Abstract
As well as generating protein isoform diversity, in some cases alternative splicing generates RNAs that harbor premature termination codons and that are subject to nonsense-mediated decay (NMD). We previously identified an apparent pseudo-exon in the rat α-tropomyosin (Tpm1) gene as a probable genuine alternatively spliced exon that causes NMD when spliced into Tpm1 RNA. Here, we report the analysis of cis-acting splicing regulatory elements within this “nonsense exon.” Guided by the data set of predicted splicing enhancer and silencer elements compiled by Zhang and Chasin, we made a series of mutations through the nonsense exon and found that like authentic exons it is densely packed with enhancer and silencer elements. Strikingly, 11 of 13 tested mutations behaved as predicted computationally. In particular, we found that a G-rich silencer at the 5′ end, which is crucial for skipping of the nonsense exon, functions by binding hnRNP-H and F.
Keywords: alternative splicing, hnRNP, nonsense-mediated decay
INTRODUCTION
Alternative splicing (AS) allows cells to regulate the inclusion or exclusion of exons, thereby generating multiple mRNA transcripts, and ultimately multiple distinct polypeptides from a single gene. Computational and microarray-based approaches suggest that the majority of human genes undergo some form of alternative splicing, and this helps to generate the rich complexity of expressed proteomes (for review, see Matlin et al. 2005; Blencowe 2006). AS may also play an important role in the regulation of transcript levels, by the inclusion of premature termination codons (PTCs), which induce nonsense-mediated decay (NMD) (for review, see Lejeune and Maquat 2005; McGlincy and Smith 2008). For a mammalian termination codon to be recognised as premature, it must lie >50–55 nucleotides (nt) upstream of an exon–exon junction (Nagy and Maquat 1998). This critical threshold distance is explained by the deposition of 350 kDa exon junction complexes (EJCs) 20–24 nt upstream of each exon–exon junction as a consequence of splicing (Le Hir et al. 2000). EJCs are displaced by the translating ribosome during the pioneer round of translation, unless they are located >∼30 nt downstream from a termination codon (Ishigaki et al. 2001). The translating ribosome stalls when the ribosome reaches a termination codon, and a termination complex is formed. Interactions between the termination complex and any remaining downstream EJCs result in the transcript being marked for degradation (Chang et al. 2007).
While NMD was originally characterized for its role in disposing of aberrant mRNAs arising from nonsense codon containing alleles, it has become clear that AS can also deliberately produce RNAs that are destined for NMD. The simple “55 nt upstream of an intron” rule to define mammalian PTCs has allowed bioinformatic analysis to investigate the extent to which AS produces mRNAs that are probable substrates for NMD. The striking finding was that between one-fifth and one-third of alternatively spliced transcripts are apparent targets of NMD (Lewis et al. 2003; Baek and Green 2005). The obvious rationale for such unproductive splicing is biological control. Verified examples include the splicing repressor PTB and the splicing activator SC35, both of which induce NMD linked AS events in their own pre-mRNAs in autoregulatory feedback loops (Sureau et al. 2001; Wollerton et al. 2004). A number of proteins that are translated only at localized sites within neurons are constitutively spliced within their 3′UTRs, rendering them NMD sensitive and thereby allowing a short pulse of protein expression after translational activation (Giorgi et al. 2007). Recent findings demonstrate that AS-NMD events are widespread among the genes for splicing regulatory proteins (Lareau et al. 2007; Ni et al. 2007) and core spliceosomal proteins (Saltzman et al. 2008), and that they can be involved in cross-regulation between different splicing regulators (Boutz et al. 2007; Makeyev et al. 2007; Spellman et al. 2007). Despite the fact that EST-based predictions are expected to underestimate the prevalence of AS-NMD, because by definition such mRNAs are underrepresented, quantitative AS microarray profiling suggested that the majority of the EST inferred cases may not be biologically significant (Pan et al. 2006). The true extent to which AS and NMD are linked in a physiologically meaningful manner is still open to question (for review, see McGlincy and Smith 2008).
We have investigated a possible example of AS-NMD in the rat α-tropomyosin gene (Tpm1), which is not even supported by EST data (Grellscheid and Smith 2006). A conserved apparent pseudo-exon was noted just downstream from an essential regulatory element (DRE) that controls selection between mutually exclusive Tpm1 exons 2 and 3 (Fig. 1A). Although not required for regulated selection of exon 2 and 3 (Gooding et al. 1998), the “pseudo-exon” is relatively well conserved, including the presence of in frame PTCs, and it can be fully activated for splicing by simple point mutations (Grellscheid and Smith 2006). The pseudo-exon can also apparently be selectively used as a “zero-length exon” (Hatton et al. 1998; Burnette et al. 2005) according to whether it splices to Tpm1 exon 2 or 3. The sequence at its 5′ end resembles a 5′ splice site and a construct in which the pseudo exon was “prespliced” to Tpm1 exon 2, exclusively used the resultant hybrid 5′ splice site (AAG|GUGGGU) for splicing to exon 4. In contrast, the sequence formed by presplicing to exon 3 (GAU|GUGGGU) was not used as a 5′ splice site, reflecting its poorer match to the consensus (Grellscheid and Smith 2006). Although originally classified as a pseudo-exon, on the basis that it has not been observed in mature spliced Tpm1 transcripts, in transgenic animals expressing a Tpm1 minigene it was observed to be spliced in a tissue-specific manner with highest levels in the heart (Ellis et al. 2004). Splicing of the 107 nt exon to exon 4 was also observed by RT-PCR of RNA from skeletal muscle, heart, and cardiomyocyte cells (Grellscheid and Smith 2006). Finally, its relative enrichment of exon splicing enhancers (ESE) over exon splicing silencers (ESS) (Grellscheid and Smith 2006) is more consistent with the profile of an authentic exon rather than a pseudo-exon (Zhang and Chasin 2004). We propose that the apparent pseudo-exon may be a genuine regulated exon that leads to the efficient destruction of mRNAs into which it is spliced. For this reason, we refer to it as a nonsense exon (NE). The role of the NE could be analogous to that of “poison” exons in the genes for SR proteins, namely quantitative post-transcriptional regulation of gene expression (Lareau et al. 2007; Ni et al. 2007).
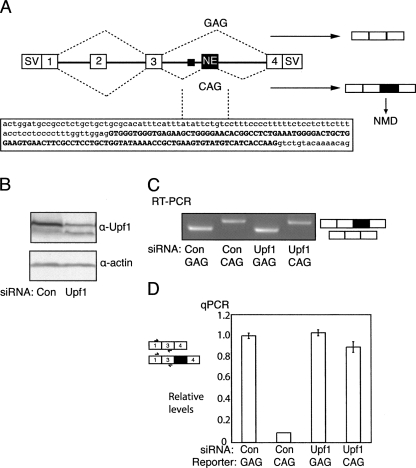
FIGURE 1.
NE inclusion leads to NMD. (A) Schematic representation of the pcDNA 1–4 splicing reporter containing rat Tpm1 exons 1 to 4. The SV40 promoter/enhancer used to express the construct and the SV40 3′ end processing signals fused to exons 1 and 4 are represented by “SV.” Exons 2 and 3 are mutually exclusive, but in most cell types exon 3 is selected predominantly or exclusively. The NE is indicated, and the black rectangle just upstream indicates its polypyrimidine tract. The sequence of the NE (upper case) and immediate flanking introns (lower case) is indicated below. In the WT pCDNA1–4–GAG minigene, the NE is skipped, but in the pCDNA1–4–CAG minigene, mutation of the NE 3′ splice site from GAG to CAG leads to its full inclusion (see C). (B) Western blot of protein from HeLa cells treated with control (con) or Upf1 targeting siRNAs, probed with antibodies against Upf1 (upper panel) or actin (lower panel). (C) RT-PCR analysis of pcDNA-1–4 splicing in HeLa cells treated with control or Upf1 targeting siRNAs. The NE is skipped in the GAG construct, but fully included in the CAG construct. (D) Quantitative real-time PCR analysis of construct RNAs from C, using primers in Tpm1 exons 1 and 3. Values are normalized to the levels of the WT GAG construct with control siRNAs. Levels of CAG transcripts were 10-fold less than those of GAG, but were largely restored by knockdown of the NMD factor Upf1.
We were interested in characterizing the sequence elements and factors that are responsible for the regulation of the NE. A number of computational approaches for global characterization of exon splicing enhancer (ESE) and silencer (ESS) motifs have been developed (Fairbrother et al. 2002; Fairbrother et al. 2004a,b; Zhang and Chasin 2004; Goren et al. 2006). We used the octamer data set of Zhang and Chasin (2004), which allows prediction both of ESEs and ESSs. Of 13 predicted elements through the length of the NE, 11 were confirmed by mutagenesis. Only in one case did the prediction suggest an ESE, whereas an ESS was confirmed. A number of potential trans-acting regulators were identified, among which hnRNP H and F were found to be crucial for repressing the NE via binding to G-rich ESSs. Our data show that the NE is regulated in a manner closely resembling that of many authentic protein-coding exons.
RESULTS
Nonsense exon induces NMD
Splicing of the Tpm1 NE into mRNA introduces an in frame stop codon 95 nt upstream of the junction between the NE and exon 4, and so is expected to lead to NMD. Consistent with this, increased levels of NE spliced to exon 4 were detected after puromycin treatment of cardiomyocytes (Grellscheid and Smith 2006). To test whether activation of the NE leads to degradation of Tpm1 RNA by NMD, we transfected Tpm1 reporter constructs into HeLa cells 24 h after treatment with siRNAs targeting Upf1, to inhibit NMD, or a control siRNA (Fig. 1B). pcDNA1–4–GAG contains wild-type (WT) Tpm1 exons 1 to 4, and the corresponding introns. The intron between exons 3 and 4 is shortened to 517 nt (compared with the WT intron of >12 kb) and includes the NE and 170 nt of downstream sequence. pcDNA1–4–CAG contains the GAG to CAG mutation of the NE 3′ splice site. Analysis of construct splicing by semiquantitative RT-PCR indicated that the CAG mutation leads to full inclusion of the NE (Fig. 1C, lanes 1,2) as reported previously (Grellscheid and Smith 2006). Expression levels of the Tpm1 constructs were next measured by qPCR relative to a cotransfected reporter construct (pCAGGPNS). Splicing of both GAG and CAG constructs in HeLa cells resulted in complete inclusion of exon 3 and skipping of exon 2 (data not shown). Therefore, qPCR measurements of exon 1–3 containing products can be used to determine overall transcript levels (Fig. 1D). Activation of NE inclusion in pcDNA1–4–CAG resulted in 90% decrease in transcript levels, compared with pcDNA1–4–GAG, where the NE is skipped (Fig. 1D). Knock-down of Upf1 had no effect upon the levels of pcDNA1–4–GAG transcript, but caused a stabilization of the pcDNA1–4–CAG transcript to almost 90% that of the original pcDNA1–4–GAG levels. These data confirm that activation of the NE results in NMD of Tpm1 transcripts.
Identification of ESEs and ESSs in the nonsense exon
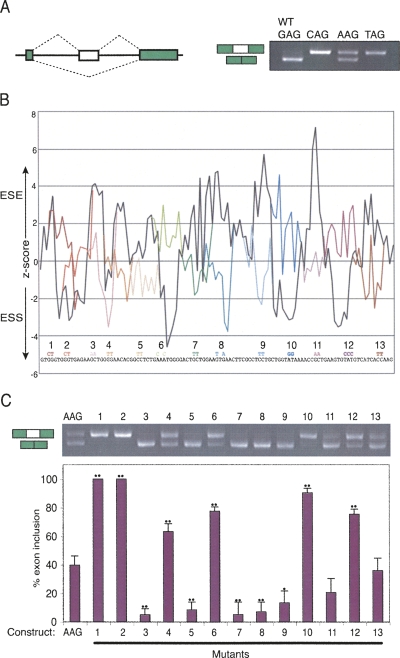
In order to systematically identify splicing regulatory sequences, the NE and flanking introns were cloned into the pCAGGPNS exon trapping vector (Fig. 2A). In this vector, the NE uses the +2 reading frame in which the first PTC is only 28 nt from the 3′ end of the NE, well beneath the 50–55 nt threshold for inducing NMD, and there are no further downstream introns in the construct. When the NE is skipped, the GFP remains in frame and its natural stop codon is in the 3′ end exon. Moreover, the sequence generated by splicing of the NE to the upstream exon (GGU|GUGGGU) has a very poor match to a 5′ splice site, so resplicing via a zero-length exon mechanism is highly unlikely. Therefore, the ratio of products corresponding to inclusion or skipping of the NE should accurately reflect the relative frequencies of the two splicing events, with no complications arising from NMD or apparent exon skipping by “resplicing.” Splicing of the vector with the WT NE (GAG 3′ss) in transfected HeLa cells involved complete exon skipping, as expected (Fig. 2A). Mutation of the 3′ss to the consensus CAG or UAG led to full exon inclusion, while mutation to AAG led to ∼40% exon inclusion. Subsequent mutagenesis to identify regulatory elements was carried out in the context of the AAG mutant construct, as its balanced splicing pattern allows identification of both ESEs and ESSs.
FIGURE 2.
Mapping ESEs and ESSs in the NE. (A) The NE and flanking introns were cloned into a GFP based exon trapping vector in either the WT form (GAG 3′ splice site) or with the 3′ splice site mutated to CAG, AAG, or TAG. The agarose gel on the right shows RT-PCR analysis of HeLa cells transfected with the four constructs. (B) Plot of ESEs and ESSs along the length of the NE predicted using the z-scores of Zhang and Chasin (2004). We found a very similar pattern of predicted elements using either the i-set or p-set of z-scores. For clarity of presentation the average of the two sets is shown here. The sequence of the NE is shown along the base of the plot, along with the base changes in mutants 1–13. Black line, WT sequence. The colored lines indicate the changes in predicted ESEs/ESSs resulting from 13 mutations introduced along the exon. Each mutation affected two or three bases, and was designed to neutralize a predicted ESE/ESS while avoiding creation of an element of opposite polarity. (C) RT-PCR analysis of HeLa cells transfected with the “WT” AAG construct, and mutants 1–13. The histogram below shows percent exon inclusion, indicating average and standard deviation of three biological repeats. Pairwise t-tests comparing each mutant to the AAG construct gave P-values <0.05 (single dots) or <0.01 (double dots).
We used Zhang and Chasin's (2004) octamer data set to predict the locations of potential ESEs and ESSs. Plotting the octamer z-scores along the length of the NE revealed the presence of numerous putative ESE and ESS elements (Fig. 2B, black line). Predicted elements were then point mutated by two base changes to inactivate them (three base changes for mutant 12), as represented by the different colored lines in Fig 2B. As far as possible, mutations were designed to neutralize elements without introducing an element of the predicted opposite polarity.
The 13 mutants and the AAG control construct were transfected into HeLa cells and their splicing analyzed by RT-PCR (Fig. 2C). Strikingly, 12 of the 13 mutations resulted in a substantial difference in splicing compared with the AAG control, and 11 of the 12 changes were consistent with the predictions of ESE or ESS activity. Only mutation 13, predicted to inactivate an ESE, resulted in no change in splicing. Of the seven other potential ESEs, mutations in six of the sites (Fig. 2C, mutants 3,5,7–9,11) resulted in a reduction in NE inclusion, confirming the predictions. Only mutation 4 in a G-rich sequence resulted in an increase in NE inclusion, contrary to the predicted ESE activity. Mutations in all five predicted ESSs (Fig. 2C, mutants 1,2,6,10,12) resulted in increased NE inclusion, consistent with the predictions. The strongest ESSs were in the G-rich region at the 5′ end of the NE; mutations 1 and 2 resulted in complete activation of splicing.
Screening of potential trans-acting factors
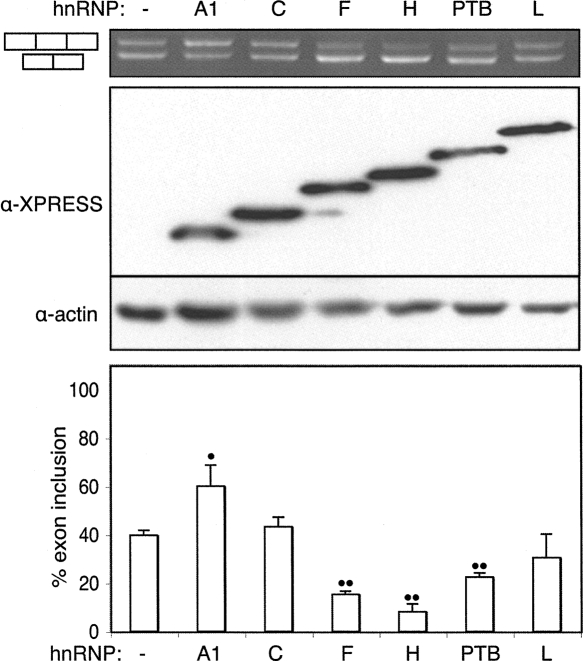
The preceding data indicated that the NE was packed with regulatory elements along its length. In order to identify potential trans-acting factors that might act via the identified elements, the effects of overexpression of a number of known splicing factors on the CAGG–AAG construct were examined. Expression vectors for the hnRNPs A1, C, F, H, PTB (I), and L were cotransfected with the CAGG–AAG reporter plasmid into HeLa cells. All six factors were expressed to comparable levels, and four of the six were shown to have substantial effects on NE splicing (Fig. 3). hnRNP F, hnRNP H, and PTB acted as splicing repressors. hnRNP H had the greatest effect, reducing NE inclusion from 40% to 10%. Overexpression of hnRNP A1, on the other hand, elevated exon inclusion to 60%, despite the fact that it most commonly acts as a splicing repressor. hnRNP C and hnRNP L had no obvious effect (the slight apparent reduction in NE splicing with hnRNP-L was not statistically significant). Transfection with a panel of 14 additional splicing regulators (SRp20, SF2/ASF, SRp30c, 9G8, SC35, SRp40, SRp55, SRp75, ETR3, CELF4, CUGBP, Sam68, hnRNPK, and SLM1) identified SRp30c and CELF4 as activators and SRp20 as a repressor of the NE (data not shown).
FIGURE 3.
Overexpressed hnRNPs affect nonsense exon splicing. Xpress-tagged hnRNP expression plasmids were cotransfected with the AAG splicing reporter. Top panel, agarose gel of RT-PCR products. Middle panel, Western blot using antibodies against the Xpress tag and actin. Lower panel, histogram indicating levels of exon inclusion from three biological repeats. Error bars indicate standard deviation. Single dots indicate P < 0.05, double dots P < 0.01 in pairwise t-test comparing control to overexpression lanes. HnRNPs F, H, and PTB repressed the nonsense exon while hnRNP A1 promoted inclusion.
hnRNP H and F repress the nonsense exon
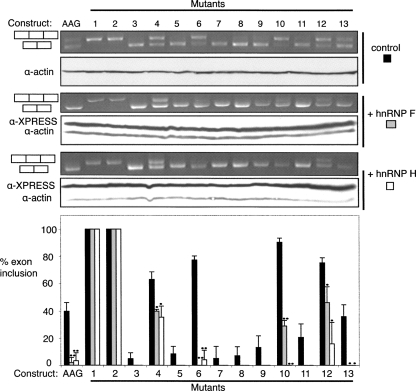
We next tested whether the effects of the candidate regulatory proteins could be associated with any of the ESEs and ESSs identified in Figure 2. In each case the regulatory protein was overexpressed with the WT construct and the 13 mutants, with a view to identifying mutations that reduce the sensitivity of the NE to the overexpressed factor. Splicing repressors were expected to decrease the amount of NE inclusion, except in the case of ESS mutations that impaired their binding. Overexpression of hnRNP F and hnRNP H led to a significant decrease in NE inclusion with mutations in ESSs 6, 10, and 12 (Fig. 4). In contrast, mutants in ESSs 1 and 2 were completely unresponsive to hnRNP F and H. This suggests that hnRNP F and H might interact with the G-rich sequences at the 5′ end of the NE, which are similar to previously identified binding sites (Matunis et al. 1994; Min et al. 1997; Chen et al. 1999; Chou et al. 1999; Fogel and McNally 2000; Caputi and Zahler 2002; Garneau et al. 2005; Han et al. 2005; Marcucci et al. 2007). Mutation of ESS 4, which is also a G-rich element, significantly diminished the extent of repression by hnRNP H and F, especially when compared with the ESS 6 mutant (Fig. 4). Of the other regulators identified by overexpression, none could be similarly associated with a particular ESE or ESS, with the exception of hnRNP A1 and CELF4, which appeared to act via ESSs 1 and 2 (data not shown). Since they are both activators, the simplest interpretation is that hnRNP A1 and CELF4 can compete with binding of repressors to ESSs 1 and 2.
FIGURE 4.
hnRNP H and F repression is mediated by ESSs 1 and 2. The “WT” AAG construct and the 13 derived ESE/ESS mutants were transfected into HeLa cells alone (top panels) or cotransfected with hnRNP F (middle panels) or hnRNP H (lower panels) expression vectors. In each set of panels, RT-PCR agarose gels are on top, and Western blots are below. The histogram below summarizes data from three biological replicates. Black bars, control, gray bars, hnRNP F overexpression, white bars, hnRNP H overexpression. Single dots indicate P < 0.05, double dots P < 0.01 in pairwise t-test comparing control to overexpression lanes. The repressive effects of hnRNP F and H were completely abrogated by mutations 1 and 2, which affected a G-rich ESS.
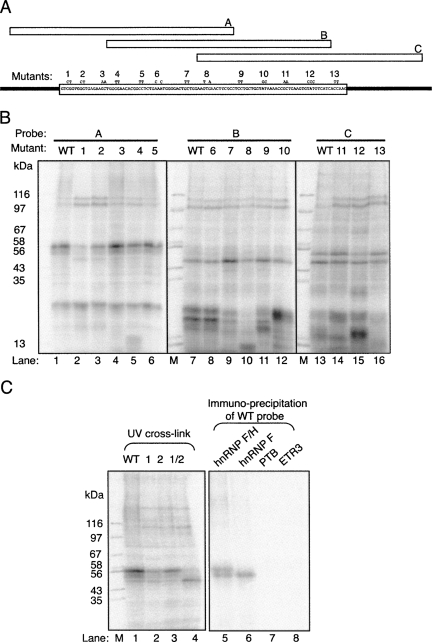
The preceding experiments allowed the activity of overexpressed proteins to be associated with a particular cis element, but did not address whether the proteins actually bound to RNA. We next used UV-crosslinking to identify proteins that bind to the NE, and to investigate whether any observed binding is affected by the ESE and ESS mutations (Fig. 5A,B). Radiolabeled 80-nt fragments A, B, and C, covering the 5′, central, and 3′ sections of the NE, respectively, were used as probes to analyze protein binding in HeLa nuclear extracts. The effects of the 13 mutations were tested within the context of the probe in which the mutation is located centrally. The three WT probes showed different patterns of crosslinking, as expected (Fig. 5B, lanes 1,7,13), and some crosslinks were affected by specific mutations (e.g., in ESSs 1,2, lanes 1–3; ESEs 7–9, lanes 7,9–11). Of particular interest, the mutations in ESSs 1 and 2, which had decreased responsiveness to hnRNP F and H (Fig. 4), caused a marked reduction in crosslinking of a pair of bands at ∼55 kDa (Fig. 5B, lanes 1–13). Combining the ESSs 1 and 2 mutations into a single RNA led to complete loss of crosslinking of the 55 kDa protein (Fig. 5C, lane 4). To test the identity of the crosslinked bands, which were of appropriate size to be hnRNP F (53 kDa) and H (56 kDa), we immunoprecipitated the crosslinked proteins (Fig. 5C). Both bands were immunoprecipitated with dual specificity anti-F/H antiserum (Fig. 5C, lane 5), while the lower band alone was precipitated with mono-specific anti-F antibodies (Fig. 5C, lane 6). Antibodies against PTB and ETR3 acted as negative controls (Fig. 5C, lanes 7,8). These data therefore indicate that hnRNP H and F bind directly to the G-rich ESSs 1 and 2.
FIGURE 5.
hnRNP H and F crosslink to the NE ESS. (A) Schematic of RNA crosslinking probes A, B, and C, indicating their positions in relationship to the NE. The positions and sequences of mutations 1–13 are indicated. (B) UV crosslinking of HeLa nuclear extract proteins to probes A, B, and C in WT or mutant versions. Protein size markers are in lanes marked “M” and sizes (kDa) are indicated to the left. (C) Left panel: UV crosslinking of probe A in WT form, and with mutations 1 and 2 individually and combined (1/2). Right panel: immunoprecipitation of proteins crosslinked to WT probe A using antibodies specific for hnRNP H + F, F, PTB, and ETR3. Size markers are in the left-hand lane, sizes in kDa are indicated to the left.
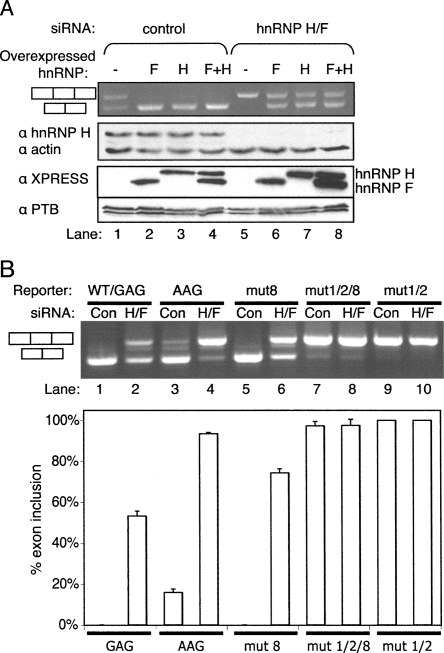
To test whether normal cellular levels of hnRNP F and H are sufficient to repress the NE we used an siRNA that targets hnRNPs H and F (Garneau et al. 2005). Substantial knockdown was achieved, as indicated by Western blots probed for hnRNP H1 (Fig. 6A, middle panel, cf. lanes 1 and 5). Knockdown of hnRNPs H and F resulted in substantially increased levels of NE inclusion in the AAG construct from 40% to 100% (Fig. 6A, upper panel, lanes 1,5). We next tested the ability of overexpressed hnRNPs H and F to reverse the effects of knockdown. For this experiment, silent mutations were introduced into the hnRNP H and F expression constructs so that they had four mismatches with the siRNA target and could thereby be readily overexpressed, as demonstrated by Western blot to detect the XPRESS epitope tag (Fig. 6A, anti-XPRESS panel). Note that the overexpressed hnRNP F and H2 are not detected by the anti-hnRNP H1 antibodies. After treatment with control siRNA, overexpression of hnRNP H and F individually and in combination, caused total repression of NE inclusion (Fig. 6A, lanes 1–4). After treatment with siRNA targeting hnRNP H and F, NE inclusion was increased to 100% but could be restored to the original levels of ∼40% exon inclusion by overexpression of hnRNP H or F (Fig. 6A, lanes 5–8). Adding back hnRNP H and F together was no more effective than addition of the individual factors. This implies that either hnRNP H or F are alone sufficient to repress the NE. We next tested the extent to which hnRNP H and F repress the NE in its WT form, with the suboptimal GAG 3′ splice site. Splicing of the WT CAGG–GAG construct was analyzed after treatment with a control or hnRNP H/F specific siRNA. Strikingly, splicing of the NE was substantially activated with levels of exon inclusion rising from undetectable to >50% (Fig. 6B, lanes 1,2). Finally, in order to test whether the silencing effects of hnRNP H and F were mediated by the G-rich ESS at the 5′ end of the NE, we combined some of the previous mutations. Mutations 1 and 2 combined, which completely impaired hnRNP H crosslinking (Fig. 4C) led to complete exon inclusion so it was not possible to test whether the mutations led to loss of responsiveness to hnRNP H/F (Fig. 6B, lanes 9,10). However, by combining mutations 1/2 with the ESE mutation 8, a low level (∼5%) of exon skipping was observed (Fig. 6B, lane 7). Knockdown of hnRNP H and F had no effect upon this residual level of exon skipping (Fig. 6B, lanes 7,8), whereas mutant 8 was still fully responsive to knockdown (Fig. 6B, lanes 5,6). These data emphasize the crucial role of hnRNPs H and F in silencing the Tpm1 NE via the G-rich ESS at its 5′ end.
FIGURE 6.
hnRNP H and F silence the nonsense exon. (A) Effects of control or hnRNP H/F targeting siRNAs and of hnRNP H and F overexpression, upon splicing of the AAG nonsense exon reporter. Four synonymous mutations were introduced into the hnRNP H and F expression constructs in the region targeted by the siRNA, thereby allowing overexpression while the endogenous hnRNP H and F are knocked down, as demonstrated by Western blot to detect the XPRESS epitope tag (anti-XPRESS panel). Upper panel, agarose gel of RT-PCR products. Lower panels, Western blots using antibodies specific to hnRNPH, actin, Xpress tag, and PTB. Knockdown of hnRNP H/F leads to full inclusion of the NE (lanes 1,5), while overexpression of siRNA resistant hnRNP H or F individually or combined can compensate for knockdown (lanes 5–8). (B) Effects of control or hnRNP H/F targeting siRNAs upon splicing of NE splicing reporters. WT (GAG, lanes 1,2), AAG (lanes 3,4), mut 8 (lanes 5,6), mut 1/2/8 (lanes 7,8), and mut 1/2 (lanes 9,10). The histogram below shows the percentage exon inclusion (mean ± SD, n = 3).
DISCUSSION
The data presented here show that the Tpm1 NE resembles a genuine exon in its complement of splicing regulatory elements. Mutations in 12 locations along the length of the NE resulted in significant changes in splicing. The sheer number of splicing regulatory sequences in the NE is perhaps initially surprising. Assuming that splicing regulatory sequences are 6–8 nt in length (Fairbrother et al. 2002), it would appear that nearly the whole NE is occupied by regulatory elements. However, similar findings have been made with exons 9 and 12 of the human CFTR gene, where numerous synonymous wobble mutations were found to influence splicing (Pagani et al. 2003; Pagani et al. 2005). Indeed, there is growing appreciation that the prevalence of such elements explains the high proportion of human diseases that at molecular level are caused by mis-splicing (Cartegni et al. 2002). The high number of ESEs, coupled with the ease with which the NE can be activated by simple mutations (Fig. 2), contrasts with the properties of a pseudoexon in the HPRT gene, which could only be activated by several combined mutations (Sun and Chasin 2000), and supports our contention that the Tpm1 NE is an authentic exon.
Of the 13 elements predicted using the Zhang and Chasin (2004) z-scores, 5/5 silencers and 6/8 enhancers were confirmed by mutation. Of the two incorrectly predicted ESEs, mutation in position 13 had no effect, while mutation 4 instead identified an ESS. An 82% validation rate had previously been reported for a set of 22 predicted ESEs (Zhang et al. 2005), but to our knowledge, predictions of ESSs have not been similarly tested. We used the PESX z-scores as a tool to guide our mutagenesis strategy, rather than setting out to rigorously validate their utility. Nevertheless, our results show that both ESEs and ESSs can be successfully predicted using this approach, even though other reports have demonstrated that the same sequence elements can have activities that depend upon their location (Goren et al. 2006; Wang et al. 2006), and the effects of local secondary structure (Muro et al. 1999; Hiller et al. 2007).
Secondary structure has been implicated as playing a major role in preventing the splicing of disease associated pseudo-exons in the human CFTR and ATM genes (Buratti et al. 2007). We noted that the Tpm1 NE contains a conserved predicted secondary structure, which would potentially “display” the ESE identified by mutation 8 in the loop of a hairpin. However, mutagenesis of the predicted hairpin was not consistent with a role for the structure in ESE activity (data not shown).
Our initial screen for proteins associated with particular regulatory elements involved first overexpression of candidate factors with the panel of ESE/ESS mutant reporters (Figs. 3, 4) and second UV crosslinking of RNA probes to identify RNA binding proteins that were sensitive to the mutations (Fig. 5). While a number of potentially interesting factors were identified, all of the approaches converged upon hnRNP H and F, which were identified to bind to, and act via, the G-rich ESSs 1 and 2. Confirmation of their repressive role was obtained by RNAi (Fig. 6). Identification of these repressor proteins also helped to explain the single incorrect ESE prediction (Fig. 2). Like ESSs 1 and 2, ESS 4 is also a G-rich element. The lack of impact of mutation 4 upon UV crosslinking of hnRNP H and F (Fig. 5B) is consistent with the milder effect of its mutation compared with ESSs 1 and 2 (Fig. 2). The sequences of ESSs 1, 2, and 4 resemble previously identified hnRNP H and hnRNP F binding sites, which contain GGG motifs (Caputi and Zahler 2001). HnRNP H and F can bind cooperatively to G-rich sequences (Alkan et al. 2006) as homo- or heterodimers (Chou et al. 1999; Kim et al. 2005). The complete activation of the NE upon mutation of either ESS 1 or 2 would be consistent with cooperative binding of a hnRNP H and/or F to both sites as a homodimer or heterodimer. The weaker G-rich ESS 4 may play a role in cooperative stabilization of hnRNP H or F binding to ESSs 1 and 2. While heterodimer formation is a possibility, the equal efficacy of either hnRNP or F alone to reverse the effects of knockdown (Fig. 6) shows that the proteins are functionally redundant in this instance.
HnRNP H and F have been shown to play a role in the activation of splicing by binding to G-rich splicing enhancers in exons (Caputi and Zahler 2002), and introns (Markovtsov et al. 2000; Garneau et al. 2005), and to enhance splicing by binding to sites at the ends of long introns (Martinez-Contreras et al. 2006). They can also repress splicing, for example, in cases where their binding site overlaps the consensus 5′ss (Buratti et al. 2004). Binding of hnRNPs H and F to ESS elements can compete with binding of SR activator proteins, such as 9G8 on Tpm1 exon 2 (Crawford and Patton 2006), and SF2/ASF on FGFR2 exon IIIc (Mauger et al. 2008). However, their first documented role in splicing was as repressors binding to a G-rich ESS in rat Tpm2 (β-tropomyosin) exon 7 (Chen et al. 1999). Similar to the NE, the G-rich ESS in Tpm2 exon 7 sits adjacent to a strong downstream ESE. HnRNP H and hnRNP F might elicit their repressive effects on these exons by interfering directly with binding of the core splicing machinery to the 3′ splice site or by disrupting binding of activators to the adjacent ESE. A further similarity between the NE and Tpm2 exon 7 is that both exons are repressed not only by hnRNP H/F, but also by PTB. Various lines of evidence support the PTB regulation of Tpm2 exon 7 (Mulligan et al. 1992; Sauliere et al. 2006; Spellman et al. 2007). Regulation of the NE was demonstrated by overexpression of PTB (Fig. 3); and the polypyrimidine tract that supports splicing of the NE is also a negative regulatory element that represses Tpm1 exon 3 by binding PTB (Perez et al. 1997; Gooding et al. 1998). Tpm2 exon 7 is selected in striated muscle cells, where it is derepressed. It is an intriguing possibility, supported in part by the high level of NE splicing in the heart of transgenic mice (Ellis et al. 2004), that the NE and Tpm2 exon 7 may be under a common program of muscle specific splicing mediated by hnRNP H/F and PTB repression in nonmuscle cells. The striking effects upon NE splicing of both overexpression (Fig. 3) or siRNA-mediated depletion of hnRNP H and F (Fig. 6) support the notion that these factors might be the primary agents of NE regulation. hnRNP F is broadly expressed in a large number of tissues in both the nuclei and cytoplasm, while hnRNP H1 and H2 expression is much higher in the nuclei than cytoplasm but more restrictive across tissue types, with most epidermal and epithelial cells showing high levels of hnRNP H1/H2 expression (Honore et al. 1999). Interestingly, cardiomyocytes show low levels of both hnRNP H1/H2 and hnRNP F, suggesting that in these cells the NE might be activated, as suggested in a transgenic mouse study (Ellis et al. 2004).
A number of other candidate factors were shown to influence NE splicing upon overexpression (Fig. 3, and data not shown), including SRp30c, hnRNP A1, and CELF4 as activators and SRp20 as a repressor. In addition, various proteins were found to crosslink to NE sequences, and some of them were sensitive to mutations that affected splicing (Fig. 5). SRp30c appeared to act via the central ESEs 7–9, as indicated by the lack of response of the mutants to SRp30c overexpression (data not shown). However, we have not been able to determine whether any of the crosslinked proteins (Fig. 5B) correspond to SRp30c. CELF4 and hnRNP A1 both activated the NE. By combining ESSs 1 and 2 mutations with ESE mutations we were able to show that both CELF4 and hnRNP A1 act via ESSs 1 and 2 (data not shown). The likeliest explanation for their activity is that when overexpressed they are able to compete with hnRNP H/F for binding. This explanation is consistent with the known antagonistic activities of hnRNP A1 and hnRNP H upon a class of neuronal specific exons (Han et al. 2005), and with the known binding preferences of the two proteins. If correct, it also indicates that protein binding at ESSs 1 and 2 is not alone sufficient for splicing repression, but that some specific feature of hnRNP H/F is required. Following the example of other splicing repressors, artificial tethering experiments might be able to identify discrete repressor domains of hnRNP H or F (Del Gatto-Konczak et al. 1999; Dauksaite and Akusjarvi 2002; Gromak et al. 2003; Expert-Bezancon et al. 2004; Rideau et al. 2006; Robinson and Smith 2006).
The best known NEs to date are in the genes for SR proteins (Lareau et al. 2007; Ni et al. 2007) where they play a possible role in direct autoregulation (Sureau et al. 2001). Inclusion of the Tpm1 NE would also lead to down-regulation of gene expression. However, since Tpm1 is a cytoskeletal protein, such regulation would necessarily be via a more complex signaling pathway. There are well-documented examples of signaling from the actin cytoskeleton to nuclear transcription factors (e.g., Miralles et al. 2003). It is an intriguing possibility that signaling from the cytoskeleton could influence Tpm1 NE splicing in a complex control circuit.
MATERIALS AND METHODS
Constructs
pcDNA1–4 constructs contains exons 1 to 4 of the Tpm1 gene, but with a large deletion in intron 3, which is >12 kb in the native gene, cloned into pcDNA-4C (Invitrogen). The pCAGGPNS vector was derived from the EGFP splicing reporter used by Ellis et al. (2004), by introducing a Pvu II, Not I, Sph I multiple cloning site into the intron between the two EGFP exons. pCAGG constructs contain the NE flanked by its native introns cloned into the Not I site of the pCAGGPNS vector. Mutations were created by PCR mediated site-directed mutagenesis using Stratagene native Pfu polymerase and verified by DNA sequencing. pCMV/PTB1: construct for in vivo expression of Xpress-tagged human PTB1 (Wollerton et al. 2001). pHNRNPH2 and pHNRNPF: constructs for in vivo expression of Xpress-tagged human hnRNPs H2 and F (Han et al. 2005). pC-XP, pA1-XP, and pL-XP: in vivo XPress-tagged expression constructs containing the ORFs of human hnRNPs C, A1, and L, respectively, cloned between the Bam HI and Eco RI sites of pcDNA-4C (Invitrogen).
Antibodies
Antibodies were obtained from the following sources: anti-PTB rabbit polyclonal antibody (Hunt and Jackson 1999); anti-ETR-3 rabbit polyclonal (Cocalico Biologicals); anti-actin rabbit polyclonal antibody (Sigma); anti-hnRNP H rabbit polyclonal supplied by F. Baralle; anti-hnRNP H/H′/F mouse monoclonal antibody (R7264), and anti-hnRNP F mouse monoclonal antibody (R7263) (Han et al. 2005); anti-XPRESS mouse monoclonal antibody (Invitrogen); anti-Upf1 rabbit polyclonal antibody (Mendell et al. 2002); donkey antimouse, antirabbit, and antirat IgG horseradish peroxidase conjugate (Jackson ImmunoReseach Laboratories).
Cell culture and transfections
HeLa cells were grown under standard conditions in Dulbecco modified Eagle-Glutamax medium (Gibco-BRL) supplemented with 10% (v/v) fetal calf serum (Sigma). Transient plasmid transfections were carried out as described previously (Wollerton et al. 2001). For hnRNP H/F and Upf1 knockdown 1 × 105 HeLa cells were seeded to a 3.5-cm well in 2 mL DMEM-Glutamax. After 24 h, 12 μL LipofectAMINE 2000 were mixed with 48 μL Opti-MEM I reduced serum medium, incubated at room temperature for 7 min and added to 24 μL (480 pmol) of siRNA duplex (20 μM), which had been mixed with 176 μL Opti-MEM I. The mixture was incubated at room temperature for 25 min, mixed with 140 μL Opti-MEM I and then added to the cells. After 48 h, effector and reporter constructs were transfected as described above. Cells were harvested for RNA and protein after 72 h. hnRNP H/F siRNA (160 pmol) (5′-GGAAGAAAUUGUUCAGUUC-3′) (Garneau et al. 2005) or Upf1 siRNA (120 pmol) (5′-GAUGCAGUUCCGCUCCAUU-3′) (Mendell et al. 2002) was used per well. Total siRNA per well was made to 480 pmol using the C2 control duplex (5′-GGUCCGGCUCCCCCAAAUG-3′) (Wagner and Garcia-Blanco 2002). siRNAs were obtained from Dharmacon in annealed form.
Analysis of cellular RNA
RNA was harvested using RNeasy columns (QIAGEN) according to the manufacturer's instructions and analyzed by reverse transcription-PCR (RT-PCR) as described (Grellscheid and Smith 2006). Transient expression of pcDNA1–4 reporter constructs was detected using reverse-transcription primer pcDNA4rev (5′-GTCGAGGCTGATCAGCGGG-3′), forward primer q1f in exon 1 (5′-ATGCTGAAGCTCGACAAAGA-3′), and reverse primer q3r in exon 3 (5′-CAGTGACACCAGCTCATCT-3′). pCAGG reporter expression was detected using reverse-transcription primer CGRT (5′-TAGTTGTACTCCAGCTT-3′), forward primer CG5′ (5′-GGCAAGAATTCGCCACCA-3′), and reverse primer CG3′ (5′-GGGTGTCGCCCTCGAACTT-3′). Digital images of agarose gels were obtained using a UV transilluminator. Band intensities were quantitated using UVP Labworks (version 4.0.0.8). Absolute Integrated ODs were determined using the “joining valleys background correlation” with the “maximum baseline slope” set to 20%. The linear response range was determined using 1 kb DNA markers over a range of concentrations.
qPCR reactions (25 μL) were assembled from 12.5 μL 2× SYBR green master mix (Applied Biosystems), 1.25 μL 10 pmol/μL forward primer, 1.25 μL 10 pmol/μL reverse primer, and 8 μL water, in a Corbett Rotor Gene 6000 series detector. Cycling conditions were 95°C for 10 min followed by 40 cycles of 95°C 10 sec, 55°C 15 sec, 72°C 20 sec. Data acquisitions were performed with Rotor Gene 6000 Series software (version 1.7). Relative quantification analysis was performed using a dynamic amplification efficiency determination for each amplification run as provided in the Comparative Quantification method with the Rotor-Gene 6000 Series software (version 1.7). Melting curve analysis (Rotor Gene 6000 version 1.7) immediately after PCR completion by heating at 95°C for 5 sec, followed by cooling to 65°C for 5 sec and continuous heating to 99°C at 0.5°C/sec under permanent fluorescence detection, was used to verify that a single product had been formed. qPCR products were also verified by gel purification and sequencing.
UV crosslinking and immunoprecipitation assays
High specific activity 32P labeled RNA fragments (20 fmol/μL) were prepared as previously described (Matlin et al. 2007), using the following pair of PCR primers to prepare templates.
Probe A:
5′-CGTAATACGACTCACTATAGGCCTCCTCCCCTTTGGTTGG-3′, and
5′-GGCGAAGTTCACTTCCAGCA-3′.
Probe B:
5′-CGTAATACGACTCACTATAGGCTGGGGAACACGGCCTCTG3′, and
5′TGATGACATACACTTCAGCG-3′.
Probe C:
5′-CGTAATACGACTCACTATAGGAAGTGAACTTCGCCTCCTG3′, and
5′-AACACTAGGGATTCTGTTTTG-3′.
32P-labeled RNA (40 fmol) was incubated at 30°C for 30 min in a 10 μL crosslinking reaction (2 μL 5× binding buffer, 4 μL HeLa nuclear extract, 2 μL buffer E, 0.5 μg rRNA nonspecific competitor, 100 mM total KCl). Samples were placed on ice and exposed twice to 960 mJ UV in a XL-1500 UV Crosslinker (Spectronics Corporation). Following addition of 4 μL RNase A and T1 mix and incubation at 37°C for 30 min, 4 μL 4× SDS loading buffer was added. Samples were heated at 90°C for 5 min, prior to separation by 12.5% SDS-PAGE. For immunoprecipitation of proteins crosslinked to radiolabeled RNA, five crosslinked samples for each RNA were pooled in a 1.5 mL centrifuge tube immediately after RNase digestion. NETS buffer (450 μL) and 5 μL antibody were then added to the tube, which was left gently rotating for 60 min at 4°C. Ten microliters of protein A sepharose slurry was then added to the tube before a further 60 min rotating at 4°C. Beads were washed three times with 500 μL NETS buffer, before heating for 5 min at 95°C in 10 μL SDS loading buffer.
ACKNOWLEDGMENTS
We thank F. Baralle, C. Milcarek, and J. Mendell for antibodies. This work was supported by Wellcome Trust Program Grant 077877 and, in part by the EC Grant EURASNET-LSHG-CT-2005-518238. J.L.C. was supported by a studentship from the Medical Research Council.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1225209.
REFERENCES
- Alkan S.A., Martincic K., Milcarek C. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem. J. 2006;393:361–371. doi: 10.1042/BJ20050538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Boutz P.L., Stoilov P., Li Q., Lin C.-H., Chawla G., Ostrow K., Shiue L., Ares M., Jr, Black D.L. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Dev. 2007;21:1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., Baralle M., De Conti L., Baralle D., Romano M., Ayala Y.M., Baralle F.E. hnRNP H binding at the 5′ splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHβ genes. Nucleic Acids Res. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E., Dhir A., Lewandowska M.A., Baralle F.E. RNA structure is a key regulatory element in pathological ATM and CFTR pseudoexon inclusion events. Nucleic Acids Res. 2007;35:4369–4383. doi: 10.1093/nar/gkm447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette J.M., Miyamoto-Sato E., Schaub M.A., Conklin J., Lopez A.J. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputi M., Zahler A.M. Determination of the RNA binding specificity of the heterogeneous nuclear ribonucleoprotein (hnRNP) H/H′/F/2H9 family. J. Biol. Chem. 2001;276:43850–43859. doi: 10.1074/jbc.M102861200. [DOI] [PubMed] [Google Scholar]
- Caputi M., Zahler A.M. SR proteins and hnRNP H regulate the splicing of the HIV-1 tev-specific exon 6D. EMBO J. 2002;21:845–855. doi: 10.1093/emboj/21.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L., Chew S.L., Krainer A.R. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- Chang Y.F., Imam J.S., Wilkinson M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- Chen C.D., Kobayashi R., Helfman D.M. Binding of hnRNP H to an exonic splicing silencer is involved in the regulation of alternative splicing of the rat β-tropomyosin gene. Genes & Dev. 1999;13:593–606. doi: 10.1101/gad.13.5.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou M.Y., Rooke N., Turck C.W., Black D.L. hnRNP H is a component of a splicing enhancer complex that activates a c-src alternative exon in neuronal cells. Mol. Cell. Biol. 1999;19:69–77. doi: 10.1128/mcb.19.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford J.B., Patton J.G. Activation of α-tropomyosin exon 2 is regulated by the SR protein 9G8 and heterogeneous nuclear ribonucleoproteins H and F. Mol. Cell. Biol. 2006;26:8791–8802. doi: 10.1128/MCB.01677-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauksaite V., Akusjarvi G. Human splicing factor ASF/SF2 encodes for a repressor domain required for its inhibitory activity on pre-mRNA splicing. J. Biol. Chem. 2002;277:12579–12586. doi: 10.1074/jbc.M107867200. [DOI] [PubMed] [Google Scholar]
- Del Gatto-Konczak F., Olive M., Gesnel M.C., Breathnach R. hnRNP A1 recruited to an exon in vivo can function as an exon splicing silencer. Mol. Cell. Biol. 1999;19:251–260. doi: 10.1128/mcb.19.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis P.D., Smith C.W., Kemp P. Regulated tissue-specific alternative splicing of enhanced green fluorescent protein transgenes conferred by α-tropomyosin regulatory elements in transgenic mice. J. Biol. Chem. 2004;279:36660–36669. doi: 10.1074/jbc.M405380200. [DOI] [PubMed] [Google Scholar]
- Expert-Bezancon A., Sureau A., Durosay P., Salesse R., Groeneveld H., Lecaer J.P., Marie J. hnRNP A1 and the SR proteins ASF/SF2 and SC35 have antagonistic functions in splicing of β-tropomyosin exon 6B. J. Biol. Chem. 2004;279:38249–38259. doi: 10.1074/jbc.M405377200. [DOI] [PubMed] [Google Scholar]
- Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- Fairbrother W.G., Holste D., Burge C.B., Sharp P.A. Single nucleotide polymorphism-based validation of exonic splicing enhancers. PLoS Biol. 2004a;2:E268. doi: 10.1371/journal.pbio.0020208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairbrother W.G., Yeo G.W., Yeh R., Goldstein P., Mawson M., Sharp P.A., Burge C.B. RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res. 2004b;32:W187–W190. doi: 10.1093/nar/gkh393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel B.L., McNally M.T. A cellular protein, hnRNP H, binds to the negative regulator of splicing element from Rous sarcoma virus. J. Biol. Chem. 2000;275:32371–32378. doi: 10.1074/jbc.M005000200. [DOI] [PubMed] [Google Scholar]
- Garneau D., Revil T., Fisette J.F., Chabot B. Heterogeneous nuclear ribonucleoprotein F/H proteins modulate the alternative splicing of the apoptotic mediator Bcl-x. J. Biol. Chem. 2005;280:22641–22650. doi: 10.1074/jbc.M501070200. [DOI] [PubMed] [Google Scholar]
- Giorgi C., Yeo G.W., Stone M.E., Katz D.B., Burge C., Turrigiano G., Moore M.J. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- Gooding C., Roberts G.C., Smith C.W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Goren A., Ram O., Amit M., Keren H., Lev-Maor G., Vig I., Pupko T., Ast G. Comparative analysis identifies exonic splicing regulatory sequences—The complex definition of enhancers and silencers. Mol. Cell. 2006;22:769–781. doi: 10.1016/j.molcel.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Grellscheid S.N., Smith C.W. An apparent pseudo-exon acts both as an alternative exon that leads to nonsense-mediated decay and as a zero-length exon. Mol. Cell. Biol. 2006;26:2237–2246. doi: 10.1128/MCB.26.6.2237-2246.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromak N., Rideau A., Southby J., Scadden A.D., Gooding C., Huttelmaier S., Singer R.H., Smith C.W. The PTB interacting protein raver1 regulates α-tropomyosin alternative splicing. EMBO J. 2003;22:6356–6364. doi: 10.1093/emboj/cdg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K., Yeo G., An P., Burge C.B., Grabowski P.J. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 2005;3:e158. doi: 10.1371/journal.pbio.00301058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton A.R., Subramaniam V., Lopez A.J. Generation of alternative Ultrabithorax isoforms and stepwise removal of a large intron by resplicing at exon–exon junctions. Mol. Cell. 1998;2:787–796. doi: 10.1016/s1097-2765(00)80293-2. [DOI] [PubMed] [Google Scholar]
- Hiller M., Zhang Z., Backofen R., Stamm S. Pre-mRNA secondary structures influence exon recognition. PLoS Genet. 2007;3:e204. doi: 10.1371/journal.pgen.0030204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honore B., Vorum H., Baandrup U. hnRNPs H, H′, and F behave differently with respect to post-translational cleavage and subcellular localization. FEBS Lett. 1999;456:274–280. doi: 10.1016/s0014-5793(99)00911-4. [DOI] [PubMed] [Google Scholar]
- Hunt S.L., Jackson R.J. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA. 1999;5:344–359. doi: 10.1017/s1355838299981414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishigaki Y., Li X., Serin G., Maquat L.E. Evidence for a pioneer round of mRNA translation: mRNAs subject to nonsense-mediated decay in mammalian cells are bound by CBP80 and CBP20. Cell. 2001;106:607–617. doi: 10.1016/s0092-8674(01)00475-5. [DOI] [PubMed] [Google Scholar]
- Kim D.H., Langlois M.A., Lee K.B., Riggs A.D., Puymirat J., Rossi J.J. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau L.F., Inada M., Green R.E., Wengrod J.C., Brenner S.E. Unproductive splicing of SR genes associated with highly conserved and ultraconserved DNA elements. Nature. 2007;446:926–929. doi: 10.1038/nature05676. [DOI] [PubMed] [Google Scholar]
- Le Hir H., Izaurralde E., Maquat L.E., Moore M.J. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J. 2000;19:6860–6869. doi: 10.1093/emboj/19.24.6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune F., Maquat L.E. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 2005;17:309–315. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Green R.E., Brenner S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makeyev E.V., Zhang J., Carrasco M.A., Maniatis T. The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci R., Baralle F.E., Romano M. Complex splicing control of the human thrombopoietin gene by intronic G runs. Nucleic Acids Res. 2007;35:132–142. doi: 10.1093/nar/gkl965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovtsov V., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Contreras R., Fisette J.F., Nasim F.U., Madden R., Cordeau M., Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biol. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A.J., Clark F., Smith C.W. Understanding alternative splicing: Toward a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Matlin A.J., Southby J., Gooding C., Smith C.W. Repression of α-actinin SM exon splicing by assisted binding of PTB to the polypyrimidine tract. RNA. 2007;13:1214–1223. doi: 10.1261/rna.219607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matunis M.J., Xing J., Dreyfuss G. The hnRNP F protein: Unique primary structure, nucleic acid-binding properties, and subcellular localization. Nucleic Acids Res. 1994;22:1059–1067. doi: 10.1093/nar/22.6.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauger D.M., Lin C., Garcia-Blanco M.A. hnRNP H and hnRNP F complex with Fox2 to silence fibroblast growth factor receptor 2 Exon IIIc. Mol. Cell. Biol. 2008;28:5403–5419. doi: 10.1128/MCB.00739-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy N.J., Smith C.W.J. Alternative splicing that leads to NMD: What is the meaning of nonsense? Trends Biochem. Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Mendell J.T., ap Rhys C.M., Dietz H.C. Separable roles for rent1/hUpf1 in altered splicing and decay of nonsense transcripts. Science. 2002;298:419–422. doi: 10.1126/science.1074428. [DOI] [PubMed] [Google Scholar]
- Min H., Turck C.W., Nikolic J.M., Black D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes & Dev. 1997;11:1023–1036. doi: 10.1101/gad.11.8.1023. [DOI] [PubMed] [Google Scholar]
- Miralles F., Posern G., Zaromytidou A.I., Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Mulligan G.J., Guo W., Wormsley S., Helfman D.M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- Muro A.F., Caputi M., Pariyarath R., Pagani F., Buratti E., Baralle F.E. Regulation of fibronectin EDA exon alternative splicing: Possible role of RNA secondary structure for enhancer display. Mol. Cell. Biol. 1999;19:2657–2671. doi: 10.1128/mcb.19.4.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E., Maquat L.E. A rule for termination-codon position within intron-containing genes: When nonsense affects RNA abundance. Trends Biochem. Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- Ni J.Z., Grate L., Donohue J.P., Preston C., Nobida N., O'Brien G., Shiue L., Clark T.A., Blume J.E., Ares M., Jr Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes & Dev. 2007;21:708–718. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani F., Stuani C., Tzetis M., Kanavakis E., Efthymiadou A., Doudounakis S., Casals T., Baralle F.E. New type of disease causing mutations: The example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- Pagani F., Raponi M., Baralle F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl. Acad. Sci. 2005;102:6368–6372. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q., Saltzman A.L., Kim Y.K., Misquitta C., Shai O., Maquat L.E., Frey B.J., Blencowe B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez I., Lin C.H., McAfee J.G., Patton J.G. Mutation of PTB binding sites causes misregulation of alternative 3′ splice site selection in vivo. RNA. 1997;3:764–778. [PMC free article] [PubMed] [Google Scholar]
- Rideau A.P., Gooding C., Simpson P.J., Monie T.P., Lorenz M., Huttelmaier S., Singer R.H., Matthews S., Curry S., Smith C.W. A peptide motif in Raver1 mediates splicing repression by interaction with the PTB RRM2 domain. Nat. Struct. Mol. Biol. 2006;13:839–848. doi: 10.1038/nsmb1137. [DOI] [PubMed] [Google Scholar]
- Robinson F., Smith C.W. A splicing repressor domain in polypyrimidine tract-binding protein. J. Biol. Chem. 2006;281:800–806. doi: 10.1074/jbc.M510578200. [DOI] [PubMed] [Google Scholar]
- Saltzman A.L., Kim Y.K., Pan Q., Fagnani M.M., Maquat L.E., Blencowe B.J. Regulation of multiple core spliceosomal proteins by alternative splicing-coupled nonsense-mediated mRNA decay. Mol. Cell. Biol. 2008;28:4320–4330. doi: 10.1128/MCB.00361-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauliere J., Sureau A., Expert-Bezancon A., Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the β-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol. Cell. Biol. 2006;26:8755–8769. doi: 10.1128/MCB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R.H., Llorian M., Smith C.W.J. Functional redundancy and cross-regulation between the splicing regulator PTB and its paralogs nPTB and ROD1. Mol. Cell. 2007;27:420–434. doi: 10.1016/j.molcel.2007.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Chasin L.A. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau A., Gattoni R., Dooghe Y., Stevenin J., Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A. RNAi-mediated PTB depletion leads to enhanced exon definition. Mol. Cell. 2002;10:943–949. doi: 10.1016/s1097-2765(02)00645-7. [DOI] [PubMed] [Google Scholar]
- Wang Z., Xiao X., Van Nostrand E., Burge C.B. General and specific functions of exonic splicing silencers in splicing control. Mol. Cell. 2006;23:61–70. doi: 10.1016/j.molcel.2006.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Robinson F., Brown E.C., Jackson R.J., Smith C.W. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Wagner E.J., Garcia-Blanco M.A., Smith C.W. Autoregulation of polypyrimidine tract binding protein by alternative splicing leading to nonsense-mediated decay. Mol. Cell. 2004;13:91–100. doi: 10.1016/s1097-2765(03)00502-1. [DOI] [PubMed] [Google Scholar]
- Zhang X.H., Chasin L.A. Computational definition of sequence motifs governing constitutive exon splicing. Genes & Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.H., Kangsamaksin T., Chao M.S., Banerjee J.K., Chasin L.A. Exon inclusion is dependent on predictable exonic splicing enhancers. Mol. Cell. Biol. 2005;25:7323–7332. doi: 10.1128/MCB.25.16.7323-7332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]