Abstract
The exon junction complex (EJC) is deposited onto spliced mRNAs and is involved in many aspects of mRNA function. We have recently reconstituted and solved the crystal structure of the EJC core made of MAGOH, Y14, the most conserved portion of MLN51, and the DEAD-box ATPase eIF4AIII bound to RNA in the presence of an ATP analog. The heterodimer MAGOH/Y14 inhibits ATP turnover by eIF4AIII, thereby trapping the EJC core onto RNA, but the exact mechanism behind this remains unclear. Here, we present the crystal structure of the EJC core bound to ADP-AIF3, the first structure of a DEAD-box helicase in the transition-mimicking state during ATP hydrolysis. It reveals a dissociative transition state geometry and suggests that the locking of the EJC onto the RNA by MAGOH/Y14 is not caused by preventing ATP hydrolysis. We further show that ATP can be hydrolyzed inside the EJC, demonstrating that MAGOH/Y14 acts by locking the conformation of the EJC, so that the release of inorganic phosphate, ADP, and RNA is prevented. Unifying features of ATP hydrolysis are revealed by comparison of our structure with the EJC–ADPNP structure and other helicases. The reconstitution of a transition state mimicking complex is not limited to the EJC and eIF4AIII as we were also able to reconstitute the complex Dbp5–RNA–ADP–AlF3, suggesting that the use of ADP–AlF3 may be a valuable tool for examining DEAD-box ATPases in general.
Keywords: EJC, AlF, transition state, DEAD-box ATPase, mRNP
INTRODUCTION
RNA helicases have been proposed to use energy from ATP hydrolysis to unwind double-stranded (ds) nucleic acid or rearrange RNA–protein complexes at virtually all steps of the gene expression pathway (Jankowsky et al. 2001; Fairman et al. 2004; Cordin et al. 2006). Helicases can be classified into five groups based on their sequences. The two largest groups are superfamilies 1 and 2 (SF1 and SF2) in which the helicases contain two RecA like domains with seven conserved motifs I, Ia, II, III, IV, V, and VI (Caruthers and McKay 2002; Singleton and Wigley 2002). The DEAD-box proteins are a ubiquitous subgroup of SF2 enzymes representing the most common type of RNA helicases. DEAD-box proteins like eIF4AIII contain the general helicase motifs and the additional Q and Ib motifs (Cordin et al. 2006). Unwinding of dsRNA and remodeling of mRNP complexes by a variety of DEAD-box ATPases have been found to require ATP hydrolysis (Ray et al. 1985; Rozen et al. 1990; Wagner et al. 1998; Svitkin et al. 2001; Bizebard et al. 2004; Fairman et al. 2004; Shu et al. 2004; Yang and Jankowsky 2005). However, the ADP-bound state of DEAD-box proteins may also be involved in RNP remodeling as Dbp5–ADP appears to be responsible for the displacement of the Nab2 protein from RNA (Tran et al. 2007).
Pre-mRNA splicing reactions in higher eukaryotes lead to the deposition of the exon junction complex (EJC) on mature mRNAs at a conserved but sequence-independent position upstream of exon junctions (Le Hir et al. 2000). The EJC constitutes a central effector of mRNA functions that provides an anchoring point successively for nuclear and cytoplasmic factors participating in mRNA transport, translation, and quality control until it most likely is displaced by ribosomes during the first round of translation (Tange et al. 2004). The core of the EJC can be reconstituted in vitro as a complex consisting of eIF4AIII bound to ATP, the most conserved part of MLN51 and the heterodimer MAGOH/Y14 on poly uracil mimicking mRNA (Ballut et al. 2005). In the presence of RNA and MLN51, eIF4AIII slowly hydrolyzes ATP while MAGOH/Y14 essentially inhibits the turnover of ATP when associating with eIF4AIII, MLN51, and RNA (Ballut et al. 2005), but the mechanism of inhibition has not been elucidated (Le Hir and Andersen 2008). At least two different mechanisms can explain the function of MAGOH/Y14 in the EJC: either it inhibits ATP hydrolysis by eIF4AIII (prehydrolysis inhibition) or ATP hydrolysis can occur, and instead, it prevents conformational changes of eIF4AIII upon ATP hydrolysis, normally leading to the release of the products, ADP, and inorganic phosphate (posthydrolysis inhibition) (Le Hir and Andersen 2008).
In order to elucidate the relationship between the stability of the EJC and the nucleotide state of its DEAD-box ATPase eIF4AIII, we have determined the structure of the EJC in complex with the transition state analog ADP–AlF3 and analyzed the hydrolysis of ATP inside assembled EJC. Additionally, we show that ADP–AlF3 can promote stable binding of DEAD-box proteins to RNA in other complexes than the EJC. Finally, by comparison with other known structures we discuss common features between SF1 and SF2 helicases.
RESULTS
The structure of EJC in complex with ADP–AlF3 mimicking the transition state
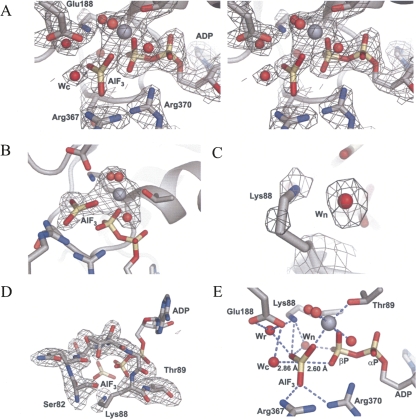
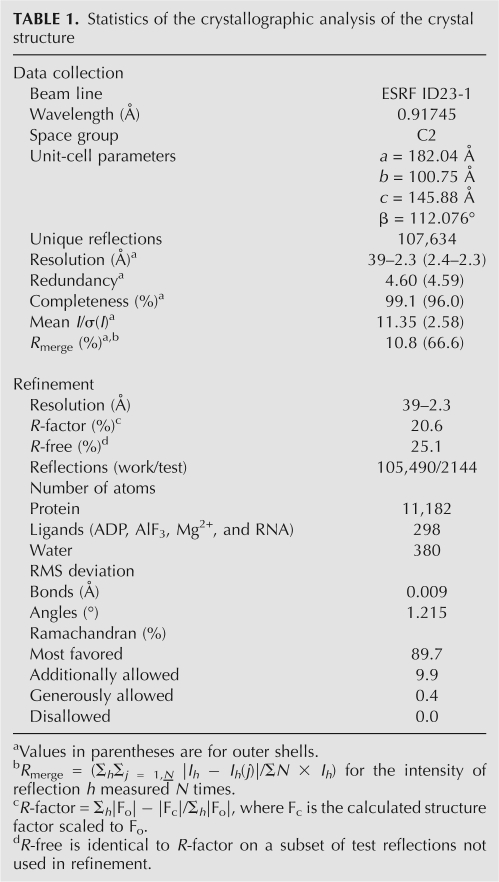
We and others recently determined the crystal structure at ∼2.3 Å resolution of a minimal reconstituted EJC in complex with a nonhydrolysable ATP analog, ADPNP, and a poly uracil oligonucleotide mRNA mimic (Andersen et al. 2006; Bono et al. 2006). To further investigate the mechanism of ATP hydrolysis within DEAD-box ATPases, we determined the crystal structure of EJC in complex with the transition state analog ADP–AlF3 (hereafter called EJC–AlF3) at the same resolution (Fig. 1; Table 1). Two EJCs are found in the asymmetric unit, and they show subtle differences. In complex II, MAGOH and Y14 are more mobile than in complex I, most likely due to crystal packing effects. Therefore, in the following, we describe only complex I. The distances between the aluminium atom and the axial oxygens (Fig. 1E) are indicative of a dissociative mechanism in which bond breaking has occurred and displays a metaphosphate-like or loose transition state (Wittinghofer 2006). The overall structure is nearly identical to the EJC–ADPNP structure except for the γ-phosphate environment, where the AlF3, mimicking the planar leaving γ-phosphate, has moved 0.7 Å toward the catalytic water (wc) that remains at the same position (Fig. 2A). The AlF3 is placed between the catalytic water and a β-phosphate oxygen in a classical transition state geometry with the leaving group coordinated by Arg367 and Arg370 in eIF4AIII (Figs. 1E, 2B). Very similar transition state geometries are seen in the UvrD helicase, the F1-ATPase and the GTPase Ras in complex with the GTPase activating protein (GAP) (Fig. 2C–E). VASA is a DEAD-box helicase from Drosophila melanogaster that regulates translation of specific mRNAs encoding factors essential for development. We previously proposed (Andersen et al. 2006), based on comparison with the structure of VASA (Sengoku et al. 2006), that coordination of the magnesium ion by Thr89 could be important in keeping the EJC in a prehydrolysis state. This is not supported, since we observe no change for Thr89 in the transition state. The main differences between the two structures with ADPNP or ADP-AlF3 occur in motif I where side chains of Ser82, Ser84, and Lys88 have changed conformations (Fig. 2A,F). In a number of other structures of G-proteins and ATPases, the equivalent of Lys88 coordinates both the β-phosphate oxygen and the leaving group (Fig. 2C–E); however, in our structure, the side chain has moved and instead a water (wn) is located between the AlF3 and the β-phosphate (Figs. 1C,E, 2F). This water may originate from the wa molecule coordinated by Ser84 in EJC-ADPNP but absent in EJC–AlF3 (Fig. 2F). The Lys88 methylene groups are mobile, thereby enabling the amino group to coordinate the γ-phosphate during its migration from the β-phosphate to the catalytic water (Figs. 1, 2). Both Ser82 and Ser84 in motif I have their alcohol group flipped compared with EJC–ADPNP (Fig. 2A,F), but single mutations of these amino acids to alanines still allow stable EJC assembly (data not shown). A stable EJC can also be assembled with eIF4AIII Lys88 mutated to alanine (Ballut et al. 2005). The C terminus of MAGOH interacts with motif I in eIF4AIII through a water bridging the backbone carbonyl of Pro145 in MAGOH with Ser82 and Thr86 in eIF4AIII in EJC–ADPNP (Andersen et al. 2006). This interaction is weakened in our new structure as Ser82 no longer coordinates this water (w) but instead the wn molecule (Fig. 2F).
FIGURE 1.
Electron density maps of the ADP–AlF3 binding site. (A) Stereo view of a SIGMAA weighted 3Fo−2Fc electron density map contoured at 1.3 σ. (B–D) Fo−Fc omit maps, using simulated annealing started at 2000 K, contoured at 3.5 σ of either (B) the ADP–AlF3 and Mg2+ with its three coordinating waters omitted. (C) Lys88 and a nearby water molecule (wn) omitted. (D) motif I (also known as the P-loop or Walker A motif) omitted. (E) Coordination of AlF3 with selected side chains, ADP, and the catalytic water (wc), the relay water (wr), wn and three Mg2+ coordinating waters. All densities are displayed on top of complex I. Carbon atoms are colored gray, nitrogen atoms blue, phosphorous atoms wheat, and oxygen atoms red. The magnesium ion is shown in gray as a sphere that is larger than the red water molecules. The AlF3 is colored as the phosphates. This color scheme is used in all figures.
TABLE 1.
Statistics of the crystallographic analysis of the crystal structure
FIGURE 2.
Surroundings of the ADP–AlF3 and comparison with the EJC–ADPNP and three other related transition state structures. (A) Overlay of EJC–ADPNP (Andersen et al. 2006) and EJC–ADP–AlF3 showing motif I as a ribbon together with selected residues and ADPNP or ADP–AlF3. The EJC–ADPNP is shown with carbon atoms and water molecules colored lime and the phosphorous atoms colored cyan. (B) Surroundings of the ADP–AlF3 and motif I with selected side chains in EJC. (C) As in (B), but of the UvrD helicase (Lee and Yang 2006), a superfamily I DNA helicase. (D) As in (B), but of the F1-ATPase (Braig et al. 2000) with β-subunit carbon atoms in gray and α-subunit carbon atoms in lime. (E) As in (B), but of the Ras–GAP complex (Scheffzek et al. 1997) with GAP carbon atoms in lime. In all structures a catalytic water (wc) is found at a similar position, while in the three ATPases a nearby water molecule (wr) is observed that may function in a proton relay during catalysis. Instead of having one “arginine finger” as in classic G-proteins (E), two “arginine fingers” are found in the ATPases. See Table 2 for comparison of distances between the two axial ligands: the catalytic water and the closest β-phosphate oxygen. (F) Close-up of motif I and its contact with MAGOH Pro145. The wn water molecule is only observed in the EJC–ADP–AlF3 complex, and the wa water molecule is only observed in the EJC–ADPNP complex.
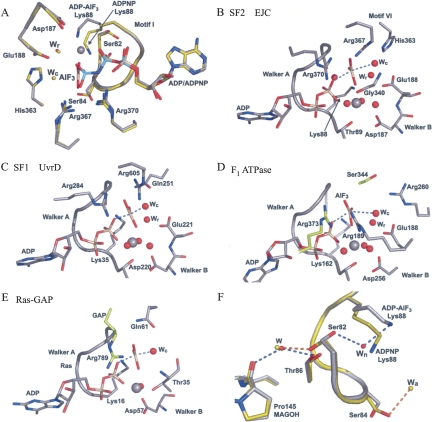
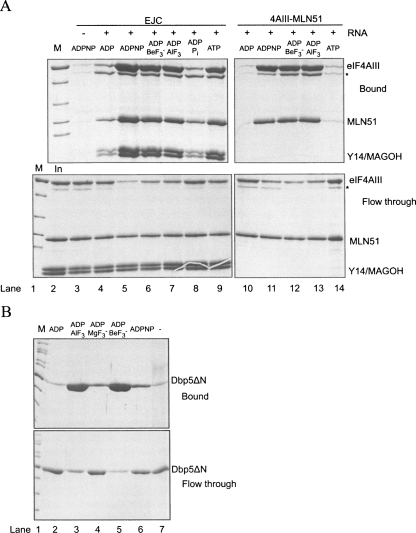
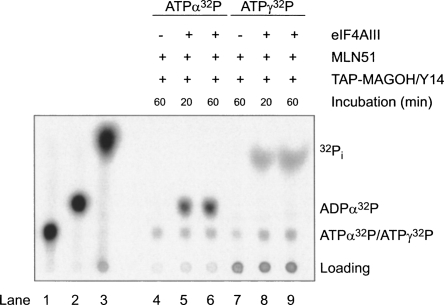
Results from Noble and Song (2007) tentatively suggested that hydrolysis can take place within EJC in vitro, but firm evidence for this has so far not been presented. To test the efficiency of EJC core assembly in the presence of different nucleotides, we performed coprecipitations with polyU ssRNA biotinylated at the 3′ end. This RNA was mixed with the EJC core recombinant proteins and incubated with various nucleotides before precipitation with streptavidin beads. We observed that the EJC is not efficiently assembled in the presence of ADP and inorganic phosphate (Pi) (Fig. 3A, cf. lanes 4 and 8). In contrast, we could detect EJC–AlF3, EJC–ADPNP, EJC–ADP–BeF3 − and EJC–ATP (Fig. 3A, lanes 5–7,9). Identical results were obtained making use of TAP–MAGOH/Y14 to precipitate formed EJC using calmodulin beads (data not shown). Within the EJC–ADP–BeF3 − complex, the ADP-BeF3 − most likely behaves as an ATP analog with an oxygen from the ADP β-phosphate acting as a ligand to the Beryllium atom (Kagawa et al. 2004). To investigate whether the EJC–ADP–Pi complex could be obtained by starting from EJC–ATP, we reconstituted the EJC with either ATPα32P or ATPγ32P, purified it, and analyzed its content of ATP and ADP or Pi, respectively, with thin-layer chromatography. Using this approach, we observed both ADP and Pi in the EJC assembled with wild type eIF4AIII (Fig. 4, lanes 6,9, respectively). As controls, only background level of ATP was observed when the EJC was not assembled (in the absence of eIF4AIII, Fig. 4, lanes 4,7). Varying the incubation times for EJC formation, 20 min versus 60 min, showed nearly identical results (Fig. 4, cf. lanes 5,6 and 8,9) and the amount of ATP was in all cases similar to the background, suggesting that the ATPase reaction occurred fast and is likely to be irreversible. These results demonstrated that the EJC is stable in the ADP–Pi state and that ATP hydrolysis was catalyzed by eIF4AIII inside the EJC because no Pi was observed when the EJC was assembled with an eIF4AIII mutant inactive for ATP hydrolysis (Ballut et al. 2005, and data not shown).
FIGURE 3.
RNA binding to DEAD-box ATPases. (A) Streptavidin pull-down of EJC components (top panels, Bound) that were incubated with the indicated nucleotides at 18°C overnight together with streptavidin–agarose beads and polyU30–biotin. An aliquot was removed (bottom panels, Flow through) before the beads were thoroughly washed and the bound protein was eluted by boiling the beads in SDS sample buffer. The concentration of inorganic phosphate was 5 mM. (B) Same as in (A), but the reaction mixtures contained Dbp5 and the indicated nucleotides and were incubated for only 1 h at room temperature. A degradation band of eIF4AIII marked * in (A) most likely corresponds to eIF4AIII without the 22 amino acids in the N-terminal known to be unstructured (Andersen et al. 2006).
FIGURE 4.
ATP hydrolysis can occur in the EJC. Thin-layer chromatography of ATP-α32P (lane 1), ATP-α32P treated with recombinant human Upf1 showing the position of ADP-α32P (lane 2) and ATP-γ32P treated with recombinant human Upf1 showing the position of the inorganic phosphate (lane 3). Purified EJC and controls (purified via TAP-MAGOH/Y14) formed with the indicated components in the presence of ATP-α32P (lanes 4–6) or in the presence of ATP-γ32P (lanes 7–9) that have been incubated for the indicated times.
The formation of stable complexes between DEAD-box proteins and RNA in the presence of ADP–AlF3 and ADP–BeF3 − is not limited to the EJC. Indeed, we observed a stable binary complex between eIF4AIII and MLN51 to RNA in the presence of either ADPNP, ADP–BeF3 −, or ADP–AlF3 (Fig. 3A), but not with ATP or ADP, as demonstrated earlier (Ballut et al. 2005). To demonstrate that the ATP analogs may be used for other DEAD-box ATPases besides eIF4AIII, we investigated the functionally unrelated Saccharomyces cerevisiae Dbp5 involved in nuclear export of mRNP (Snay-Hodge et al. 1998) and which has been demonstrated to bind single-stranded RNA in the presence of ADPNP (Weirich et al. 2006). A crucial step in export is the release of mRNP at the cytosolic face of the nuclear pore complex (NPC), making the export process directional. Dbp5 is thought to participate in this step by remodeling the mRNP to remove the export factor Mex67 from the mRNP. This facilitates the passage through the NPC (Lund and Guthrie 2005) and prevents the return of the mRNP to the nucleus. An N terminally truncated Dbp5 formed highly stable complexes with poly uracil in the presence of ADP–AlF3 or ADP–BeF3 − (Fig. 3B, lanes 3,5), less stable complexes with ADPNP or ADP–MgF3 − (Fig. 3B, lanes 4,6), while no complex was observed with ADP (Fig. 3B, cf. lanes 2 and 7).
Our results demonstrate how ADP–AlF3 can be used as a transition state analog for DEAD-box ATPases, and that MAGOH/Y14 locks EJC in a conformation that allows ATP hydrolysis but prevent the release of the products.
DISCUSSION
Stabilization of EJC by MAGOH/Y14
A unique feature of the EJC is its stable association with mature mRNAs from splicing to the first cycle of translation. The stability of the EJC core is conferred by the association of the heterodimer MAGOH/Y14 with the otherwise unstable eIF4AIII–MLN51–RNA–ATP complex (Ballut et al. 2005). The results presented in this study suggest that the EJC stabilization ensured by MAGOH/Y14 is not due to an inhibition of the catalytic activity of eIF4AIII per se (prehydrolysis inhibition). Indeed, ATP hydrolysis can occur inside the EJC (Fig. 4) strongly suggesting that the tight association with MAGOH/Y14 prevents release of ADP, Pi, and RNA from eIFAIII (posthydrolysis inhibition) that would make hydrolysis irreversible (Le Hir and Andersen 2008). Our results are in agreement with the original observation that MAGOH/Y14 inhibit the turnover ATPase activity of eIF4AIII as part of the EJC core (Ballut et al. 2005). Our demonstration of a stable EJC in the presence of ADP–Pi is in excellent agreement with a recent kinetic study of the Escherichia coli DEAD-box ATPase DbpA associated with ribosome biogenesis (Henn et al. 2008). This study showed that both ATP hydrolysis and Pi release limits the ATPase cycle, and that the DbpA–ATP–rRNA and DbpA–ADP–Pi–rRNA complexes occur at frequencies of 53% and 33%, respectively, at steady state (Henn et al. 2008). Hence, for both the EJC and DbpA–rRNA complexes, a large fraction appears to exist in the ADP–Pi state.
The transition state of helicases
In most crystal structures of G-proteins and ATPases the distance between the two axial oxygens coordinating the planar γ-phosphate analog (here AlF3) is shorter than 4.5 Å (Table 2), whereas we observe a distance of 5.3 Å. Although we cannot rule out that this may be slightly shorter in the transition state of DEAD-box ATPases in general, we believe that our structure provides a very good approximation. The distance between the catalytic water and the β-phosphate oxygen is 4.9 Å in the ADPNP bound state of VASA (Sengoku et al. 2006). In addition, the small GTPase Rac1, which has a distance of 5.4 Å in the transition mimicking state (Table 2), has been demonstrated to be active in ATP hydrolysis (Stebbins and Galan 2000).
TABLE 2.
Distances between the two axial ligands, the catalytic water, and the closest β-phosphate oxygen, of the γ-phosphate mimic (AlF3, AlF4 − or MgF3 −)
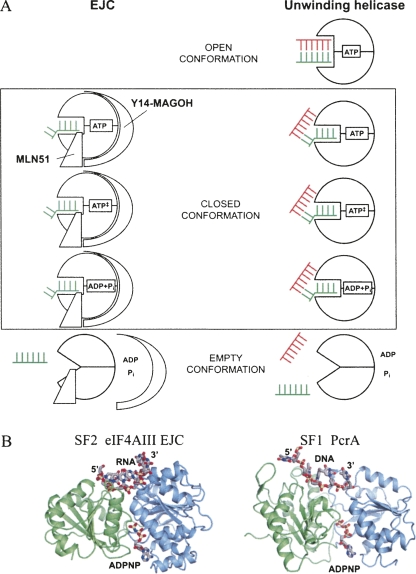
The differences between the structures of the EJC in the ADPNP state and in the transition state occur only in the vicinity of the γ-phosphate, and this potentially has implications for the dsRNA unwinding activity of DEAD-box helicases. To our knowledge, unwinding has not been observed with nonhydrolysable ATP analogs or mutants unable to hydrolyze ATP for any DEAD-box ATPase (Ray et al. 1985; Rozen et al. 1990; Svitkin et al. 2001; Bizebard et al. 2004; Shu et al. 2004; Yang and Jankowsky 2005). VASA, DedI, eIF4AI and other helicases can unwind blunt end dsRNA (Rogers et al. 1999; Cordin et al. 2006; Sengoku et al. 2006; Yang and Jankowsky 2006) but ATP hydrolysis is required. Therefore, these proteins must be able to bind dsRNA in the ATP state with the RNA binding site between the two helicase core domains more open than in the VASA–ADPNP–RNA structure. We envision that in DEAD-box helicases an open conformation in the ATP state with room for dsRNA must exist. This changes to a closed conformational with ATP bound (as observed in the VASA structure). Unwinding occurs during formation of the closed conformation. In this conformation the nucleotide can be present in the ATP state, the transitions state, or the ADP + Pi state. In the EJC, hydrolysis appears to be irreversible, but this may not be the case for other DEAD-box proteins. Dissociation of ADP, Pi, and RNA from the helicase forms the empty conformation (Fig. 5A, unwinding helicase). For the EJC, eIF4AIII is in the closed conformation with ssRNA bound in both the ATP state, transition state, and post-hydrolysis state that predominate. The post-hydrolysis state is stable due to Y14/MAGOH, and the empty conformation is presumable induced by an outer force like the ribosome, which triggers release of MAGOH/Y14 followed by release of RNA, ADP, and Pi (Fig. 5A, EJC).
FIGURE 5.
(A) A model for the role of ATP hydrolysis in relation to an unwinding helicase (right side) or the EJC (left side). Three states are shown, the open conformation, where the DEAD-box protein is bound to ATP and dsRNA, the closed conformation, where eIF4AIII and the helicase display a very similar structure and finally the empty conformation after dissociation. The ssRNA is depictured as a green comb, while the complementary strand in the dsRNA is a red comb. The kink in the ssRNA induced by the DEAD-box protein is indicated as a kink in the comb. See text for further discussion. (B) Comparison of the closed ATP–RNA bound conformation of eIF4AIII within the EJC (left panel) and the closed conformation of PcrA in the ATP–DNA complex (right panel) (pdb entry 3PJR). For clarity, only the two RecA domains (N-terminal blue, C-terminal green) are shown for both proteins, and only the singlestranded region of the DNA is shown for PcrA.
To our knowledge, this is the first demonstration of a stable DEAD-box-RNA trapped in a complex mimicking the ATPase transition state. The finding that ADP–AlF3 allows complex formation with two different DEAD-box ATPases (eIF4AIII and Dbp5; Fig. 3) suggests that ADP–AlF3 could be an important tool for analyzing DEAD-box proteins in general, not only for structural investigations but also when studying the enzymatic properties of DEAD-box proteins.
Unifying aspects of ATP hydrolysis between SF1 and SF2 ATPases
Crystal structures of the SF1 helicases PcrA (Velankar et al. 1999) and UvrD (Lee and Yang 2006) in complex with ADPNP and ds DNA with a 3′ single-stranded region have been determined. Importantly, the structure of UvrD in complex with ADP and MgF3 − mimicking the ATPase transition state (Fig. 2C) in a manner similar to that of EJC–AlF3 gave the first structural insight into the mechanism of ATP hydrolysis in SF1 helicases. In UvrD, only minimal changes are observed in the transition state at the nucleotide binding site compared with the ATP state; however, these are amplified into movement of a “gating” helix at the binding pocket for the 3′ end of the ssDNA (Lee and Yang 2006). The recent structures of UvrD were determined at a resolution, allowing observation of water molecules in the vicinity of the γ-phosphate, in contrast to the structure of PcrA. Comparison of PcrA and UvrD structures with those of the SF2 DEAD-box proteins VASA (Sengoku et al. 2006) and eIF4AIII (Andersen et al. 2006; Bono et al. 2006) in complex with RNA and ADPNP, reveals several highly conserved key structural features beyond their common use of the seven conserved motifs for DNA/RNA and ATP binding. First, the packing of the two RecA domains 1 and 2 in eIF4AIII are quite similar, not only to that observed in VASA (Andersen et al. 2006), but also to the packing of the two domains in PcrA (Fig. 5B) and UvrD (data not shown). As an example, eIF4AIII and PcrA can be superimposed with a root-mean-square deviation of 3.4 Å over 284 common Cα positions from both RecA domains. Second, the direction of the single-stranded region is conserved in all four structures, the 5′ end is always at the second RecA domain, and the 3′ end at the first RecA domain. Third, the transition state geometry is rather similar in EJC–AlF3 and UvrD–MgF3 − with two arginine “fingers” in contact with the β-phosphate and the planer γ-phosphate mimic. In addition, both structures contain a relay water wr (Fig. 2B–C) located in between the glutamate of motif II, the γ-phosphate mimic, and the Mg2+ coordinating water. In contrast to both the EJC–ADPNP and VASA–ADPNP structures, the relay water is not observed in the UvrD–ADPNP structure; however, since there is sufficient space for it, this may be due to the lower resolution of the structure (2.6 Å) compared with that of the UvrD–MgF3 − structure (2.2 Å). In summary, we predict that details of the mechanism of ATP hydrolysis are conserved between the two superfamilies of helicases. Many of these features are likely to be shared with the F1-ATPase, as described earlier (Andersen et al. 2006).
MATERIALS AND METHODS
Plasmids and purifications
Dbp5 was expressed from a pET30 vector (Novagen) that was modified by having a TEV protease cleavage site inserted after the N-terminal 6xHis tag. Plasmids encoding either eIF4AIII, MAGOH, and Y14 residues 51–176 (Y14Δ50), or the MLN51 fragment (MLN51-S), were identical to those previously described (Andersen et al. 2006). The plasmids were transformed into the BL21 Rosetta E. coli strain and cultures were grown at 37°C until an OD600 of ∼0.8 and then induced overnight at 20°C with 1 mM IPTG. Cells were resuspended in lysis buffer containing 20 mM Tris HCl at pH 7.6, 500 mM KCl (eIF4AIII and MLN51), 300 mM KCl (MAGOH/Y14), or 200 mM NaCl (Dbp5), 20 mM Imidazole, 5 mM MgCl2, 0.5 mM β-mercaptoethanol (β-ME), 1 mM PMSF, complete protease inhibitor (Roche Diagnostics GmbH), and 10% glycerol. The cells were opened by sonication and then centrifuged at 14,000 rpm for 30 min. Prior to loading on the Ni2+ charged IMAC column (GE Healthcare) the samples were filtered through a 0.45-μm filter. The protein was eluted from the Ni2+ affinity column by running a gradient from 20–500 mM imidazole in a buffer containing 20 mM Tris–HCl at pH 7.6, 500 mM KCl (eIF4AIII and MLN51), 300 mM KCl (MAGOH/Y14), or 200 mM NaCl (Dbp5), 5 mM MgCl2, 0.5 mM β-ME, 0.5 mM PMSF, and 10% glycerol. The affinity tag was removed with 1/300 (w/w) TEV protease digestion on ice for 2 h. The cleaved products were dialyzed overnight against 20 mM Tris–HCl at pH 7.6, 500 mM KCl (eIF4AIII and MLN51), 300 mM KCl (MAGOH/Y14), or 200 mM NaCl (Dbp5), 5 mM MgCl2, 0.5 mM β-ME, and 10% glycerol. The sample was loaded on the Ni2+ affinity column again and the flow-through containing essentially pure protein was collected. Dbp5 was diluted four times in water and further purified on a Source 15Q using the following buffer, 20 mM Tris–HCl at pH 8, 10% glycerol, 50 mM NaCl, 0.5 mM dithiotretiol (DTT), and eluted by increasing the NaCl concentration in a linear gradient to 200 mM.
Reconstitution of EJC
For reconstitution of the EJC, the eIF4AIII and MLN51 were mixed in an equimolar ratio, while the MAGOHY/14 heterodimer was added in excess, in a buffer containing 100 mM Tris–HCl at pH 7.6, 5 mM MgCl2, 1 mM DTT, 150 mM KCl, 1 mM ADP (A2754, Sigma), 1 mM AlCl3 (21,747, Acros Organics), 10 mM NaF (71,519, Fluka BioChemika), and polyU (P9528, Sigma; 1/3 wrna/wprotein). The mixture was incubated at 18°C overnight and then treated with RNase A (0.1 mg RNase A (R5500, Sigma)/10 mg EJC) for 30 min at 18°C to cleave unprotected RNA. Ammonium sulphate (AmS) was added to 1.4 M and the sample loaded on a Source isopropyl column (GE Healthcare). The complex was eluted by running a gradient from 1.4-0 M AmS in a buffer containing 20 mM Tris HCl at pH 7.6, 5 mM MgCl2, and 1 mM DTT. Fractions containing purified complex were precipitated with AmS and resuspended in crystallization buffer (20 mM Tris at pH 7.6, 150 mM KCl, 5 mM MgCl2, and 1 mM DTT) to 5 mg/mL. Crystals were grown in hanging drops against a reservoir containing 7% PEG3350, 50 mM Tris HCl at pH 8.8, 200 mM NaAcetate. The crystals were transferred to a cryoprotection buffer (25% Ethylene Glycol, 4% PEG3350, 50 mM Tris HCl at pH 8.8) and frozen in liquid nitrogen.
Structure determination and refinement
Diffraction data were processed and scaled using the XDS package (Kabsch 2001) (Table 1). The structure of the EJC–AlF3 was solved by molecular replacement using PHASER (Storoni et al. 2004) with the EJC (Andersen et al. 2006) as a search model. Model building and averaging of density maps were done in the program “O” (Jones et al. 1991) and the fitted models were refined with CNS (Brünger et al. 1998) and PHENIX.REFINE (Adams et al. 2002). Molecular graphics figures were prepared with PYMOL (DeLano 2002).
Streptavidin pull-down assay
The streptavidin agarose beads (High Capacity Streptavidin Agarose Resin, Pierce), 20 μL 50% slurry per reaction, were washed in 0.5 mL binding buffer (20 mM HEPES–NaOH at pH 7.2, 125 mM NaCl, 5 mM MgCl2, 2,5% glycerol, 1 mM DTT for EJC, and 20 mM HEPES–NaOH at pH 7.2, 100 mM NaCl, 5 mM MgCl2, 1 mM β-ME for Dbp5) three times using an Eppendorf centrifuge to pellet the beads by spinning for 2 min at 3k rpm. The beads were aliquoted and either 0.2 nmol for EJC or 0.1 nmol for Dbp5 of polyU30-biotin (Dharmacon) per reaction together with the proteins and nucleotides of interest were mixed and incubated as described in the figure legend after which the beads were washed three times using 0.5 mL binding buffer. The bound protein was eluted by boiling the beads in 2.5xSDS sample buffer followed by separation on a 12% SDS gel, which was Coomassie stained. The concentrations of nucleotides/analogs were 1 mM for ADPNP, ADP, ATP, BeSO4, and AlCl3. NaF was added to a final concentration of 10 mM and KH2PO4/K2HPO4 to a final concentration of 5 mM.
Thin-layer chromatography
EJC assembled with either [α-32P] or [γ-32P] ATP was purified on calmodulin beads by using TAP-MAGOH/Y14 as bait as follows: 2 μg of each EJC proteins were mixed in binding buffer BB-125 (20 mM HEPES at pH 7.4, 5 mM MgCl2, 2,5% glycerol, 1 mM DTT, 125 mM NaCl) supplemented with 1 mM cold ATP, 5 μCi of [α-32P] or [γ-32P] ATP (3000 Ci mmol−1, Perkin-Elmer), 170 nM 47-mer ssRNA (3′ end biotinylated RNA purchased from Dharmacon), and incubated for 20 min or 60 min at 30°C in a final volume of 25 μL. To the mix was added 12 μL of Calmodulin Resin (50% slurry, Stratagen) and 250 μL of BB-250 supplemented with 0.1 mM of cold ATP to eliminate non specific binding of ATP to the beads. After incubation for 1 h at 4°C, the resin was washed four times with 500 μL BB-250, 0.1 mM ATP. Proteins were eluted with 20 μL of 10 mM Tris at pH 7.5, 20 mM EGTA. SDS was added to a final concentration of 0.1% and proteins were denatured at 65°C, and 5 μL of the resulting solution was spotted on PEI-cellulose. The TLC plate was developed in 0.3 M K2HPO4 at pH 7.6, and the radioactivity was visualized and quantified with a phosphorimager (Molecular Dynamics).
ACKNOWLEDGMENTS
We are grateful to G. Hartvigsen for technical assistance, E. Jankowsky for discussion and suggestions, and the staffs at ESRF and SLS for help with data collection. K.H.N. was supported by the Danish National Research Foundation and the Alfred Benzon Foundation. G.R.A. was supported by FNU and the Danish National Research Foundation. H.C. and H.L.H. were supported by the CNRS and the Agence Nationale de la Recherche. Coordinates and structure factors have been deposited at the RCSB protein data bank with the ID code 3EX7.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1283109
REFERENCES
- Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Andersen C.B.F., Ballut L., Johansen J.S., Chamieh H., Nielsen K.H., Oliveira C., Pedersen J.S., Seraphin B., Le Hir H., Andersen G.R. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- Ballut L., Marchadier B., Baguet A., Tomasetto C., Seraphin B., Le Hir H. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 2005;12:861–869. doi: 10.1038/nsmb990. [DOI] [PubMed] [Google Scholar]
- Bizebard T., Ferlenghi I., Iost I., Dreyfus M. Studies on three E. coli DEAD-box helicases point to an unwinding mechanism different from that of model DNA helicases. Biochemistry. 2004;43:7857–7866. doi: 10.1021/bi049852s. [DOI] [PubMed] [Google Scholar]
- Bono F., Ebert J., Lorentzen E., Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Braig K., Menz R.I., Montgomery M.G., Leslie A.G., Walker J.E. Structure of bovine mitochondrial F(1)-ATPase inhibited by Mg2+ ADP and aluminium fluoride. Structure. 2000;8:567–573. doi: 10.1016/s0969-2126(00)00145-3. [DOI] [PubMed] [Google Scholar]
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.-S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. doi: 10.1016/s0959-440x(02)00298-1. [DOI] [PubMed] [Google Scholar]
- Coleman D.E., Berghuis A.M., Lee E., Linder M.E., Gilman A.G., Sprang S.R. Structures of active conformations of Gi α 1 and the mechanism of GTP hydrolysis. Science. 1994;265:1405–1412. doi: 10.1126/science.8073283. [DOI] [PubMed] [Google Scholar]
- Cordin O., Banroques J., Tanner N.K., Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. The PyMOL user's manual. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Fairman M.E., Maroney P.A., Wang W., Bowers H.A., Gollnick P., Nilsen T.W., Jankowsky E. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- Graham D.L., Lowe P.N., Grime G.W., Marsh M., Rittinger K., Smerdon S.J., Gamblin S.J., Eccleston J.F. MgF3 − as a transition state analog of phosphoryl transfer. Chem. Biol. 2002;9:375–381. doi: 10.1016/s1074-5521(02)00112-6. [DOI] [PubMed] [Google Scholar]
- Henn A., Cao W., Hackney D.D., De La Cruz E.M. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J. Mol. Biol. 2008;377:193–205. doi: 10.1016/j.jmb.2007.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E., Gross C.H., Shuman S., Pyle A.M. Active disruption of an RNA–protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Cowan S., Zou J.-Y., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. In: Rossmann M.G., Arnold E., editors. International tables for crystallography. Kluwer Academic Publishers; Dordrecht, The Netherlands: 2001. chap. 25.22.29. [Google Scholar]
- Kagawa R., Montgomery M.G., Braig K., Leslie A.G., Walker J.E. The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 2004;23:2734–2744. doi: 10.1038/sj.emboj.7600293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Yang W. UvrD helicase unwinds DNA one base pair at a time by a two-part power stroke. Cell. 2006;127:1349–1360. doi: 10.1016/j.cell.2006.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H., Andersen G.R. Structural insights into the exon junction complex. Curr. Opin. Struct. Biol. 2008;18:112–119. doi: 10.1016/j.sbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde E., Maquat L.E., Moore M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund M.K., Guthrie C. The DEAD-box protein Dbp5p is required to dissociate Mex67p from exported mRNPs at the nuclear rim. Mol. Cell. 2005;20:645–651. doi: 10.1016/j.molcel.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Menz R.I., Walker J.E., Leslie A.G. Structure of bovine mitochondrial F(1)-ATPase with nucleotide bound to all three catalytic sites: Implications for the mechanism of rotary catalysis. Cell. 2001;106:331–341. doi: 10.1016/s0092-8674(01)00452-4. [DOI] [PubMed] [Google Scholar]
- Nassar N., Hoffman G.R., Manor D., Clardy J.C., Cerione R.A. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat. Struct. Biol. 1998;5:1047–1052. doi: 10.1038/4156. [DOI] [PubMed] [Google Scholar]
- Noble C.G., Song H. MLN51 stimulates the RNA-helicase activity of eIF4AIII. PLoS One. 2007;2:e303. doi: 10.1371/journal.pone.0000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Eathiraj S., Munson M., Lambright D.G. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature. 2006;442:303–306. doi: 10.1038/nature04847. [DOI] [PubMed] [Google Scholar]
- Ray B.K., Lawson T.G., Kramer J.C., Cladaras M.H., Grifo J.A., Abramson R.D., Merrick W.C., Thach R.E. ATP-dependent unwinding of messenger RNA structure by eukaryotic initiation factors. J. Biol. Chem. 1985;260:7651–7658. [PubMed] [Google Scholar]
- Rogers G.W., Jr, Richter N.J., Merrick W.C. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- Rozen F., Edery I., Meerovitch K., Dever T.E., Merrick W.C., Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian M.R., Kabsch W., Wiesmuller L., Lautwein A., Schmitz F., Wittinghofer A. The Ras–RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science. 1997;277:333–338. doi: 10.1126/science.277.5324.333. [DOI] [PubMed] [Google Scholar]
- Scrima A., Wittinghofer A. Dimerization-dependent GTPase reaction of MnmE: How potassium acts as GTPase-activating element. EMBO J. 2006;25:2940–2951. doi: 10.1038/sj.emboj.7601171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengoku T., Noreki O., Nakamura A., Kobayashi S., Shigeyuki Y. Structural basis for RNA unwinding by the DEAD-box protein Drosophila vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Shu Z., Vijayakumar S., Chen C.F., Chen P.L., Lee W.H. Purified human SUV3p exhibits multiple-substrate unwinding activity upon conformational change. Biochemistry. 2004;43:4781–4790. doi: 10.1021/bi0356449. [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Wigley D.B. Modularity and specialization in superfamily 1 and 2 helicases. J. Bacteriol. 2002;184:1819–1826. doi: 10.1128/JB.184.7.1819-1826.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snay-Hodge C., Colot H., Goldstein A., Cole C. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J. 1998;17:2663–2676. doi: 10.1093/emboj/17.9.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins C.E., Galan J.E. Modulation of host signaling by a bacterial mimic: Structure of the Salmonella effector SptP bound to Rac1. Mol. Cell. 2000;6:1449–1460. doi: 10.1016/s1097-2765(00)00141-6. [DOI] [PubMed] [Google Scholar]
- Storoni L.C., McCoy A.J., Read R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D Biol. Crystallogr. 2004;60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Svitkin Y.V., Pause A., Haghighat A., Pyronnet S., Witherell G., Belsham G.J., Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tange T.O., Nott A., Moore M.J. The ever-increasing complexities of the exon junction complex. Curr. Opin. Cell Biol. 2004;16:279–284. doi: 10.1016/j.ceb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Tran E.J., Zhou Y., Corbett A.H., Wente S.R. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Velankar S.S., Soultanas P., Dillingham M.S., Subramanya H.S., Wigley D.B. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Wagner J.D., Jankowsky E., Company M., Pyle A.M., Abelson J.N. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich C.S., Erzberger J.P., Flick J.S., Berger J.M., Thorner J., Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat. Cell Biol. 2006;8:668–676. doi: 10.1038/ncb1424. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A. Phosphoryl transfer in Ras proteins, conclusive or elusive? Trends Biochem. Sci. 2006;31:20–23. doi: 10.1016/j.tibs.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Wurtele M., Wolf E., Pederson K.J., Buchwald G., Ahmadian M.R., Barbieri J.T., Wittinghofer A. How the Pseudomonas aeruginosa ExoS toxin downregulates Rac. Nat. Struct. Biol. 2001;8:23–26. doi: 10.1038/83007. [DOI] [PubMed] [Google Scholar]
- Yang Q., Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- Yang Q., Jankowsky E. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat. Struct. Mol. Biol. 2006;13:981–986. doi: 10.1038/nsmb1165. [DOI] [PubMed] [Google Scholar]