Abstract
The bacterial signal recognition particle (SRP) binds to ribosomes synthesizing inner membrane proteins and, by interaction with the SRP receptor, FtsY, targets them to the translocon at the membrane. Here we probe the conformation of SRP and SRP protein, Ffh, at different stages of targeting by measuring fluorescence resonance energy transfer (FRET) between fluorophores placed at various positions within SRP. Distances derived from FRET indicate that SRP binding to nontranslating ribosomes triggers a global conformational change of SRP that facilitates binding of the SRP receptor, FtsY. Binding of SRP to a signal-anchor sequence exposed on a ribosome-nascent chain complex (RNC) causes a further change of the SRP conformation, involving the flexible part of the Ffh(M) domain, which increases the affinity for FtsY of ribosome-bound SRP up to the affinity exhibited by the isolated NG domain of Ffh. This indicates that in the RNC–SRP complex the Ffh(NG) domain is fully exposed for binding FtsY to form the targeting complex. Binding of FtsY to the RNC–SRP complex results in a limited conformational change of SRP, which may initiate subsequent targeting steps.
Keywords: trigger factor, SRP, SRP receptor, FtsY, membrane targeting
INTRODUCTION
Ribosomes synthesizing inner membrane proteins in bacteria are targeted to the plasma membrane by the signal recognition particle (SRP) pathway (Keenan et al. 2001; Nagai et al. 2003; Doudna and Batey 2004). According to current models, SRP is recruited to translating ribosomes when an SRP-specific signal-anchor sequence (SAS) emerges from the peptide exit tunnel of the ribosome-nascent chain (RNC) complex, leading to high-affinity binding of SRP. The RNC–SRP complex is directed to the membrane by the interaction with the SRP receptor, FtsY, which is associated with the membrane and bound to the translocon (Angelini et al. 2005). Subsequently, the RNC is transferred to the translocation pore through which the nascent protein passes during further elongation. In the bacterial system, the early steps of targeting can take place when the nascent chains are still contained within the peptide exit tunnel (Bornemann et al. 2008). In such a case, the RNCs are sorted at the membrane after the nascent peptide has emerged from the ribosome, and the targeting complex is maintained only with RNCs that expose an SAS, whereas ribosomes exposing other sequences are released from the membrane (Bornemann et al. 2008).
In Escherichia coli, the SRP consists of 4.5S RNA (114 nucleotides [nt]) and a single protein, Ffh. Ffh is responsible for signal sequence recognition (Krieg et al. 1986; Kurzchalia et al. 1986), ribosome binding (Gu et al. 2003), and the transfer of the nascent peptide chain to the translocation machinery through the SRP receptor, FtsY (Connolly et al. 1991). Ffh is a 48-kDa GTPase that consists of three domains, the N-terminal N domain, the G domain, comprising the GTP binding site, and the C-terminal M domain where the major RNA-binding site is located (Bernstein et al. 1989; Poritz et al. 1990). The bacterial SRP receptor, FtsY, another GTPase, is a 52-kDa protein that has an acidic N-terminal A domain followed by an NG domain (Gill and Salmond 1990). The NG domains of Ffh and FtsY are homologous in sequence, and the crystal structures of the NG domains of Ffh from Thermus aquaticus and of FtsY from E. coli revealed a high degree of structural similarity (Freymann et al. 1997; Montoya et al. 1997). The NG domains of Ffh and FtsY form a pseudosymmetric heterodimeric complex featuring a combined GTP binding site that accommodates two GTP molecules (Egea et al. 2004; Focia et al. 2004).
Structural models of SRP complexed with bacterial translating ribosomes were obtained by cryo-electron microscopy (cryo-EM) (Halic et al. 2006a; Schaffitzel et al. 2006). According to these models, SRP binds to the large ribosomal subunit close to the peptide exit. The 4.5S RNA projects away from the peptide exit and is kinked by about 30° in the vicinity of nucleotide 72. The M domain of Ffh binds both 4.5S RNA and the exposed signal-anchor sequence. Consistent with cross-linking results (Gu et al. 2003), the NG domain interacts through the N domain with the ribosomal protein L23, while the G domain does not contact the ribosome (Halic et al. 2006a; Schaffitzel et al. 2006). Although the two cryo-EM reconstructions indicate a similar orientation of SRP, details of the SRP structures are different. The structure of the bacterial SRP–RNC complex with the receptor, FtsY, is not known; the cryo-EM reconstruction of the homologous eukaryotic complex indicated a gross rearrangement of SRP upon binding the receptor (Halic et al. 2006b). In general, Ffh and 4.5S RNA appear to undergo conformational changes upon interaction with one another (Rosendal et al. 2003; Buskiewicz et al. 2005a,b), with FtsY (Buskiewicz et al. 2005a; Spanggord et al. 2005), or with the ribosome (Gu et al. 2005; Halic et al. 2006a; Schaffitzel et al. 2006). Affinity measurements have revealed that SRP binding to the ribosome and of FtsY to ribosome-bound SRP is enhanced substantially when an SAS is exposed, both on eukaryotic (Flanagan et al. 2003) and E. coli (Bornemann et al. 2008) ribosomes, compared to nontranslating ribosomes, implying structural changes. It is not clear, however, to what extent SRP binding to the ribosome or to the signal peptide exposed on the ribosome induced the structural changes of SRP. Furthermore, the influence of the binding of the SRP receptor FtsY on the structure of ribosome-bound SRP is not clear.
In the present article, we used distances within SRP measured by fluorescence resonance energy transfer (FRET) to derive the conformation of SRP in the complex with vacant 70S ribosomes, with RNCs carrying 50 N-terminal amino acids of leader peptidase and exposing the SRP-specific SAS, and with RNC and FtsY. The influence of binding the chaperone trigger factor (TF) on the structure of RNC-bound SRP was also assessed. Multiple distances between residues in the M and NG domains of Ffh and between residues in Ffh and a defined position in 4.5S RNA were measured and used to build models of SRP on the ribosome. The results provide information about structural changes of SRP passing through various stages of targeting, indicating that the NG domain of Ffh becomes more accessible upon SRP binding to the ribosome. These results are corroborated by an increase of the affinity of FtsY binding to SRP, ribosome-bound SRP, RNC-bound SRP, or the isolated Ffh(NG) domain, as determined by equilibrium fluorescence titrations.
RESULTS
Experimental approach
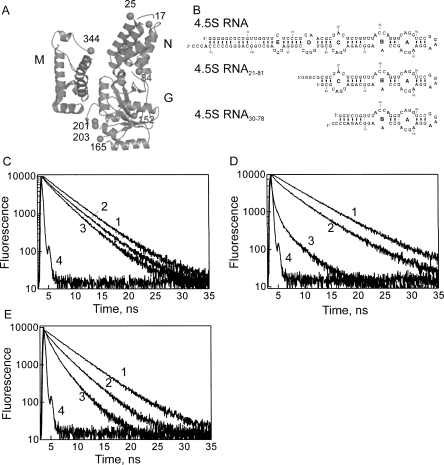
Ffh mutants were constructed with one or two cysteine residues at nonconserved surface positions (Fig. 1A; Buskiewicz et al. 2005b). In the NG domain, cysteines were introduced in the N domain (positions 17, 25), at the junction of N and G domains (position 84), and in the G domain (positions 152, 165, 201, 203). In the M domain, the intrinsic cysteine residue (position 406) was replaced with serine, while cysteine was introduced at position 344 at the junction between helix αM1 and the finger loop (Keenan et al. 1998). Cysteine residues were labeled pairwise (position 344 and one of the other positions) with either Oregon Green 488 (OG) or BODIPY FL (Bpy) for measuring FRET between either two OG or two Bpy fluorophores (“homo-FRET”). For control measurements, the respective single-labeled proteins were also prepared. The labeled proteins were fully active in binding 4.5S RNA, ribosome, and FtsY (Buskiewicz et al. 2005b). Furthermore, SRP containing labeled Ffh was bound to RNC exposing the signal-anchor sequence of Lep with the same high affinity as SRP with unmodified Ffh. Thus, fluorophores at the indicated positions did not interfere with signal peptide binding to Ffh (data not shown).
FIGURE 1.
FRET measurements. (A) Positions of engineered cysteine residues in Ffh. N, G, and M domains are arranged as described previously (Buskiewicz et al. 2005a,b). Positions of cysteine residues at which fluorescent dyes were introduced are indicated by spheres and numbers. (B) 4.5S RNA constructs: full-length 4.5S RNA and the truncated constructs 4.5S RNA21–81 and 4.5S RNA30–78. (C–E) Fluorescence decay of OG attached to Ffh at positions 17 (C), 201 (D), or 344 (E) with Alx555 at the 3′ end of 4.5S RNA21–81. (1) RNC–SRP, donor alone, (2) SRP, donor+acceptor, (3) RNC–SRP, donor+acceptor, (4) excitation pulse.
When two identical fluorophores with overlapping excitation and emission spectra are sufficiently close to one another, the fluorescence anisotropy is lowered as a result of FRET (Lakowicz 1999). The present protein-bound fluorophore pairs exhibited R 0(2/3) values (50% FRET efficiency assuming freely rotating fluorophores) of 43 Å (OG–OG) and 54 Å (Bpy–Bpy). Fluorescence anisotropy was measured for double-labeled proteins and compared to the anisotropy of equimolar mixtures of the respective single-labeled proteins; from the difference, the efficiency of FRET between the two fluorophores was calculated (Supplementary Information). From the FRET efficiencies, the distances between position 344 in the M domain and the respective positions in the N or G domains were calculated. For positions measured with both fluorophores, the same distances to C344 were obtained.
Another set of distances was obtained by measuring FRET (“hetero-FRET”) between one donor fluorophore (OG or Bpy) attached to Ffh and an acceptor (Alexa Fluor 555 or 647, Alx) attached to the 3′ end (G81) of truncated 4.5S RNA21–81 comprising residues 21–81 of 4.5S RNA (Fig. 1B). The construct contained the complete Ffh binding site, including internal loops A, B, and C, and bound Ffh with the same affinity (∼50 pM) as full-length 4.5S RNA (Buskiewicz et al. 2005a). The structures of Ffh bound to 4.5S RNA21–81 or to full-length 4.5S RNA were the same, based on distances measured by FRET (Buskiewicz et al. 2005b). SRP formed with 4.5S RNA21–81 had the same ribosome-binding affinity as SRP formed with full-length 4.5S RNA (Bornemann et al. 2008). A shorter fragment, 4.5S RNA30–78, was also used; it binds Ffh with the same affinity as full-length 4.5S RNA (Batey et al. 2001), but lacks internal loop C, which comprises the binding site for the NG domain of Ffh (Buskiewicz et al. 2005a; Hainzl et al. 2007).
The three different donor–acceptor couples used for hetero-FRET measurements covered an extended distance range, with R 0(2/3) values of 66 Å (OG/Alx555), 50 Å (OG/Alx647), and 44 Å (Bpy/Alx647). FRET efficiencies were determined from the difference between donor fluorescence intensities of Ffh–RNA complexes in which only Ffh was labeled (donor alone) and of the complexes of labeled Ffh with labeled RNA (donor plus acceptor); the signal of the acceptor-alone control was negligible. Additionally, donor fluorescence lifetimes were measured in the absence and presence of acceptor (Fig. 1C–E), and FRET efficiencies were determined from the decrease of the lifetimes due to FRET. Results of fluorescence and fluorescence lifetime measurements and the respective distances calculated from FRET efficiencies are summarized in the Supplementary Information. Essentially identical distances were obtained when different donor–acceptor pairs were used to measure the same distance.
Structure of SRP bound to RNC
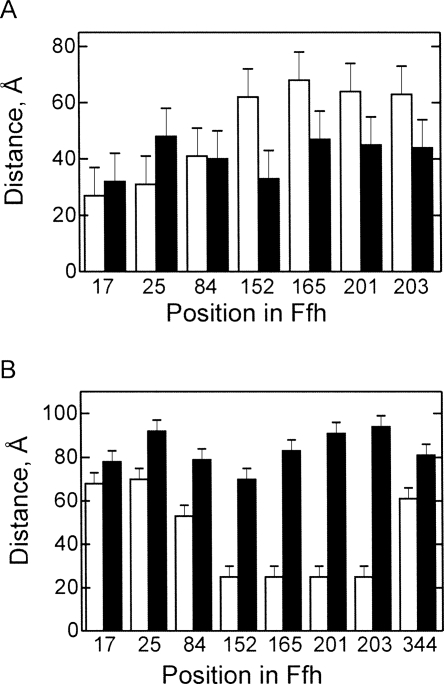
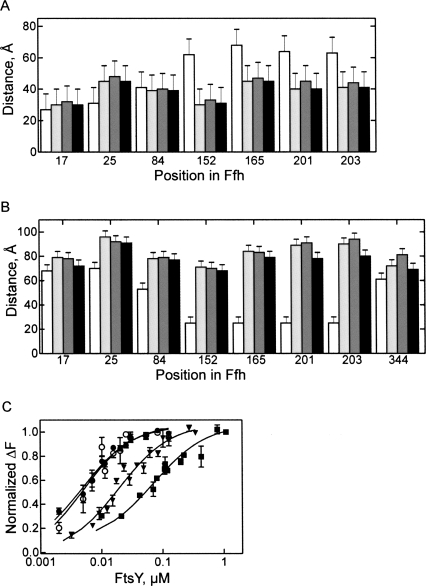
The Ffh domain arrangement in SRP bound to RNC was studied with ribosomes stalled at the end of truncated mRNA and carrying as a nascent chain the first 50 amino acids of leader peptidase. The length of the nascent peptide was sufficient to expose the signal-anchor sequence of Lep for the interaction with SRP (Angelini et al. 2005). Upon binding of SRP to the RNC, the distances between positions 344 in the M domain and 17 and 25 in the N domain of Ffh increased, while those between 344 and the positions in the G domain decreased (Fig. 2A), suggesting a conformational change that moves the N domain away from, and the G domain closer to, the M domain. Identical FRET distances between M and NG domains were found for Ffh bound to full-length 4.5S RNA or to 4.5S RNA21–81. No FRET was observed between labels placed on the 3′ end of full-length 4.5S RNA and any of the Ffh positions, suggesting that the distances were too long for FRET to take place. The distances from almost all positions in Ffh to the 3′ end of 4.5S RNA21–81 (position G81) increased in the SRP–RNC complexes, in particular from positions in the G domain (Fig. 2B, 152, 165, 201, 203). Interestingly, also the distance between position 344 in the M domain and G81 in 4.5 S RNA21–81 increased from ∼60 Å in free SRP to 80 Å in the SRP–RNC complex. This may indicate a change in the RNA conformation, i.e., assuming a somewhat less pronounced kink at loop C or flexibility of the part of the M domain where position 344 is located.
FIGURE 2.
FRET distances in free SRP and the SRP–RNC complex. (A) Distances between position 344 in the M domain of Ffh and the indicated positions in the N domain (17, 25) and G domain (84, 152, 165, 201, 203). (B) Distances between the 3′ end of 4.5S RNA21–81 and the indicated positions in Ffh. (White bars) Free SRP; (black bars) SRP–RNC complex.
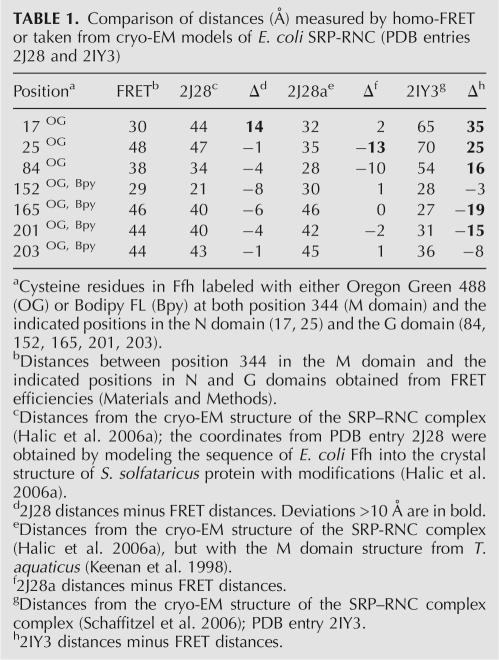
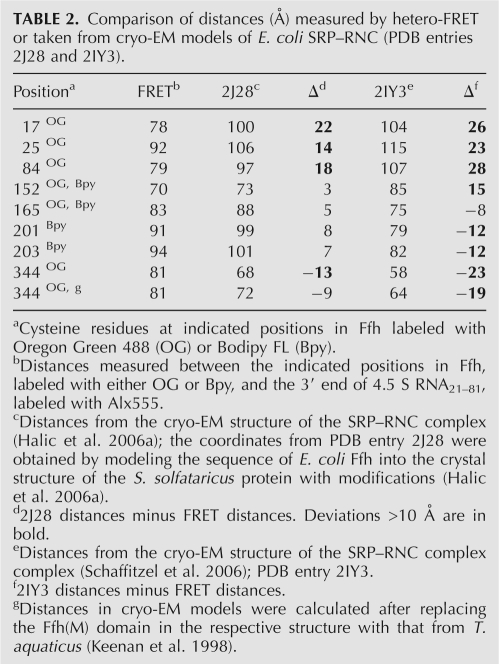
The FRET distances between the NG and M domains (Table 1) and between positions in Ffh and 4.5S RNA21–81 (Table 2) were compared to those derived from two cryo-EM models published recently (Halic et al. 2006a; Schaffitzel et al. 2006). Most FRET distances were consistent with the respective distances derived from one model (Halic et al. 2006a), but deviated considerably from those derived from the other (Schaffitzel et al. 2006). Deviations from the former model included distances between position 344 in the M domain and G81 in 4.5S RNA21–81 as well as between positions in the M and N domains. As the structure of the E. coli full-length M domain is not available, the cryo-EM model of the M domain (Halic et al. 2006b) apparently was obtained by building the sequence of the E. coli Ffh(M) domain into the structure of SRP54 from Sulfolobus solfataricus (Rosendal et al. 2003) with additional adjustments for the position of the finger loop. However, the part of the M domain that comprises helix αM1, the finger loop, and, possibly, the linker between G and M domains vary considerably in SRP structures from different organisms (Keenan et al. 1998; Clemons et al. 1999; Batey et al. 2000; Rosendal et al. 2003; Hainzl et al. 2007). Thus, we tested whether fitting other known structures would give a better agreement with the distances measured by FRET. When the structure of the T. aquaticus M domain (Keenan et al. 1998) was superimposed on helices αM3, αM4, and αM5 of the cryo-EM model (Halic et al. 2006a), the position of residue 344 changed such that the predicted distance to G81 in 4.5S RNA21–81 became 72 Å, closer to the value measured by FRET, 81 Å (Table 2). This suggests that the orientation of the flexible finger loop on the ribosome is more accurately reflected by the T. aquaticus structure of isolated Ffh than by the modified S. sulfataricus structure.
TABLE 1.
Comparison of distances (Å) measured by homo-FRET or taken from cryo-EM models of E. coli SRP-RNC (PDB entries 2J28 and 2IY3)
TABLE 2.
Comparison of distances (Å) measured by hetero-FRET or taken from cryo-EM models of E. coli SRP–RNC (PDB entries 2J28 and 2IY3).
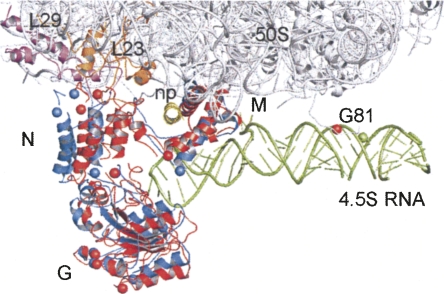
To account for other differences between the present FRET distances and those from the cryo-EM model (Halic et al. 2006a), we modeled the orientation of the N and G domains relative to 4.5S RNA and the M domain of Ffh using the FRET distances as constraints. To transform FRET distances into three-dimensional models of SRP, we used a program, FRETsg (Schröder and Grubmüller 2004), that generates an ensemble of configurations of residues in space that are consistent with the experimentally determined distances between these positions. This ensemble was used for aligning the labeled residues onto the respective residues in the cryo-EM model (Fig. 3; Halic et al. 2006a). The overall position of the G domain in our model agrees well with that in the cryo-EM model (Halic et al. 2006a), while the tip of the N domain occupies a somewhat different position on protein L23, further away from L29 compared to the cryo-EM model. However, this position of the N domain is consistent with the observed strong cross-links to ribosomal protein L23 from a cross-linker attached to N-domain positions 17 or 25 and the lack of cross-links to L29 (Gu et al. 2003). As described above, the flexible part of the M domain is moved in the model. The position of G81 is slightly shifted, compared to the cryo-EM structure, which may reflect the position of the dye that is attached to G81 by a flexible linker.
FIGURE 3.
Model of SRP bound to RNC. The position of Ffh domains N, G, and M derived from FRET is in red, the arrangement of the RNA and Ffh domains in the cryo-EM model (Halic et al. 2006a) are in green and blue, respectively. Red spheres label positions used for FRET and placed by pairwise alignment using FRETsg (Supplementary Information); blue spheres label positions according to the cryo-EM model; green sphere indicates position 81 of 4.5S RNA in the cryo-EM model. The 50S subunit is indicated in gray, proteins L23 and L29 in orange and purple, respectively. (np) Nascent peptide from the cryo-EM model.
Conformation of SRP on the ribosome at different stages of targeting
Next, we analyzed the overall structure of SRP bound to vacant ribosomes, RNCs, and RNCs in the presence of FtsY, the latter complex representing the complete targeting complex poised to transfer the RNC to the translocon. The FRET distances within SRP were quite similar in the SRP complexes with vacant ribosomes and RNCs (Fig. 4A,B). The only noticeable distance change was observed for positions 344 in the M domain and G81 of 4.5S RNA21–81, which may be caused by a movement of part of the M domain upon binding the SAS exposed on the RNC. Thus, the overall domain orientation of SRP bound to vacant ribosomes or RNCs exposing the SAS is probably quite similar, indicating that any structural change resulting from recognition of the SAS is limited to a local rearrangement. This suggests that the SRP–ribosome interaction alone already induces a conformation of SRP in which FtsY binding does not require much further conformational change. The prediction from this scenario is that the affinity of FtsY for ribosome-bound SRP should be higher than for free SRP. This prediction was examined by the fluorescence titration experiments described in the following.
FIGURE 4.
FRET distances in SRP at different stages of targeting. FRET distances were measured in free SRP (white bars), SRP-70S (light gray), SRP-RNC (dark gray), or SRP-RNC-FtsY (black) complexes. (A) Distances between the indicated positions and position 344 in the M domain. (B) Distances from the indicated positions to the 3′ end of 4.5S RNA21–81. (C) Affinity of FtsY binding to SRP in different functional stages. Fluorescence titrations were carried out with SRP formed from Ffh(OG84) and full-length 4.5S RNA. Free SRP (▪); SRP bound to vacant 70S ribosomes (▼); SRP bound to Lep50-RNC (●); isolated Ffh(NG)(OG84) domain (○). Nonlinear fitting (continuous lines; see Materials and Methods) yielded the following K d values: FtsY-SRP, (70 ± 9) nM; FtsY-SRP-70S, (20 ± 3) nM; FtsY-SRP-RNC, (3 ± 0.1) nM; FtsY-Ffh(NG), (3 ± 0.2) nM.
Binding affinities were determined by equilibrium titration, monitoring the fluorescence change of OG attached to position 84 of Ffh. The observed affinity of FtsY binding to free SRP was 70 nM (Fig. 4C), comparable to previous values (Peluso et al. 2000; Bornemann et al. 2008). When SRP was bound to vacant ribosomes, the affinity to FtsY was increased to 20 nM. Binding to RNCs increased the affinity of SRP for FtsY to 4 nM, i.e., almost 20 times compared to the binding to free SRP. Strikingly, FtsY bound equally strongly to the isolated NG domain of Ffh labeled with OG at position 84 (Fig. 4C), in which the interaction surface for binding the FtsY NG domain is freely accessible, as to SRP bound to RNC, indicating that also in the latter complex the NG domain interface of Ffh is fully exposed.
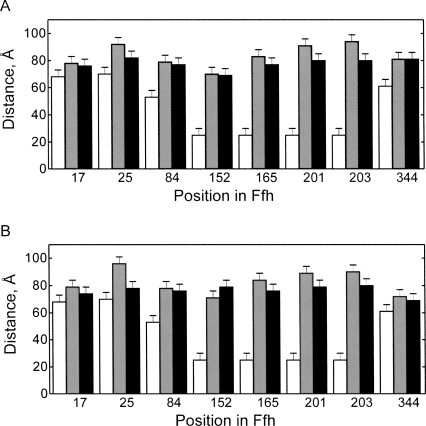
Upon FtsY binding to SRP–RNC, the position of part of the M domain changes, as indicated by the decrease in the distance between position 344 in the M domain and G81 of 4.5S RNA21–81. Furthermore, the G domain moves in the direction of G81, as suggested by shorter distances between the label in 4.5S RNA21–81 and positions 201 and 203 in Ffh, while the distances to other positions in the NG domain remained unchanged. Thus, FtsY induced local alterations in the conformation of Ffh on the ribosome, although the overall Ffh domain arrangement was similar in SRP–RNC and SRP–RNC–FtsY complexes.
The chaperone TF binds in the vicinity of the peptide exit tunnel of the ribosome, sharing with SRP L23 as a common binding partner. Upon binding of TF to the SRP–RNC complex, several distances changed (Fig. 5A). The distances between the 3′ end of 4.5S RNA21–81 and position 25, 201, and 203 decreased by 10–14 Å. Nevertheless, the overall SRP conformation remained close to that in the SRP–RNC complex and was quite different from the conformation of free SRP. The distance between position 344 in the M domain and G81 in 4.5S RNA21–81 did not change upon TF binding. Very similar results were obtained with vacant 70S ribosomes (Fig. 5B). These results suggest that binding of TF induces a local rearrangement of SRP, in keeping with observations from cross-linking experiments (Ullers et al. 2003; Buskiewicz et al. 2004).
FIGURE 5.
Influence of TF on the conformation of ribosome-bound SRP. Changes of distances upon binding of TF to the complex of SRP with RNC (A) or vacant ribosomes (B) were measured by FRET. Distances between the indicated positions in M and NG domains, labeled with OG, and the 3′ end of 4.5S RNA21–81, labeled with Alx 647, were measured in free SRP (white bars), SRP-RNC or SRP-ribosome (gray bars), and SRP-RNC-TF or SRP-ribosome-TF (black) complexes.
Role of 4.5S RNA
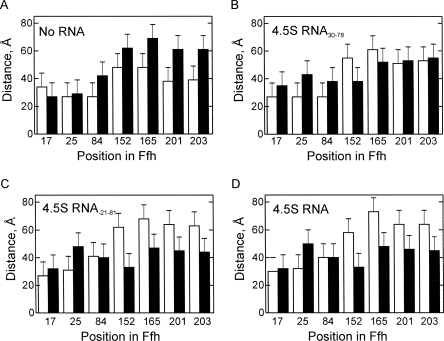
Because the cellular function of Ffh requires binding to 4.5S RNA, we tested how the domain arrangement of Ffh on the ribosome is affected by the presence of 4.5S RNA. Distances were measured between position 344 in the M domain and positions in the N and G domains, because only these distances can be compared for complexes with and without 4.5S RNA. The domain arrangement of free Ffh is quite different from that in the SRP complex, as indicated by characteristically different distances between M domain and G domain (Fig. 6A–D; Buskiewicz et al. 2005b). Upon addition of Ffh to RNC, the distances between the NG and M domains changed considerably, suggesting that Ffh can bind to RNCs even in the absence of 4.5S RNA. However, the structure of the resulting Ffh–RNC complex appeared strikingly different from that formed with SRP. The largest deviations were found in the distances between the M domain and positions 152, 165, 201, and 203 in the G domain, which were in the range of 61–70 Å in the complex without RNA, 38–55 Å in the complex with 4.5S RNA30–78, and 33–48 Å with full-length 4.5S RNA or the 4.5S RNA21–81 fragment. These results indicate that 4.5S RNA strongly affects the conformation of Ffh bound to ribosome and that the magnitude of the effect depends on the presence of the C loop in 4.5S RNA. Analogous results were obtained when Ffh and different SRP complexes with ribosomes or RNCs and/or FtsY were compared (Supplementary Information). Thus, Ffh in ribosome-bound SRP assumes the specific domain arrangement that facilitates subsequent steps of membrane targeting.
FIGURE 6.
Effect of 4.5S RNA. Distances from position 344 in the Ffh(M) domain to the indicated positions in the Ffh(NG) domain were measured in the absence of ribosomes (white bars) and with RNCs (black bars). (A) Ffh, (B) Ffh–4.5S RNA30–78, (C) Ffh–4.5S RNA21–81, (D) Ffh–4.5S RNA.
DISCUSSION
Structural changes of SRP during targeting
The present results reveal that SRP upon binding to the ribosome undergoes a gross conformational change, indicating that the NG and M domains move apart and the G domain is released from the binding interaction with 4.5S RNA. Unlike SRP54 from S. solfataricus, for which an open structure has been reported both in the unbound and the RNA-bound state (Rosendal et al. 2003), Ffh from E. coli is present in a closed conformation in which NG and M domains strongly interact with each other (Buskiewicz et al. 2005a,b), similar to the arrangement of the domains in the first molecule (A/A) of the asymmetric unit in the crystal structure of T. aquaticus Ffh (Keenan et al. 1998). Binding of 4.5S RNA induced a movement of the Ffh(NG) and Ffh(M) domains away from one another, but overall a closed arrangement was retained (Buskiewicz et al. 2005b). Footprinting and biochemical studies indicated that in SRP both M and G domains are bound to 4.5S RNA (Buskiewicz et al. 2005a), albeit with grossly different affinities (∼50 pM and ∼0.5 μM, respectively), the M domain binding at loops A and B (Batey et al. 2000) and the G domain at loop C (Buskiewicz et al. 2005a). The crystal structure of Methanococcus jannaschii SRP54 in complex with the S domain fragment of 7S RNA and SRP19 protein (Hainzl et al. 2007) suggested the existence of two conformations of SRP54, a closed arrangement with the NG domain interacting with the RNA in a way similar to that proposed for E. coli SRP (Buskiewicz et al. 2005a,b), and an open conformation that appeared when the contact between the NG domain of SRP54 and 7S RNA was sterically hindered.
The rearrangement of SRP induced by binding to the ribosome exposes the interaction surface on the NG domain of Ffh for binding of FtsY, which—according to biochemical, biophysical (Buskiewicz et al. 2005a,b), and structural (Hainzl et al. 2007) evidence—is occluded in free SRP. The details of SRP placement on RNC, particularly of the NG domain, are different in the two published cryo-EM reconstructions (Halic et al. 2006a; Schaffitzel et al. 2006). Overall, the FRET data are consistent with one of the cryo-EM models (Halic et al. 2006a), with only minor adjustments in the position of the N domain, and are incompatible with the other (Schaffitzel et al. 2006), some distances deviating as much as 35 Å. One interesting difference between the models derived from FRET and cryo-EM (Halic et al. 2006a) concerns the distance between position 344 in the Ffh(M) domain and G81 in 4.5S RNA. The comparison of different M domain structures suggests that the orientation of the αM1 helix and the following finger loop varies in SRP54/Ffh proteins from different organisms (Keenan et al. 1998; Clemons et al. 1999; Batey et al. 2000; Rosendal et al. 2003). These structural differences may reflect sequence and structure variations of the orthologous proteins. Alternatively, they could suggest that the position of helix αM1 and the finger loop can vary, depending on the functional state of SRP.
Another question concerns the conformation of SRP bound to ribosomes that do not expose an SAS. Cross-linking results indicated that the contact between the N domain of Ffh and protein L23 at the peptide exit is independent of the presence of an SAS and may be established early during targeting (Gu et al. 2003). On the other hand, the cross-link between 4.5S RNA and 23S rRNA was observed only when the signal sequence was exposed (Rinke-Appel et al. 2002). In cryo-EM reconstructions of SRP in the complex with the vacant ribosome, there was no density for the M domain and 4.5S RNA (Schaffitzel et al. 2006). This prompted the suggestion that early in targeting, i.e., before the signal sequence has emerged from the ribosome, the M domain of Ffh and 4.5S RNA are mobile, the contact of 4.5S RNA with the ribosome is not yet established, and the only contact between SRP and the ribosome is the contact between the Ffh(N) domain and protein L23 (Schaffitzel et al. 2006). Mobile M domain and 4.5S RNA implies that SRP can be present in different domain arrangements on different ribosome–SRP complexes, which would be expected to result in different average FRET distances in SRP on vacant ribosomes compared to SRP bound to RNCs. According to the FRET data, the overall orientation of Ffh on the ribosome appears rather similar without and with exposed signal sequence, arguing against models implying an extensive mobility of parts of SRP or gross conformational changes upon signal sequence recognition. On the other hand, local rearrangements are likely, such as adjustments of the contacts between the Ffh(N) domain and L23, as indicated by an altered cross-linking pattern (Gu et al. 2003) and changes in the position of elements of the M domain (present data). The latter may involve the finger loop alone or, additionally, the αM1 helix in the M domain, which constitute part of the hydrophobic groove where nascent signal peptides are thought to bind. In the absence of the SAS, the part of the M domain comprising the αM1 helix-finger loop appears to be in a conformation compatible with that observed in the structure from S. solfataricus. Upon exposure of the SAS, this part of the M domain moves, probably to accommodate the nascent peptide in the hydrophobic groove presumed to be (part of) the binding site of the signal peptide. Upon interaction with FtsY, the movement is reversed, suggesting that FtsY induces the release of the signal peptide from the M domain, thereby probably initiating transfer of the nascent peptide to the translocon. Binding of FtsY to the SRP–RNC complex results in a small movement of the G domain away from the RNA and a reversal of the changes in the arrangement of the αM1 helix-finger loop region in the M domain. The rearrangements may play a role in communicating the transfer of the signal-anchor sequence to the GTPase sites in Ffh and FtsY, e.g., through L23, which is contacted by both the finger loop of the M domain (Halic et al. 2006a) and the N domain of Ffh (Gu et al. 2003).
Besides SRP, also TF binds in the vicinity of the peptide exit of the ribosome. SRP and TF can bind simultaneously (Buskiewicz et al. 2004; Raine et al. 2004); both use L23 as a contact site at the ribosome and occupy adjacent regions on the ribosomal surface (Kramer et al. 2002; Buskiewicz et al. 2004; Ferbitz et al. 2004; Baram et al. 2005; Schlunzen et al. 2005). SRP and TF can bind to the ribosome before the signal sequence is exposed (Kramer et al. 2002; Buskiewicz et al. 2004). The present data show that, upon binding of TF to the ribosomes, the overall Ffh domain arrangement does not change, but there are slight adjustments in the positioning of both N domain (position 25) and G domain (positions 201 and 203). The conformation of SRP in the complex with vacant ribosomes or RNCs in the presence of TF is very similar, except for a different orientation of the αM1 helix-finger loop part. Although the nascent chain was modeled to be tightly packed to the hydrophobic grooves of TF on one side and the M domain on the other (Schlunzen et al. 2005), the distance between position 344 in the M domain and G81 in 4.5S RNA21–81 is identical in the SRP–RNC complexes in the presence and absence of TF, suggesting that TF does not hinder the movement of the finger loop as induced by the binding of the signal-anchor sequence.
Affinity of FtsY binding to SRP at different stages of targeting
SRP can bind to vacant ribosomes or to RNCs with short nascent chains that are fully contained in the ribosome exit tunnel (Bornemann et al. 2008). When the length of the nascent peptide in the tunnel reaches ∼30 amino acids, i.e., is still completely contained within the tunnel and unable to bind to SRP (Angelini et al. 2006), the affinity of SRP binding to RNC is increased ∼50-fold compared to RNCs with shorter nascent peptides (<26 amino acids) or nontranslating ribosomes. The affinity increase is mediated by protein L23 (Bornemann et al. 2008), which reaches into the tunnel and forms part of the SRP binding site. Exposure of the SAS, which presumably binds to SRP with micromolar affinity (Zheng and Gierasch 1997), did not increase the affinity of SRP binding to the RNC (Bornemann et al. 2008). The likely explanation for this finding is that the binding energy gained from interactions of Ffh with the SAS is consumed by the conformational rearrangements indicated by the present FRET data. These include a movement of the αM1-finger loop region and a local rearrangement of the NG domain.
The structural changes in SRP upon binding to the ribosome at different targeting stages are correlated with the affinity of FtsY binding. Binding to RNCs increased the affinity of SRP for FtsY almost 20 times compared to the binding to free SRP. Interestingly, FtsY binds equally strongly to the SRP–RNC and to the isolated NG domain of Ffh in which the interaction surface is exposed without any conformational change. This indicates that the conformational change of Ffh in SRP required for FtsY binding is promoted by SRP binding to the ribosome. While SRP showed a similar global structure on vacant ribosomes and RNCs, with the binding site for FtsY on the Ffh(NG) domain exposed, the affinity for FtsY increased only 3.5-fold when SRP was bound to vacant ribosomes, compared to the 20-fold increase observed with the SRP–RNC complex. One clear structural difference between those complexes is the location of the αM1 helix-finger loop of the M domain. It seems likely, therefore, that the M domain in the complex of SRP with vacant ribosomes sterically hinders FtsY binding. Given that the A domain of FtsY was lacking in the heterodimeric NG–NG domain complexes for which crystal structures were determined (Egea et al. 2004; Focia et al. 2004), a clash between the A domain of FtsY and the M domain of Ffh is not excluded. In addition, the position of the αM1 helix-finger loop region may affect the G domain, e.g., through repositioning of the linker peptide that connects M and G domains or of the N domain through interactions with protein L23. Thus, although the surface of the Ffh(NG) domain appears to be more easily accessible for FtsY binding in the SRP–ribosome complex, compared to free SRP, additional adjustments, as induced by the binding of the signal peptide, appear to be necessary to reach the high affinity for FtsY binding observed for the SRP-RNC complex.
Role of 4.5S RNA
The present data show that the conformations of ribosome-bound Ffh are quite different in the presence or absence of 4.5S RNA. Given that the N domain of Ffh was cross-linked to the protein L23 independent of 4.5S RNA (Gu et al. 2003), the position of the NG domain is likely to be similar in the ribosome complexes of Ffh and SRP. Then, in order to account for the measured distances, the position of the Ffh(M) domain must be different in the two complexes. The simplest explanation for this is that the contacts of 4.5S RNA with the M domain and/or the ribosome stabilize the observed open conformation of SRP on the ribosome. The structure of Ffh in ribosome-bound SRP was the same for full-length 4.5S RNA or the 4.5S RNA21–81 fragment which contains loops A–C. Although the distances in Ffh induced by the 4.5S RNA30–78 fragment were not identical to those induced by the longer 4.5S RNA constructs, the direction of changes in Ffh conformation was similar with all three constructs. This is consistent with the observation that 4.5S RNA30–78 could replace full-length 4.5S RNA in vivo (Batey et al. 2000), despite the differences in the conformations of free SRP formed with the 4.5S RNA30–78 fragment compared to full-length 4.5S RNA (Fig. 6B; Buskiewicz et al. 2005b). 4.5S RNA may act as a tether for positioning the M domain at the exit tunnel, e.g., through contacts between loop C of 4.5S RNA and the ribosomal protein L18 (and possibly L17), as indicated in the cryo-EM reconstructions (Halic et al. 2006a; Schaffitzel et al. 2006). An influence of 4.5S RNA on the conformation of the M domain has been observed (Zheng and Gierasch 1997). Thus, a major effect of 4.5S RNA may be to stabilize a configuration of Ffh that is favorable for the interaction with FtsY and the SAS by separating NG and M domains and by stabilizing the M domain conformation. This is in line with previous observations indicating that 4.5S RNA facilitates the interaction of Ffh with FtsY (Peluso et al. 2000). Thus, 4.5S RNA appears to act as a guide for arranging Ffh on the ribosome and stabilizing the SRP conformation that is most favorable for the interaction with the SRP receptor FtsY and subsequent steps of targeting.
MATERIALS AND METHODS
Materials
Buffer A is composed of 25 mM HEPES (pH 7.5), 70 mM ammonium acetate, 30 mM potassium acetate, 7 mM magnesium acetate. Alexa Fluor 555/647 hydrazide, Oregon Green 488 maleimide, and BODIPY-FL maleimide were from Molecular Probes. Ni-NTA agarose was from Qiagen. All other chemicals were obtained from Sigma or Merck. Proteins and RNAs were purified and fluorescence modifications were conducted as described previously (Buskiewicz et al. 2005b). TF with an oligohistidine tag at the C terminus was a gift of Elke Deuerling (University of Konstanz, Germany).
E. coli strains and plasmids
BL21 (DE3) pLysS strain was used for expressing FtsY and Ffh from pET9-FtsY and pET24-Ffh plasmids, respectively (Jagath et al. 2000). The plasmid pT7-4.5S was used to prepare full-length 4.5S RNA and 4.5S RNA20–81 fragment by in vitro transcription using T7 RNA polymerase (Lentzen et al. 1994; Buskiewicz et al. 2005b). The 4.5S RNA30–78 fragment was purchased from Dharmacon. Replacement of residues at positions 17, 25, 84, 152, 165, 201, 203, or 344 from Ffh with cysteine was performed by PCR mutagenesis by the QuickChange method using Pfu polymerase (Promega) as described (Gu et al. 2003; Buskiewicz et al. 2005b). Mutations were generated in plasmid pET24-Ffh coding for Ffh extended by six histidines at the C terminus. Mutations were confirmed by DNA sequencing.
Ribosome–nascent chain complexes
Ribosomes from E. coli MRE600 and purified components of the translation system were prepared as described (Rodnina and Wintermeyer 1995; Rodnina et al. 1999). Truncated mRNA coding for 50 N-terminal amino acids of leader peptidase (Lep) was translated and RNCs were purified as described (Gu et al. 2003; Bornemann et al. 2008). A total of 75% or more of the ribosomes present in the RNC preparation carried a nascent peptide, as determined from the amount of f[3H]Met in trichloroacetic acid (TCA)-precipitable peptides after alkaline hydrolysis of peptidyl–tRNA. The peptide, which was labeled by the incorporation of [14C]Leu, was homogeneous on SDS gel electrophoresis and migrated at the expected position (Bornemann et al. 2008).
FRET measurements
SRP was formed by mixing equimolar amounts of Ffh and full-length or truncated 4.5S RNA in buffer A with GDPNP (0.2 mM). SRP-RNC complexes were prepared by mixing SRP (0.015 μM) with purified RNC (0.015 μM) in the same buffer for 5 min at room temperature (RT). When indicated, FtsY (0.015–0.1 μM) or TF (0.5–1.0 μM) were added and incubated for another 15 min at RT. For measurements with vacant ribosomes SRP (0.15 μM), 70S ribosomes (0.15 μM) and, where indicated, FtsY (0.15 μM) or TF (0.5–1.5 μM) were added. FRET efficiencies were determined from both steady-state fluorescence (intensity or anisotropy) and fluorescence lifetime measurements, carried out as described in Supplementary Information. Measurements were performed in triplicate, and standard deviations were <5%. Distances between fluorophore couples, R, were estimated from FRET efficiencies, E, based on R 0 values as previously described (Lakowicz 1999; Buskiewicz et al. 2005b; for details, see Supplemental Material). Ranges of R 0 values were estimated from κ2 values determined from anisotropy measurements with the respective single-labeled SRP (Lakowicz 1999; Buskiewicz et al. 2005b). Due to the limited rotational mobility of the protein-bound dyes, as indicated by anisotropy values >0.3, the uncertainty range of the distances derived from homo-FRET, included in the range of values determined for R 0 min and R 0 max, was ∼±10 Å. The uncertainty ranges of the distances derived from FRET between labels in Ffh and RNA were ±5 Å or less, owing to a high rotational mobility of the fluorophores at the 3′ end of 4.5S RNA21–81.
Modeling
Structural models of SRP on RNC were built using FRETsg (Schröder and Grubmüller 2004), as described previously (Buskiewicz et al. 2005b; see Supplemental Material).
Affinity measurements
Fluorescence titrations were performed by adding increasing amounts of FtsY to Ffh(OG84) or Ffh(Bpy165) (2 nM) alone or bound to 4.5S RNA (2 nM) and vacant 70S ribosomes (100, 150, 200, or 500 nM), or to Ffh(NG) alone (2 nM) in buffer A with GDPNP (0.2 mM). FtsY titrations with RNCs were performed at different concentrations of SRP and RNC (SRP/RNC: 0.5 nM/1 nM; 0.5 nM/4 nM; 1 nM/4 nM; 1 nM/10 nM, 2 nM/10 nM). After correction for dilution, the data were evaluated by nonlinear fitting (Wilden et al. 2006) using Table Curve software (Jandel Scientific). The same K d values were obtained from titrations performed at various concentrations.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
Time-resolved measurements were performed at the Center for Fluorescence Spectroscopy, University of Maryland at Baltimore, School of Medicine, Baltimore, MD, USA. We thank Ignacy Gryczynski for help with the time-resolved fluorescence measurements and discussions; Elke Deuerling for providing trigger factor; Frank Peske for cysteine constructs of Ffh; and Astrid Böhm, Carmen Schillings, Simone Möbitz, and Petra Striebeck for expert technical assistance. The work was supported by the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, and the Deutsche Akademische Austauschdienst (DAAD).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1285609.
REFERENCES
- Angelini S., Deitermann S., Koch H.G. FtsY, the bacterial signal-recognition particle receptor, interacts functionally and physically with the SecYEG translocon. EMBO Rep. 2005;6:476–481. doi: 10.1038/sj.embor.7400385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelini S., Boy D., Schiltz E., Koch H.G. Membrane binding of the bacterial signal recognition particle receptor involves two distinct binding sites. J. Cell Biol. 2006;174:715–724. doi: 10.1083/jcb.200606093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baram D., Pyetan E., Sittner A., Auerbach-Nevo T., Bashan A., Yonath A. Structure of trigger factor binding domain in biologically homologous complex with eubacterial ribosome reveals its chaperone action. Proc. Natl. Acad. Sci. 2005;102:12017–12022. doi: 10.1073/pnas.0505581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R.T., Rambo R.P., Lucast L., Rha B., Doudna J.A. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Batey R.T., Sagar M.B., Doudna J.A. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- Bernstein H.D., Poritz M.A., Strub K., Hoben P.J., Brenner S., Walter P. Model for signal sequence recognition from amino-acid sequence of 54K subunit of signal recognition particle. Nature. 1989;340:482–486. doi: 10.1038/340482a0. [DOI] [PubMed] [Google Scholar]
- Bornemann T., Jöckel J., Rodnina M.V., Wintermeyer W. Signal sequence-independent membrane targeting of ribosomes containing short nascent peptides within the exit tunnel. Nat. Struct. Mol. Biol. 2008;15:494–499. doi: 10.1038/nsmb.1402. [DOI] [PubMed] [Google Scholar]
- Buskiewicz I., Deuerling E., Gu S.Q., Jockel J., Rodnina M.V., Bukau B., Wintermeyer W. Trigger factor binds to ribosome-signal-recognition particle (SRP) complexes and is excluded by binding of the SRP receptor. Proc. Natl. Acad. Sci. 2004;101:7902–7906. doi: 10.1073/pnas.0402231101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Kubarenko A., Peske F., Rodnina M.V., Wintermeyer W. Domain rearrangement of SRP protein Ffh upon binding 4.5S RNA and the SRP receptor FtsY. RNA. 2005a;11:947–957. doi: 10.1261/rna.7242305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskiewicz I., Peske F., Wieden H.J., Gryczynski I., Rodnina M.V., Wintermeyer W. Conformations of the signal recognition particle protein Ffh from Escherichia coli as determined by FRET. J. Mol. Biol. 2005b;351:417–430. doi: 10.1016/j.jmb.2005.06.023. [DOI] [PubMed] [Google Scholar]
- Clemons W.M., Jr, Gowda K., Black S.D., Zwieb C., Ramakrishnan V. Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 Å resolution: Evidence for the mechanism of signal peptide binding. J. Mol. Biol. 1999;292:697–705. doi: 10.1006/jmbi.1999.3090. [DOI] [PubMed] [Google Scholar]
- Connolly T., Rapiejko P.J., Gilmore R. Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science. 1991;252:1171–1173. doi: 10.1126/science.252.5009.1171. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Batey R.T. Structural insights into the signal recognition particle. Annu. Rev. Biochem. 2004;73:539–557. doi: 10.1146/annurev.biochem.73.011303.074048. [DOI] [PubMed] [Google Scholar]
- Egea P.F., Shan S.O., Napetschnig J., Savage D.F., Walter P., Stroud R.M. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- Ferbitz L., Maier T., Patzelt H., Bukau B., Deuerling E., Ban N. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 2004;431:590–596. doi: 10.1038/nature02899. [DOI] [PubMed] [Google Scholar]
- Flanagan J.J., Chen J.C., Miao Y., Shao Y., Lin J., Bock P.E., Johnson A.E. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 2003;278:18628–18637. doi: 10.1074/jbc.M300173200. [DOI] [PubMed] [Google Scholar]
- Focia P.J., Shepotinovskaya I.V., Seidler J.A., Freymann D.M. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freymann D.M., Keenan R.J., Stroud R.M., Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- Gill D.R., Salmond G.P. The identification of the Escherichia coli ftsY gene product: An unusual protein. Mol. Microbiol. 1990;4:575–583. doi: 10.1111/j.1365-2958.1990.tb00626.x. [DOI] [PubMed] [Google Scholar]
- Gu S.Q., Peske F., Wieden H.J., Rodnina M.V., Wintermeyer W. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 2003;9:566–573. doi: 10.1261/rna.2196403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S.Q., Jockel J., Beinker P., Warnecke J., Semenkov Y.P., Rodnina M.V., Wintermeyer W. Conformation of 4.5S RNA in the signal recognition particle and on the 30S ribosomal subunit. RNA. 2005;11:1374–1384. doi: 10.1261/rna.7219805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T., Huang S., Sauer-Eriksson A.E. Interaction of signal-recognition particle 54 GTPase domain and signal-recognition particle RNA in the free signal-recognition particle. Proc. Natl. Acad. Sci. 2007;104:14911–14916. doi: 10.1073/pnas.0702467104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M., Blau M., Becker T., Mielke T., Pool M.R., Wild K., Sinning I., Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006a;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- Halic M., Gartmann M., Schlenker O., Mielke T., Pool M.R., Sinning I., Beckmann R. Signal recognition particle receptor exposes the ribosomal translocon binding site. Science. 2006b;312:745–747. doi: 10.1126/science.1124864. [DOI] [PubMed] [Google Scholar]
- Jagath J.R., Rodnina M.V., Wintermeyer W. Conformational changes in the bacterial SRP receptor FtsY upon binding of guanine nucleotides and SRP. J. Mol. Biol. 2000;295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- Keenan R.J., Freymann D.M., Walter P., Stroud R.M. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- Keenan R.J., Freymann D.M., Stroud R.M., Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- Kramer G., Rauch T., Rist W., Vorderwulbecke S., Patzelt H., Schulze-Specking A., Ban N., Deuerling E., Bukau B. L23 protein functions as a chaperone docking site on the ribosome. Nature. 2002;419:171–174. doi: 10.1038/nature01047. [DOI] [PubMed] [Google Scholar]
- Krieg U.C., Walter P., Johnson A.E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T.V., Wiedmann M., Girshovich A.S., Bochkareva E.S., Bielka H., Rapoport T.A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Lakowicz J.R. Principles of fluorescence spectroscopy. Kluwer Academic/Plenum Publishers; New York: 1999. pp. 305–309. [Google Scholar]
- Lentzen G., Dobberstein B., Wintermeyer W. Formation of SRP-like particle induces a conformational change in E. coli 4.5S RNA. FEBS Lett. 1994;348:233–238. doi: 10.1016/0014-5793(94)00599-0. [DOI] [PubMed] [Google Scholar]
- Montoya G., Svensson C., Luirink J., Sinning I. Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature. 1997;385:365–368. doi: 10.1038/385365a0. [DOI] [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Kuglstatter A., Menichelli E., Isel C., Jovine L. Structure, function and evolution of the signal recognition particle. EMBO J. 2003;22:3479–3485. doi: 10.1093/emboj/cdg337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso P., Herschlag D., Nock S., Freymann D.M., Johnson A.E., Walter P. Role of 4.5S RNA in assembly of the bacterial signal recognition particle with its receptor. Science. 2000;288:1640–1643. doi: 10.1126/science.288.5471.1640. [DOI] [PubMed] [Google Scholar]
- Poritz M.A., Bernstein H.D., Strub K., Zopf D., Wilhelm H., Walter P. An E. coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science. 1990;250:1111–1117. doi: 10.1126/science.1701272. [DOI] [PubMed] [Google Scholar]
- Raine A., Ivanova N., Wikberg J.E., Ehrenberg M. Simultaneous binding of trigger factor and signal recognition particle to the E. coli ribosome. Biochimie. 2004;86:495–500. doi: 10.1016/j.biochi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Rinke-Appel J., Osswald M., von Knoblauch K., Mueller F., Brimacombe R., Sergiev P., Avdeeva O., Bogdanov A., Dontsova O. Crosslinking of 4.5S RNA to the Escherichia coli ribosome in the presence or absence of the protein Ffh. RNA. 2002;8:612–625. doi: 10.1017/s1355838202020095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Wintermeyer W. GTP consumption of elongation factor Tu during translation of heteropolymeric mRNAs. Proc. Natl. Acad. Sci. 1995;92:1945–1949. doi: 10.1073/pnas.92.6.1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina M.V., Savelsbergh A., Matassova N.B., Katunin V.I., Semenkov Y.P., Wintermeyer W. Thiostrepton inhibits the turnover but not the GTPase of elongation factor G on the ribosome. Proc. Natl. Acad. Sci. 1999;96:9586–9590. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendal K.R., Wild K., Montoya G., Sinning I. Crystal structure of the complete core of archaeal signal recognition particle and implications for interdomain communication. Proc. Natl. Acad. Sci. 2003;100:14701–14706. doi: 10.1073/pnas.2436132100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffitzel C., Oswald M., Berger I., Ishikawa T., Abrahams J.P., Koerten H.K., Koning R.I., Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- Schlunzen F., Wilson D.N., Tian P., Harms J.M., McInnes S.J., Hansen H.A., Albrecht R., Buerger J., Wilbanks S.M., Fucini P. The binding mode of the trigger factor on the ribosome: Implications for protein folding and SRP interaction. Structure. 2005;13:1685–1694. doi: 10.1016/j.str.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Schröder G.F., Grubmüller H. FRETsg: Biomolecular structure model building from multiple FRET experiments. Comput. Phys. Commun. 2004;158:150–157. [Google Scholar]
- Spanggord R.J., Siu F., Ke A., Doudna J.A. RNA-mediated interaction between the peptide-binding and GTPase domains of the signal recognition particle. Nat. Struct. Mol. Biol. 2005;12:1116–1122. doi: 10.1038/nsmb1025. [DOI] [PubMed] [Google Scholar]
- Ullers R.S., Houben E.N., Raine A., ten Hagen-Jongman C.M., Ehrenberg M., Brunner J., Oudega B., Harms N., Luirink J. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 2003;161:679–684. doi: 10.1083/jcb.200302130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilden B., Savelsbergh A., Rodnina M.V., Wintermeyer W. Role and timing of GTP binding and hydrolysis during EF-G-dependent tRNA translocation on the ribosome. Proc. Natl. Acad. Sci. 2006;103:13670–13675. doi: 10.1073/pnas.0606099103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N., Gierasch L.M. Domain interactions in E. coli SRP: Stabilization of M domain by RNA is required for effective signal sequence modulation of NG domain. Mol. Cell. 1997;1:79–87. doi: 10.1016/s1097-2765(00)80009-x. [DOI] [PubMed] [Google Scholar]