Abstract
The essential transcriptional repressor REST (repressor element 1-silencing transcription factor) plays central roles in development and human disease by regulating a large cohort of neural genes. These have conventionally fallen into the class of known, protein-coding genes; recently, however, several noncoding microRNA genes were identified as REST targets. Given the widespread transcription of messenger RNA-like, noncoding RNAs (“macroRNAs”), some of which are functional and implicated in disease in mammalian genomes, we sought to determine whether this class of noncoding RNAs can also be regulated by REST. By applying a new, unbiased target gene annotation pipeline to computationally discovered REST binding sites, we find that 23% of mammalian REST genomic binding sites are within 10 kb of a macroRNA gene. These putative target genes were overlooked by previous studies. Focusing on a set of 18 candidate macroRNA targets from mouse, we experimentally demonstrate that two are regulated by REST in neural stem cells. Flanking protein-coding genes are, at most, weakly repressed, suggesting specific targeting of the macroRNAs by REST. Similar to the majority of known REST target genes, both of these macroRNAs are induced during nervous system development and have neurally restricted expression profiles in adult mouse. We observe a similar phenomenon in human: the DiGeorge syndrome-associated noncoding RNA, DGCR5, is repressed by REST through a proximal upstream binding site. Therefore neural macroRNAs represent an additional component of the REST regulatory network. These macroRNAs are new candidates for understanding the role of REST in neuronal development, neurodegeneration, and cancer.
Keywords: REST, NRSF, RE1, ncRNA, noncoding RNA, macroRNA, DGCR5, neural stem cell, transcription, regulation
INTRODUCTION
Despite recent technological and conceptual advances in genomics, the full functional content of mammalian genomes remains uncertain. In addition to well-annotated protein-coding exons, there is evidence for extensive functionality in noncoding DNA: (1) the small variation in numbers of known genes between species of vastly different complexity; (2) extensive negative selection outside of protein-coding exons (Lunter et al. 2006); and (3) the existence of tens of thousands of non-protein-coding transcripts (noncoding RNAs or ncRNAs) in mammalian cells (Maeda et al. 2006). These observations have led to the hypothesis that uncharacterized ncRNAs may underlie unexplained aspects of organismal complexity, evolution, and human disease (Mattick 2004). Discovering and assigning function to ncRNAs is a major challenge to contemporary genomics.
Clues about the functions of genes are often gleaned from their spatiotemporal expression patterns and the regulatory factors controlling them. One such transcriptional regulator is the repressor element 1-silencing transcription factor (REST), which represses a large cohort of neurally restricted genes with roles in nervous system development and neuronal function (Chong et al. 1995; Schoenherr and Anderson 1995). REST is recruited to the regulatory regions of target genes by its cognate DNA recognition motif, the repressor element 1 (RE1). Gene repression is thence affected by the nucleation of co-repressor complexes containing various histone-modifying and chromatin-remodeling activities (Ooi and Wood 2007). During development, REST is a key member of regulatory networks governing embryonic stem cell (ESC) pluripotency (Ballas et al. 2005; Boyer et al. 2005; Loh et al. 2006) and neural differentiation (Chen et al. 1998; Ballas et al. 2001; Greenway et al. 2007). In the adult, REST plays key roles in several disease-related pathways, including neuronal death following ischemia (Calderone et al. 2003), cardiac hypertrophy (Kuwahara et al. 2003), vascular smooth muscle proliferation (Cheong et al. 2005), and neurodegeneration in Huntington's disease (Zuccato et al. 2003). In these cases, de novo identification of REST target genes by sequence analysis has greatly facilitated our understanding of the disease pathway. Importantly, this understanding relies on our ability to accurately predict all classes of REST target genes—the focus of this study.
Since its discovery, REST has been the subject of bioinformatics approaches to identify its target genes by searching for RE1s in genomic sequence. This was facilitated by the unusual length of the RE1: at 21 base pairs (bp), it is considerably longer than most transcriptional regulatory motifs, enabling the accurate identification of REST-binding sites by sequence analysis. We, and others, have employed consensus sequence (Schoenherr et al. 1996; Lunyak et al. 2002; Bruce et al. 2004), position-specific scoring matrix (Johnson et al. 2006), and sequence conservation (Mortazavi et al. 2006; Wu and Xie 2006) to create genome-wide maps of REST-binding sites. Target genes were then inferred by simply comparing RE1 locations to catalogs of known (predominantly protein-coding) genes and identifying the most proximal gene to each RE1. These studies agree on the approximate number of REST target genes and their enrichment for neural-related function. But while recent genome-wide experiments have validated the accuracy of later bioinformatics RE1 predictions, it has not been possible to comprehensively test resultant REST target gene predictions. Moreover, recent microarray gene expression studies in our laboratory indicate that REST only regulates a small fraction of annotated, protein-coding target genes, despite recruitment to the majority of predicted RE1 sites (Johnson et al. 2008a). This leads us to ask, do new classes of noncoding REST target genes exist, which are not included in previous annotations?
In fact, the first non-protein-coding target genes of REST were recently discovered among the class of small, regulatory RNAs, the microRNAs (Bartel 2003). A cohort of neurally expressed microRNA genes is repressed through conserved RE1s in nonneural and immature neural cell types (Conaco et al. 2006; Wu and Xie 2006; Johnson et al. 2008b). Interestingly, we were able to show that at least one of these microRNAs, mir-132 (a positive regulator of neurite growth) (Vo et al. 2005), is aberrantly repressed in the striatum of Huntington's disease patients, similar to protein-coding REST targets like BDNF (Zuccato et al. 2003). This supports the notion that new classes of noncoding target genes remain to be discovered, and could participate in disease processes involving REST.
Among the noncoding RNAs, the biological roles and molecular mechanism of microRNAs are relatively well understood, and their distinctive structural characteristics mean that they are accurately annotated (although new examples are still being discovered) (Griffiths-Jones 2004). In contrast, little is known about the numerous, but rather amorphous, class of so-called “macroRNAs” (Furuno et al. 2006): frequently spliced, mRNA-like transcripts with few defining structural or sequence characteristics. The FANTOM3 Transcriptome Consortium discovered 34,030 distinct macroRNAs in the mouse genome (Maeda et al. 2006). The significance of this pervasive macroRNA transcription is the subject of some debate; their generally low expression levels (Imanishi et al. 2004) and the small number discovered in functional screens before now support the argument that they represent “transcriptional noise” (Ponjavic et al. 2007). However, diverse evidence supports their important biological role: (1) nonneutral sequence conservation of promoters and splice sites that is comparable to that of protein-coding genes (Ponjavic et al. 2007); (2) cell- and tissue-specific expression patterns (Ravasi et al. 2006; Mercer et al. 2008); (3) disease associations (Louro et al. 2007; Pasmant et al. 2007; Khalil et al. 2008; Perez et al. 2008); and (4) experimentally demonstrated functions for an increasing number of examples, including NRON (Willingham et al. 2005), trafficking of the transcription factor NFAT; Xist (Nesterova et al. 2001), X-chromosome inactivation; HOTAIR (Rinn et al. 2007), transcriptional silencing of the HOXD locus; and Evf, coactivator to the homeodomain protein Dlx-2 (Feng et al. 2006).
In light of this, we asked whether REST can regulate expression of macroRNAs. To discover potential macroRNA targets of REST, we devised an unbiased annotation strategy that considers both protein-coding and noncoding transcripts as potential targets. Reannotation of RE1 catalogs from mouse and human suggests that REST regulates macroRNA expression from approximately one-quarter of its binding sites. Experimental validation of a focused set of candidate targets yields two neural-specific macroRNAs that are directly regulated by REST in mouse. In human, the DiGeorge syndrome-associated macroRNA DGCR5 (Sutherland et al. 1996) is a REST target. Our findings indicate that neural macroRNAs constitute a new class of REST target gene. Future large-scale identification of such macroRNAs should provide new candidates for understanding the role of REST in development and in diseases such as cancer, cardiac hypertrophy, and neurodegeneration.
RESULTS
An unbiased annotation pipeline for REST target gene discovery
We previously generated genome-wide maps of REST-binding sites (RE1s) in mouse and human, using a position-specific scoring matrix (PSSM) (Johnson et al. 2006). In that initial study, the sole target gene of each RE1 was defined as the nearest known gene to, and residing within 100 kb of, that RE1, almost invariably a protein-coding gene given the small numbers of known noncoding RNAs in curated gene catalogs. To investigate whether REST can regulate “macroRNAs” (defined as messenger RNA-like transcripts with no protein-coding capacity, which do not belong to well-defined, small ncRNA categories), we reannotated target genes for the same RE1 maps using a novel, unbiased pipeline that relies on experimental evidence for transcription, rather than on catalogs of known genes. Accordingly, the definition of potential targets was widened to include all high-quality mRNAs and ESTs, in addition to known protein-coding genes in Refseq and UCSC KnownGene catalogs. To provide increased flexibility in the annotation process, we collapsed potential target transcripts into transcriptional units (TUs): a TU is defined as the collection of all mRNAs and ESTs that overlap by at least one exonic base pair and are transcribed from the same genomic strand (Engstrom et al. 2006). A TU may represent a member of known Refseq or UCSC KnownGene catalogs, or may be an unannotated gene with high-quality mRNA or EST support. We downloaded 1783 mouse and 1301 human RE1s from the RE1 Database (Johnson et al. 2006). Of these, 1769 and 1283 could be unambiguously mapped to current mouse and human genomes, respectively, and were annotated (details of the annotation procedure can be found in Materials and Methods; full-annotation data sets can be found in Supplemental File 1). For every RE1, the nearest TU was assigned as the target gene. Although target gene assignment was based on proximity as in previous studies (Johnson et al. 2006, 2007; Mortazavi et al. 2006; Wu and Xie 2006; Otto et al. 2007), we modified the annotation strategy to account for possible regulation of multiple nearby target genes and for the possibility that an RE1 can regulate a nearby gene that is not the most proximal. Thus, we annotated all TUs within a 20-kb window centered on the RE1 as targets; if none were found, then the single most proximal TU within a 200-kb window was assigned as the target.
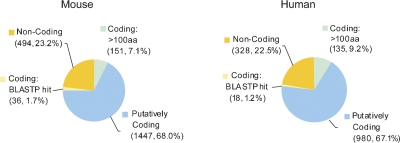
Using this methodology, we found that 77% and 78% of putative REST target genes encode protein in mouse and human, respectively (Fig. 1). As in previous studies (Johnson et al. 2006, 2007; Mortazavi et al. 2006; Wu and Xie 2006), these sets are significantly enriched for Gene Ontology terms relating to neuronal development and function (data not shown). Comparison of the two species' known protein-coding targets showed that at least 452 (31.2% and 46.1% of all protein-coding targets in mouse and human, respectively) are orthologous targets in both, reflecting evolutionary conservation of the REST regulatory network. In addition to known protein-coding targets, we found that ∼10% of targets (mouse 8.8%, human 10.4%) were not classified in known catalogs, but are likely to encode protein based on (1) BLASTP matches to other known proteins or (2) the existence of open reading frames (ORFs) greater than 100 amino acids, a crude but effective indicator of protein-coding capacity (Frith et al. 2006). Subsequent BLASTP analysis showed that a substantial minority of the >100 amino acids set are likely to be unannotated protein-coding genes (see Supplemental File 2). We found no substantive ORFs in the remaining 23% of target TUs (mouse 23.2%, human 22.5%), making them candidate macroRNA targets of REST.
FIGURE 1.
Protein-coding capacity of REST targets. The protein-coding capacity of every annotated target transcriptional unit (TU) was assessed: They were classified as “putatively coding” (annotated as such in public databases), “coding: BLASTP hit” (having significant homology with another known protein coding sequence, evidenced by BLASTP), “coding: >100 amino acids” (having an open reading frame greater than 100 amino acids on the positive strand), or “noncoding” (satisfying none of these criteria). See Materials and Methods for more details of the RE1 annotation pipeline.
Experimental validation of novel macroRNA targets of REST in neural stem cells
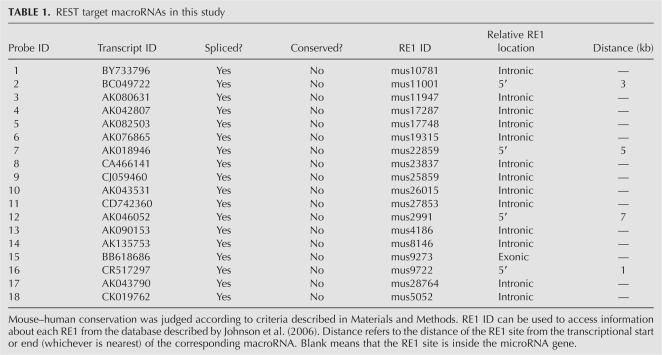
Reannotation suggested that REST may regulate expression of macroRNAs from almost one-quarter of its binding sites. In order to test whether such regulation takes place, we selected a short list of 18 RE1–macroRNA pairs for experimental validation (Table 1). These pairs were selected on the basis of good experimental support for both occupancy at the RE1 and expression of the macroRNA, and for the likelihood of a regulatory relationship between the two (see Materials and Methods for a detailed description of the selection process). All the candidate macroRNAs are spliced, and none are conserved in human by our conservation criteria (see Materials and Methods). First, we screened this panel of macroRNAs for REST binding at their associated RE1 sites. We already had evidence that REST physically interacts with the macroRNA genes in vivo: the 18 RE1s of our study set were selected on the basis of highly significant enrichment (P < 0.01) in multiple biological replicates of mouse embryonic stem (ES) cells by chromatin immunoprecipitation microarrays (ChIP-chip) (Supplemental File 3; Johnson et al. 2008a). We independently validated these findings by ChIP-qPCR (Fig. 2), detecting statistically significant (P < 0.05) recruitment of REST at all 18 macroRNAs.
TABLE 1.
REST target macroRNAs in this study
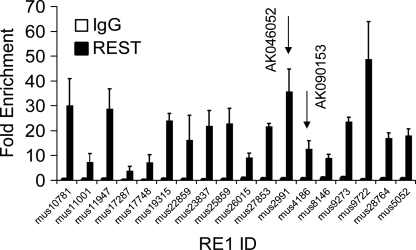
FIGURE 2.
REST is recruited to candidate target macroRNAs in vivo. To confirm the recruitment of REST to candidate macroRNAs, chromatin immunoprecipitation (ChIP) was carried out on the mouse embryonic stem cell line, E14. Experiments were carried out on at least three independent biological replicates, using a REST antibody, or negative control, nonspecific IgG. Resulting ChIP DNAs were interrogated by quantitative real-time PCR with primers flanking candidate REST binding sites (RE1s) and a distal non-RE1 locus. Fold enrichments were calculated with reference to the non-RE1 amplicon, using the ΔΔCT normalization method. Error bars indicate the standard error of the mean. RE1s of validated REST target macroRNAs are shown by arrows (see Figs. 3, 4).
In addition to ChIP evidence for recruitment, transcriptional responsiveness to REST inhibition indicates that a gene is a direct target of REST. Consequently, we used intron-spanning Taqman probes to profile macroRNA expression in response to the widely used dominant-negative REST construct, DN:REST (Chen et al. 1998). We expected that target macroRNAs would display elevated levels in the presence of DN:REST. REST is an important regulator of many neural genes in neural stem cells (NSCs); consequently, we carried out gene expression studies in the mouse NSC line, NS5 (Conti et al. 2005). We identified two candidate macroRNAs that displayed elevated levels in response to overexpression of DN:REST: AK046052 (Fig. 3) and AK090153 (Fig. 4). These observed differences in expression for both were statistically significant. We conclude, therefore, that REST negatively regulates transcription of some macroRNAs.
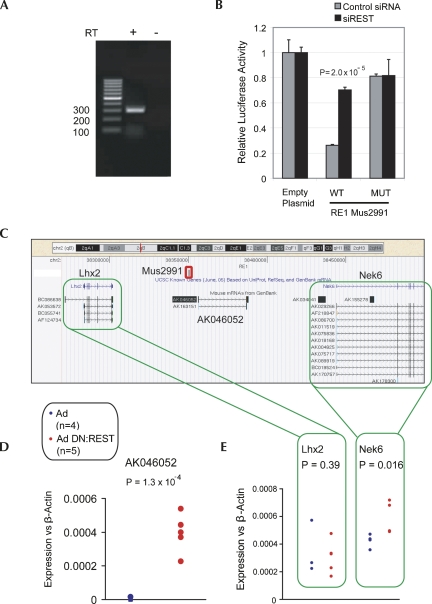
FIGURE 3.
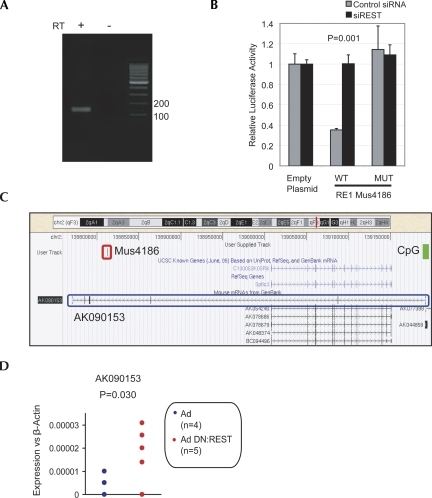
Noncoding transcript AK046052 is targeted by REST. (A) Expression of AK046052 in NS5 was validated by RT-PCR. A control reverse transcription (RT) reaction was carried out without RTase enzyme (−RT). (B) RE1 mus2991 is capable of repressing transcription of a reporter gene in a REST-dependent manner. The RE1 sequence (WT), or a mutated version (MUT), was tested in luciferase assay. The luciferase plasmid, pGL4TK, containing RE1 sequence constructs upstream of the TK promoter was transfected into HEK293 cells in the presence of siRNA targeting REST mRNA (siREST), or control nontargeting siRNA. Luciferase intensity was normalized to that of a control, Renilla plasmid. Ratios are expressed relative to pGL4TK containing no RE1 (Empty). Experiments were carried out on three biological replicates and two technical replicates. Error bars represent the standard error of the mean (SEM), and statistical significance was judged by a Student's t-test. (C) AK046052 resides on mouse Chromosome 2, flanked by known protein-coding genes Lhx2 and Nek6. The validated REST binding site (RE1) mus2991 is located ∼7 kb upstream of AK046052 (red rectangle). (D) The adenovirally delivered dominant-negative REST construct (Ad DN:REST) activated transcription of AK046052 in the neural stem cell line, NS5. Transcript levels of AK046052 were measured using intron-spanning Taqman probes, and normalized to the housekeeping gene, β-actin. Assays were carried out on four control replicates (adenovirus only, Ad) and five dominant-negative replicates (Ad DN:REST). (E) Nek6 was weakly de-repressed by DN:REST, while Lhx2 was unaffected. Expression assays using intron-spanning primers with SYBR green qPCR were carried out on the same samples as in D. Statistical significance was calculated using Student's t-test.
FIGURE 4.
Noncoding transcript AK090153 is targeted by REST. (A) Expression of AK090153 in NS5 was validated by RT-PCR. A control reverse transcription (RT) reaction was carried out without RTase enzyme (−RT). (B) The ability of RE1 mus4186 to repress reporter gene expression in a REST-dependent manner was demonstrated as in Figure 3B by luciferase assay. (C) AK090153 resides on mouse Chromosome 2 and encompasses the protein-coding gene Sptlc3 on the opposite strand. The validated REST binding site (RE1) mus4186 lies toward the 3′ intronic region of AK090153 (red rectangle). A CpG island encompasses the transcriptional start site (green rectangle). (D) Adenovirally delivered dominant-negative REST (Ad DN:REST) de-repressed transcription of AK090153 in the neural stem cell line, NS5. Transcript levels were measured by means of intron-spanning Taqman probes and normalized to the housekeeping gene β-actin. Assays were carried out on four control replicates (adenovirus only, Ad) and five dominant-negative replicates (Ad DN:REST). No expression of Sptlc3 could be detected using SYBR green qPCR with the same cDNA samples. Statistical significance was calculated using Student's t-test.
We validated the expression of macroRNA AK046052 in NS5 cells by conventional RT-PCR (Fig. 3A). It is regulated via the evolutionarily conserved RE1, mus2991, located 7 kb upstream of its non-CpG transcriptional start site (Fig. 3C; Supplemental File 4). In addition to recruiting REST in vivo, mus2991 is capable of repressing transcription of a luciferase reporter gene in a REST-dependent manner (Fig. 3B). The transcript mainly consists of various types of repeat elements (Supplemental File 5). Silent in control NS5, AK046052 was consistently activated by the presence of DN:REST (Fig. 3D). AK046052 is distally flanked by two protein-coding genes: Lhx2, a homeobox transcription factor involved in neurodevelopment, and Nek6, encoding a serine/threonine protein kinase cell cycle regulator. The ability of REST to simultaneously regulate multiple genes from a single binding site remains controversial, although based on its known role as an epigenetic regulator, it has been suggested that REST can repress extended intervals of chromatin and associated genes (Lunyak et al. 2002). We tested this in the context of AK046052 and found that Nek6 was weakly, but significantly, repressed in NS5 (Lhx2 was unaffected) (Fig. 3E). Subsequent inspection of whole-genome, chromatin immunoprecipitation coupled with deep sequencing revealed evidence for weak REST recruitment to the first intron of Nek6 in multiple cell types, making it unclear whether repression of Nek6 is effected through mus2991 (at ∼100 kb from the Nek6 promoter) or a weaker, intronic binding site (Johnson et al. 2008a). Nevertheless, these results indicate that REST silences AK046052 via recruitment to an upstream RE1.
Similarly, expression of the second REST target macroRNA, AK090153, could be validated in NS5 (Fig. 4A). Its associated RE1, mus4186, repressed luciferase reporter gene expression (Fig. 4B). This element is evolutionarily conserved and lies in a 3′-intron of AK090153, which initiates within an annotated CpG island (Fig. 4C; Supplemental File 4). Inhibition of REST function by DN:REST resulted in significant de-repression of AK090153 (Fig. 4D), indicating, together with the ChIP and luciferase data, that AK090153 is a direct REST target gene. AK090153 lies antisense to and encompasses Sptlc3, encoding a serine palmitoyltransferase. The gene was silent in both control and DN:REST-expressing NS5 (data not shown).
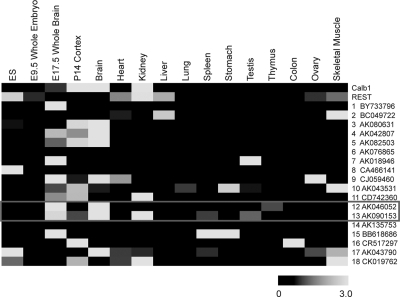
Many macroRNAs have been found to have highly tissue-specific expression patterns (Ravasi et al. 2006; Mercer et al. 2008); most recently, the group of John Mattick used data from the Allen Brain Atlas to demonstrate regionalized expression of noncoding transcripts in mouse brain (Mercer et al. 2008). Consistent with our target macroRNA set being preferentially selected from brain-derived cDNA libraries, we found that AK046052 and AK090153 are largely restricted to the brain, and induced after embryonic day 9.5 (E9.5) (Fig. 5). Interestingly, levels of both are higher in whole-brain RNA samples compared to dissected cortex, suggesting that they may also have region-specific expression within the brain (unfortunately, neither transcript is included in the Allen Brain Atlas). Similarly, the other macroRNAs from the study set displayed (1) largely heterogeneous expression levels in the organs and tissues of the adult mouse (principally, but not exclusively, highest in the brain), and (2) induction during neurodevelopment (although some exceptions are principally expressed in undifferentiated ES cells).
FIGURE 5.
MacroRNAs are developmentally regulated and have tissue-specific expression profiles. The expression levels of candidate REST-target macroRNAs were measured using intron-spanning Taqman probes. Assays were carried out in duplicate on cDNAs from the developing mouse nervous system, as well as from a panel of tissues from the adult. mRNAs for protein-coding genes Calb1 and Rest were also measured. All levels were normalized to the housekeeping gene β-actin. Data are displayed as a heat map, where black indicates no detectable signal, and the intensity of gray indicates varying degrees of expression. For every macroRNA transcript, levels are expressed relative to the mean from all tissue samples. (ES) E14 mouse embryonic stem cell.
A human macroRNA target of REST: The DiGeorge syndrome-associated DGCR5
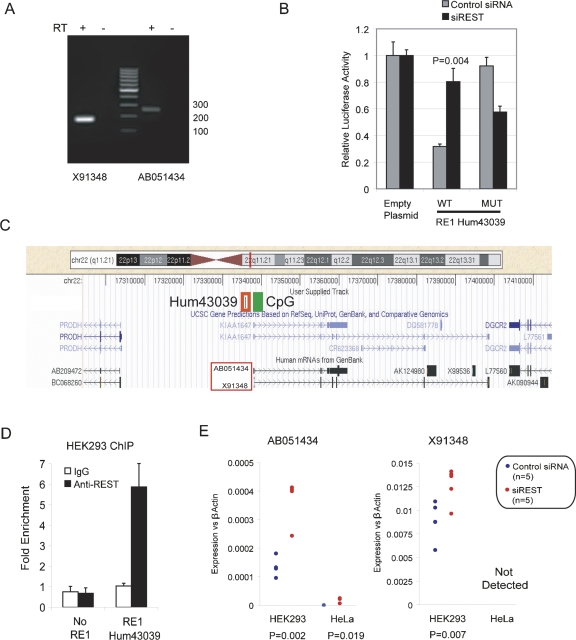
For a relatively small number of macroRNAs, functional roles and disease associations have been experimentally demonstrated and are included in Refseq and UCSC KnownGene catalogs. We searched our annotation data sets of RE1-associated genes for any previously identified macroRNAs belonging to such catalogs (microRNAs were included in the annotation pipeline, but are not discussed further). Among those we identified was DGCR5 (DiGeorge Critical Region 5), a spliced macroRNA that was found to be disrupted in a DiGeorge syndrome patient by a balanced translocation (Sutherland et al. 1996). DGCR5 is principally expressed in the brain and has two distinct splice isoforms: AB051434 (5427 nucleotides [nt], six exons) and X91348 (1284 nt, six exons) (named here for the accession number of their exemplar mRNA) (Fig. 6C). Neither has a credible open reading frame (ORF). Both were detected in human HEK293 cells using conventional RT-PCR (Fig. 6A). A high-scoring, evolutionarily conserved RE1, hum43039, is located 2160 bp upstream of the DGCR5 transcription start site, which itself lies within a CpG island (Fig. 6C; Supplemental File 4). Hum43039 repressed a luciferase reporter gene in HEK293 cells, in a manner dependent on REST (Fig. 6B). REST is recruited to this region in HEK293, as assayed by ChIP (Fig. 6D). Moreover, both isoforms of DGCR5 were responsive to knockdown of REST by siRNA; this regulatory effect was observed in both HEK293 and in the cervical carcinoma cell line HeLa (Fig. 6E). We could find no such evidence for regulation of DGCR5-flanking genes PRODH (a proline oxidase) and DGCR2 (a predicted adhesion molecule), confirming the specificity of DGCR5 regulation by REST (Supplemental File 6).
FIGURE 6.
The DGCR5 macroRNA from the human DiGeorge Syndrome locus is a target of REST. (A) DGCR5 isoforms AB051434 and X91348 were both detected in HEK293 by specific primers. A control reverse transcription (RT) reaction was carried out without RTase enzyme (−RT). (B) The ability of RE1 hum43039 to repress reporter gene expression in a REST-dependent manner was demonstrated as in Figure 3B by luciferase assay. (C) The DGCR5 promoter lies within a CpG island (green rectangle). The RE1 hum43039 lies in the proximal 5′ region (red rectangle). DGCR5 is flanked by the protein-coding genes PRODH and DGCR2. (D) Chromatin immunoprecipitation (ChIP) was carried out to validate recruitment of REST to the DGCR5 RE1. ChIP DNAs were prepared using an anti-REST antibody and nonspecific IgGs, from three independent biological samples of HEK293 cells. DNAs were interrogated in SYBR green qPCR using primers flanking the DGCR5 RE1, as well as with two negative control, non-RE1 proximal primer sets (No RE1). Fold enrichments were calculated by normalizing to the signal from a non-RE1 region, using the ΔΔCT method. (E) DGCR5 levels were assayed in the human cell lines HEK293 and HeLa. Cells were either transfected with nontargeting (control) siRNA, or an siRNA construct targeting REST mRNA. cDNA was prepared from five independent biological samples. Expression levels of both DGCR5 splice isoforms were independently measured using intron-spanning Taqman probes designed to hybridize to unique exons in each transcript. Statistical significance was calculated by Student's t-test.
Conclusions
By a combination of genome annotation and focused experimental validation, we have demonstrated that neural macroRNAs can be repressed by REST in mouse and human. From a set of 18 candidate macroRNA targets in mouse, we identified two that responded to a loss of REST binding, a similar validation rate to predicted protein-coding targets: 8% (96/1242) of detectable, protein-coding genes within 10 kb of a REST-binding site were responsive to DN:REST in the same cells (Johnson et al. 2008a). Additionally, we identified a disease-associated macroRNA from the DiGeorge locus, DGCR5, as a target in human. These three macroRNAs are regulated in cis by evolutionarily conserved, proximal REST-binding sites. All have brain-restricted expression profiles, in common with other REST target genes. These findings continue a trend beginning with the discovery that REST regulates members of another major class of noncoding RNAs, the microRNAs (Conaco et al. 2006; Johnson et al. 2006; Wu and Xie 2006). Although our validation experiments were carried out on a small set of candidates to prove the principle, they suggest that macroRNAs may comprise a substantial fraction of REST target genes: given that (1) various bioinformatics (Johnson et al. 2006; Mortazavi et al. 2006; Wu and Xie 2006) and genomic (Johnson et al. 2007; Otto et al. 2007) studies find 1000–5000 REST-binding sites in a given cell; (2) our annotation pipeline indicated that 23% of RE1s may target a macroRNA; and (3) our rate of ncRNA target validation was 11% (2/18), we estimate that the number of macroRNA targets of REST lies in the range 25–1150. Future studies will expand the REST macroRNA target list, while also assigning function to those already discovered. Given their demonstrated regulatory potential and their highly regulated transcription in the brain, we expect that the repression of macroRNA expression by REST is an important means of regulating neural phenotype in the nervous system and elsewhere. Furthermore, these findings help explain the highly neural-specific expression profile of many macroRNAs. (Mercer et al. 2008).
MATERIALS AND METHODS
Genomic mapping of RE1s
We downloaded 1781 mouse and 1301 human RE1 sites, identified by our previous position-specific scoring matrix (PSSM) study, from the RE1 Database (http://www.bioinformatics.leeds.ac.uk/RE1db_mkII/) (Johnson et al. 2006). For each, 200 bp of flanking DNA was downloaded in addition to the core RE1 sequence. The unique identifier (RE1 ID) for each site was used throughout the study. The UCSC Genome Browser, version 145 (Kent et al. 2002) and BLAT (Kent 2002) were used to map and visualize alignments of each RE1-binding site query (221 bp) to the mouse mm7 assembly (NCBI build 35) or the human hg18 assembly (NCBI build 36.1). Queries that yielded (1) no BLAT hits, (2) equal-scoring BLAT hits to multiple genomic locations, or (3) had >75% repetitive DNA coverage (as defined by RepeatMasker) (Kent et al. 2002) were excluded. Otherwise, the best-scoring full-length BLAT alignment was used for annotation.
Unbiased annotation of REST targets
The complete mouse and human annotation data sets from this study can be found in the Annotation Data Set (Supplemental File 1). Importantly, we did not limit annotation to RefSeq and UCSC KnownGene entries, instead considering all transcriptional units (TUs) supported by any EST or mRNA evidence. TUs are defined as any collection of ESTs and cDNAs that represent transcripts in the same orientation and share at least one base pair of shared exonic sequence (Engstrom et al. 2006). Every known gene is therefore a TU but may have ESTs or cDNAs extending beyond the KnownGene or RefSeq boundaries. However, not every TU corresponds to a known gene: some TUs are novel, unannotated, and/or non-protein-coding.
ESTs and cDNAs with non-GT/AG splice junctions, <96% identity to the genome, ORESTES/RAGE/MAGE library origins, nonlinear splicing patterns indicative of cDNA library chimerism, and/or ambiguous genomic mappings, were not considered of sufficient quality and were excluded. Our initial annotation of each RE1 site cataloged all TUs within a 20-kb window centered on the RE1 site. If no such TUs existed, then we considered a 200-kb window centered on the RE1 site and annotated the single most proximal TU as the target, excluding any other less proximal TUs. The distance of each putative target TU from its RE1 was calculated: intronic/exonic RE1s are defined as 0, otherwise, distance refers to the most proximal point of the TU to the RE1. We also classified each RE1 site as intronic, exonic, 5′ proximal (<20 kb), 3′ proximal, 5′ distal (20–100 kb), or 3′ distal relative to its target TUs. RE1 sites >200 kb away from any TUs were classified as gene-desert sites and were not assigned to any target genes. RE1 sites mapped to genome assembly fragments separated by assembly gaps from any TU-bearing assembly fragments were also not assigned to any target genes.
Annotated TUs corresponding to RefSeq or KnownGene entries were given those entries' descriptive names (from literature or HUGO nomenclature). Wherever both descriptive names and clone numbers (e.g., KIAA/FLJ/LOC/MGC designations) were available, descriptive names had priority. Other TUs were named using GenBank accession numbers of their representative transcripts. In general, a representative transcript of a TU was chosen based on the greatest number of exons and/or the greatest genomic distance covered. In a small number of cases, TUs did not have RefSeq or KnownGene support, but could be related to a putative ortholog in another species; these TUs were named after the ortholog.
Occasionally, genomic landscape complexity was reflected in overlapping targets, including but not limited to targets cis-antisense to one another, targets with in tronic overlaps, and targets initiating at potential bidirectional promoters. Intercalation of such targets was noted during annotation.
Protein-coding assessment of REST targets
All RefSeq- or KnownGene-supported targets were annotated as “putatively coding.” Remaining targets were tested with ORF Finder from NCBI (http://www.ncbi.nlm.nih.gov/projects/gorf/): targets having a longest positive-strand ORF >100 amino acids were annotated as “coding due to >100 amino acids”; otherwise, the longest positive-strand ORF was compared to all known proteins by NCBI BLASTP (Altschul et al. 1990). Resultant hits were carefully evaluated to eliminate low-complexity and short-fragment results from evolutionarily distant organisms. Such BLASTP hits were defined as “coding due to BLASTP hit.” All RE1-target TUs that had <100 amino acids ORFs not supported by credible BLASTP hits were classified as “noncoding.” The >100 amino acids set was also tested by BLASTP: for each, the highest-scoring, nonself, mammalian hit can be found in Supplemental File 2.
Conservation of TUs
We used the other-species mRNA and 17-species MultiZ conservation tracks (Blanchette et al. 2004) to determine which noncoding RNA TUs had exons conserved in multiple species. Targets with MultiZ peaks overlapping every exon and targets with exon–intron structures corresponding to those of other-species mRNAs were annotated as conserved.
REST target macroRNAs
The following strategy was used to create a short list of 18 potential macroRNA targets of REST for experimental study. From the manual annotation of 1781 mouse RE1s, all those associated with target genes identified as “noncoding” were collected. All such TUs that did not have mRNA support were removed. This set was then cross-referenced to REST chromatin immunoprecipitation microarray (ChIP-chip) data from three mouse cell lines (Johnson et al. 2008a), to identify those sites that recruit REST in vivo (ChIP-chip data are provided as Supplemental File 3). From the resulting set of 174 REST-bound macroRNA targets, 18 promising macroRNA target genes were manually selected with reference to the following criteria: (1) presence of splicing, (2) proximal upstream or intronic RE1, and (3) expression in embryonic- or brain-derived cDNA libraries.
Cell culture, siRNA transfection, and adenoviral infection
HEK293 and HeLa cells were cultured at 37°C in 5% CO2, in high glucose DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, and 1% nonessential amino acids. E14 and NS5 cells were cultured following procedures described by Lim et al. (2007) and Conti et al. (2005), respectively. For transfection, 0.05 nMol of REST Smartpool siRNA or nontargeting, scrambled siRNA (Dharmacon) was transfected into subconfluent cells grown in 24-well plates, using Dharmafect 2 reagent. RNA was harvested 3 d post-transfection. NS5 were infected with recombinant adenoviruses: (Ad-) control empty adenovirus, or (Ad DN:REST-) dominant-negative construct (Chen et al. 1998). Purified adenovirus was added to subconfluent cells at the minimum titer such that >90% of cells became GFP-fluorescent with minimal cell death. RNA was harvested 48 h post-infection. The efficacy of DN:REST overexpression in de-repressing target gene expression and of siRNAs in knocking down REST mRNA was validated experimentally (Supplemental File 7).
Chromatin immunoprecipitation (ChIP)
ChIP was carried out using the Hinxton protocol (ENCODE 2007). Subconfluent cells were cross-linked in situ with 1% formaldehyde for 10 min at room temperature, then quenched with excess glycine. Cell pellets were resuspended in Cell Lysis Buffer on ice. Resultant nuclei were pelleted and resuspended in Nuclei Lysis Buffer on ice, such that there was 100 μL of NLB for every 1 × 107 cells. Sonication on ice produced DNA fragments of ∼600 bp. Chromatin was diluted 10-fold with Dilution Buffer and pre-cleared with nonspecific IgG and BSA-blocked protein G Sepharose. Chromatin from 1 × 107 cells was immunoprecipitated overnight with 10 μg of anti-REST IgG (Upstate 07-579), or nonspecific IgG, then retrieved with 50 μL of BSA-blocked protein G Sepharose beads. Beads were washed twice with Wash Buffer 1, then once with Wash Buffer 2, then twice with TE. Immune complexes were eluted twice with 225 μL of Elution Buffer. De-cross-linked and proteinase-treated DNA was purified using the QIAGEN PCR Purification kit. Control, nonimmunoprecipitated Input DNA samples were also prepared. ChIP enrichments were measured by qPCR using primers to RE1s, or control non-RE1 regions. Enrichment values were calculated with respect to the latter using the ΔΔCT method (Livak and Schmittgen 2001).
Luciferase assay
Fifty-one base-pair oligonucleotides encompassing test RE1s were cloned into the pGL4 luciferase reporter plasmid, upstream of the constitutive thymidine kinase (TK) promoter. Mutant RE1s were also prepared, where the essential CC and GG dinucleotides were swapped to abolish REST binding. All sequences can be found in Supplemental File 8. Plasmids were transfected using Dharmafect 2 reagent into subconfluent HEK293 cells grown in 24-well plates (0.5 μg per well), along with 5 ng of the control Renilla plasmid, pSV40-Ren, and relevant siRNAs. After 48 h, cells were harvested and subjected to luciferase assay using the Promega Dual Luciferase Kit according to the manufacturer's protocol. Three biological and two technical replicates were carried out in each case. Luciferase readings were normalized to Renilla, then compared to ratios from cells transfected with pGL4TK plasmid lacking any RE1 insert.
Mouse tissue samples
Mouse adult tissue RNA samples were obtained from BD Biosciences. Embryonic mouse tissue RNA samples were the kind gift of Dr. Philip Gaughwin (Genome Institute of Singapore).
Gene expression analysis
One microgram of RNA was reverse-transcribed using the ABI cDNA Archive kit, along with a control reaction containing no reverse transcriptase. cDNA was interrogated in either SYBR green (coding transcripts and DGCR5) or Taqman (mouse noncoding transcripts) real-time quantitative PCR reaction in an ABI Prism 7900HT unit. For all spliced transcripts, intron-spanning primers or Taqman probes were used. Gene expression was normalized to that of the housekeeping gene β-actin by the ΔΔCT method (Livak and Schmittgen 2001). We observed no change in expression of another housekeeping gene, β2 microglobulin, upon siRNA or DN:REST treatment (data not shown). Specificity of all qPCR amplifications was gauged by the presence of a unique peak in a melt curve analysis. Genes were considered undetectable when >50% of samples showed no amplification of correct template, as evidenced by melt curve analysis. PCR products were cloned and sequenced to confirm correct amplification of AK046052, AK090153, AB051434, and X91348, in their respective cell lines. RT-PCR primer sequences can be found in Supplemental File 8.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Jameelah Sheik Mohamed for technical assistance with the Taqman assay; Galih Kunarso for bioinformatics help; Dr. Philip Gaughwin and Kee Yew Wong for providing mouse RNA samples; and Dr. Andrew Hutchins for critically reading the manuscript. L.L., R.J., N.J.B., and L.W.S. designed the project. T.P. and Z.-H.L. curated REST target genes. J.H. created automatically curated target gene sets. R.J., C.H.-L.T., and R.R.V. performed experiments. The article was written by L.L. and R.J. with contributions from the C.H.-L.T., J.H., R.R.V., T.P., Z.-H.L., N.J.B., and L.W.S.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1127009.
REFERENCES
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ballas N., Battaglioli E., Atouf F., Andres M.E., Chenoweth J., Anderson M.E., Burger C., Moniwa M., Davie J.R., Bowers W.J., et al. Regulation of neuronal traits by a novel transcriptional complex. Neuron. 2001;31:353–365. doi: 10.1016/s0896-6273(01)00371-3. [DOI] [PubMed] [Google Scholar]
- Ballas N., Grunseich C., Lu D.D., Speh J.C., Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2003;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Lee T.I., Cole M.F., Johnstone S.E., Levine S.S., Zucker J.P., Guenther M.G., Kumar R.M., Murray H.L., Jenner R.G., et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce A.W., Donaldson I.J., Wood I.C., Yerbury S.A., Sadowski M.I., Chapman M., Gottgens B., Buckley N.J. Genome-wide analysis of repressor element 1 silencing transcription factor/neuron-restrictive silencing factor (REST/NRSF) target genes. Proc. Natl. Acad. Sci. 2004;101:10458–10463. doi: 10.1073/pnas.0401827101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderone A., Jover T., Noh K.-M., Tanaka H., Yokota H., Lin Y., Grooms S.Y., Regis R., Bennett M.V.L., Zukin R.S. Ischemic insults derepress the gene silencer REST in neurons destined to die. J. Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-F., Paquette A.J., Anderson D.J. NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat. Genet. 1998;20:136–142. doi: 10.1038/2431. [DOI] [PubMed] [Google Scholar]
- Cheong A., Bingham A., Li J., Kumar B., Sukumar P., Munsch C., Buckley N., Neylon C., Porter K., Beech D., et al. Downregulated REST transcription factor is a switch enabling critical potassium channel expression and cell proliferation. Mol. Cell. 2005;20:45–52. doi: 10.1016/j.molcel.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Chong J., Tapia-Ramirez J., Kim S., Toledo-Aral J., Zheng Y., Boutros M., Altshuller Y., Frohman M., Kraner S., Mandel G. REST: A mammalian silencer protein that restricts sodium channel gene expression to neurons. Cell. 1995;80:949–957. doi: 10.1016/0092-8674(95)90298-8. [DOI] [PubMed] [Google Scholar]
- Conaco C., Otto S., Han J.-J., Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc. Natl. Acad. Sci. 2006;103:2422–2427. doi: 10.1073/pnas.0511041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.-L., Cattaneo E., et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engstrom P.G., Suzuki H., Ninomiya N., Akalin A., Sessa L., Lavorgna G., Brozzi A., Luzi L., Tan S.L., Yang L., et al. Complex loci in human and mouse genomes. PLoS Genet. 2006;2:e47. doi: 10.1371/journal.pgen.0020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Bi C., Clark B.S., Mady R., Shah P., Kohtz J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith M.C., Bailey T.L., Kasukawa T., Mignone F., Kummerfeld S.K., Madera M., Sunkara S., Furuno M., Bult C.J., Quackenbush J., et al. Discrimination of non-protein-coding transcripts from protein-coding mRNA. RNA Biol. 2006;3:40–48. doi: 10.4161/rna.3.1.2789. [DOI] [PubMed] [Google Scholar]
- Furuno M., Pang K.C., Ninomiya N., Fukuda S., Frith M.C., Bult C., Kai C., Kawai J., Carninci P., Hayashizaki Y., et al. Clusters of internally primed transcripts reveal novel long noncoding RNAs. PLoS Genet. 2006;2:e37. doi: 10.1371/journal.pgen.0020037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway D.J., Street M., Jeffries A., Buckley N.J. RE1 silencing transcription factor maintains a repressive chromatin environment in embryonic hippocampal neural stem cells. Stem Cells. 2007;25:354–363. doi: 10.1634/stemcells.2006-0207. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanishi T., Itoh T., Suzuki Y., O'Donovan C., Fukuchi S., Koyanagi K.O., Barrero R.A., Tamura T., Yamaguchi-Kabata Y., Tanino M., et al. Integrative annotation of 21,037 human genes validated by full-length cDNA clones. PLoS Biol. 2004;2:e162. doi: 10.1371/journal.pbio.0020162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Gamblin R.J., Ooi L., Bruce A.W., Donaldson I.J., Westhead D.R., Wood I.C., Jackson R.M., Buckley N.J. Identification of the REST regulon reveals extensive transposable element-mediated binding site duplication. Nucleic Acids Res. 2006;34:3862–3877. doi: 10.1093/nar/gkl525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D., Mortazavi A., Myers R., Wold B. Genome-wide mapping of in vivo protein–DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Johnson R., Teh C.H.-l., Kunarso G., Wong K.Y., Srinivasan G., Cooper M.L., Volta M., Chan S.S.-l., Lipovich L., Pollard S.M., et al. REST regulates distinct transcriptional networks in embryonic and neural stem cells. PLoS Biol. 2008a;6:e256. doi: 10.1371/journal.pbio.0060256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R., Zuccato C., Belyaev N.D., Guest D.J., Cattaneo E., Buckley N.J. A microRNA-based gene dysregulation pathway in Huntington's disease. Neurobiol. Dis. 2008b;29:438–445. doi: 10.1016/j.nbd.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The Human Genome Browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil A.M., Faghihi M.A., Modarresi F., Brothers S.P., Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwahara K., Saito Y., Takano M., Arai Y., Yasuno S., Nakagawa Y., Takahashi N., Adachi Y., Takemura G., Horie M., et al. NRSF regulates the fetal cardiac gene program and maintains normal cardiac structure and function. EMBO J. 2003;22:6310–6321. doi: 10.1093/emboj/cdg601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.S., Loh Y.-H., Zhang W., Li Y., Chen X., Wang Y., Bakre M., Ng H.-H., Stanton L.W. Zic3 is required for maintenance of pluripotency in embryonic stem cells. Mol. Biol. Cell. 2007;18:1348–1358. doi: 10.1091/mbc.E06-07-0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loh Y.-H., Wu Q., Chew J.-L., Vega V.B., Zhang W., Chen X., Bourque G., George J., Leong B., Liu J., et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Louro R., Nakaya H.I., Amaral P.P., Festa F., Sogayar M.C., da Silva A.M., Verjovski-Almeida S., Reis E.M. Androgen responsive intronic noncoding RNAs. BMC Biol. 2007;5:4. doi: 10.1186/1741-7007-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G., Ponting C.P., Hein J. Genome-wide identification of human functional DNA using a neutral indel model. PLoS Comput. Biol. 2006;2:e5. doi: 10.1371/journal.pcbi.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunyak V.V., Burgess R., Prefontaine G.G., Nelson C., Sze S.H., Chenoweth J., Schwartz P., Pevzner P.A., Glass C., Mandel G., et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- Maeda N., Kasukawa T., Oyama R., Gough J., Frith M., Engstrom P.G., Lenhard B., Aturaliya R.N., Batalov S., Beisel K.W., et al. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet. 2006;2:e62. doi: 10.1371/journal.pgen.0020062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick J.S. RNA regulation: A new genetics? Nat. Rev. Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mercer T.R., Dinger M.E., Sunkin S.M., Mehler M.F., Mattick J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A., Thompson E.C.L., Garcia S.T., Myers R.M., Wold B. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesterova T.B., Slobodyanyuk S.Y., Elisaphenko E.A., Shevchenko A.I., Johnston C., Pavlova M.E., Rogozin I.B., Kolesnikov N.N., Brockdorff N., Zakian S.M. Characterization of the genomic Xist locus in rodents reveals conservation of overall gene structure and tandem repeats but rapid evolution of unique sequence. Genome Res. 2001;11:833–849. doi: 10.1101/gr.174901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi L., Wood I.C. Chromatin crosstalk in development and disease: Lessons from REST. Nat. Rev. Genet. 2007;8:544–554. doi: 10.1038/nrg2100. [DOI] [PubMed] [Google Scholar]
- Otto S.J., McCorkle S.R., Hover J., Conaco C., Han J.-J., Impey S., Yochum G.S., Dunn J.J., Goodman R.H., Mandel G. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 2007;27:6729–6739. doi: 10.1523/JNEUROSCI.0091-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmant E., Laurendeau I., Heron D., Vidaud M., Vidaud D., Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- Perez D.S., Hoage T.R., Pritchett J.R., Ducharme-Smith A.L., Halling M.L., Ganapathiraju S.C., Streng P.S., Smith D.I. Long, abundantly expressed noncoding transcripts are altered in cancer. Hum. Mol. Genet. 2008;17:642–655. doi: 10.1093/hmg/ddm336. [DOI] [PubMed] [Google Scholar]
- Ponjavic J., Ponting C.P., Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T., Suzuki H., Pang K.C., Katayama S., Furuno M., Okunishi R., Fukuda S., Ru K., Frith M.C., Gongora M.M., et al. Experimental validation of the regulated expression of large numbers of noncoding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J., Kertesz M., Wang J., Squazzo S., Xu X., Brugmann S., Goodnough L., Helms J., Farnham P., Segal E., et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;127:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenherr C., Anderson D. The neuron-restrictive silencer factor (NRSF): A coordinate repressor of multiple neuron-specific genes. Science. 1995;5202:1360–1363. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- Schoenherr C.J., Paquette A.J., Anderson D.J. Identification of potential target genes for the neuron-restrictive silencer factor. Proc. Natl. Acad. Sci. 1996;93:9881–9886. doi: 10.1073/pnas.93.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland H.F., Wadey R., McKie J.M., Taylor C., Atif U., Johnstone K.A., Halford S., Kim U.J., Goodship J., Baldini A., et al. Identification of a novel transcript disrupted by a balanced translocation associated with DiGeorge syndrome. Am. J. Hum. Genet. 1996;59:23–31. [PMC free article] [PubMed] [Google Scholar]
- Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H., Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl. Acad. Sci. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham A., Orth A., Batalov S., Peters E., Wen B., Aza-Blanc P., Hogenesch J., Schultz P. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wu J., Xie X. Comparative sequence analysis reveals an intricate network among REST, CREB, and miRNA in mediating neuronal gene expression. Genome Biol. 2006;7:R85. doi: 10.1186/gb-2006-7-9-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B.R., Hayden M.R., Timmusk T., et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. [DOI] [PubMed] [Google Scholar]