Abstract
The major facilitator superfamily (MFS) represents the largest group of secondary active membrane transporters, and its members transport a diverse range of substrates. Recent work shows that MFS antiporters, and perhaps all members of the MFS, share the same three-dimensional structure, consisting of two domains that surround a substrate-translocation pore. The advent of crystal structures of three MFS antiporters sheds light on their fundamental mechanism; they operate via a single-binding site, alternating-access mechanism, involving a rocker-switch type movement of the two halves of the protein. In the sn-glycerol-3-phosphate transporter (GlpT) from E. coli, the substrate-binding site is formed by several charged residues and a histidine that can be protonated. Salt bridge formation and breakage is involved in the conformational changes of the protein during transport. In this review, we attempt to set forth a set of mechanistic principles that characterize all MFS antiporters.
Keywords: Secondary membrane transporter proteins, rocker-switch mechanism, GlpT, UhpT, OxlT, EmrD
INTRODUCTION
Control of substrate movement across cytoplasmic or internal membranes is vital to the long-term stability of a cell. To achieve this, nature has evolved a diverse system of channel and transporter proteins that effect the translocation of ions and small hydrophilic molecules across these membranes. Bacteria employ three main types of transporter to fulfill this function: (i) the so-called “primary” transporters, such as P-type ATPases and ATP-binding cassette (ABC) transporters, which use the energy released from ATP hydrolysis to drive ions or solutes across the membrane; (ii) the “secondary” transporters, which drive substrate translocation by exploiting the free energy stored in the ion or solute gradients generated by primary transporters; and (iii) “group translocation” systems, that couple translocation of a substrate to its chemical modification, resulting in release of a modified substrate at the opposite side of the membrane.
About 25% of all known membrane transport proteins in prokaryotes belong to the major facilitator superfamily (MFS) (67), the largest and most diverse superfamily of secondary active transporters. The MFS contains 58 distinct families (www.tcdb.org), with about 5000 sequenced members identified to date (www.membranetransport.org) (13, 53, 58, 63, 64, 67), a number that expected to swell as more genomes are sequenced. This superfamily is ubiquitous in all kingdoms of life and in all biological cells, and includes members of direct medical and pharmaceutical significance. Mechanistically, MFS transporters display three distinct kinetic mechanisms: there are (i) uniporters, which transport only one type of substrate and are energized solely by the substrate gradient; (ii) symporters, which translocate two or more substrates in the same direction simultaneously, making use of the electrochemical gradient of one of them as the driving force; and (iii) antiporters, which transport two or more substrates, but in opposite directions across the membrane. Individual members within the MFS show stringent specificity, yet as a group the superfamily accepts an enormous diversity of substrate types (ions, sugars, sugar phosphates, drugs, neurotransmitters, nucleosides, amino acids and peptides, to list a few). As a result, mechanistic studies of the MFS allows one to focus on fundamental questions related to how proteins recognize substrates and transport them across the membrane. Our understanding of these issues is well advanced in some model systems. This is especially true of the Escherichia coli lactose:H+ permease (LacY), an MFS symporter that has been the subject of intense experimental scrutiny for many decades by a number of laboratories (1, 21, 38, 39). By contrast, insight into the molecular details of mechanism is less advanced for antiporters, which are the main topic of this review. Here, we focus on studies of bacterial MFS antiporters, in particular the closely related sn-glycerol-3-phosphate/phosphate (GlpT) and hexosephosphate:phosphate (UhpT) antiporters from E. coli, and the oxalate:formate transporter (OxlT) from Oxalobacter formigenes. We try to provide a snapshot of contemporary understanding of structural and mechanistic aspects of these important and fascinating integral membrane proteins.
AN ABBREVIATED HISTORY
From the time of Hippocrates and Galen, who believed substances were absorbed through orifices in blood vessels in the intestinal wall, hypotheses to account for the translocation of even large substrates across biological membranes invoked processes such as osmosis and simple diffusion (20). But as it stands today, that sector of membrane biology concerned with solute transport more properly owes its identity to theoretical and practical studies dating from the early 1950s. Until that time, those interested in transport of sugars, amino acids, or similar metabolites were constrained by a conceptual framework focused on the flow and distribution of ions (Na, K, Cl) across membranes, with findings evaluated in terms of coefficients of diffusion or permeability, flux ratio measurements, and whether or not a system could be considered at equilibrium (33). This all changed in the early 1950's, when W.F. Widdas offered a new perspective in his study of glucose transport by the sheep placenta (80) and human erythrocytes (81), and when others applied this same view to sugar transport by rat small intestine (19), and phosphate transport across bacterial membranes (54, 55). Widdas' particular contribution was to posit an intermediate - a complex reflecting the direct and stoichiometric combination of substrate and a ‘carrier’, much like the intermediate complexes then being invoked in the context of enzymatic catalysis. Accordingly, to frame the process of transport, he proposed that descriptions of substrate (glucose) transport must recognize terms relevant to ligand binding (which, by the way, legitimized use of biochemistry as a practical tool); he also imagined that the liganded carrier diffused across the membrane, so that a kinetic formulation would include terms describing mobility of the carrier/substrate complex. At the opposite surface, the bound passenger would be released, and the unoccupied carrier would return to the original membrane surface to complete a cycle - one cycle (turnover) leading to net transfer of one molecule of substrate (80). Of course, we no longer believe that protein carriers literally diffuse across the membrane, although this is true of peptide carrier ionophores (e.g., valinomycin). Instead, in the spirit of Occam's razor, we accept the concept of a single-binding site, alternating-access mechanism, and variations thereof, in which the transporter can possess two major alternating conformations, inward-facing (Ci) and outward-facing (Co) (36, 72, 73, 76, 79). In this case, translocation of substrate across the membrane is catalyzed by the interconversion of these two conformations. Indeed, much of the current emphasis in the field is dedicated to developing experimental approaches that describe this conformational change, as noted in this review. Such efforts now appear to validate the two general kinds of models discussed over the years. On the one hand, structural insights provided by study of MFS family members are nicely summarized by the “rocker-switch” mechanism (35), a term meant to suggest that substrate might be fixed in space at its binding site, while alternating-access is provided as protein conformational changes alternately generate pathways of access to either surface (51). As well, one might imagine a “gated-pore” (59), in which substrate moves as through an ion channel, but with gates that alternately open and close at either end. This is a view more suited to structural features revealed by certain non-MFS transporters. The now accelerating pace of structural work should provide enough examples to populate a continuum between these two extremes, and perhaps even define new axes to consider.
STRUCTURAL CONSIDERATIONS
General Structure of MFS Proteins
Until the recent arrival of high-resolution three-dimensional (3D) structures of representatives of the MFS, the field relied on indirect methods, such as phylogeny and sequence analysis, as a guide to structure and function of these proteins (13). Hydropathy analysis of protein sequences and topological studies with gene-fusion constructs predicted that almost all MFS proteins possess a uniform topology of 12 transmembrane a-helices (TMs) connected by hydrophilic loops, with both their N- and C-termini located in the cytoplasm (58, 66, 67). Interestingly, the N-terminal half of the protein (TM1-TM6) displays weak sequence homology to the C-terminal half (TM7-TM12), suggesting that the molecule may have arisen following a gene duplication/fusion event (49). This has implications, now confirmed, regarding an underlying structural symmetry, as noted later. Exceptions to the 12 TM rule do exist - a few MFS families have 14 TMs, one representative has only six and yet another 24 (58). The extra two helices in the 14 TM members probably arose via insertion of the central cytoplasmic loop into the membrane, whereas the 24 TM member is likely a consequence of a gene fusion event (66); one suspects that the lone example with six TMs functions as a homodimer.
MFS proteins typically consist of 400-600 amino acids, and analysis of their primary sequences revealed that within any single family, sequence similarity is highly significant (58). By contrast, at the level of the superfamily, individual MFS members share low sequence identity or similarity, and are united only by a pair of conserved signature sequences, DRXXRR, at equivalent positions in the N- and C-terminal halves of the proteins, in loops that join TM2 to TM3 and TM8 to TM9, respectively (49). Along with the conserved transmembrane topology and the internal sequence homology between the two halves of the proteins, this duplicated signature sequence suggests a common ancestral gene for the MFS.
The importance of these earlier studies to a structural perspective was underlined with the publication of high-resolution 3D crystal structures of E. coli GlpT (35), LacY (2) and EmrD (83), and a lower resolution structure of O. formigenes OxlT (30, 32). This revealed that all these MFS proteins - despite their sequence divergence - indeed share almost the same 3D structures. Essentially all existing biochemical and biophysical data for these and other MFS proteins are in agreement with these structures (45, 47, 56). This leads to the notion of a shared fold that acts as a scaffold for all MFS proteins, irrespective of their particular function as a symporter, uniporter or antiporter.
Structure of MFS Antiporters
Detailed 3D structural information is currently available for three MFS antiporters, all from prokaryotes (30, 35, 83). The structure of GlpT (35), along with that of the related LacY symporter (2, 22), has been determined in the inward facing or Ci conformation. Two additional structures - those of the oxalate exchanger, OxlT, and the multidrug transporter, EmrD - may represent transporters in an occluded state (30, 32, 83), but there is as yet no structure of an MFS transporter in the opposite, outward facing or Co conformation. Nevertheless, the available structures emphasize the general conservation of fold and architecture, consisting of two domains with a pore between them.
Publication of these structures, in combination with previous mutagenesis, biochemical and biophysical studies (6-8, 17, 18), has enabled a dramatic enrichment of our understanding of transporter mechanism. Furthermore, as we summarize later in this review, such structural information has been invaluable for the design of new studies to test mechanistic aspects of these proteins.
Structure of GlpT in the CiConformation
As a member of the organophosphate:phosphate antiporter (OPA) family of the MFS, GlpT functions to couple an outward flow of internal inorganic phosphate (Pi) to the uptake of G3P into the cell (26). G3P is an important intermediate in both glycolysis and phospholipid biosynthesis, and it can act as the sole energy source for bacterial growth (48, 62). In GlpT and other antiporters that utilize Pi-linked exchange, the net reaction is enabled by the outwardly directed inorganic phosphate gradient. In addition to bacterial OPA proteins, homologues of GlpT have been identified in a range of eukaryotes, including plants (12), fruit flies (3), mice (71) and humans (11). The 3.3 Å structure of GlpT showed the molecule to have a silhouette reminiscent of a Mayan temple (35). The periplasmic side of the protein is plateau-like and protrudes only slightly into the external surface. In contrast, several TMs extend beyond the membrane interface at the cytoplasmic side.
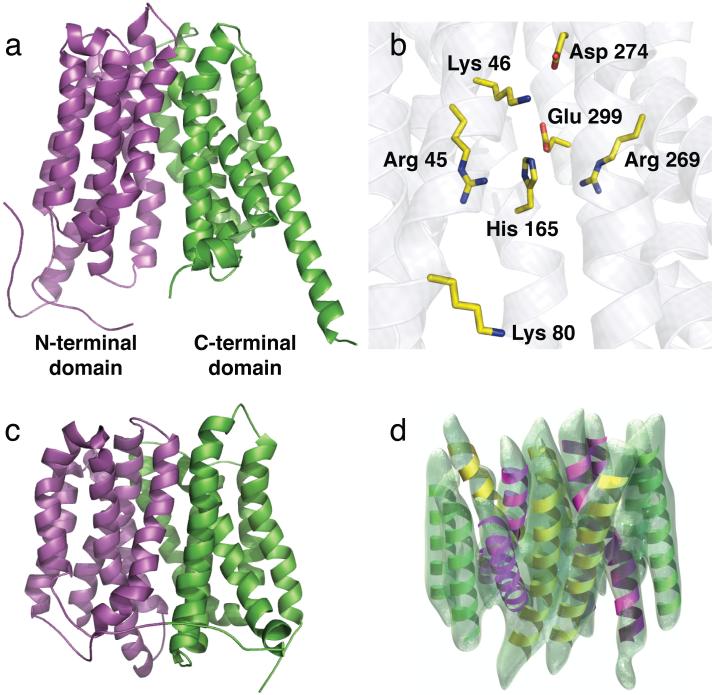
GlpT consists of N- and C-terminal domains related by a pseudo two-fold symmetry (Figure 1a). Each domain is composed of six transmembrane α-helices, consisting of two 3-helix bundles inserted into the membrane in opposite orientations. These TMs are packed loosely in the membrane, presumably to facilitate helix and domain mobility that occurs during the substrate translocation process (28, 29). A long cytoplasmic loop (L6-7) of 45 residues, which is disordered in the crystal structure, links the N- and C-terminal domains (35). While this loop acts as a constraint for movement of the N- and C-terminal domains, it likely permits a relatively large interdomain movement; this has implications for the substrate translocation mechanism discussed later.
Figure 1.
3D structures of GlpT, EmrD and OxlT viewed parallel to the membrane. (a) The 3.3 Å structure of GlpT in the Ci conformation with the transmembrane α-helices of the N-terminal domain colored magenta, and those of the C-terminal domain in green. The substrate-translocation pore is situated between the two domains (35). (b) The GlpT substrate-binding site, depicting the basic residues intimately involved in binding (Lys 80, Arg 45, His 165 and Arg 269) and those that participate in intra- and interhelical salt bridge formation (Lys 46, Asp 274 and Glu 299). (c) The 3.5 Å structure of EmrD in a compact, occluded state (83). (d) The 6.5 Å density map of OxlT, also in an occluded state, with 12 TMs modeled into it (30). In all of the above, the periplasmic side of each transporter is at the top.
Although MFS proteins such as the lactose transporter LacS from Streptococcus thermophilus (75) and the Bacillus subtilis tetracycline transporter TetL (65) are dimeric, GlpT and its close homologue, UhpT, both function as monomers (5, 10). For this reason, the permeation pathway is presumed to reside within the monomer itself, and the 3D GlpT structure immediately suggests that the substrate translocation pathway is the pore saddled by the GlpT N- and C-terminal domains (Figure 1a) (35). When viewed perpendicular to the membrane, the pore is corralled by a rectangular fence composed of eight of the twelve TMs (two on each of the two sides, and two each at the front and back), which act as a scaffolding for the remaining four TMs. The TMs that form the front and back of the fence (TM2 and TM11, and TM5 and TM8, respectively) are curved like bananas, so that each helix pair is shaped like an hourglass, in which the helices make contact with one another at the periplasmic side of the membrane but are separated from each other in the cytosol. The curvature of these helices holds significance for the rocker-switch type of movement that the GlpT N- and C-terminal domains undergo during the transport reaction cycle (35).
In the crystal structure of GlpT, the substrate translocation pore is open only to the cytoplasmic side of the membrane (Figure 1a) (35). The pore is prevented from forming a portal through the membrane by portions of TM1 and TM7, which fills gaps in the barrier with the side chains of nine aromatic residues. This arrangement ensures there is no communication between opposite sides of the membrane when the transporter is in the Ci conformation, an important structural feature consistent with the substrate-translocation mechanism. From the middle of the membrane, the substrate-translocation pore opens out to form a well about 30 Å deep, with a funnel-shaped outer section and a more cylindrical inner section. The base of this inner section is lined by side chains that yield a positive surface electrostatic potential, and this is likely to enhance binding of GlpT to its negatively charged, oxyanionic substrates (10). The interior surface of the funnel section, however, is greasy, and lined with mostly hydrophobic residues that would prevent adhesion of ions and water molecules and thereby direct hydrophilic substrates down to the electropositive binding site. The upper and outermost sides of the pore are formed by the cytoplasmic ends of the N-terminal domain TM4 and TM5 helices, their connecting loop L4-5, and the corresponding C-terminal helices and loop - TM10 and TM11, and L10-11 (35). The cytoplasmic ends of TM1 and TM7 make contacts with these loops (L4-5 and L10-11, respectively), and it has been pointed out that this arrangement appears optimal for transmitting substrate-induced conformational changes throughout the protein (35).
The information provided by the 3D structure of GlpT (Figure 1a) has been invaluable to efforts to unravel general principles of substrate binding and protein architecture in the MFS. But when considered together with the structures of two other MFS antiporters - the O. formigenes oxalate/formate transporter OxlT (30, 32) and E. coli drug/H+ antiporter EmrD (83) - it is also possible to derive important clues as to the conformational change that MFS antiporters undergo during the transport cycle.
Structures of OxlT and EmrD in an Occluded Conformation
In contrast to the overall Mayan temple-shape of GlpT in the Ci conformation, the OxlT and EmrD structures show these proteins in a more compact, occluded conformation. A projection map at 6 Å (27) and a 3D map of OxlT at 6.5 Å (Figure 1d) (30) were determined using electron crystallography of two-dimensional crystals grown in the presence of saturating concentrations of substrate (oxalate), giving us a first glimpse of what MFS proteins look like on the molecular level in terms of helix organization. The overall topology of the transporter revealed 12 TMs arranged around a central pore that is closed to both sides of the membrane (30). As does GlpT, the OxlT protein consists of two 6-helix bundles representing the N- and C-terminal domains. As might be expected if this structure reflects a different conformation than does GlpT, there are subtle differences due to the relative displacement of the OxlT helices compared to those of GlpT. Although the low resolution of the OxlT structure did not permit visualization of bound substrate, the overall structure is much more compact and closed to both sides of the membrane, suggesting it was crystallized in an occluded state (30). This conformation likely corresponds to a key intermediate state in the reaction pathway somewhere between the Ci and Co conformations (40). Using the GlpT structure as a template, the 6.5 Å OxlT density map was used subsequently to model a structure of the latter in the substrate-bound occluded state (32). This, too, suggested that substrate binding initiates a global conformational change imperative for function (31). Similarly, biochemical and biophysical studies have also provided evidence of a more compact form of GlpT upon substrate-binding (10, 35).
The other MFS protein whose 3D structure in an occluded state is EmrD (83). With respect to substrate specificity, EmrD differs significantly from GlpT and OxlT in that it can export a broad range of hydrophobic substrates from the cell (58). Nevertheless, the 3.5 Å crystal structure of EmrD (Figure 1c) showed that, like other MFS proteins, it consists of 12 TMs organized as a pair of six-helix domains resembling those found in OxlT and GlpT. The pore surrounded by these helices is closed to both sides of the membrane. Although the relative orientation of some of the helices (TM3, TM6, TM9 and TM12) is similar to that of their GlpT counterparts, the others deviate substantially from the observed positions of their GlpT equivalents. The EmrD structure also shows two long helical regions (composed of TM4 and TM5, and TM10 and TM11, and the loops that connect them) on the cytoplasmic side of the membrane that are arranged much closer to the substrate-translocation pore, and that extend farther into the cytoplasm than their GlpT counterparts (83). It was suggested that these regions and a run of positively charged residues located at the end of TM4, act as a substrate-specificity filter (83). Akin to the OxlT structure, the EmrD helices form a compact structure of ~50 Å in the plane of the membrane by ~45 Å along the membrane normal. Consistent with its function of catalyzing the translocation of hydrophobic compounds across the bacterial inner membrane, the substrate-translocation pore of EmrD is much more hydrophobic than that of GlpT, with stacked aromatic side chains lining the pore surface, probably to aid in substrate binding (83). That this transporter has been crystallized with its substrate-translocation pore closed to both sides of the membrane makes it likely that it has been captured in an occluded state (83). It is surprising, then, that no substrate is visible in this structure, since in the absence of substrate an occluded state is a high energy, unstable state that would, in principle be refractory to crystallization. The absence of substrate, therefore, is likely a consequence of insufficient resolution.
In summary, we now have access to three 3D structures of MFS antiporters, one in the substrate-free Ci conformation (35), and the others in a substrate-bound, occluded state (30, 83).
SUBSTRATE SPECIFICITY OF MFS ANTIPORTERS
One intriguing aspect of MFS transporters as a whole is their ability to differentiate between vast selections of often very similar substrates. Indeed, there is probably an individual MFS transporter for each small or medium-sized hydrophilic molecule of biological relevance in the cell. The simplicity of the MFS fold design, that of two symmetrical domains saddling a substrate-specific pathway, has been used repeatedly by nature, so clearly it is the presence of only a few amino acid residues at the substrate-binding site that determines the specificity of each transporter for its cognate substrate.
Among MFS antiporters, substrate specificity determinants have been well studied in UhpT (23-25). An intrahelical ion pair in UhpT formed between an asparagine (D388) and lysine (K391) of TM11 was found to be essential for normal UhpT function (23). Because TM11 lines the substrate translocation pore in UhpT, it was suggested that residues that compose sections of it could function as determinants of and regulate substrate specificity of the transporter (24). Furthermore, an uncompensated cationic charge at position 388 or 391 in UhpT resulted in gain-of-function mutants that preferred divalent phosphoenolpyruvate as a substrate over the monovalent hexosephosphates normally transported by UhpT (24). In contrast, an uncompensated anionic charge at position 388 increased the preference of UhpT for monovalent hexosephosphates over divalent sugar species (23). A related antiporter from Salmonella typhimurium, PgtP, which transports phosphoenolpyruvate, lacks the equivalent D388-K391 salt bridge but does possess an uncompensated R391, indicating that residues at positions 388 and 391 in these transporters act as determinants for substrate selectivity (23).
More recent work has used structural, computational and biochemical analyses to describe a more detailed mechanism for substrate binding by GlpT (43). GlpT, like other MFS transporters, operates via a single-binding site, alternating-access mechanism (76) involving a rocker-switch type movement of the protein (35, 44). As the published structure of GlpT is of the molecule in the absence of substrate, in the inward-facing or Ci conformation, the nature of the substrate-binding site could only be inferred (Figure 1b) (35). Nevertheless, strong arguments attest to the validity of this inference. Two conserved, positively charged residues, arginine 45 (R45) from TM1 and arginine 269 (R269) from TM7, are located at the inner end of the substrate-translocation pore visualized in the GlpT structure, and these basic residues are strategically placed to interact with substrate as central components of the binding site (35). In the crystal structure, the shortest distance between the R45 and R269 side chains is 9.9 Å. For both side chains to form a hydrogen bond of 2.9 Å in length with the negatively charged oxygen atoms of the Pi or G3P substrate, they must move 1.4 Å closer to each other. This could occur if substrate binding pulls TM1 and TM7 closer together (35). Molecular dynamics (MD) simulations support the notion that R45 and R269 directly bind to substrates (43). As expected, mutation of either arginine into a lysine eliminates transport activity of GlpT reconstituted into proteoliposomes. Both these residues are conserved between GlpT and its close homologue from E. coli, UhpT, and also in G6PT, the human microsomal glucose-6-phosphate transporter (4, 17). Importantly, the two equivalent arginine residues (R46 and R275) in UhpT have also been identified as substrate-binding elements, since only these two among all UhpT arginines are essential (17). This is entirely consistent with the binding chemistry suggested by the GlpT crystal structure. Similarly, in human G6PT, mutation of R28 (equivalent to GlpT R45) to cysteine or histidine abolishes transport activity; indeed, such mutations cause glycogen storage disease type Ib (4, 14). Notably, paired basic residues are often essential for function in other MFS anion transporters such as the nitrate transporter, NrtA, from Aspergillus nidulans (74), the putative E. coli nitrate:nitrite antiporter, NarU (37), and OxlT (78).
Other residues are also involved directly in substrate binding in OPA family proteins. Studies on UhpT have implicated a lysine (K82) and histidine residue (H168) as important to substrate binding (23, 25), and equivalent residues in GlpT (K80 and H165) are found lining the substrate-translocation pathway (Figure 1b). In a recent analysis of the GlpT structure, it has been suggested that K80, in partnership with R45, R269 and H165, plays a vital role in substrate binding (43). This proposal was supported by MD simulations (43) suggesting that H165 is involved in substrate coordination only when it is protonated. The maintenance of a positive charge on the H165 residue during substrate binding presumably enables stronger binding to the GlpT anionic substrates. While changes in the ionization state of histidine are common in many enzyme-catalyzed reactions (68), observations in LacY (57), UhpT (25) and now GlpT (43), highlight its importance in membrane transporter proteins in general. In GlpT, H165 is surrounded by several conserved aromatic residues (Y38, Y42, Y76, W138, W161, Y362 and Y393) (35, 46). As it is known that aromatic residues such as tyrosine stabilize positive charges within the membrane electric field (16) it is possible the primary function of the conserved tyrosines and other aromatic residues surrounding the binding site of GlpT is not in substrate binding, but rather in stabilizing the basicity of the binding site. Preservation of the basicity of the substrate-binding site has been shown to be vital for function of OxlT (78, 82), and a membrane-embedded positive charge is proposed to be absolutely essential for multidrug transport by the E. coli MFS protein MdfA (69). Moreover, several aromatic residues are required for substrate transport in the human organic anion transporter hOAT1 (61), and both histidine and tyrosine residues appear to participate in substrate binding by another MFS protein, the mammalian H+/peptide transporter PepT1 (15).
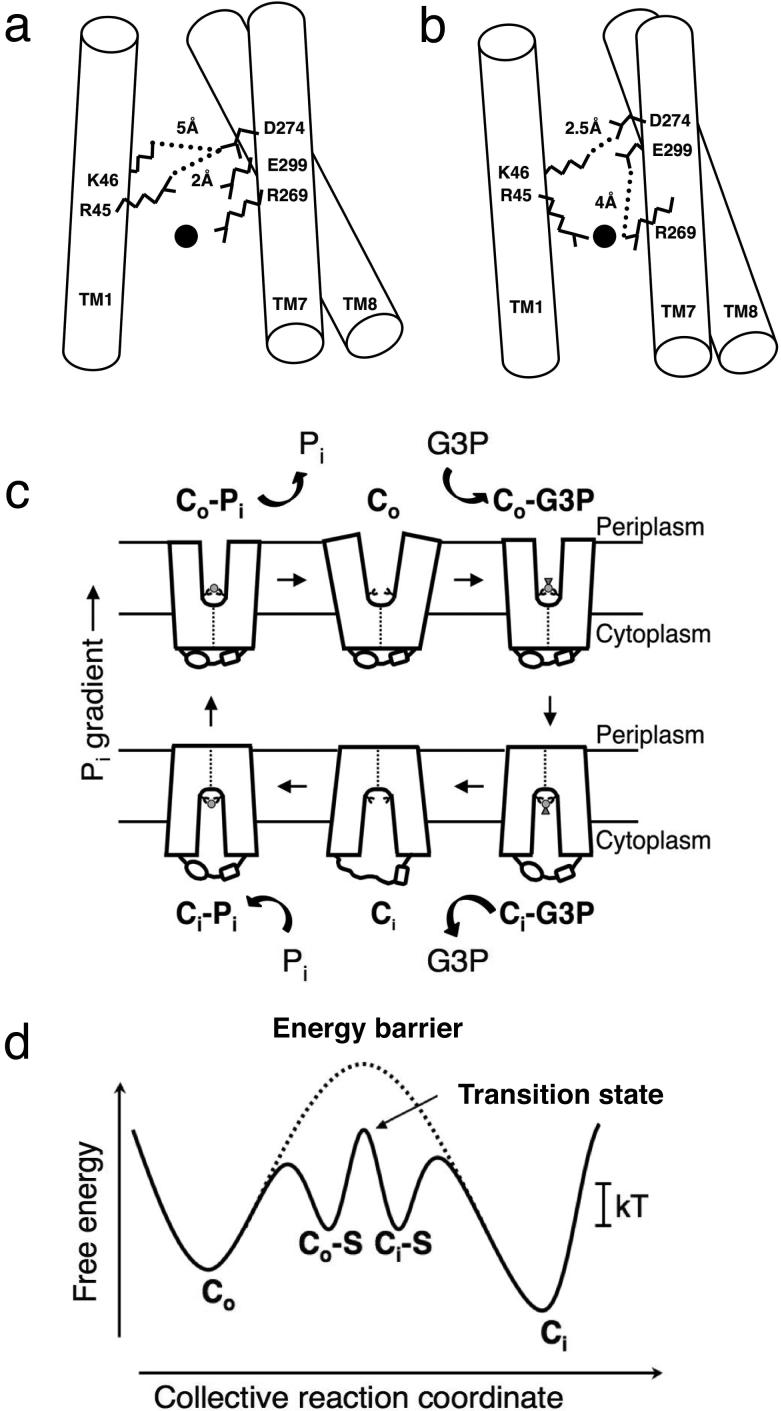
Taken together, a reasonably detailed mechanism for substrate binding by the inward-facing Ci conformation of GlpT can be summarized as follows: (1) an initial weak binding of substrate occurs near the center of the substrate-binding site, involving three basic residues, K80, R45 and R269 (Figure 1b). At this stage H165 is unprotonated and does not participate strongly in binding; (2) H165 then undergoes protonation at the δ-nitrogen position, perhaps facilitated by the proximity of Pi, and its side chain moves closer toward and interacts more strongly with the substrate. At the same time, the R45 and R269 side chains also move 1.4 Å nearer to each other, pulling TM1 and TM7 closer together on the cytoplasmic side (35). Coupled with a reorientation of the substrate (see below), this elicits stronger, more stable interactions between the substrate and K80, R45, R269 and protonated H165, and thus tighter binding to the transporter. The intrinsic binding energy released by this stronger interaction is utilized to overcome the energy barrier to conformational change of the unloaded transporter (Figure 2d); (3) subsequent deprotonation of H165 weakens the interactions with the substrate, now bound in the Co conformation, allowing it to be released into the periplasm.
Figure 2.
Substrate-binding and transport cycle of GlpT. (a) Schematic diagram illustrating the salt bridges that are formed and broken upon initial loose and subsequent tight binding of substrate to GlpT. TM1, 7 and 8 are depicted as cylinders, and the amino acid side chains that participate in salt bridge formation are depicted as stick models. The substrate molecule is represented as a filled circle. When substrate binds loosely to GlpT in the Ci conformation, and H165 is unprotonated, interhelical salt bridges are formed between R45-D274, and K46-D274 (44). (b) Protonation of H165 elicits tighter substrate binding, and R45 and R269 move closer to each other, pulling TM1 and TM7 closer together. The R45-D274 interhelical salt bridge breaks and a new intrahelical one forms between R269 and E299. The existing interhelical salt bridge formed between K46 and D274 becomes stronger, and the transporter takes on a more compact conformation (44). (c) Schematic diagram of the single binding site, alternating access mechanism with a rocker-switch type of movement for the GlpT-mediated G3P-Pi exchange reaction. The diagram describes the proposed conformational changes that the transporter undergoes during the reaction cycle. Co represents the protein in the outward facing conformation and Ci the inward facing one. The G3P substrate is represented by a small disk and triangle, and Pi is represented by a small disk (35). (d) A schematic free energy diagram illustrating the energy levels of the different conformations of GlpT that occur during the transport reaction cycle under physiological conditions. In the absence of substrate-binding the energy barrier prevents the conformational interconversion between the Co and Ci states of the transporter. Substrate binding lowers the energy barrier sufficiently to allow Brownian motion (kT) to drive the conformational interconversion. The energy barrier is represented by a dotted line. S denotes substrate (43).
Such a mechanism agrees with earlier energetics considerations of membrane transport. It has been suggested before for membrane transporter systems that reduction in the energy barrier to conformational change - paid for by intrinsic binding energy between substrate and its binding site - is dependent on formation of an initial loose complex followed by a tight complex that forms in the transition state (41, 42). This mechanism is probably conserved amongst all OPA family antiporters.
ROCKER-SWITCH MECHANISM
A fundamental problem encountered in the study of membrane transport concerns the mechanism of substrate translocation; following substrate binding, how can a membrane-embedded protein catalyze movement of the substrate from one side of the membrane to the other? In both MFS antiporters and symporters, it is clear that the substrate-binding site has access to each side of the membrane in alternating fashion. This can now be seen as a direct consequence of domain movement that allows the N- and C-terminal portions of the transporter to rock back and forth against each along an axis that runs along the domain interface in the membrane. This “rocker-switch” mechanism, implied from the crystal structures of GlpT (35) and LacY (2), is the contemporary version of what Mitchell referred to as the ‘mobile barrier’ model, a name intended to replace the idea (and implications) of the ‘mobile carrier’ (51). Recent work on LacY has supported the notion of a global conformational change during substrate translocation (50, 70).
Salt Bridge Formation and Breakage during Substrate Translocation
The current model of GlpT-mediated transport invokes a single-binding site, alternating-access mechanism accompanied by rocker-switch type movement of the N- and C-terminal domains of the transporter; in kinetic terms (see below), this reflects that interconversion between the Ci and Co (and vice versa) conformations is favored upon substrate binding. Recent work using a combination of MD simulations and biochemical experiments suggests that formation and breaking of inter- and intradomain salt bridges is important in controlling the helical motions necessary for the large scale conformational change between Ci and Co (43). Examination of the crystal structure of GlpT implicated three conserved, charged residues - K46, D274 and E299 - that line the periplasmic end of the substrate-binding pore as of potential importance to function (Figure 1b). MD simulations and biochemical experiments then suggested that two interdomain salt bridges formed by D274 with two separate basic residues (K46 and R45) act in concert as “springs” to stabilize the different conformations during the transport reaction cycle (Figures 2a and 2b). Initially, when H165 is unprotonated and Pi binds loosely to the binding site, there is only a weak interaction between K46 and D274, and between R269 and E299 (Figure 2a). At the same time, there is a strong salt bridge formed between R45 and D274. This would maintain the transporter in the Ci conformation in which the binding site allows substrate approach from the cytoplasmic side of the substrate translocation pore. Subsequent tighter binding of substrate, accompanied by protonation of H165, pulls R45 of TM1 and R269 of TM7 closer together. Tight substrate binding, because it competes for interaction with R45 and R269, would then be followed by disruption of the interdomain R45-D274 salt bridge, and the formation of stronger inter-and intradomain ones between K46 and D274, and subsequently R269 and E299, respectively, resulting in an interdomain movement and relative rotation of the helices in each domain (Figure 2b). Therefore, substrate binding effectively introduces a ‘competing’ salt bridge, weakening interactions that stabilize the Ci conformation. This enables new interactions involved in domain rotation, possibly involving yet other salt bridges. In this model, the salt bridge dynamics are part of the spring that permits the delicately poised GlpT molecule to flip - via the rocker-switch mechanism (35, 47) -from the substrate-bound inward-facing Ci-S conformation to the outward-facing Co-S conformation. The salt bridges may even act as the “pivot” upon which the “teeter-totter” motion of the two domains of the protein is centered. It is pertinent to note here that the E. coli outer membrane protein OmpA uses the formation and breaking of salt bridges to control the gating of its substrate-translocation pore (34).
Evidence for conformational changes during substrate translocation by antiporters is available from the structure of GlpT (35), and from its comparisons with the OxlT (30, 31) and EmrD (83) structures. As noted before, a model of OxlT in substrate-bound form gave insight into the conformational changes that occur upon substrate binding to antiporters (32), suggesting that the main consequence of transition from the unloaded, inward-facing Ci state to the closed, substrate-bound Ci-S state is a closing of the cytoplasmic ends of the N- and C-terminal domains, involving a swiveling movement of each domain relative to the other (Figure 2c). Further, it was suggested the continuation of this movement brings the protein into the Co-S state, in which the cytoplasmic ends of the domains closer together than the periplasmic ones, thereby revealing the binding site to the periplasm and exposing the substrate to it. Due to symmetry of the global architecture of OxlT, the Co state is likely to mirror that of the Ci state (31, 32).
Kinetic Considerations
Recent kinetic studies on GlpT provide further evidence for the rocker-switch mechanism, and this has allowed the reaction cycle to be described in detailed physical chemistry terms (44). The rate of the complete reaction cycle is associated with, and dictated by, large conformational changes in the protein. An occluded state of the protein is also expected during substrate translocation (40), although whether this state is as stable in GlpT as it is in OxlT and EmrD is unknown. As the entire antiport process must occur at a physiologically realistic rate and be responsive to changes in substrate concentrations on both sides of the membrane, reasonable values for the rate constants are needed for the individual stages of the reaction cycle (Figure 2c). Like other antiporters, GlpT obeys Michaelis-Menten kinetics and operates via a Ping-Pong mechanism (79). This allows one to conceptually distinguish six discrete steps, with three steps dedicated to the binding, translocation and release of one of the transported substrates and the other three describing the same events for transport of the countersubstrate. This represents the minimum number of steps required for antiport. The half-reaction for translocation of one substrate (Pi in the case of GlpT) can therefore be written as,
| (Scheme 1) |
The three stages of the half-reaction are: (i) binding of substrate to the protein in the Ci conformation; (ii) switching of the substrate-bound form of the protein from the Ci-Pi to Co-Pi state, which allows the pathway to face the opposing surface; and, (iii) release of the translocated substrate into the periplasm. A detailed description of the antiport reaction mechanism begins with GlpT in the Ci conformation, with its substrate-binding site accessible only to Pi on the cytoplasmic side of the membrane (Figure 2c). In this state, the antiporter is in a stable configuration of low energy. The conformational change from the Ci to Co state (or vice versa) of unloaded, substrate-free transporter over the energy barrier is extremely slow compared to the substrate-bound form (Figure 2c). Thus, one may derive from Scheme 1,
| (Eqn. 1) |
where Kd = (k-1/k1), and Km is dependent on the ratios of (k-1/k1) and (k3/k-1). Biochemical experiments that exploit the quenching of intrinsic tryptophan fluorescence of GlpT upon binding of G3P substrate in detergent solution gives an apparent substrate binding dissociation constant of ~0.8 μM (44). Significantly, the binding of substrate to GlpT is independent of temperature, as is the Michaelis constant for transport, Km, calculated from assays performed using both E. coli whole cells and purified GlpT reconstituted into proteoliposomes (44). The next kinetic event (the k2/k-2 step) represents the interconversion of the protein from the Ci-Pi to the Co-Pi state, which is accompanied by exposure of the substrate-binding site to the opposite side of the membrane (Figure 2c). This step encompasses the transition state and is the rate-limiting step of the whole transport process (18). Crucially, transport assays showed that this step is highly temperature-dependent, with the maximal velocity of transport, Vmax, increasing with temperature (44). This led to the conclusion that release of binding energy alone is insufficient to drive the Ci-Pi to Co-Pi transition, and that an energy input of roughly 35 kJ mol-1 (at 37°C) is derived as thermal energy from the environment (Figure 2d). The final step of the half-reaction described in (Scheme 1) involves release of bound substrate into the periplasmic compartment, and presentation of the binding site to countersubstrate (G3P in the case of GlpT) to allow the antiport reaction to recur by reversal of the three steps just described (Figure 2c). In the periplasm, the relatively low affinity of the transporter for Pi (10, 26) allows its replacement by G3P, whereas in the cytoplasm Pi replaces G3P at the binding site due its much higher cellular concentration (4 mM under non-growing conditions) (77), thus driving the entire transport reaction. This mechanism effectively allows substrate-bound GlpT to randomly sample the Ci and Co states via simple Brownian motion; the net direction of transport is the determined by substrate association/dissociation reactions in accordance with the rules of mass action at either surface. We believe that all antiporters of the MFS likely utilize this same mechanistic strategy.
Substrate Exchange Stoichiometry
Even today, ambiguity exists as to the actual stoichiometry of exchange in MFS antiporters. For technical reasons discussed by Maloney et al. (53), investigations into the stoichiometry of glucose-6-phosphate(G6P)/phosphate exchange were performed on membrane vesicles of S. lactis. A striking finding from this work was the effect of pH on the substrate-exchange process (9, 52). First, a reduction in pH slowed the reaction due to a velocity effect. Dropping the pH from 7 to 5.2 reduced the Vmax 10-fold, whereas the Km remained constant for both heterologous and homologous exchange reactions, implying that monobasic and dibasic sugars were equally effective as substrates. Second, the pH had a direct impact on stoichiometry, with a 2:1 Pi:sugar phosphate exchange measured at pH 7, and a 1:1 exchange measured at pH 5.2 (9). Based on these findings, along with the presumption that monovalent Pi but not divalent Pi is the preferred exchange substrate, a model was forwarded that incorporated a bifunctional substrate binding site that accepted either two monovalent sugar phosphates (2HG6P1-) or a single divalent anion (1G6P2-), but not both (53). This seemed to reconcile the observed 2:1 (2Pi vs 1G6P) stoichiometry at alkaline pH, and a 1:1 (2P 1-i vs 2G6P1-) ratio at the acid pH. The arrival of the crystal structure of GlpT casts some doubt on these interpretations since there does not appear to be sufficient room in the translocation pore for two molecules of Pi, much less G3P. Nevertheless, because the GlpT structure has no bound substrate (35), the issue remains unsettled, and it will be of great interest to reexamine the issue of how these transporters select among the various ionic forms of their substrates. The finding that H165 in GlpT is likely to accept a proton during the binding of Pi (43) immediately raises the question of whether the proton is abstracted from the medium or from Pi and how this might affect selectivity among mono- and dibasic forms of Pi.
FINAL CONSIDERATIONS
As a final point in this review, it is worth noting that redundancy with respect to membrane transporters is built into the cell. Often, the cell contains both primary and secondary active transporters for the same substrate. As the largest primary transporter superfamily in the cell (60), the ABC proteins, like their MFS counterparts, display a diverse substrate specificity. Although secondary active transporters do not establish as steep a substrate gradient as primary transporters do, they may be viewed as energetically more frugal. This allows the cell to modulate expression of each type of transporter for a particular substrate according to cellular and environmental needs (46).
SUMMARY POINTS
We hope this review has outlined to the reader a set of principles that apply equally to all MFS antiporters.
All MFS proteins share essentially the same 3D structure, with two domains that saddle the substrate-translocation pore.
Conservation of the blueprint for the fold of MFS antiporters implies that the diversity of substrate preference seen in the MFS is a result of changes in only a few residues in the substrate-binding site and translocation pathway.
Release of intrinsic binding energy upon binding of substrate lowers the activation energy barrier sufficiently to allow Brownian motion to drive substrate translocation across the membrane.
Substrate translocation, via the rocker-switch mechanism, is accompanied by large, global conformational change of the protein that involves formation and breaking of inter- and intrahelical salt bridges.
FUTURE ISSUES
Although steady progress has been made in illuminating the biology of MFS antiporters, significant questions remain.
How many conformational states of the transporter are there? There is an urgent need to define the transition states in conformational space. Characterization of these states and the ability to achieve an individual state in vitro is the main hurdle to realizing a full structural characterization of any secondary active transporter.
We eagerly await the arrival of high-resolution structures of an MFS antiporter in the outward-facing Co conformation, and in the Ci conformation with substrate bound.
Are the same residues involved in binding substrate in both the Ci and Co conformations, and what are the exact determinants of substrate specificity and ionic selectivity?
ACKNOWLEDGEMENTS
C.J.L and D.N.W would like to thank past members of the lab; Jonas Almqvist, Manfred Auer, Yafei Huang, Myon Jin Kim, Joanne Lemieux, Xiaodan Li, Jinmei Song, Céline Soudant and Xiaorong Zhang for their dedicated and enthusiastic work on GlpT. The authors' work was supported in part by NIH Grants DK-053973 (to D.N.W) and GM-24195 (to P.C.M).
Mini-Glossary of Terms
- Antiporter
a membrane transport protein that transports two or more substrates in opposite directions across the cell membrane using the gradient of one to drive movements of the other.
- Ci
inward- or cytoplasmic-facing conformation of bacterial antiporters.
- Co
outward- or periplasmic-facing conformation of bacterial antiporters.
- Alternating-access
general mechanism by which the single-binding site of MFS antiporters is sequentially exposed to the cytoplasmic and periplasmic sides of the membrane.
- Rocker-switch mechanism
the motion of the N- and C-terminal domains of MFS transporters that results in alternating-access of the substrate-binding site.
Abbreviations and Acronyms
- ABC transporters
ATP-binding cassette transporters
- G3P
glycerol-3-phosphate
- G6P
glucose-6-phosphate
- GlpT
sn-glycerol-3-phosphate transporter from E. coli
- MD
molecular dynamics
- MFS
major facilitator superfamily
- OPA
organophosphate:phosphate antiporter
- OxlT
oxalate:formate antiporter from Oxalobacter formigenes
- Pi
inorganic phosphate
- TMs
transmembrane α-helices
- UhpT
hexose phosphate:phosphate antiporter from E. coli
LITERATURE CITED
- 1.Abramson J, Kaback HR, Iwata S. Structural comparison of lactose permease and the glycerol-3-phosphate antiporter: members of the major facilitator superfamily. Curr. Opin. Struct. Biol. 2004;14:413–9. doi: 10.1016/j.sbi.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–15. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 3.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 4.Almqvist J, Huang Y, Hovmöller S, Wang DN. Homology modeling of the human microsomal glucose-6-phosphate transporter explains the mutations that cause the glycogen storage disease type Ib. Biochemistry. 2004;43:9289–97. doi: 10.1021/bi049334h. [DOI] [PubMed] [Google Scholar]
- 5.Ambudkar SV, Anantharam V, Maloney PC. UhpT, the sugar phosphate antiporter of Escherichia coli, functions as a monomer. J. Biol. Chem. 1990;265:12287–92. [PubMed] [Google Scholar]
- 6.Ambudkar SV, Larson TJ, Maloney PC. Reconstitution of sugar phosphate transport systems of Escherichia coli. J. Biol. Chem. 1986;261:9083–86. [PubMed] [Google Scholar]
- 7.Ambudkar SV, Maloney PC. Characterization of phosphate:hexose 6-phosphate antiport in membrane vesicles of Streptococcus lactis. J. Biol. Chem. 1984;259:12576–85. [PubMed] [Google Scholar]
- 8.Ambudkar SV, Maloney PC. Reconstitution of anion exchange from Streptococcus lactis. Fed. Proc. 1985;44:1806–06. [Google Scholar]
- 9.Ambudkar SV, Sonna LA, Maloney PC. Variable stoichiometry of phosphate-linked anion exchange in Streptococcus lactis: implications for the mechanism of sugar phosphate transport by bacteria. Proc. Natl. Acad. Sci. USA. 1986;83:280–4. doi: 10.1073/pnas.83.2.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auer M, Kim MJ, Lemieux MJ, Villa A, Song J, et al. High-yield expression and functional analysis of Escherichia coli glycerol-3-phosphate transporter. Biochemistry. 2001;40:6628–35. doi: 10.1021/bi010138+. [DOI] [PubMed] [Google Scholar]
- 11.Bartoloni L, Wattenhofer M, Kudoh J, Berry A, Shibuya K, et al. Cloning and characterization of a putative human glycerol 3-phosphate permease gene (SLC37A1 or G3PP) on 21q22.3: Mutation analysis in two candidate phenotypes, DFNB10 and a glycerol kinase deficiency. Genomics. 2000;70:190–200. doi: 10.1006/geno.2000.6395. [DOI] [PubMed] [Google Scholar]
- 12.Bevan M, Bancroft I, Bent E, Love K, Goodman H, et al. Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature. 1998;391:485–8. doi: 10.1038/35140. [DOI] [PubMed] [Google Scholar]
- 13.Chang AB, Lin R, Studley WK, Tran CV, Saier MH. Phylogeny as a guide to structure and function of membrane transport proteins. Mol. Membr. Biol. 2004;21:171–81. doi: 10.1080/09687680410001720830. [DOI] [PubMed] [Google Scholar]
- 14.Chen LY, Pan CJ, Shieh JJ, Chou JY. Structure-function analysis of the glucose-6-phosphate transporter deficient in glycogen storage disease type Ib. Hum. Mol. Genet. 2002;11:3199–207. doi: 10.1093/hmg/11.25.3199. [DOI] [PubMed] [Google Scholar]
- 15.Chen XZ, Steel A, Hediger MA. Functional roles of histidine and tyrosine residues in the H+-peptide transporter PepT1. Biochem. Biophy. Res. Comm. 2000;272:726–30. doi: 10.1006/bbrc.2000.2851. [DOI] [PubMed] [Google Scholar]
- 16.Dougherty DA. Cation-π interactions in chemistry and biology: a new view of benzene, Phe, Tyr, and Trp. Science. 1996;271:163–8. doi: 10.1126/science.271.5246.163. [DOI] [PubMed] [Google Scholar]
- 17.Fann M, Davies AH, Varadhachary A, Kuroda T, Sevier C, et al. Identification of two essential arginine residues in UhpT, the sugar phosphate antiporter of Escherichia coli. J. Membr. Biol. 1998;164:187–95. doi: 10.1007/s002329900404. [DOI] [PubMed] [Google Scholar]
- 18.Fann MC, Maloney PC. Functional symmetry of UhpT, the sugar phosphate transporter of Escherichia coli. J. Biol. Chem. 1998;273:33735–40. doi: 10.1074/jbc.273.50.33735. [DOI] [PubMed] [Google Scholar]
- 19.Fisher RB, Parsons DS. Glucose movements across the wall of the rat small intestine. J. Physiol. 1953;119:210–23. doi: 10.1113/jphysiol.1953.sp004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldschmidt S. On the mechanism of absorption from the intestine. Physiol. Rev. 1921;1:421–53. [Google Scholar]
- 21.Guan L, Kaback HR. Lessons from lactose permease. Ann. Rev. Biophys. Biomol. Struct. 2006;35:67–91. doi: 10.1146/annurev.biophys.35.040405.102005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan L, Mirza O, Verner G, Iwata S, Kaback HR. Structural determination of wild-type lactose permease. Proc. Natl. Acad. Sci. USA. 2007;104:15294–98. doi: 10.1073/pnas.0707688104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall JA, Fann MC, Maloney PC. Altered substrate selectivity in a mutant of an intrahelical salt bridge in UhpT, the sugar phosphate carrier of Escherichia coli. J. Biol. Chem. 1999;274:6148–53. doi: 10.1074/jbc.274.10.6148. [DOI] [PubMed] [Google Scholar]
- 24.Hall JA, Maloney PC. Transmembrane segment 11 of UhpT, the sugar phosphate carrier of Escherichia coli, is an alpha-helix that carries determinants of substrate selectivity. J. Biol. Chem. 2001;276:25107–13. doi: 10.1074/jbc.M102017200. [DOI] [PubMed] [Google Scholar]
- 25.Hall JA, Maloney PC. Altered oxyanion selectivity in mutants of UhpT, the Pi-linked sugar phosphate carrier of Escherichia coli. J. Biol. Chem. 2005;280:3376–81. doi: 10.1074/jbc.M409965200. [DOI] [PubMed] [Google Scholar]
- 26.Hayashi S, Koch JP, Lin ECC. Active transport of L-a-glycerophosphate in Escherichia coli. J. Biol. Chem. 1964;239:3098–105. [PubMed] [Google Scholar]
- 27.Heymann JA, Sarker R, Hirai T, Shi D, Milne JL, et al. Projection structure and molecular architecture of OxlT, a bacterial membrane transporter. EMBO J. 2001;20:4408–13. doi: 10.1093/emboj/20.16.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hildebrand P, Guenther S, Goede A, Forrest L, Frommel C, Preissner R. Hydrogen-bonding and packing features of membrane proteins: functional implications. Biophys. J. 2008 doi: 10.1529/biophysj.107.110395. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hildebrand PW, Rother K, Goede A, Preissner R, Frommel C. Molecular packing and packing defects in helical membrane proteins. Biophys. J. 2005;88:1970–77. doi: 10.1529/biophysj.104.049585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirai T, Heymann JA, Shi D, Sarker R, Maloney PC, Subramaniam S. Three-dimensional structure of a bacterial oxalate transporter. Nat. Struct. Biol. 2002;9:597–600. doi: 10.1038/nsb821. [DOI] [PubMed] [Google Scholar]
- 31.Hirai T, Heymann JAW, Shi D, Subramaniam S. Comparative structural analysis of the "symmetric", substrate-bound state of the oxalate transporter OxlT with the "cytoplasmically-open" state of the MFS transporters GlpT and LacY. Biophys. J. 2004;86:611–11. [Google Scholar]
- 32.Hirai T, Subramaniam S. Structure and transport mechanism of the bacterial oxalate transporter OxlT. Biophys. J. 2004;87:3600–07. doi: 10.1529/biophysj.104.049320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hodgkin AL, Huxley AF. Currents carried by sodium and potassium ions through the membrane of the giant axon of Loligo. J. Physiol. 1952;116:449–72. doi: 10.1113/jphysiol.1952.sp004717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong H, Szabo G, Tamm LK. Electrostatic couplings in OmpA ion-channel gating suggest a mechanism for pore opening. Nat. Chem. Biol. 2006;2:627–35. doi: 10.1038/nchembio827. [DOI] [PubMed] [Google Scholar]
- 35.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 2003;301:616–20. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 36.Jencks WP. The utilization of binding energy in coupled vectorial processes. Adv. Enzymol. Relat. Areas Mol. Biol. 1980;51:75–106. doi: 10.1002/9780470122969.ch2. [DOI] [PubMed] [Google Scholar]
- 37.Jia W, Cole JA. Nitrate and nitrite transport in Escherichia coli. Biochem. Soc. Trans. 2005;33:159–61. doi: 10.1042/BST0330159. [DOI] [PubMed] [Google Scholar]
- 38.Kaback HR. Structure and mechanism of the lactose permease. Comptes Rendus Biologies. 2005;328:557–67. doi: 10.1016/j.crvi.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 39.Kaback HR, Wu J. From membrane to molecule to the third amino acid from the left with a membrane transport protein. Q. Rev. Biophys. 1997;30:333–64. doi: 10.1017/s0033583597003387. [DOI] [PubMed] [Google Scholar]
- 40.Klingenberg M. Ligand-protein interaction in biomembrane carriers. The induced transition fit of transport catalysis. Biochemistry. 2005;44:8563–70. doi: 10.1021/bi050543r. [DOI] [PubMed] [Google Scholar]
- 41.Krupka RM. Role of substrate binding forces in exchange-only transport systems: I. Transition-state theory. J. Membr. Biol. 1989;109:151–8. doi: 10.1007/BF01870854. [DOI] [PubMed] [Google Scholar]
- 42.Krupka RM. Role of substrate binding forces in exchange-only transport systems: II. Implications for the mechanism of the anion exchanger of red cells. J. Membr. Biol. 1989;109:159–71. doi: 10.1007/BF01870855. [DOI] [PubMed] [Google Scholar]
- 43.Law CJ, Almqvist J, Bernstein A, Goetz RM, Soudant C, Wang DN. Salt bridge dynamics upon substrate binding to GlpT participate in conformational change during transport. 2008. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Law CJ, Yang Q, Soudant C, Maloney PC, Wang DN. Kinetic evidence is consistent with the rocker-switch mechanism of membrane transport by GlpT. Biochemistry. 2007;46:12190–97. doi: 10.1021/bi701383g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lemieux MJ. Eukaryotic major facilitator superfamily transporter modeling based on the prokaryotic GlpT crystal structure. Mol. Membr. Biol. 2007;24:333–41. doi: 10.1080/09687680701496507. [DOI] [PubMed] [Google Scholar]
- 46.Lemieux MJ, Huang Y, Wang DN. Glycerol-3-phosphate transporter of Escherichia coli: structure, function and regulation. Res. Microbiol. 2004;155:623–29. doi: 10.1016/j.resmic.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 47.Lemieux MJ, Huang Y, Wang DN. Structural basis of substrate translocation by the Escherichia coli glycerol-3-phosphate transporter: a member of the major facilitator superfamily. Curr. Opin. Struct. Biol. 2004;14:405–12. doi: 10.1016/j.sbi.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 48.Lin ECC. Glycerol dissimilation and its regulation in bacteria. Ann. Rev. Microbiol. 1976;30:535–78. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- 49.Maiden MCJ, Davis EO, Baldwin SA, Moore DCM, Henderson PJF. Mammalian and bacterial sugar-transport proteins are homologous. Nature. 1987;325:641–43. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- 50.Majumdar DS, Smirnova I, Kasho V, Nir E, Kong XX, et al. Single-molecule FRET reveals sugar-induced conformational dynamics in LacY. Proc. Natl. Acad. Sci. USA. 2007;104:12640–45. doi: 10.1073/pnas.0700969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maloney PC. Bacterial transporters. Curr. Opin. Cell. Biol. 1994;6:571–82. doi: 10.1016/0955-0674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 52.Maloney PC, Ambudkar SV. Anion exchange in bacteria. Variable stoichiometry of phosphate: sugar 6-phosphate antiport. Ann. NY. Acad. Sci. 1985;456:245–7. doi: 10.1111/j.1749-6632.1985.tb14871.x. [DOI] [PubMed] [Google Scholar]
- 53.Maloney PC, Ambudkar SV, Anatharam V, Sonna LA, Varadhachary A. Anion-exchange mechanisms in bacteria. Microbiol. Rev. 1990;54:1–17. doi: 10.1128/mr.54.1.1-17.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitchell P. Transport of phosphate across the surface of Micrococcus pyogenes; nature of the cell inorganic phosphate. J. Gen. Microbiol. 1953;9:273–87. doi: 10.1099/00221287-9-2-273. [DOI] [PubMed] [Google Scholar]
- 55.Mitchell P. Transport of phosphate across the osmotic barrier of Micrococcus pyogenes; specificity and kinetics. J. Gen. Microbiol. 1954;11:73–82. doi: 10.1099/00221287-11-1-73. [DOI] [PubMed] [Google Scholar]
- 56.Mueckler M, Makepeace C. Transmembrane segment 12 of the Glut1 glucose transporter is an outer helix and is not directly involved in the transport mechanism. J. Biol. Chem. 2006;281:36993–98. doi: 10.1074/jbc.M608158200. [DOI] [PubMed] [Google Scholar]
- 57.Padan E, Sarkar HK, Viitanen PV, Poonian MS, Kaback HR. Site-specific mutagenesis of histidine residues in the lac permease of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1985;82:6765–8. doi: 10.1073/pnas.82.20.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pao SS, Paulsen IT, Saier MH. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patlak C. Contributions to the theory of active transport: II. The gate type non-carrier mechanism and generalizations concerning tracer flow, efficiency, and measurement of energy expenditure. Bull. Math. Biophys. 1957;19:209–35. [Google Scholar]
- 60.Paulsen IT, Nguyen L, Sliwinski MK, Rabus R, Saier MH. Microbial genome analyses: Comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 2000;301:75–100. doi: 10.1006/jmbi.2000.3961. [DOI] [PubMed] [Google Scholar]
- 61.Perry JL, Dembla-Rajpal N, Hall LA, Pritchard JB. A three-dimensional model of human organic anion transporter 1: aromatic amino acids required for substrate transport. J. Biol. Chem. 2006;281:38071–9. doi: 10.1074/jbc.M608834200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rao NN, Roberts MF, Torriani A, Yashphe J. Effect of glpT and glpD mutations on expression of the phoA gene in Escherichia coli. J. Bacteriol. 1993;175:74–9. doi: 10.1128/jb.175.1.74-79.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ren Q, Paulsen IT. Comparative analyses of fundamental differences in membrane transport capabilities in prokaryotes and eukaryotes. PLoS Comput. Biol. 2005;1:e27. doi: 10.1371/journal.pcbi.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ren Q, Paulsen IT. Large-scale comparative genomic analyses of cytoplasmic membrane transport systems in prokaryotes. J. Mol. Microbiol. Biotechnol. 2007;12:165–79. doi: 10.1159/000099639. [DOI] [PubMed] [Google Scholar]
- 65.Safferling M, Griffith H, Jin J, Sharp J, De Jesus M, et al. The TetL tetracycline efflux protein from Bacillus subtilis is a dimer in the membrane and in detergent solution. Biochemistry. 2003;42:13969–76. doi: 10.1021/bi035173q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saier MH. Tracing pathways of transport protein evolution. Mol. Micro. 2003;48:1145–56. doi: 10.1046/j.1365-2958.2003.03499.x. [DOI] [PubMed] [Google Scholar]
- 67.Saier MH, Jr., Beatty JT, Goffeau A, Harley KT, Heijne WH, et al. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1999;1:257–79. [PubMed] [Google Scholar]
- 68.Schneider F. Histidine in enzyme active centers. Angew. Chem. Int. Ed. Engl. 1978;17:583–92. doi: 10.1002/anie.197805831. [DOI] [PubMed] [Google Scholar]
- 69.Sigal N, Vardy E, Molshanski-Mor S, Eitan A, Pilpel Y, et al. 3D model of the Escherichia coli multidrug transporter MdfA reveals an essential membrane-embedded positive charge. Biochemistry. 2005;44:14870–80. doi: 10.1021/bi051574p. [DOI] [PubMed] [Google Scholar]
- 70.Smirnova I, Kasho V, Choe JY, Altenbach C, Hubbell WL, Kaback HR. Sugar binding induces an outward-facing conformation of LacY. Proc. Natl. Acad. Sci. USA. 2007;104:16504–09. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takahashi Y, Miyata M, Zheng P, Imazato T, Horwitz A, Smith JD. Identification of cAMP analogue inducible genes in RAW264 macrophages. Biochim. Biophys. Acta. 2000;1492:385–94. doi: 10.1016/s0167-4781(00)00133-0. [DOI] [PubMed] [Google Scholar]
- 72.Tanford C. Simple model for the chemical potential change of a transported ion in active transport. Proc. Natl. Acad. Sci. USA. 1982;79:2882–84. doi: 10.1073/pnas.79.9.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tanford C. Mechanism of free energy coupling in active transport. Annu. Rev. Biochem. 1983;52:379–409. doi: 10.1146/annurev.bi.52.070183.002115. [DOI] [PubMed] [Google Scholar]
- 74.Unkles SE, Rouch DA, Wang Y, Siddiqi MY, Glass AD, Kinghorn JR. Two perfectly conserved arginine residues are required for substrate binding in a high-affinity nitrate transporter. Proc. Natl. Acad. Sci. USA. 2004;101:17549–54. doi: 10.1073/pnas.0405054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Veenhoff LM, Heuberger EH, Poolman B. The lactose transport protein is a cooperative dimer with two sugar translocation pathways. EMBO J. 2001;20:3056–62. doi: 10.1093/emboj/20.12.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vidavar GA. Inhibition of parallel flux and augmentation of counter flux shown by transport models not involving a mobile carrier. J. Thoer. Biol. 1966;10:301–06. doi: 10.1016/0022-5193(66)90128-7. [DOI] [PubMed] [Google Scholar]
- 77.Vink R, Bendall MR, Simpson SJ, Rogers PJ. Estimation of H+ to adenosine 5'-triphosphate stoichiometry of Escherichia coli ATP synthase using 31P NMR. Biochemistry. 1984;23:3667–75. doi: 10.1021/bi00311a015. [DOI] [PubMed] [Google Scholar]
- 78.Wang X, Sarker RI, Maloney PC. Analysis of substrate-binding elements in OxlT, the oxalate:formate antiporter of Oxalobacter formigenes. Biochemistry. 2006;45:10344–50. doi: 10.1021/bi060746v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.West IC. Ligand conduction and the gated-pore mechanism of transmembrane transport. Biochim. Biophys. Acta. 1997;1331:213–34. doi: 10.1016/s0304-4157(97)00007-5. [DOI] [PubMed] [Google Scholar]
- 80.Widdas WF. Inability of diffusion to account for placental glucose transfer in the sheep and consideration of the kinetics of a possible carrier transfer. J. Physiol. 1952;118:23–39. doi: 10.1113/jphysiol.1952.sp004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Widdas WF. Kinetics of glucose transfer across the human erythrocyte membrane. J. Physiol. 1953;120:23–24. [Google Scholar]
- 82.Yang Q, Wang X, Ye L, Mentrikoski M, Mohammadi E, et al. Experimental tests of a homology model for OxlT, the oxalate transporter of Oxalobacter formigenes. Proc. Natl. Acad. Sci. USA. 2005;102:8513–18. doi: 10.1073/pnas.0503533102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli. Science. 2006;312:741–44. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]