Abstract
Previous reports have demonstrated that the mtDNA of mouse common inbred strains (CIS) originated from a single female ancestor and that mtDNA mutations occurred during CIS establishment. This situation provides a unique opportunity to investigate the impact of individual mtDNA variations on complex traits in mammals. In this study, we compiled the complete mtDNA sequences of 52 mouse CIS. Phylogenetic analysis demonstrated that 50 of the 52 CIS descended from a single female Mus musculus domesticus mouse, and mtDNA mutations have accumulated in 26 of the CIS. We then generated conplastic strains on the C57BL/6J background for 12 mtDNA variants with one to three functional mtDNA mutations. We also generated conplastic strains for mtDNA variants of the four M. musculus subspecies, each of which contains hundreds of mtDNA variations. In total, a panel of conplastic strains was generated for 16 mtDNA variants. Phenotypic analysis of the conplastic strains demonstrated that mtDNA variations affect susceptibility to experimental autoimmune encephalomyelitis and anxiety-related behavior, which confirms that mtDNA variations affect complex traits. Thus, we have developed a unique genetic resource that will facilitate exploration of the biochemical and physiological roles of mitochondria in complex traits.
The mammalian mitochondrial genome (mtDNA) is a closed, circular, double-stranded DNA with genes encoding mitochondrial oxidative phosphorylation (OXPHOS) enzyme complexes that are essential for ATP production (Anderson et al. 1981). In humans, mtDNA variations can be rare pathogenic mutations or deletions that cause maternally inherited mtDNA disorders or common mtDNA variants that lead to functional changes and thus predispose individuals for polygenic diseases (Wallace et al. 1988; Taylor and Turnbull 2005). However, unique genetic characteristics of mammalian mtDNA, for example, the lack of recombination and the transmission as one haplotype, hamper efforts to identify precisely the variations responsible for traits and effects and to define the biochemical and physiological consequences of individual variations. The same characteristics complicate the genetic manipulation of mtDNA. Although several transmitochondrial mice have been produced, a mouse with a single mtDNA mutation has not yet been generated (Pinkert and Trounce 2002).
An unexpected observation provided a potential solution to the limitations of mtDNA. In 1982, Ferris et al. (1982) demonstrated that most mouse common inbred strains (CIS) had the same mtDNA RFLP pattern, which suggested that mouse CIS were descended from a single female. This conclusion was confirmed by sequencing the mtDNA of CIS (Johnson et al. 2001; Bayona-Bafaluy et al. 2003; Mathews et al. 2005; Goios et al. 2007), which also demonstrated that several mutations had accumulated during the establishment of the strains, with a higher ratio of nonsynonymous/synonymous mutations than in wild mice (Goios et al. 2007). Those mutations are of potential functional importance, as has been demonstrated in recent studies (Johnson et al. 2001; Mathews et al. 2005; Moreno-Loshuertos et al. 2006). These findings suggest that the systematic production of conplastic strains (mitochondrial substitution strains) with CIS mtDNA carrying one or more functional mutations would constitute a genetic resource to investigate the roles of individual mtDNA variations in complex traits.
In the present study, we report the generation of a unique public resource of 16 conplastic strains that carry potential functional mtDNA mutations on a C57BL/6J genetic background. In humans, mtDNA variations are associated with many neurological diseases such as Alzheimer’s disease, Parkinson’s disease, multiple sclerosis (MS), and bipolar disorder (Shoffner et al. 1993; Kalman and Alder 1998; Kato et al. 2001; van der Walt et al. 2003). Recently, we reported that the mtDNA allele nt13708A increases susceptibility to MS (Yu et al. 2008). To test the usefulness of this genetic resource, we investigated the conplastic strains with regard to nervous system phenotypes, in particular experimental autoimmune encephalomyelitis (EAE), a mouse model of MS, and anxiety-like behavior as an aspect of bipolar disorder. We show that mtDNA variations affect both phenotypes, which suggests that conplastic strains are a useful genetic resource for investigating the role of mtDNA in complex traits.
Results
Sequence and phylogeny of mtDNA in mouse inbred strains
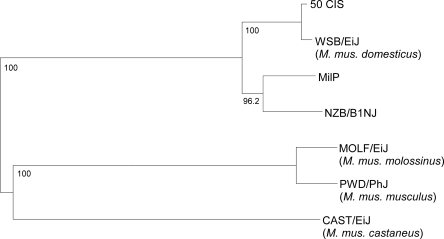
We sequenced the complete mtDNA of 27 common inbred strains: 129P3/J, BPH/2J, BPL/1J, BPN/3J, BUB/BnJ, C57BL/10J, CE/J, I/LnJ, LG/J, LP/J, LT/SvEi, MA/MyJ, MRL/MpJ, NOR/LtJ, P/J, PL/J, RF/J, RIIIS/J, SB/LeJ, SEA/GnJ, SJL/J, SM/J, SOD1/EiJ, STR/ort, SWR/J, TALLYHO/JngJ, and YBR/EiJ. Including the previously published sequences of 29 inbred strains (Loveland et al. 1990; Johnson et al. 2001; Bayona-Bafaluy et al. 2003; Mathews et al. 2005; Goios et al. 2007), the complete mtDNA sequences of 56 inbred mouse strains were collected. The strains consisted of 52 CIS and four wild-derived inbred strains (WIS). A phylogenetic tree based on the mtDNA sequences demonstrated that all 52 CIS, with the exception of NZB/B1NJ and MilP, shared a sub-branch related to WSB/EiJ, a WIS belonging to the Mus musculus domesticus subspecies (Fig. 1). This confirmed the previous conclusion that most CIS were descended from one M. musculus domesticus female mouse (Ferris et al. 1982).
Figure 1.
Phylogenetic tree of mtDNA from mouse inbred strains. The tree was generated with ClustalW software on the basis of the mtDNA sequences of 56 mouse inbred strains, including 52 classic inbred strains (CIS) and four wild-derived inbred strains (WIS). For the four WIS, the taxonomic classification is indicated. Numbers correspond to bootstrap values (percentage of 1000 total bootstrap replicates).
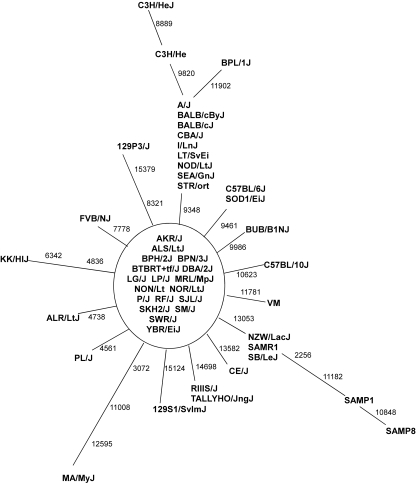
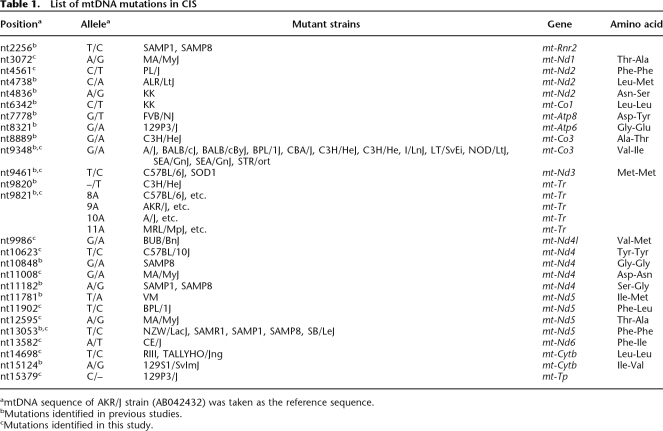
To further characterize the 50 CIS with the same maternal origin, we generated a parsimonious tree for all mtDNA variations in these strains. The mtDNA sequences of the 50 mouse strains formed a star-like network, with a main group of 18 strains in the center (Fig. 2). The other 32 strains were one to three variations apart from the central group. Four mtDNA variations—nt9348 G/A, nt9416 C/T, nt13053 T/C, and nt14698 T/C—were carried by at least two strains, which indicates that they occurred in the early stage of development of CIS. Each of the other variations was carried by only one strain, which suggests that most mtDNA variations in CIS were recent mutations. In total, 26 mtDNA sequence variations were identified in the 52 CIS. Four of the variations were located in tRNA- or rRNA-coding regions, and 22 were in protein-coding regions, including 15 nonsynonymous and seven synonymous variations (Table 1).
Figure 2.
The most parsimonious tree for all mouse mtDNA variations found in 50 common inbred strains. The mtDNA sequence of the AKR/J strain (AB042432) was taken as the reference sequence, with the positions of the mtDNA variations indicated. nt9821, an A-repeat variation, and some other heteroplasmic variations were not included.
Table 1.
List of mtDNA mutations in CIS
amtDNA sequence of AKR/J strain (AB042432) was taken as the reference sequence.
bMutations identified in previous studies.
cMutations identified in this study.
Generation of mouse conplastic strains
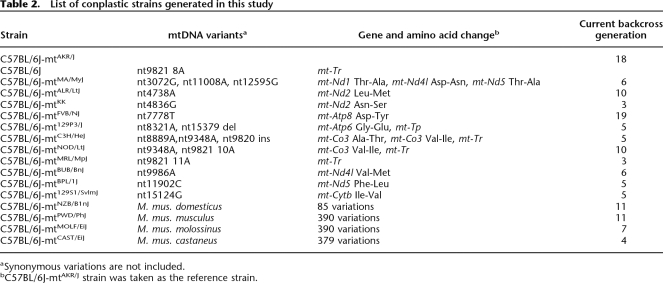
Next, we generated conplastic strains for potentially functional mtDNA mutations by introducing the mtDNA variant from the donor into recipient strains with successive backcrossing for 10 or more generations. The 15 nonsynonymous and four RNA variations were considered to be potentially functional mtDNA mutations. We generated conplastic strains for mtDNA variants from 12 CIS, each of which differs from the reference mtDNA by only one to three polymorphisms. Those 12 mtDNA variants encompass 14 potentially functional mtDNA variations, including 12 nonsynonymous polymorphisms and two polymorphisms in tRNA genes. Owing to the unavailability of mtDNA donor strains, we did not generate conplastic strains for the other five potentially functional mtDNA mutations. We also generated conplastic strains for the mtDNA variants of four M. musculus subspecies, each of which carries hundreds of sequence variations compared with the reference sequence. The strain C57BL/6J was used as the recipient strain for all conplastic strains. Altogether, 16 conplastic strains on the C57BL/6J genetic background were generated or are in production (Table 2).
Table 2.
List of conplastic strains generated in this study
aSynonymous variations are not included.
bC57BL/6J-mtAKR/J strain was taken as the reference strain.
Effects of mtDNA variants on EAE
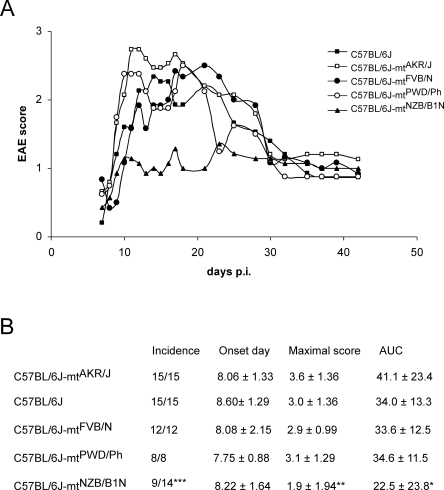
To evaluate the usefulness of this unique genetic resource, we performed some phenotypic analyses on a subset of the available conplastic strains. The C57BL/6J strain is used widely for induction of EAE by immunization with MOG 53-55 peptides. In humans, it has been suggested that mtDNA variations affect susceptibility to MS (Yu et al. 2008). Therefore, we investigated the effect of mtDNA variants on EAE. For the EAE model, we used five conplastic strains: C57BL/6J, C57BL/6J-mtAKR/J, C57BL/6J-mtFVB/N, C57BL/6J-mtPWD/Ph, and C57BL/6J-mtNZB/B1N. Male and female mice from each conplastic strain were used equally in the experiments. No difference in the clinical score between males and females was observed. Thus, to perform the analysis, we combined data from both sexes for each conplastic strain. After EAE induction, the disease began on day 3, reached a peak on days 11–17, then started to subside and reached the chronic phase on day 30 after immunization. All five conplastic strains showed a similar course of disease. However, the C57BL/6J-mtNZB/B1N strain was more resistant to EAE than were the other four conplastic strains (Fig. 3A). Further analysis demonstrated that the resistance was due to a lower disease incidence in C57BL/6J-mtNZB/B1N strain (∼64%) than in the other four strains (100%) (Fig. 3B). Therefore, we concluded that mtDNA variations affect the susceptibility to EAE.
Figure 3.
mtDNA variations affect susceptibility to EAE. (A) Development of EAE in five conplastic strains. The x-axis presents the days post-immunization, and the y-axis indicates the EAE score. (B) Comparison of EAE clinical phenotypes, including disease incidence, onset day (only the disease mice were included), maximal disease score, and area under the curve (AUC). The C57BL/6J-mtAKR/J strain was taken as the reference strain, and the statistical significance was calculated for each strain compared with the C57BL/6J-mtAKR/J strain. Asterisks indicate significant differences: *P < 0.05; **P < 0.01; ***P < 0.001.
Effects of mtDNA variants on behavior
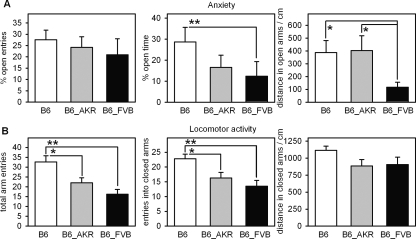
The C57BL/6J mtDNA differs from AKR/J mtDNA only in nucleotide 9821 (nt9821; an A-repeat variation), and the FVB/N mtDNA differs from AKR/J mtDNA only in nt7778 (G/T variation), the latter of which lies in the mitochondrial ATPase subunit-8 gene. In humans, mutations in the ATPase subunit-6 and subunit-8 genes were found in patients with a family history of bipolar disorder with manic and depressive episodes, for which maternal transmission was suspected (Kato et al. 2001). Therefore, C57BL/6J, C57BL/6J-mtAKR/J, and C57BL/6J-mtFVB/N strains were used to investigate the role of single mtDNA polymorphisms in such complex traits as anxiety-like behavior and related neurophysiological markers. We examined anxiety by testing the mice in the elevated plus maze. Interestingly, C57BL/6J-mtFVB/N mice were significantly more anxious than C57BL/6J-mtAKR/J and C57BL/6J mice (Fig. 4A). They spent less time in the open arms of the maze than C57BL/6J mice and walked shorter distances in open arms than C57BL/6J-mtAKR/J and C57BL/6J mice.
Figure 4.
Behavior of C57BL/6J (B6), C57BL/6J-mtAKR/J (B6_AKR), and C57BL/6J-mtFVB/N (B6_FVB) mice on the elevated plus maze. (A) Anxiety was measured by the percentage of entries into open arms, the percentage of time spent in open arms, and the distance walked in open arms. (B) Locomotor activity was recorded as total entries into open and closed arms, the number of times mice entered closed arms, and the distance walked in closed arms. Data represent means and SEM of nine to 12 mice. Asterisks indicate significant differences between strains; : *P < 0.05; **P < 0.01; Mann-Whitney test.
To test whether this effect was due to overall reduced locomotor activity, we counted total arm entries, entries into closed arms, and distances walked in closed arms. C57BL/6J-mtFVB/N showed less total arm entries and entries into closed arms but no difference in distances walked in closed arms (Fig. 4B). The C57BL/6J-mtFVB/N mice did not leave closed arms as often as the other strains, not even via the central platform to the opposite closed arms (data not shown), and thus rarely re-entered closed arms. These differences cannot be explained by reduced mobility of the C57BL/6J-mtFVB/N mice because the distance walked in closed arms was not affected by the mtDNA mutation.
To determine whether behavioral differences were reflected by stress hormones or brain activity, serum corticosterone and brain neurotransmitter levels were analyzed. There were no differences between mouse strains, which indicated that the behavioral differences were based on different pathways (Supplemental Tables 1, 2). Taken together, these results indicate that single mtDNA variations can affect anxiety-related behavior.
Discussion
In this study, we generated a panel of conplastic strains on the C57BL/6J genetic background. Compared with the control strain, the 16 new strains carried either few or numerous mtDNA variations. We showed that the mtDNA variations could affect susceptibility to EAE. This observation is consistent with those in humans, in which mtDNA variations are associated with susceptibility to MS (Yu et al. 2008). Our results provide confirmatory evidence for the role of mitochondria in MS. Furthermore, we provide a mouse model for investigating the mechanism of the mitochondrial contribution to MS.
Evidence exists for the effects of mtDNA variations on central nervous system functions. mtDNA deletion affects the regulation of diurnal rhythms in humans and mice and the estrus cycle in mice. A single study has searched for associations between mtDNA variations and cognitive functions in humans (Skuder et al. 1995). A mouse study associated mtDNA variations with differences in cognitive functions and exploratory behavior but not with anxiety-like behavior (Roubertoux et al. 2003). In the present study, we show that a single mtDNA variation can affect anxiety-like behavior. This discrepancy with the prior study can be explained by the fact that Roubertoux and colleagues used different mouse strains than those employed here. In particular, their mice did not have nonsynonymous variations in the genes mt-Atp6 and mt-tp8, which are the genes that have been associated with bipolar disorder in humans (Kato et al. 2001). The C57BL/6J-mtFVB/N mice carry a mutation in the mt-Atp8 gene that could be responsible for the increased anxiety-like behavior observed in these mice. Thus, our mitochondrial substitution strains represent a promising resource to study anxiety as an aspect of bipolar disorder.
In humans, a growing body of evidence suggests that mtDNA polymorphisms are associated with complex diseases or traits such as diabetes, Alzheimer’s disease, Parkinson’s disease, MS, cancer, aging, and bipolar disorder (De Benedictis et al. 1999; McMahon et al. 2000; Kato et al. 2001; Poulton et al. 2002; van der Walt et al. 2003, 2004; Canter et al. 2005; Mohlke et al. 2005; Kalman 2006; Kazuno et al. 2006). However, dissection of the role of individual mt gene variants has been hampered by the lack of appropriate experimental models. The conplastic strains generated in this study provide at least a partial solution to that difficulty because the C57BL/6J strain is the most widely used mouse strain and has been used as an animal model for many diseases and traits. The conplastic strains also could be used to investigate interactions between mtDNA and nuclear genomes with available transgenes that carry known disease-linked mutations. Mitochondrial–nuclear DNA epistasis is a common phenomenon that profoundly affects complex traits (Johnson et al. 2001; Roubertoux et al. 2003; Del Giudice et al. 2005; Capri et al. 2006; Gusdon et al. 2007).
In conclusion, we present a novel public genetic resource for investigating mtDNA variations. Using this resource, we demonstrated that mtDNA variations can affect both susceptibility to EAE and behavior. Therefore, these conplastic strains will be useful for investigating the role of mtDNA variations in mammals and will facilitate exploration of the biochemical and physiological roles of mitochondria in complex traits.
Methods
Mice
All inbred mouse strains used in this study were obtained from the Jackson Laboratory. To generate the conplastic strains, we crossed mtDNA donor strain females with male C57BL/6J and then backcrossed the female offspring to the male C57BL/6J. In each generation, four female mice were selected randomly and backcrossed to two C57BL/6J males (one male with two females). Such backcrosses were performed for 10 subsequent generations, and the offspring after the tenth generation were regarded as conplastic strains that carried the nuclear genome from the recipient strain and the mitochondrial genome from the donor strain. To check the purity of the nuclear genome of conplastic strains, we performed SNP analysis with Illumina Mouse SNP medium-density panels, which contain 1449 SNPs that cover the entire mouse genome. Animals were housed at the animal facility at the University of Rostock, and all procedures were pre-approved by the local Animal Care Committee.
DNA and sequencing
mtDNA sequences of 29 inbred mouse strains were available from http://www.ncbi.nlm.nih.gov or from the literature (Loveland et al. 1990; Johnson et al. 2001; Bayona-Bafaluy et al. 2003; Mathews et al. 2005; Goios et al. 2007). We sequenced mtDNA from an additional 27 inbred mouse strains. DNA samples from all strains in this study were obtained from the Jackson Laboratory, with the exception of DNA from the STR/ort strain, which was prepared from liver with the QIAamp DNA Mini Kit (QIAGEN). We amplified 14 overlapping fragments of ∼1.3–2.5 kb covering the entire mtDNA molecule (Supplemental Table 3). PCR reactions contained 1.5 mM MgCl2, 0.25 mM dNTP, 50 ng of template DNA, 1 μM each primer, and 1 unit of Gold AmpliTaq DNA polymerase (PE Applied Biosystems) in a final volume of 50 μL. Amplification conditions were as follows: 95°C for 5 min; two cycles of 94°C for 30 sec, 60°C for 1 min, and 72°C for 1.5 min; 35 cycles of 94°C for 30 sec, 55°C for 1 min, and 72°C for 2 min; and a final extension at 72°C for 7 min. PCR was conducted with the GeneAmp PCR System 9700 cycler (Applied Biosystems, Inc.). PCR products were purified with the QIAquick PCR Purification Kit (QIAGEN) according to the manufacturer’s protocol, and the purified PCR products were directly sequenced at the QIAGEN genome service center. Sequences were compared using C57BL/6J as the reference sequence. We confirmed all mtDNA variations identified in CIS by sequencing a second PCR product.
Phylogenetic analysis
Phylogenetic analyses were performed with ClustalW software. We constructed a phylogenetic tree based on the mtDNA sequences of 58 inbred strains, and the bootstrap values (percentage of 1000 total bootstrap replicates) were calculated. For the WIS, the taxonomic classifications were retrieved from the Jackson Laboratory or literature as follows: Mus musculus musculus, PWD/PhJ; Mus musculus domesticus, WSB/EiJ; Mus musculus molossinus, MOLF/EiJ; and Mus musculus castaneus, CAST/EiJ.
Induction and evaluation of EAE
To induce EAE in C57BL/6 mice, 16-wk-old mice were immunized at the base of the tail with 120 μg of rat myelin-oligodendrocyte glycoprotein (MOG) 35-55 peptide dissolved in 50 μL of PBS and mixed with an equal volume (50 μL) of CFA (IFA with 4 mg/mL Mycobacterium tuberculosis). Two hundred nanograms of pertussis toxin dissolved in 100 μL of PBS was injected intraperitoneally immediately after immunization and 48 h later. The clinical scoring of EAE commenced at day 7 after immunization, and animals were monitored for signs of EAE every day during the acute phase and three times per week during the chronic phase. The EAE score was determined as follows: 0, no detectable sign of EAE; 1, tail weakness; 2, tail paralysis; 3, paralysis in one hind leg; 4, paralysis in two hind legs but can move; 5, paralysis in two hind legs and cannot move; and 6, moribund.
Behavioral test
The elevated plus maze tests the conflict between the tendency to explore a novel environment and the aversive properties of brightly lit, open, and elevated areas. The elevated plus maze consists of four arms, each 30 cm in length and 5 cm in width. Two opposing arms are enclosed in opaque, 15-cm-high side and end walls. The structure is elevated by 60 cm. Six-week-old male mice of the strains C57BL/6J, C57BL/6J-mtAKR/J, or C57BL/6J-mtFVB/N were either received from Harlan-Winkelmann (Borchen, Germany) at 3 wk of age (C57BL/6J) or reared as siblings (C57BL/6J-mtAKR/J or C57BL/6J-mtFVB/N) in our facility. We tested anxiety on the basis of the following parameters: (1) percentage of entries into open arms, (2) percentage of time spent in open arms, and (3) distance traveled in open arms. Locomotor activity was determined as (1) total entries into open and closed arms, (2) entries into closed arms, and (3) distance traveled in closed arms. Mice were put into the central position of the maze and left undisturbed for 10 min. Their behavior was recorded by a videocamera and later analyzed by a blinded observer. For all mice, testing was performed between 10 a.m. and 12 p.m. to minimize the influence of the time of day.
Corticosterone and neurotransmitter analyses
Because the elevated plus maze might induce stress in the tested mice, corticosterone and neurotransmitter levels were analyzed in age-matched male mice that were not tested for their behavior. After cervical dislocation, mice were rapidly decapitated, and their trunk blood was collected. Brains were isolated, snap-frozen in liquid nitrogen, and stored at −80°C until neurotransmitter analysis. For all mice, sampling was performed between 9:30 and 9:45 a.m. to minimize the influence of the time of day. Blood was allowed to clot for 2 h and then centrifuged at 2000g for 10 min. Sera were stored frozen at −20°C until corticosterone analysis.
Serum corticosterone concentrations were measured in duplicate with a commercially available double antibody rat corticosterone 125I-RIA kit (DRG Diagnostics) according to the manufacturer’s instructions. Cross-reactivities of the antibody to potential competing plasma steroids were <0.5%. The test sensitivity was 7.2 ng/mL, and intra- and interassay coefficients of variation (CV) were 6.9% and 8.1%, respectively.
Whole brain concentrations of noradrenalin (NA), its metabolite 3-methoxy-4-hydroxyphenylglycol (MHPG), dopamine (DA), its metabolites 3,4-dihydroxyphenylacetic acid (DOPAC) and homovanillic acid (HVA), 5-hydroxytryptamine (5-HT), and its metabolite 5-hydroxyindole-3-acetic acid (5-HIAA) were determined in duplicate by high-performance liquid chromatography (HPLC) with electrochemical detection. Brains were weighed and homogenized on ice for 5 min with a hand homogenizer in 4 mL of 0.2 M perchloric acid, followed by centrifugation at 45,000g for 10 min at 4°C. After collection of the supernatants, the procedure was repeated. Pooled supernatants of the repeated extractions were again centrifuged at 45,000g for 10 min at 4°C. Twenty microliters of each sample was then injected directly in the HPLC system equipped with a 125 × 4-mm reversed-phase column packed with Prontosil C18 AQ (Bischoff Analysentechnik). As the mobile phase, 58 mM sodium hydrogen phosphate buffer containing 1.2 mM octansulfonic acid, 0.3 mM EDTA, 0.2 mM potassium chloride, and 6% methanol (pH 3.8) was used at a flow rate of 1.2 mL/min. Electrochemical detection was achieved by an ISAAC cell with a glassy carbon working electrode set at a potential of 600 mV (Shimadzu). Dihydroxybenzylamine (DHBA) was used as an internal standard for quantification, and tissue contents of the above neurotransmitters and their metabolites were expressed as nanograms per gram of wet tissue weight. As indices of NA, DA, and 5-HT turnover, the MHPG/NA, DOPAC/DA, HVA/DA, and 5-HIAA/5-HT ratios were calculated.
Statistical analysis
Quantitative traits of mouse strains were compared with the nonparametric Mann-Whitney test. Qualitative traits, for example, the incidence of EAE, were compared with the Fisher’s exact test. In all statistical tests, P < 0.05 was considered to be statistically significant.
Acknowledgments
We thank Rica Waterstadt, Ilona Klamfuss, and Claudia Voigt for assistance in DNA sequencing, animal care, and cell culture. This study was supported by the Hertie foundation (1.01.1/07/001) and the EU-FP6 (MRTN-CT-2004-005693 EURO-RA).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.078865.108.
References
- Anderson S., Bankier A.T., Barrell B.G., de Bruijn M.H., Coulson A.R., Drouin J., Eperon I.C., Nierlich D.P., Roe B.A., Sanger F., et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Bayona-Bafaluy M.P., Acin-Perez R., Mullikin J.C., Park J.S., Moreno-Loshuertos R., Hu P., Perez-Martos A., Fernandez-Silva P., Bai Y., Enriquez J.A. Revisiting the mouse mitochondrial DNA sequence. Nucleic Acids Res. 2003;31:5349–5355. doi: 10.1093/nar/gkg739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canter J.A., Kallianpur A.R., Parl F.F., Millikan R.C. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005;65:8028–8033. doi: 10.1158/0008-5472.CAN-05-1428. [DOI] [PubMed] [Google Scholar]
- Capri M., Salvioli S., Sevini F., Valensin S., Celani L., Monti D., Pawelec G., De Benedictis G., Gonos E.S., Franceschi C. The genetics of human longevity. Ann. N. Y. Acad. Sci. 2006;1067:252–263. doi: 10.1196/annals.1354.033. [DOI] [PubMed] [Google Scholar]
- De Benedictis G., Rose G., Carrieri G., De Luca M., Falcone E., Passarino G., Bonafe M., Monti D., Baggio G., Bertolini S., et al. Mitochondrial DNA inherited variants are associated with successful aging and longevity in humans. FASEB J. 1999;13:1532–1536. doi: 10.1096/fasebj.13.12.1532. [DOI] [PubMed] [Google Scholar]
- Del Giudice L., Massardo D.R., Pontieri P., Wolf K. Interaction between yeast mitochondrial and nuclear genomes: Null alleles of RTG genes affect resistance to the alkaloid lycorine in rho0 petites of Saccharomyces cerevisiae. Gene. 2005;354:9–14. doi: 10.1016/j.gene.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Ferris S.D., Sage R.D., Wilson A.C. Evidence from mtDNA sequences that common laboratory strains of inbred mice are descended from a single female. Nature. 1982;295:163–165. doi: 10.1038/295163a0. [DOI] [PubMed] [Google Scholar]
- Goios A., Pereira L., Bogue M., Macaulay V., Amorim A. mtDNA phylogeny and evolution of laboratory mouse strains. Genome Res. 2007;17:293–298. doi: 10.1101/gr.5941007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusdon A.M., Votyakova T.V., Reynolds I.J., Mathews C.E. Nuclear and mitochondrial interaction involving mt-Nd2 leads to increased mitochondrial reactive oxygen species production. J. Biol. Chem. 2007;282:5171–5179. doi: 10.1074/jbc.M609367200. [DOI] [PubMed] [Google Scholar]
- Johnson K.R., Zheng Q.Y., Bykhovskaya Y., Spirina O., Fischel-Ghodsian N. A nuclear-mitochondrial DNA interaction affecting hearing impairment in mice. Nat. Genet. 2001;27:191–194. doi: 10.1038/84831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman B. Role of mitochondria in multiple sclerosis. Curr. Neurol. Neurosci. Rep. 2006;6:244–252. doi: 10.1007/s11910-006-0012-0. [DOI] [PubMed] [Google Scholar]
- Kalman B., Alder H. Is the mitochondrial DNA involved in determining susceptibility to multiple sclerosis? Acta Neurol. Scand. 1998;98:232–237. doi: 10.1111/j.1600-0404.1998.tb07301.x. [DOI] [PubMed] [Google Scholar]
- Kato T., Kunugi H., Nanko S., Kato N. Mitochondrial DNA polymorphisms in bipolar disorder. J. Affect. Disord. 2001;62:151–164. doi: 10.1016/s0165-0327(99)00173-1. [DOI] [PubMed] [Google Scholar]
- Kazuno A.A., Munakata K., Nagai T., Shimozono S., Tanaka M., Yoneda M., Kato N., Miyawaki A., Kato T. Identification of mitochondrial DNA polymorphisms that alter mitochondrial matrix pH and intracellular calcium dynamics. PLoS Genet. 2006;2:e128. doi: 10.1371/journal.pgen.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loveland B., Wang C.R., Yonekawa H., Hermel E., Lindahl K.F. Maternally transmitted histocompatibility antigen of mice: A hydrophobic peptide of a mitochondrially encoded protein. Cell. 1990;60:971–980. doi: 10.1016/0092-8674(90)90345-f. [DOI] [PubMed] [Google Scholar]
- Mathews C.E., Leiter E.H., Spirina O., Bykhovskaya Y., Gusdon A.M., Ringquist S., Fischel-Ghodsian N. mt-Nd2 allele of the ALR/Lt mouse confers resistance against both chemically induced and autoimmune diabetes. Diabetologia. 2005;48:261–267. doi: 10.1007/s00125-004-1644-8. [DOI] [PubMed] [Google Scholar]
- McMahon F.J., Chen Y.S., Patel S., Kokoszka J., Brown M.D., Torroni A., DePaulo J.R., Wallace D.C.2000Mitochondrial DNA sequence diversity in bipolar affective disorder Am. J. Psychiatry 1571058–1064. .Comment . 2001158 1169–1170. [DOI] [PubMed] [Google Scholar]
- Mohlke K.L., Jackson A.U., Scott L.J., Peck E.C., Suh Y.D., Chines P.S., Watanabe R.M., Buchanan T.A., Conneely K.N., Erdos M.R., et al. Mitochondrial polymorphisms and susceptibility to type 2 diabetes-related traits in Finns. Hum. Genet. 2005;118:245–254. doi: 10.1007/s00439-005-0046-4. [DOI] [PubMed] [Google Scholar]
- Moreno-Loshuertos R., Acin-Perez R., Fernandez-Silva P., Movilla N., Perez-Martos A., Rodriguez de Cordoba S., Gallardo M.E., Enriquez J.A. Differences in reactive oxygen species production explain the phenotypes associated with common mouse mitochondrial DNA variants. Nat. Genet. 2006;38:1261–1268. doi: 10.1038/ng1897. [DOI] [PubMed] [Google Scholar]
- Pinkert C.A., Trounce I.A. Production of transmitochondrial mice. Methods. 2002;26:348–357. doi: 10.1016/S1046-2023(02)00041-5. [DOI] [PubMed] [Google Scholar]
- Poulton J., Luan J., Macaulay V., Hennings S., Mitchell J., Wareham N.J. Type 2 diabetes is associated with a common mitochondrial variant: Evidence from a population-based case-control study. Hum. Mol. Genet. 2002;11:1581–1583. doi: 10.1093/hmg/11.13.1581. [DOI] [PubMed] [Google Scholar]
- Roubertoux P.L., Sluyter F., Carlier M., Marcet B., Maarouf-Veray F., Cherif C., Marican C., Arrechi P., Godin F., Jamon M., et al. Mitochondrial DNA modifies cognition in interaction with the nuclear genome and age in mice. Nat. Genet. 2003;35:65–69. doi: 10.1038/ng1230. [DOI] [PubMed] [Google Scholar]
- Shoffner J.M., Brown M.D., Torroni A., Lott M.T., Cabell M.F., Mirra S.S., Beal M.F., Yang C.C., Gearing M., Salvo R., et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- Skuder P., Plomin R., McClearn G.E., Smith D.L., Vignetti S., Chorney M.J., Chorney K., Kasarda S., Thompson L.A., Detterman D.K., et al A polymorphism in mitochrondrial DNA associated with IQ? Intelligence. 1995;21:1–11. [Google Scholar]
- Taylor R.W., Turnbull D.M. Mitochondrial DNA mutations in human disease. Nat. Rev. Genet. 2005;6:389–402. doi: 10.1038/nrg1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt J.M., Nicodemus K.K., Martin E.R., Scott W.K., Nance M.A., Watts R.L., Hubble J.P., Haines J.L., Koller W.C., Lyons K., et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am. J. Hum. Genet. 2003;72:804–811. doi: 10.1086/373937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Walt J.M., Dementieva Y.A., Martin E.R., Scott W.K., Nicodemus K.K., Kroner C.C., Welsh-Bohmer K.A., Saunders A.M., Roses A.D., Small G.W., et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci. Lett. 2004;365:28–32. doi: 10.1016/j.neulet.2004.04.051. [DOI] [PubMed] [Google Scholar]
- Wallace D.C., Singh G., Lott M.T., Hodge J.A., Schurr T.G., Lezza A.M., Elsas L.J., Nikoskelainen E.K. Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science. 1988;242:1427–1430. doi: 10.1126/science.3201231. [DOI] [PubMed] [Google Scholar]
- Yu X., Koczan D., Sulonen A., Akkad D., Kroner A., Comabella M., Costa G., Corongiu D., Goertsches R., Camina-Tato M., et al. mtDNA nt13708A variant increases the risk of multiple sclerosis. PLoS One. 2008;3:e1530. doi: 10.1371/journal.pone.0001530. [DOI] [PMC free article] [PubMed] [Google Scholar]