOSC10 remains to be determined.
A revised map of mammalian pre-rRNA processing
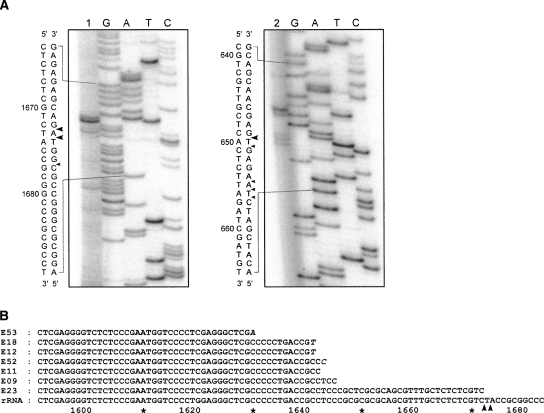
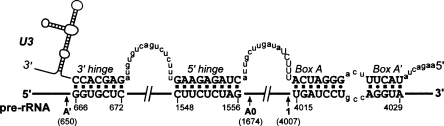
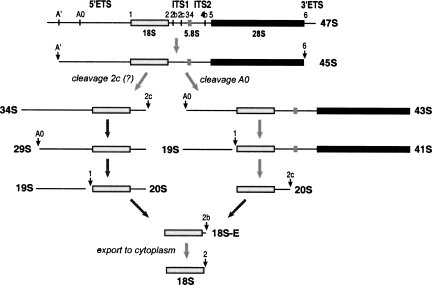
The sequence of the mouse 5′ETS contains two elements that are complementary to the single-stranded regions of the U3 snoRNA called 3′ hinge and 5′ hinge (Fig. 3). The 3′ hinge in the mouse is complementary to a region 16 nt downstream from the primary cleavage site. The complementarity between 5′ETS and the U3 3′ hinge is a conserved feature in metazoans and is required for 18S formation, as suggested by mutation analysis in X. laevis (Borovjagin and Gerbi 2000). The 5′ hinge of mouse U3 can base-pair with 5′ETS 118 nt upstream of the second cleavage site we describe here. In other organisms studied in this respect, the A0 cleavage in 5′ETS was observed 50–230 nt downstream from the 5′-hinge binding site (see Discussion). Base-pairing between pre-rRNA and the hinge regions is thought to mediate docking of U3-containing complexes on pre-rRNA (Borovjagin and Gerbi 2005). For consistency with the nomenclature used in Xenopus and T. brucei (Hartshorne et al. 2001; Borovjagin and Gerbi 2005), we propose to designate the newly identified site at nucleotide 1673/1674 as A0 and rename the primary cleavage site at nucleotide 650 (formerly site 0) as A′. Taken together, our findings suggest a new picture of the processing pathway that leads to the synthesis of mouse 18S rRNA (Fig. 4).
FIGURE 3.
A summary of the proposed interactions between mouse U3 snoRNA and pre-rRNA. Two regions (5′ hinge and 3′ hinge) in U3 snoRNA can base-pair with two sequences in the 5′ETS. Box A and A′ in U3 exhibit conserved complementarity to the 5′ region in 18S rRNA. According to the model of Borovjagin and Gerbi (2005), base-pairing between the U3 hinges and 5′ETS may be required for the spatial reorganization of the complex allowing binding of boxes A and A′ to the 18S sequences in pre-rRNA.
FIGURE 4.
A revised map of pre-rRNA processing leading to 18S rRNA formation in mouse cells. The major parts on the primary transcript (47S) and cleavage sites are indicated at the top. The 47S pre-rRNA is normally short-lived in cells due to the rapidly occurring processing at sites A′ in 5′ETS and 6 in 3′ETS that generate 45S pre-rRNA. Subsequent cleavages may first occur either in ITS1 (probably at site 2c) or in 5′ETS (site A0). The end result of both pathways is the removal of the remaining portion of 5′ETS yielding 20S pre-rRNA, which is converted to 18S-E and finally processed to the mature 18S rRNA in the cytoplasm. Conventional names for the precursors are based on the nomenclature of Bowman et al. (1981) and Eichler and Craig (1994).
DISCUSSION
In this study, we identified a novel processing site within mouse pre-rRNA. The existence of this site in the 5′ETS is supported by hybridizations, primer extensions, and sequencing analysis of 5′ETS-derived precursors. These results provide a corrected picture of pre-rRNA processing in the mouse and support the view that U3-dependent processing steps that involve the molecular interaction between this snoRNA and 5′ETS have been conserved throughout eukaryotes.
Mammalian A0 site
In all studied organisms, the conserved elements within the 5′ domain of U3 can base-pair with 18S sequences within pre-rRNA, and this interaction is critical for 18S rRNA formation (Hughes 1996; Sharma and Tollervey 1999). In addition, phylogenetic and mutational studies suggest that the 5′ hinge and 3′ hinge of U3 snoRNA base-pair with 5′ETS and help to correctly position U3 on pre-rRNA (Borovjagin and Gerbi 2000; Borovjagin and Gerbi 2005). In X. laevis, the 5′ hinge interaction site has been located 46 nt upstream of the A0 cleavage (Borovjagin and Gerbi 2001). The corresponding region in T. brucei (termed site 3) lies 162 nt upstream of A0 (Hartshorne and Toyofuku 1999). The 5′ hinge region in S. cerevisiae U3 can be crosslinked to the 5′ETS sequence ∼140 nt upstream of the A0 cleavage (Beltrame and Tollervey 1992).
Our data provide experimental evidence in support of the model that predicts that processing of the 5′ETS in all eukaryotic pre-rRNA involves common mechanisms. The mouse U3 3′ hinge elements can base-pair to a short sequence adjacent to site A′, while the newly discovered processing site at position 1673/1674 is located ∼120 nt downstream from the region that can base-pair with the mouse U3 5′ hinge sequence (Fig. 3). The arrangement of processing sites in the mouse 5′ETS is thus similar to that in X. laevis and T. brucei. These data also extend the results of a recent study that showed accumulation of a precursor analogous to 29S pre-rRNA after treatment of human HeLa cells with leptomycin B (Rouquette et al. 2005). In this study, the putative human A0 site was mapped at position G1643, which is located ∼230 nt downstream from the region that exhibits complementarity to the 5′ hinge of U3 snoRNA according to the phylogenetic analysis of Borovjagin and Gerbi (2000). Thus, both human and mouse cells appear to possess the A0 processing site within 5′ETS located a short distance downstream from the region capable of base-pairing with the 5′ hinge of U3.
Role of Rrp6 homologs in degradation of 5′ETS spacers
EXOSC10 is a member of the RNase D family of exonucleases, and is associated with the exosome complex (Raijmakers et al. 2004). We found that depletion of mouse EXOSC10 leads to a dramatic accumulation of the excised A′–A0 fragment in mouse cells (Fig. 1C, lane 6). Previous studies in S. cerevisiae demonstrated that depletion of exosome components leads to accumulation of a 5′–A0 spacer fragment (de la Cruz et al. 1998; Allmang et al. 1999). Recently, it was shown that a 450-nt fragment derived from 5′ETS accumulates upon knockdown of RRP6L2, one of RRP6 homologs in Arabidopsis thaliana (Lange et al. 2008). Thus, degradation of 5′-proximal fragments generated by A0 cleavage is a conserved function of the Rrp6-like exonucleases. Notably, the exosome-associated helicase Mtr4p (Dob1p) is also required for the efficient spacer degradation (de la Cruz et al. 1998). What is a possible reason for these observations? Computer analysis of the 5′ regions preceding A0 shows no apparent secondary structure that could explain the requirement for the exosome in spacer degradation. Recently, several proteins have been shown to associate directly with nascent pre-rRNA transcripts (Gallagher et al. 2004; Segerstolpe et al. 2008). These proteins might facilitate interactions between ETS and U3 snoRNA or participate in cleavage reactions, but could also impede degradation of the excised fragments when exosome function is compromised. We speculate that the exosome and an associated helicase may be required to displace such proteins that bind to pre-rRNA upstream of the A0 site.
Variability in processing events
Although pre-rRNA processing is often depicted as a linear pathway, many cleavages in mammalian rRNA do not follow a strict temporal order (Bowman et al. 1981). The observed steady-state levels of “major” and “minor” precursors likely reflect the different kinetics of their synthesis and conversion to daughter species. Our analysis of mouse rRNA precursors shows that cleavage at the newly identified site A0 can occur either before or after processing in ITS1 that separates 18S from the 5.8S/28S rRNA (Fig. 4). One notable observation, however, is that the 5′ETS fragment with the 3′ end generated by cleavage at site 1 (19S RNA) appears to have its 5′ end derived by cleavage A0, whereas 24S RNA extending from site A′ to site 1 (see Fig. 1A) was not readily detectable in our assays. This result suggests that processing at sites 1 and A0 may be coupled to some extent, possibly reflecting the requirement for the binding of U3 snoRNA to both 5′ETS and 18S sequences. The exact mechanism of these cleavages and the enzymes that catalyze these processing events require further investigation.
MATERIALS AND METHODS
Cell culture, protein, and RNA analyses
NIH 3T3 cells were maintained in DMEM (Gibco) supplemented with 10% calf serum and penicillin/streptomycin. EXOSC10 knockdowns were carried out by calcium phosphate-mediated transfections with an siRNA Smartpool (Dharmacon). Protein levels were monitored by immunoblotting using antibodies against PM-Scl100 (Sigma P4124) and beta-tubulin (Sigma T4026). RNA was isolated using Trizol (Invitrogen). Hybridization analysis of rRNA was performed as described (Pestov et al. 2008) using probes listed in Table 1.
Analysis of the 5′ ends of RNA
A total of 4 pmol of primers A0prEx4 or A′prEx2 were [32P]-labeled with T4 polynucleotide kinase, mixed with 1 μg RNA, incubated at 90°C for 5 min, and annealed at 60°C for 5 min. Primer extensions were performed in a 20 μL reaction mixture containing 50 U MonsterScript reverse transcriptase (Epicentre), 1×first strand buffer supplied with the enzyme and 4.5% formamide for 1 h at 60°C. Reactions were stopped by the addition of 0.3 M sodium acetate, 2 mM EDTA, and 0.1% SDS. Reaction products were precipitated with ethanol, dried, and dissolved in 3 μL of loading buffer (95% formamide, 10 mM EDTA, 0.1% xylene cyanol, and 0.1% bromophenol blue, at final pH 11). Mouse cDNA clones obtained from ATCC (GenBank IDs: CF899888 and AA546819) were used as reference templates for sequencing with SequiTherm EXCEL II (Epicentre). Sequencing reactions were diluted 10 times and resolved next to primer extension products on a 6% polyacrylamide/urea gel.
Analysis of the 3′ ends of RNA
Total RNA was isolated from cells transfected with EXOSC10 siRNA, dephosphorylated with alkaline phosphatase, purified by ethanol precipitation, and ligated with 5′-phosphorylated oligonucleotide Lig1_1 that has a blocked 3′ end (Table 1) using T4 RNA ligase (NEB). Reaction products were precipitated, dissolved in 10 mM Tris–HCl at pH 8, annealed with primer Lig1com2 at 90°C for 5 min, followed by 5 min at 60°C, and used for cDNA synthesis with MonsterScript reverse transcriptase. PCR amplification of 5′ETS-derived sequences was performed using primers A0st2 and Lig1com2 with the Taq Master Mix (NEB) supplemented with 5% DMSO. PCR products were purified from an agarose gel and cloned into the pGEM-T vector (Promega).
ACKNOWLEDGMENTS
We thank Natalia Shcherbik for critical reading of the manuscript and helpful suggestions. This work was supported by NIH Grant GM074091 to D.G.P.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1384709.
REFERENCES
- Allmang C., Petfalski E., Podtelejnikov A., Mann M., Tollervey D., Mitchell P. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes & Dev. 1999;13:2148–2158. doi: 10.1101/gad.13.16.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrame M., Tollervey D. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J. 1992;11:1531–1542. doi: 10.1002/j.1460-2075.1992.tb05198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovjagin A.V., Gerbi S.A. The spacing between functional cis-elements of U3 snoRNA is critical for rRNA processing. J. Mol. Biol. 2000;300:57–74. doi: 10.1006/jmbi.2000.3798. [DOI] [PubMed] [Google Scholar]
- Borovjagin A.V., Gerbi S.A. Xenopus U3 snoRNA GAC-Box A′ and Box A sequences play distinct functional roles in rRNA processing. Mol. Cell. Biol. 2001;21:6210–6221. doi: 10.1128/MCB.21.18.6210-6221.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borovjagin A.V., Gerbi S.A. An evolutionary intra-molecular shift in the preferred U3 snoRNA binding site on pre-ribosomal RNA. Nucleic Acids Res. 2005;33:4995–5005. doi: 10.1093/nar/gki815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman L.H., Rabin B., Schlessinger D. Multiple ribosomal RNA cleavage pathways in mammalian cells. Nucleic Acids Res. 1981;9:4951–4966. doi: 10.1093/nar/9.19.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Kressler D., Tollervey D., Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae . EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon F., Gallagher J.E.G., Compagnone-Post P.A., Mitchell B.M., Porwancher K.A., Wehner K.A., Wormsley S., Settlage R.E., Shabanowitz J., Osheim Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler D.C., Craig N. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1994;49:197–239. doi: 10.1016/s0079-6603(08)60051-3. [DOI] [PubMed] [Google Scholar]
- Gallagher J.E.G., Dunbar D.A., Granneman S., Mitchell B.M., Osheim Y., Beyer A.L., Baserga S.J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P., Rybin V., Bassler J., Petfalski E., Strauss D., Marzioch M., Schäfer T., Kuster B., Tschochner H., Tollervey D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Hartshorne T., Toyofuku W. Two 5′-ETS regions implicated in interactions with U3 snoRNA are required for small subunit rRNA maturation in Trypanosoma brucei . Nucleic Acids Res. 1999;27:3300–3309. doi: 10.1093/nar/27.16.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T., Toyofuku W., Hollenbaugh J. Trypanosoma brucei 5′ETS A′-cleavage is directed by 3′-adjacent sequences, but not two U3 snoRNA-binding elements, which are all required for subsequent pre-small subunit rRNA processing events. J. Mol. Biol. 2001;313:733–749. doi: 10.1006/jmbi.2001.5078. [DOI] [PubMed] [Google Scholar]
- Henras A.K., Soudet J., Gérus M., Lebaron S., Caizergues-Ferrer M., Mougin A., Henry Y. The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell. Mol. Life Sci. 2008;65:2334–2359. doi: 10.1007/s00018-008-8027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J., LaCava J., Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- Hughes J.M. Functional base-pairing interaction between highly conserved elements of U3 small nucleolar RNA and the small ribosomal subunit RNA. J. Mol. Biol. 1996;259:645–654. doi: 10.1006/jmbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Hughes J.M., Ares M.J. Depletion of U3 small nucleolar RNA inhibits cleavage in the 5′ external transcribed spacer of yeast pre-ribosomal RNA and impairs formation of 18S ribosomal RNA. EMBO J. 1991;10:4231–4239. doi: 10.1002/j.1460-2075.1991.tb05001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Craig N., Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol. Cell. Biol. 1987;7:2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S., Tyc K., Steitz J.A., Sollner-Webb B. The U3 small nucleolar ribonucleoprotein functions in the first step of pre-ribosomal RNA processing. Cell. 1990;60:897–908. doi: 10.1016/0092-8674(90)90338-f. [DOI] [PubMed] [Google Scholar]
- Lange H., Holec S., Cognat V., Pieuchot L., Le Ret M., Canaday J., Gagliardi D. Degradation of a polyadenylated rRNA maturation by-product involves one of the three RRP6-like proteins in Arabidopsis thaliana . Mol. Cell. Biol. 2008;28:3038–3044. doi: 10.1128/MCB.02064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski E., Shevchenko A., Mann M., Tollervey D. The exosome: A conserved eukaryotic RNA processing complex containing multiple 3′ → 5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- Pestov D.G., Lapik Y.R., Lau L.F. Assays for ribosomal RNA processing and ribosome assembly. In: Bonifacino J.S., et al., editors. Current protocols in cell biology. Wiley; New York: 2008. pp. 22.11.1–22.11.16. [DOI] [PubMed] [Google Scholar]
- Raijmakers R., Schilders G., Pruijn G.J.M. The exosome, a molecular machine for controlled RNA degradation in both nucleus and cytoplasm. Eur. J. Cell Biol. 2004;83:175–183. doi: 10.1078/0171-9335-00385. [DOI] [PubMed] [Google Scholar]
- Rouquette J., Choesmel V., Gleizes P. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–2872. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segerstolpe A., Lundkvist P., Osheim Y.N., Beyer A.L., Wieslander L. Mrd1p binds to pre-rRNA early during transcription independent of U3 snoRNA and is required for compaction of the pre-rRNA into small subunit processomes. Nucleic Acids Res. 2008;36:4364–4380. doi: 10.1093/nar/gkn384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., Tollervey D. Base pairing between U3 small nucleolar RNA and the 5′ end of 18S rRNA is required for pre-rRNA processing. Mol. Cell. Biol. 1999;19:6012–6019. doi: 10.1128/mcb.19.9.6012. [DOI] [PMC free article] [PubMed] [Google Scholar]