Abstract
To identify genes that are regulated by cis-acting functional elements in acute lymphoblastic leukemia (ALL) we determined the allele-specific expression (ASE) levels of 2529 genes by genotyping a genome-wide panel of single nucleotide polymorphisms in RNA and DNA from bone marrow and blood samples of 197 children with ALL. Using a reproducible, quantitative genotyping method and stringent criteria for scoring ASE, we found that 16% of the analyzed genes display ASE in multiple ALL cell samples. For most of the genes, the level of ASE varied largely between the samples, from 1.4-fold overexpression of one allele to apparent monoallelic expression. For genes exhibiting ASE, 55% displayed bidirectional ASE in which overexpression of either of the two SNP alleles occurred. For bidirectional ASE we also observed overall higher levels of ASE and correlation with the methylation level of these sites. Our results demonstrate that CpG site methylation is one of the factors that regulates gene expression in ALL cells.
Acute lymphoblastic leukemia (ALL) is a malignant disease originating from disturbed development of blood progenitor cells that are committed to differentiate in the B-cell or T-cell pathway. ALL can be subdivided into cytogenetically distinct subtypes including B-cell progenitor leukemias with chromosomal translocations t(12;21), t(1;19), and t(9;22), rearrangements on chromosome 11q23, and hyperdiploid and hypodiploid karyotypes (Greaves and Wiemels 2003). These chromosomal aberrations are considered to be important in the initiation of leukemia, but most likely other genetic factors are also required to induce acute leukemia (Pui et al. 2008). Although additional mutations have been identified in some ALL cases (Weng et al. 2004; Mullighan et al. 2007), the complete spectrum of specific genes and their functional variants that lead to ALL remain to be elucidated. The challenge now is to identify and understand how genetic variation at higher resolution than the chromosomal aberrations affects the functions of molecular pathways that alter proliferation, differentiation, and survival of lymphocyte progenitor cells, leading to their conversion into leukemia.
During the past decade a large number of genome-wide gene expression studies using microarray-based methods have identified genes that allow classification of ALL subtypes or might be of predictive value for the outcome of treatment of ALL patients (Willenbrock et al. 2004; Cheok and Evans 2006; Flotho et al. 2007). However, they have not been able to identify the specific functional elements that regulate the expression of individual genes in ALL, which is important for the understanding of the inherited and epigenetic changes that result in ALL.
The Encyclopedia of DNA Elements (ENCODE) project has documented that the expression of protein-coding genes is regulated by both inherited genetic and epigenetic mechanisms (International Human Genome Sequencing Consortium 2004; The ENCODE Project Consortium 2007). Recent genome-wide association studies using single nucleotide polymorphism (SNP) markers predict that the expression of a large proportion of human genes is regulated by cis-acting regulatory SNPs located outside protein-coding regions of genes (Dixon et al. 2007; Goring et al. 2007; Stranger et al. 2007; Emilsson et al. 2008). At the same time, the Human Epigenome Project (HEP) is working toward the identification of DNA methylation that regulates the expression of human genes in multiple tissues on a genome-wide scale (Eckhardt et al. 2006).
Determination of the allele-specific expression (ASE) levels of genes by quantitative genotyping of heterozygous SNPs on the RNA level, using genomic DNA as reference (Pastinen and Hudson 2004) can be used as a guide for identifying cis-acting genetic and epigenetic variation that regulate gene expression. In the ASE approach the relative expression levels of the two alleles of a gene are measured in the same sample, and therefore environmental or trans-acting regulatory factors that might affect the expression levels of the genes are controlled for (Bray et al. 2003; Pastinen et al. 2003; Pastinen et al. 2005; Mahr et al. 2006; Serre et al. 2008). Many studies on ASE have been performed in immortalized lymphoblastoid cell lines from the Center d’Etude du Polymorphisme Humain collection (CEPH) (Pastinen et al. 2003; Pastinen et al. 2005; Gimelbrant et al. 2007; Pollard et al. 2008; Serre et al. 2008), and ASE has also been detected in cultured cancer cell lines (Milani et al. 2007; Serre et al. 2008).
Methylation of CpG dinucleotides in the proximity of the transcription start site frequently silences gene expression. It is also recognized that hypermethylation of tumor suppressor genes, as well as hypomethylation of oncogenes may lead to various forms of cancer (Jones and Baylin 2007). Aberrant methylation of CpG sites in the promoter regions of genes has been identified in leukemic cell lines or primary ALL cells, and correlated with the expression of individual genes, but such studies have been hampered by a limited representation of the studied genomic regions and/or by a small number of cell samples included in the analysis (Taylor et al. 2007; Figueroa et al. 2008; Kuang et al. 2008). Although smaller studies have clearly shown that DNA methylation in promoter regions affects the expression of individual genes (Eckhardt et al. 2006; Kerkel et al. 2008), comprehensive studies of DNA methylation and its effect on gene expression are lagging behind reports on cis-acting regulatory SNPs.
To identify genes that are regulated by cis-acting functional elements in ALL we performed a genome-wide survey of ASE of 8000 genes in 197 bone marrow and peripheral blood samples from children diagnosed with ALL in the five Nordic countries. We also determined the methylation levels of 1306 CpG sites located in the promoter regions of 400 genes that displayed ASE and correlated the methylation levels at the CpG sites with ASE of these genes.
Results
Detection of ASE in leukemic cells
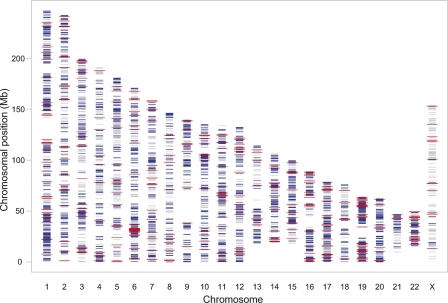
To identify genes that display differential expressions of the two alleles in leukemic cells, we screened 8000 genes distributed over all human autosomes and the X chromosome in bone marrow or peripheral blood cells of 197 children with ALL. The samples were collected at the time of ALL diagnosis. According to microscopic analysis all samples included in the study contained >90% leukemic cells. We used the Infinium I assay and HumanNS-12 Genotyping BeadChip to genotype 13,917 SNPs in RNA and DNA extracted from the cells to detect ASE. The NS-12 BeadChips assay genes with a good coverage of the human genome, with 80% of the SNPs located in annotated exons or untranslated regions of mRNA. To be informative for the detection of ASE, a SNP has to be heterozygous in DNA, and expressed at a detectable level in RNA. Of the SNPs included on the BeadChip, 3531 SNPs (32%) distributed over 2529 genes were informative in the 197 samples genotyped in our study (Fig. 1).
Figure 1.
Genome-wide distribution of 8000 genes included on the NS-12 BeadChips (gray), 2529 genes, which contained heterozygous SNPs and were expressed in the ALL cell samples included in the study (blue), and 400 genes for which we detected allele-specific gene expression (red). The chromosome numbers are given on the x-axis and the chromosomal positions (Mb) on the y-axis.
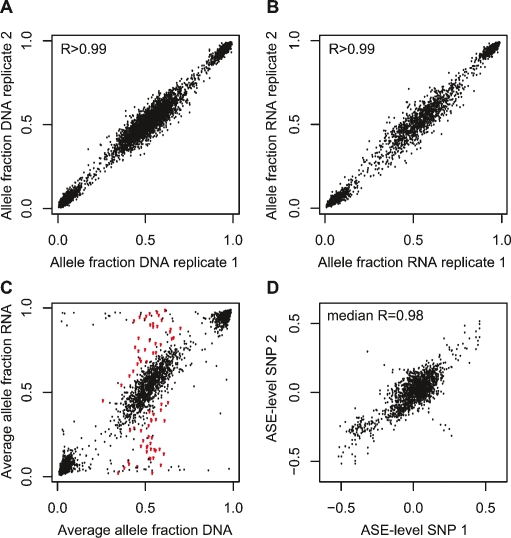
To detect ASE we measured the average fluorescence signals from the two SNP alleles (A1 and A2) in triplicate RNA and DNA samples and determined the allele fraction [A1/(A1 + A2)] for each SNP by dividing the mean fluorescence signal from one allele (A1) by the sum of the fluorescence signals of both alleles (A1 + A2). The Infinium I assay performed robustly in the genotyping, as evidenced by an excellent correlation of >0.99 between the allele fraction determined in replicate RNA and DNA samples from each individual (Fig. 2A,B). We then compared the allele fraction [A1/(A1 + A2)] in RNA with the corresponding allele fraction in genomic DNA from the same sample, using a stringent significance threshold of P < 0.001 for the difference between the allele fractions for a SNP in RNA and DNA for scoring ASE in each individual sample (Fig. 2C). We also required that ASE was observed in at least eight samples. (See Supplemental Fig. 1 for examples of genotype scatter plots for three SNPs with different patterns of ASE.) To obtain a quantitative measure for the differential allelic expression we subtracted the allele fractions [A1/(A1 + A2)] determined in DNA from that in RNA and refer to this difference as the ASE level. The high median correlation (0.98) between the ASE level determined using SNPs located in the same exon of a gene provided additional evidence for the robust performance of quantitative genotyping by the Infinium I assay (Fig. 2D; Supplemental Fig. 2). We also validated the ASE levels determined by the NS-12 BeadChips by quantitative Sanger sequencing of nine genes, and observed a high correlation (0.86) between these two independent methods (Supplemental Fig. 3).
Figure 2.
Genotyping by the NS-12 BeadChips to detect allele-specific gene expression. (A) Correlation between the allele fractions determined in replicate DNA samples for 3531 expressed SNPs in one ALL sample. The median correlation between the allele fraction obtained in replicate assays in all 197 samples was 0.9969 (range 0.9934–0.9986). (B) Correlation between the allele fractions determined by genotyping the same 3531 SNPs in replicate RNA samples from the same sample as in A. The median correlation between the allele fraction obtained in replicate assays in all 197 samples was 0.9956 (range 0.9779–0.9984). (C) Average allele fractions from triplicate assays of 3531 SNPs in RNA and DNA from the same sample as above. The red dots represent the allele fraction in RNA for SNPs that display allele-specific expression, i.e., SNPs that are heterozygous in DNA and show a significant difference (P < 0.001) in the mean allele fraction between RNA and DNA from the same cell sample as in A and B. (D) Pairwise correlation between allele-specific expression (ASE) levels determined using pairs of informative SNPs located in the same exon of 16 different genes. The ASE level for each SNP is given as the average difference in allele fraction between triplicate DNA and triplicate RNA samples. Shown are the results from 16 genes, of which 11 genes had two SNPs in the same exon, two genes had three SNPs in the same exon, and three genes had more than three SNPs in the same exon and were heterozygous in 9–112 samples, totaling 1658 observations. The pairwise correlation between ASE-levels determined with these SNPs ranged from 0.68 to 0.99 (median 0.98), with the exception of three SNPs in the FPR1 gene, between which there was an obvious inverse correlation between the ASE levels in a subset of the samples. As can be seen in Supplemental Figure 2 these SNPs are located outside the main linkage disequilibrium (LD) block of the FPR1 gene.
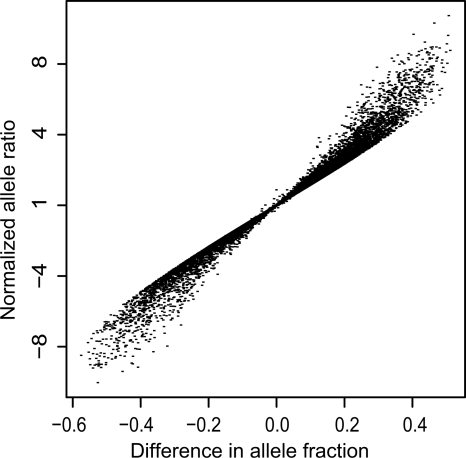
For comparison, we determined the differential allelic expression by calculating the ratio between average fluorescence signal intensities measured for the two alleles of a SNP (A1/A2), and normalized this allele ratio in RNA by dividing it with the allele ratio in DNA for the same SNP in the same sample. Figure 3 shows the correlation between ASE determined according to the difference in allele fraction between RNA and DNA and according to the normalized allele ratio in RNA for all informative SNPs. Particularly for SNPs with a large overexpression of one of the alleles, the allele fraction showed lower variability between replicates and was less affected by differences in expression levels between the genes than the allele ratio. The ASE level based on the allele fraction provided better resolution for scoring ASE than the normalized fold-expression level of one allele and was therefore applied as a measure for differential allelic expression throughout the study.
Figure 3.
Correlation between ASE determined using allele fractions and normalized allele ratios. The ASE levels determined according to the difference in allele fraction [A1/(A1 + A2)] between SNPs in RNA and DNA are shown on the x-axis. The fold overexpression of one allele according to the allele ratios (A1/A2) for SNPs in RNA normalized against the allele ratio for SNPs in DNA are shown on the y-axis. Mean values from triplicate assays of 3531 informative SNPs in 197 ALL samples are shown (∼700,000 data points).
We detected ASE for 470 SNPs located in 400 genes, which corresponds to 16% of the genes with informative SNPs on the NS-12 BeadChips. The genes that displayed ASE contained 1–12 SNPs per gene (average 1.4). As can be seen in Figure 1, the genes that are subject to ASE are evenly distributed across the autosomal chromosomes, indicating that ASE is a common phenomenon also in primary ALL cells. The higher density of SNPs in the MHC region on chromosome 6 and on chromosome 19 is reflected by a larger number of genes with ASE in these regions. According to KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis, genes involved in cell communication, extracellular matrix receptor interactions, β-alanine metabolism, antigen processing and presentation, cell adhesion and type 1 diabetes mellitus were significantly overrepresented among the genes for which we detected ASE (Supplemental Table 1).
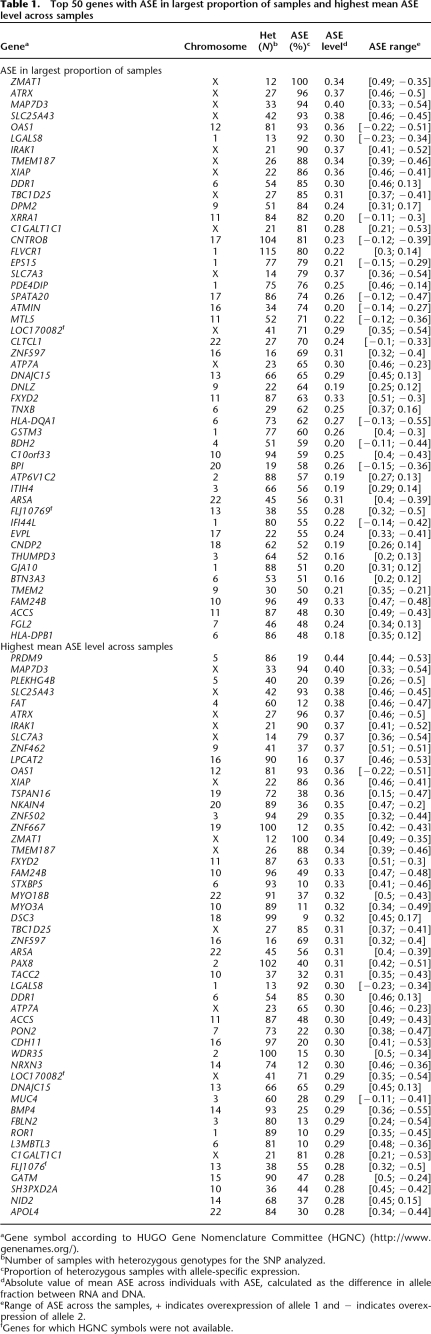
Table 1 shows a list of the 50 genes for which we observed ASE in the largest proportion of the samples with informative SNPs and of the 50 genes that exhibited the highest ASE levels, and Supplemental Table 2 provides the complete list of the 400 genes for which we detected ASE. The ASE levels calculated according to allele fractions in RNA and DNA varied among the genes and among the samples from 0.09 to 0.58. These ASE values correspond to 1.4- to >14-fold overexpression of one of the alleles (Fig. 3). For as many as 222 of the genes we observed monoallelic expression according to an allele fraction in RNA that was indistinguishable from that in a DNA sample with a homozygous genotype in at least one sample. For 67 of the genes we observed monoallelic expression in five or more samples. Expectedly, all heterozygous SNPs with ASE on the X chromosome indicate monoallelic expression.
Table 1.
Top 50 genes with ASE in largest proportion of samples and highest mean ASE level across samples
Interestingly, the same allele was overexpressed in all samples for about 45% of the genes, while for 55% of the genes the overexpressed allele differed between samples. As can be seen in Figure 4A, there is a substantial overrepresentation of genes with large ASE levels among the genes with bidirectional ASE (P = 1.4 × 10−12; Fisher’s exact test). This result suggests that randomly occurring epigenetic alterations rather than cis-acting inherited genetic variants might regulate the expression of genes with bidirectional ASE.
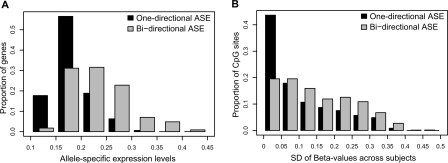
Figure 4.
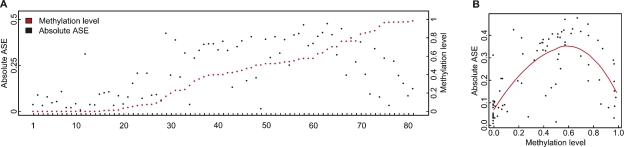
Variation of ASE and CpG site methylation levels in genes with one-directional and bidirectional ASE. (A) Bins of average absolute values for allele-specific expression of all genes (n = 400) are shown on the x-axis, and the proportion of genes in each bin of ASE values are shown on the y-axis for genes with one-directional (black bars) and bidirectional (gray bars) ASE. The graph illustrates significantly larger ASE values in genes with bidirectional ASE than in genes with one-directional ASE (P = 1.4 × 10−12). (B) The variation in CpG site methylation for all CpG sites (n = 1306) is shown on the x-axis as bins of standard deviations (SD) for the methylation levels (beta-values from the GoldenGate assay) across samples for each individual CpG site. The proportion of CpG sites in each bin of SDs shown on the y-axis were obtained by dividing the number of CpG sites in each bin by the total number of CpG sites in genes with one-directional (black bars) and bidirectional (gray bars) ASE, respectively. The graph shows that genes with bidirectional ASE according to data from 267 SNPs display a larger variation in methylation levels than genes with one-directional ASE according to data from 203 SNPs (P = 2.2 × 10−5).
Effect of CpG methylation on allele-specific gene expression
To investigate the correlation between DNA methylation and ASE, we searched for differential methylation of CpG sites in the promoters and first introns of the genes for which we had observed ASE. Two to 10 CpG sites per gene (average 3.7) were included in a panel of 1536 CpG sites for methylation analysis, with a preference for the genes with high ASE levels or apparent monoallelic expression. The methylation levels at these CpG sites were determined by the GoldenGate methylation assay in the same 197 samples from ALL patients that were analyzed for detection of ASE. This assay provides a quantitative measure of the methylation levels (beta-value) for each analyzed CpG site, with beta-values ranging from 0 to 1.0, corresponding to no methylation on either allele to complete methylation of both alleles.
We found that most of the analyzed CpG sites (72%) showed little variability, with consistent low (average beta-value < 0.25) or high methylation levels (average beta-value > 0.75) in all samples. A general observation was that CpG sites in CpG islands, i.e., regions of at least 200 bp in size with increased GC content and at CpG sites located within 800 bp regions upstream of the transcription start site of a gene displayed low levels of methylation (Supplemental Fig. 3). About one-fourth of the analyzed CpG sites located in 96 genes displayed variation in the methylation levels with a standard deviation >0.20 for the beta-values across samples. These differentially methylated CpG sites are most informative for the identification of a quantitative correlation between methylation of CpG sites and allele-specific gene expression. Because we had observed larger ASE levels in genes with bidirectional ASE (Fig. 4A), we first tested for differences in variability of CpG site methylation between genes that exhibit one-directional and genes that exhibit bidirectional ASE. Comparison of the variation in CpG site methylation between these two groups of genes showed that the genes with bidirectional ASE displayed a significantly larger variability in methylation of CpG sites than those with one-directional ASE (P = 2.2 × 10−5; Fisher’s exact test) (Fig. 4B). This finding is consistent with random methylation of the two alleles of a gene, and suggests that methylation has a strong effect on regulation of gene expression.
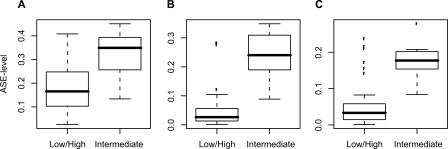
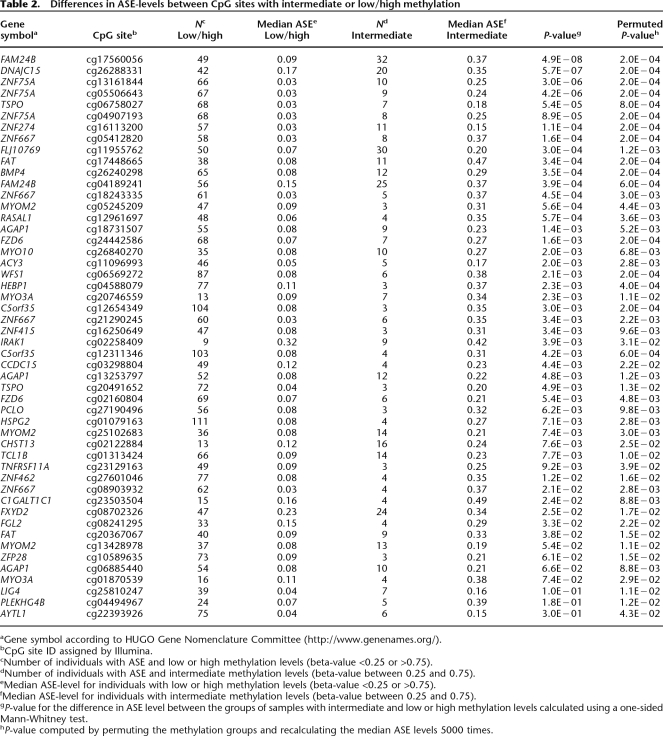
Next, we tested for a correlation between ASE levels and variability in CpG site methylation for 312 genes with differentially methylated CpG sites. We compared the ASE levels between the group of samples containing CpG sites that displayed intermediate (0.25–0.75) methylation levels, where variability in CpG site methylation could cause ASE and the group of samples that contained CpG sites with high (>0.75) or low (<0.25) levels of methylation, for which CpG site methylation is not expected to result in ASE. Figure 5 shows three examples of genes with a significant correlation between variability in CpG site methylation and ASE level. For 50 of the CpG sites located in regulatory regions of 35 genes we observed significantly higher ASE levels for the group of genes with variability in CpG site methylation with permuted P-values <0.05 and median ASE difference >0.1 (Table 2). The permuted P-values for the correlation between variability in CpG site methylation and ASE for all CpG sites are provided in Supplemental Table 4.
Figure 5.
Correlation between ASE and CpG site methylation. Comparison of ASE levels between samples with low or high methylation levels (beta-value <0.25 or >0.75) and samples with intermediate methylation levels (beta-value 0.25–0.75) exemplified by three genes. ASE levels for DNAJC15 in samples with low or high beta-values (n = 42) and intermediate beta-values (n = 20) at the CpG site cg26288331 (unadjusted P = 5.7 × 10−7; permuted P = 2.0 × 10−4) (A), ZNF75A in samples with low or high beta-values (n = 67) and intermediate beta-values (n = 9) at the CpG site cg05506643 (unadjusted P = 4.2 × 10−6; permuted P = 2.0 × 10−4) (B), and TSPO in samples with low or high beta-values (n = 68) and intermediate beta-values (n = 7) at the CpG site cg06758027 (unadjusted P = 5.4 × 10−5; permuted P = 8.0 × 10−4) (C).
Table 2.
Differences in ASE-levels between CpG sites with intermediate or low/high methylation
aGene symbol according to HUGO Gene Nomenclature Committee (http://www.genenames.org/).
bCpG site ID assigned by Illumina.
cNumber of individuals with ASE and low or high methylation levels (beta-value <0.25 or >0.75).
dNumber of individuals with ASE and intermediate methylation levels (beta-value between 0.25 and 0.75).
eMedian ASE-level for individuals with low or high methylation levels (beta-value <0.25 or >0.75).
fMedian ASE-level for individuals with intermediate methylation levels (beta-value between 0.25 and 0.75).
gP-value for the difference in ASE level between the groups of samples with intermediate and low or high methylation levels calculated using a one-sided Mann-Whitney test.
hP-value computed by permuting the methylation groups and recalculating the median ASE levels 5000 times.
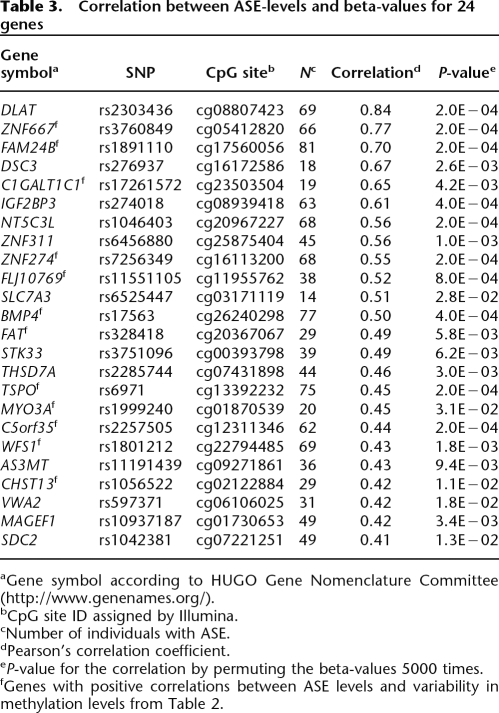
For 282 genes with differentially methylated CpG sites, we tested for a quantitative correlation between the ASE level and the beta-value for CpG site methylation in individual samples with informative SNPs. When data for more than one SNP and/or CpG site were available for a gene, we used the most variable ASE- and beta-values to minimize the number of tests performed. This analysis identified 24 genes with a suggestive quantitative correlation (Pearson’s R > 0.4 and P < 0.05) between the ASE level and CpG site methylation (Table 3). Twelve out of the 35 genes with a significant difference in ASE-levels between the groups of samples with CpG sites that displayed variability in methylation levels and high or low methylation levels shown in Table 2 also showed a correlation between ASE-level and CpG site methylation in individual samples. A clear correlation (Pearson’s R = 0.7) is exemplified by the data for FAM24B in Figure 6. The methylation levels for FAM24B range from 0 to 1.0 and the ASE-levels range from 0 to 0.5. The shape of the regression curve for the ASE-level as a function of the methylation level, which has a maximum at a beta-value of 0.59, is consistent with increased methylation of one allele until complete methylation. As the methylation levels increase further to above 0.59, the ASE-levels decrease, which is consistent with increased methylation of the other allele of FAM24B. Thus we have demonstrated a correlation between ASE and CpG site methylation using three different approaches.
Table 3.
Correlation between ASE-levels and beta-values for 24 genes
aGene symbol according to HUGO Gene Nomenclature Committee (http://www.genenames.org/).
bCpG site ID assigned by Illumina.
cNumber of individuals with ASE.
dPearson’s correlation coefficient.
eP-value for the correlation by permuting the beta-values 5000 times.
fGenes with positive correlations between ASE levels and variability in methylation levels from Table 2.
Figure 6.
Correlation between ASE levels and CpG site methylation levels for the FAM24B gene in individual samples. (A) The methylation levels of the CpG site cg17560056 (red dots, left y-axis) and the absolute values for the ASE-levels (SNP rs1891110) (black dots, right y-axis) for FAM24B in individual heterozygous samples listed on the horizontal axis (n = 81). (B) The ASE-levels (y-axis) plotted against the methylation levels (x-axis) for FAM24B (black dots). The regression curve (R = 0.7; permuted P = 2.0 × 10−4) fitted to these data points is shown in red.
Discussion
The study presented here is the first systematic survey of ASE in primary cancer cells, and it is, to our knowledge, the largest survey of ASE carried out to date, with respect to number of samples and number of genes included. We determined the ASE-levels of 2529 genes in 197 lymphoblast samples collected from children with newly diagnosed ALL prior to therapy. Because ALL cells are characterized by chromosomal aberrations (Pui et al. 2008), of which those that alter gene copy numbers could cause genotyping errors on the level of DNA and affect gene expression levels, we designed our study to avoid this problem. In the 165 pre-B ALL samples included in our study most of the known chromosomal aberrations have been found to occur in only a minority of the samples (Forestier and Schmiegelow 2006). Based on cytogenetic information of the ALL samples and the chromosomal distribution of our observations of ASE we estimate that less than 10% of our ASE observations originate from a duplicated or amplified chromosome. In our study we made over 8000 observations of ASE, and for each chromosome, the number of ASE observations from chromosomes with a normal copy number was substantially larger than that from a duplicated or amplified chromosome. To avoid possible detection of individual genes with ASE due to rare copy-number alterations in the ALL cells, we scored ASE only for genes where we detected ASE in eight or more samples. Moreover, by requiring statistical significance for the difference between the allele fraction measured in DNA and RNA from each individual sample, we circumvented gene dosage effects caused by the expression of several gene copies in hyperdiploid cells or amplified genomic regions. Owing to this study design, possible confounding effects of chromosomal amplifications or other unknown genomic copy-number variations on the overall ASE data presented in our study would remain minor. Thus we were able to use ASE to identify genes with true cis-acting regulatory elements that affect gene expression in ALL. By applying stringent criteria for scoring ASE, we found that 16% of the genes with informative heterozygous SNPs displayed ASE in our collection of ALL cells.
For genotyping we used the HumanNS-12 BeadChips which are based on the Infinium I assay, which avoids PCR-amplification for sample treatment and uses robust allele-specific primer extension of biotinylated dNTPs for discrimination of the two SNP alleles (Gunderson et al. 2005). Both alleles of each SNP are scored with an identical PCR-free single-color detection procedure, which reduces variation in the fluorescence signals between the two SNP alleles, and hence distortion of the allele ratios depending on the amount of target nucleic acid subjected to genotyping. This is a particularly important advantage for quantitative genotyping of allele-specific RNA transcripts, which are expressed at different levels in the cells. Using the Infinuim I assay we were able to detect and quantify ASE over a wide range, from 1.4-fold to about 14-fold overexpression of one allele in each individual sample. Our quantitative data on ASE contrast the qualitative data on ASE presented in previous studies that have used PCR for sample preparation and microarray-based hybridization with allele-specific oligonucleotide (ASO) probes for genotyping (Pant et al. 2006; Gimelbrant et al. 2007; Bjornsson et al. 2008), although ASE was scored semi-quantitatively as 2-, 4-, and 10-fold overexpression of one allele using the same procedure in a recent study (Pollard et al. 2008). Presumably, amplification biases between the alleles expressed at different levels caused by PCR and experimental noise from the hybridization arrays prohibit quantification of ASE. A recent study used the GoldenGate assay, which like the Infinium I assay, is based on allele-specific primer extension, but employs a two-color PCR-based detection procedure for detection of ASE (Serre et al. 2008). In this study 1.5-fold ASE was detectable in groups of samples, but not in individual samples.
The large number of samples analyzed in our study in combination with accurate and highly reproducible quantitative SNP genotyping using the HumanNS-12 BeadChips, which contain a representative set of human genes, allowed us to detect several interesting features of allele-specific gene expression in the ALL cells. We found that for 55% of the genes, including 12 genes on the X chromosome, the ASE was bidirectional with either of the SNP alleles as the overexpressed one. We detected a substantially larger proportion of genes with bidirectional ASE in primary lymphoblasts from ALL patients than a recent study on cultured lymphoblast cell lines, in which bidirectional ASE (flipping) was observed only for 14% of the informative SNPs (69 SNPs out of 469 with twofold ASE) (Pollard et al. 2008). The explanation for this discrepant result may be that our study included a larger number of samples and hence a larger number of informative SNPs, and employed sensitive detection of ASE using the Infinium I assay. Obviously, there could also be genuine differences in the gene expression patterns between cultured lymphocytes and primary ALL cells. The ASE-levels measured in our study showed a large variation between individual ALL samples and between genes, from 1.4-fold overexpression of one allele to apparent monoallelic expression, which was indistinguishable from the allelic expression in samples of homozygous genotype. We also noted that bidirectional ASE was more prevalent among genes with high ASE-levels, including the genes with apparent monoallelic expression.
In our study, we identified only two autosomal genes, PAX8 and OAS1, that exhibited consistent monoallelic expression in all samples with ASE, while for most of the genes samples with monallelic expression represented a minority. A recent study reported that 10% of analyzed human autosomal genes (371 out of 3939 genes) show stable monoallelic expression in cultured clonal B-lymphoblast cell lines (Gimelbrant et al. 2007). But this study included a low number of samples and did not appear to recognize the possibility of differential allelic expression. Consequently, all genes with ASE might have been classified as being monoallelically expressed in this study. It is technically difficult to assign monoallelic expression to individual genes and samples unequivocally, because the incidence of monoallelic expression depends on the sensitivity of the genotyping method used for detecting a minority allele and on the algorithm used for defining monoallelic expression. These technical differences between our study and the study by Gimelbrant et al. (2007), who used ASO hybridization on Affymetrix 500K SNP arrays for genotyping are the likely reasons for the large differences in incidence of monoallelic expression between the two studies.
To examine to what extent methylation causes ASE in the ALL cells, we determined the methylation levels of 1306 CpG sites in the promoter regions and first introns of the genes that exhibited ASE in our ALL samples. Using three different approaches, we found a clear correlation between CpG site methylation and ASE. First, we observed a significantly larger variability in methylation of CpG sites in promoter regions of the genes that displayed bidirectional ASE. This finding suggests that bidirectional ASE occurs as a consequence of CpG site methylation, which is randomly distributed between the two chromosomes and causes allele-specific silencing of the expression of one of the alleles. We speculate that one-directional ASE could more commonly be caused by inherited regulatory polymorphisms that affect binding of transcription factors or enhancers in an allele-specific manner, although an early study on ASE also suggests that bidirectional ASE could be caused by regulatory SNPs that are not linked with the SNPs used to detect ASE (Pastinen et al. 2003). In our study only three known imprinted genes, ATP10A, SLC22A18, and SPON2, exhibited bidirectional ASE, and surprisingly, we did not observe monoallelic expression in all our ALL samples for any of these genes. Expression of both alleles of imprinted genes in a subset of the samples could possibly be due to loss of imprinting in cancer cells as previously shown for the IGF2 gene in ALL (Vorwerk et al. 2003). Second, we detected higher levels of ASE for 35 genes with CpG sites that displayed variation in methylation levels between the samples, which indicates a quantitative correlation between ASE and CpG site methylation. These genes include several genes, like FAM24B, ZNF75A, ZNF274, ZNF667, FLJ10769, and FAT, for which little is known about their functions, but also some genes that are interesting because of their potential role in ALL. Silencing of DNAJC15 by methylation has been associated with increased chemotherapeutic resistance in ovarian cancer (Shridhar et al. 2001) and TSPO is the ligand for several anticancer agents (Santidrian et al. 2007) and has been shown to be overexpressed in chronic lymphocytic leukemia (CLL) cells (Carayon et al. 1996).
Third, the quantitative data for ASE obtained by the Infinium I assays and for the methylation levels of CpG sites by genotyping of bisulfite-modified DNA by the Golden Gate assay allowed detection of a direct quantitative correlation between ASE and CpG site methylation in individual samples. This correlation was particularly striking for the FAM24B gene, but also evident for several other genes, including DLAT, ZNF667, DSC3, C1GALT1C1, and IFG2BP3. DSC3 is a member of the cadherin superfamily of cell adhesion molecules and has been shown to be silenced by aberrant DNA methylation in primary breast tumor specimens (Oshiro et al. 2005). Considering that we included only 1500 CpG sites out of the total 50,000 CpG sites in the promoters and first introns of the genes for which we detected ASE in the ALL cells, it can be expected that the expression of additional genes may be regulated in allele-specific manner by methylation of CpG sites.
We conclude that the identification of a large set of genes that exhibit ASE in primary ALL cells and identification of a subset of these genes for which gene expression seems to be regulated by methylation opens up new perspectives for more detailed studies on the molecular events that lead to ALL and affect the response to therapy and clinical outcome in patients with ALL.
Methods
Patients and samples
This study included bone marrow or peripheral blood samples from 197 children diagnosed with acute lymphoblastic leukemia at centers for pediatric oncology in the five Nordic countries and enrolled on the Nordic Society of Pediatric Hematology and Oncology (NOPHO) ALL 1992 or NOPHO ALL 2000 treatment protocol during 1998–2006 (Gustafsson et al. 2000). The distribution of samples between the five Nordic countries was: Sweden, n = 109; Denmark, n = 36; Norway, n = 29; Finland, n = 17; and Iceland, n = 6. The median age of the patients was 5.3 yr, range 0.1–17.7, and 165 of them were of B-cell precursor and 29 of T-cell phenotype (three were difficult to classify). Samples were collected in heparinized tubes prior to treatment and shipped to the laboratory in Uppsala within 24–36 h. Leukemic cells were isolated from the samples by 1.077 g/mL Ficoll-Isopaque (Pharmacia) density-gradient centrifugation. The proportion of leukemic cells was estimated on May-Grünwald-Giemsa-stained cytocentrifugate preparations, using light microscopy. The cell samples selected for analysis contained at least 90% lymphoblasts after separation. Pellets of 2–10 million cells were immediately frozen and stored at −70°C in established tissue banks at Uppsala University Hospital following institutional guidelines.
DNA and RNA extraction
DNA and RNA was extracted from samples with 2–10 million cells using the AllPrep DNA/RNA Mini Kit (Qiagen), including the optional on-column DNase digestion step using the RNase-Free DNase Set (Qiagen) to ensure complete removal of carry over DNA from the RNA samples. Absence of DNA in the RNA samples was verified by PCR amplification of at least 100 ng of RNA with intragenic primers for the GAPDH gene. The DNA and RNA samples were quantified using the NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies) and the integrity of the RNA was examined by capillary electrophoresis with a Bioanalyzer using RNA 6000 Nano Labchips (Agilent). For the pure and intact samples 1 μg of RNA was reverse transcribed into double stranded cDNA using the Illumina TotalPrep RNA Amplification kit (Ambion) stopping after the double stranded cDNA purification step and stored at −70°C.
Genotyping
Allele-specific gene expression levels were measured by genotyping 13,917 SNPs in DNA and RNA (cDNA) from the patient cells using the Infinium I assay (Gunderson et al. 2005) and HumanNS-12 BeadChip (Illumina). The NS-12 BeadChips contain over 11,000 SNPs in annotated exons and untranslated mRNA regions of 6310 genes, which were all known SNPs with a minor allele frequency >1% at the time when the original BeadChip was designed (Evans et al. 2008). In addition to the genome-wide coverage of SNPs in coding regions of genes, the BeadChips contain about 2000 SNPs in introns and flanking regions of genes. Reagents and protocols supplied by the manufacturer were used throughout the genotyping process. The format of the NS-12 BeadChips allowed genotyping of triplicate DNA and RNA samples from two cell samples per BeadChip. An equivalent of one-fourth of the reverse transcribed RNA (1 μg of RNA) or 250 ng of genomic DNA from each sample were processed according to standard Infinium protocols. In brief, DNA was amplified by whole-genome amplification and fragmented to several hundred bases by enzymatic digestion. Purified DNA was resuspended in hybridization buffer, denatured, and hybridized to the Human NS-12 BeadChip overnight at 48°C, After an overnight hybridization, the BeadChips were assembled into a Te Flow Through Chamber (Illumina) followed by washing, allele-specific primer extension with biotinylated dNTPs, and streptavidin-phycoerythrin sandwich staining (Gunderson et al. 2005). The BeadChips were then washed with low salt wash buffer, coated with a protective agent, and imaged on an Illumina BeadStation GX scanner.
Interpretation of genotyping data
The raw fluorescence signal intensities measured from the BeadChips were analyzed using the BeadStudio software (Illumina). The cluster file supplied by Illumina was initially used for genotype assignment, followed by manual adjustment of the clusters. The average genotype call rate was 96% in the DNA samples. RNA samples with a total fluorescence signal intensity (A1 + A2) below 600 were considered not to be expressed in the cells and were excluded from further analysis. The statistical significance for the difference in the allele fraction [A1/(A1 + A2)] in RNA compared to that in DNA was tested with the limma software (Smyth et al. 2005), which applies linear models and empirical Bayes methods to assess differential gene expression (Smyth 2004). The significance threshold for scoring ASE was set to P < 0.001. After applying these automatic filters, genes flagged with ASE were again inspected visually in BeadStudio, after which 3531 SNPs were finally scored for ASE analysis and all samples passed the quality control. The frequency of ASE scored in ALL samples from the five Nordic countries was similar.
Quantitative sequencing
DNA fragments spanning nine SNPs in nine genes (ARSA, BZRP, DLAT, FAAH, FLJ10769, LGALS8, NKAIN4, OAS1, and RNF168) from the NS-12 BeadChip were amplified by PCR from genomic DNA and RNA (cDNA) from eight ALL cell samples. The same primers were used for each SNP for genomic DNA and RNA, except for ARSA and NKAIN4 where the SNP was located close to an exon/intron boundary. The PCR products were sequenced using BigDye Terminator v3.1 chemistry and an Applied Biosystems 3730XL DNA sequencer. The success rate of sequencing was 97%. The sequence traces were analyzed using the “Peak Picker” software specifically developed for quantitative determination of allelic expression levels (Ge et al. 2005). The normalized allele ratios in RNA determined by the PeakPicker software were converted to ASE levels, which were compared with the corresponding ASE levels measured in the same samples using the NS-12 genotyping BeadChips.
Analysis of DNA methylation
A custom designed GoldenGate methylation analysis panel (Illumina) including 1536 CpG sites was used for the analysis of the CpG sites upstream or in the first intron of 386 of the genes with ASE. On average 3.7 CpG sites were selected per gene. Between 600 and 750 ng of DNA from the cell samples was treated with sodium bisulfite using reagents and protocols from the EZ-96 DNA Methylation Kit (Zymo Research). The DNA samples were first chemically denatured followed by overnight (16h) incubation in sodium bisulfite reagent at 50°C, which converts unmethylated cytosines into uracils. After this treatment, the samples were incubated at 4°C for 10 min and then purified with a desulfonation reagent and a clean-up reagent in reaction columns. A whole-genome amplified (WGA) DNA sample, where methylated C-residues are not replicated and the CpG sites remain unmethylated was used as a negative control for the bisulfite treatment and subsequent genotyping procedure. A DNA sample treated with SssI methyltransferase to methylate all CpG sites was used as a positive control for the methylation assay.
After bisulfite treatment of the DNA samples, the cytosines in the CpG sites were genotyped as C/T polymorphisms. The GoldenGate assay uses two allele-specific oligonucleotides and two locus-specific oligonucleotides for each CpG site. Briefly, the bisulfite treated DNA was biotinylated and immobilized on paramagnetic beads. The allele- and locus-specific oligonucleotides were hybridized to the immobilized DNA, the allele-specific primers were extended with dNTPs and the extension products were ligated to the locus-specific oligonucleotides according to protocols from Illumina. The DNA templates created by primer extension and ligation were amplified by PCR with fluorescently labeled universal primers complementary to sequences in the allele- and locus-specific oligonucleotides. The subsequent steps are identical to those for the standard GoldenGate genotyping assay (Shen et al. 2005). The fluorescence signals were measured from the BeadArrays using an Illumina BeadStation GX scanner. The fluorescence data were then analyzed using the BeadStudio software (Illumina). The software assigns a beta-value for each CpG site, which corresponds to the ratio between the fluorescence signal from the methylated allele (C) and the sum of the fluorescent signals of the methylated (C) and unmethylated (T) alleles (Bibikova et al. 2006).
CpG sites with a detection P-value above 0.05 in more than 50 samples according to the BeadStudio software, which indicates a less robust signal, were excluded from further analysis (n = 230), leaving 1306 CpG sites for the final analysis of the 197 samples. The unmethylated WGA-sample that served as a negative control had a median beta-value of 0.07 across all CpG sites and the methylated SssI methyltransferase-treated DNA sample that served as positive control for the assay had a median beta-value of 0.80 across all CpG sites, with detection P-values below 0.05.
Statistical analyses
The quality controlled genotype and methylation data from BeadStudio (Illumina) were further exported as text files for analysis in Microsoft Excel or using the R-software package (The R Development Core Team 2008; http://www.r-project.org). The significance of the difference in the allele fraction in RNA compared to that in DNA was determined using the limma software (Smyth et al. 2005). Genes with ASE were examined for biologically relevant associations using the WebGestalt (Zhang et al. 2005) tool (http://genereg.ornl.gov/webgestalt/). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were assessed for enrichment of genes with ASE using a hypergeometric test to compare the proportion of genes with ASE with the proportion of genes that were classified as informative by RNA genotyping with the NS-12 BeadChips.
Correlations between ASE and CpG site methylation were assessed by linear regression using Pearson’s correlation. P-values for the correlation were computed by permuting the beta-values 5000 times. A one-sided Mann-Whitney test, where the median ASE values for samples with beta-values of <0.25 or >0.75 (“low/high”) were compared to median ASE values for samples with beta-values 0.25–0.75 (“intermediate”). Adjusted P-values were computed by permuting the methylation groups and recalculating the median ASE-levels 5000 times. Fisher’s exact test was used to compare the number of genes with absolute ASE-values ≥ 0.25 in the groups of genes with one- and bidirectional ASE. To compare the variation in methylation levels between genes with one- and bidirectional ASE, the samples were grouped according to “low/high” and “intermediate” methylation levels as above. The genes were then divided into groups based on the methylation status for the majority of the samples with ASE. Fisher’s exact test was used to compare the number of genes with “intermediate,” i.e., more variable methylation levels, between the groups of genes with one- and bidirectional ASE.
Acknowledgments
The ASE and methylation analyses were performed using equipment at the SNP technology platform in Uppsala (www.genotyping.se) with the assistance of Torbjörn Öst and Marie Lindersson. We thank all colleagues in the Nordic Society of Pediatric Hematology and Oncology who provided the patient samples. Financial support for the study was provided by the Swedish Cancer Foundation (A.-C.S.), the Swedish Research Council for Science and Technology (A.-C.S.), the Knut and Alice Wallenberg Foundation (to the SNP technology platform), the Marcus Borgström and Anna Maria Lundin Foundations (L.M.), the Nordic Center of Excellence in Disease Genetics (A.K.) and the Swedish Childhood Cancer Foundation (G.L.). Kjeld Schmiegelow holds the Danish Childhood Cancer Foundation Research Professorship.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083931.108.
References
- Bibikova M., Lin Z., Zhou L., Chudin E., Garcia E.W., Wu B., Doucet D., Thomas N.J., Wang Y., Vollmer E., et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson H.T., Albert T.J., Ladd-Acosta C.M., Green R.D., Rongione M.A., Middle C.M., Irizarry R.A., Broman K.W., Feinberg A.P. SNP-specific array-based allele-specific expression analysis. Genome Res. 2008;18:771–779. doi: 10.1101/gr.073254.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray N.J., Buckland P.R., Owen M.J., O'Donovan M.C. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- Carayon P., Portier M., Dussossoy D., Bord A., Petitpretre G., Canat X., Le Fur G., Casellas P. Involvement of peripheral benzodiazepine receptors in the protection of hematopoietic cells against oxygen radical damage. Blood. 1996;87:3170–3178. [PubMed] [Google Scholar]
- Cheok M.H., Evans W.E. Acute lymphoblastic leukaemia: A model for the pharmacogenomics of cancer therapy. Nat. Rev. Cancer. 2006;6:117–129. doi: 10.1038/nrc1800. [DOI] [PubMed] [Google Scholar]
- Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Eckhardt F., Lewin J., Cortese R., Rakyan V.K., Attwood J., Burger M., Burton J., Cox T.V., Davies R., Down T.A., et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat. Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.M., Barrett J.C., Cardon L.R. To what extent do scans of non-synonymous SNPs complement denser genome-wide association studies? Eur. J. Hum. Genet. 2008;16:718–723. doi: 10.1038/sj.ejhg.5202011. [DOI] [PubMed] [Google Scholar]
- Figueroa M.E., Reimers M., Thompson R.F., Ye K., Li Y., Selzer R.R., Fridriksson J., Paietta E., Wiernik P., Green R.D., et al. An integrative genomic and epigenomic approach for the study of transcriptional regulation. PLoS One. 2008;3:e1882. doi: 10.1371/journal.pone.0001882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho C., Coustan-Smith E., Pei D., Cheng C., Song G., Pui C.H., Downing J.R., Campana D. A set of genes that regulate cell proliferation predicts treatment outcome in childhood acute lymphoblastic leukemia. Blood. 2007;110:1271–1277. doi: 10.1182/blood-2007-01-068478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forestier E., Schmiegelow K. The incidence peaks of the childhood acute leukemias reflect specific cytogenetic aberrations. J. Pediatr. Hematol. Oncol. 2006;28:486–495. doi: 10.1097/01.mph.0000212972.90877.28. [DOI] [PubMed] [Google Scholar]
- Ge B., Gurd S., Gaudin T., Dore T., Lepage P., Harmsen E., Hudson T.J., Pastinen T. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584–1591. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Greaves M.F., Wiemels J. Origins of chromosome translocations in childhood leukaemia. Nat. Rev. Cancer. 2003;3:639–649. doi: 10.1038/nrc1164. [DOI] [PubMed] [Google Scholar]
- Gunderson K.L., Steemers F.J., Lee G., Mendoza L.G., Chee M.S. A genome-wide scalable SNP genotyping assay using microarray technology. Nat. Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- Gustafsson G., Schmiegelow K., Forestier E., Clausen N., Glomstein A., Jonmundsson G., Mellander L., Makipernaa A., Nygaard R., Saarinen-Pihkala U.M. Improving outcome through two decades in childhood ALL in the Nordic countries: The impact of high-dose methotrexate in the reduction of CNS irradiation. Nordic Society of Pediatric Haematology and Oncology (NOPHO) Leukemia. 2000;14:2267–2275. doi: 10.1038/sj.leu.2401961. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V.V., Schupf N., Vilain E., et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kuang S.Q., Tong W.G., Yang H., Lin W., Lee M.K., Fang Z.H., Wei Y., Jelinek J., Issa J.P., Garcia-Manero G. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia. 2008;22:1529–1538. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- Mahr S., Burmester G.R., Hilke D., Gobel U., Grutzkau A., Haupl T., Hauschild M., Koczan D., Krenn V., Neidel J., et al. Cis- and trans-acting gene regulation is associated with osteoarthritis. Am. J. Hum. Genet. 2006;78:793–803. doi: 10.1086/503849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani L., Gupta M., Andersen M., Dhar S., Fryknas M., Isaksson A., Larsson R., Syvanen A.C. Allelic imbalance in gene expression as a guide to cis-acting regulatory single nucleotide polymorphisms in cancer cells. Nucleic Acids Res. 2007;35:e34. doi: 10.1093/nar/fk11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–764. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- Oshiro M.M., Kim C.J., Wozniak R.J., Junk D.J., Munoz-Rodriguez J.L., Burr J.A., Fitzgerald M., Pawar S.C., Cress A.E., Domann F.E., et al. Epigenetic silencing of DSC3 is a common event in human breast cancer. Breast Cancer Res. 2005;7:R669–R680. doi: 10.1186/bcr1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant P.V., Tao H., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T., Hudson T.J. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Pastinen T., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics. 2003;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- Pastinen T., Ge B., Gurd S., Gaudin T., Dore C., Lemire M., Lepage P., Harmsen E., Hudson T.J. Mapping common regulatory variants to human haplotypes. Hum. Mol. Genet. 2005;14:3963–3971. doi: 10.1093/hmg/ddi420. [DOI] [PubMed] [Google Scholar]
- Pollard K.S., Serre D., Wang X., Tao H., Grundberg E., Hudson T.J., Clark A.G., Frazer K. A genome-wide approach to identifying novel-imprinted genes. Hum. Genet. 2008;122:625–634. doi: 10.1007/s00439-007-0440-1. [DOI] [PubMed] [Google Scholar]
- Pui C.H., Robison L.L., Look A.T. Acute lymphoblastic leukaemia. Lancet. 2008;371:1030–1043. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- The R Development Core Team . R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2008. [Google Scholar]
- Santidrian A.F., Cosialls A.M., Coll-Mulet L., Iglesias-Serret D., de Frias M., Gonzalez-Girones D.M., Campas C., Domingo A., Pons G., Gil J. The potential anticancer agent PK11195 induces apoptosis irrespective of p53 and ATM status in chronic lymphocytic leukemia cells. Haematologica. 2007;92:1631–1638. doi: 10.3324/haematol.11194. [DOI] [PubMed] [Google Scholar]
- Serre D., Gurd S., Ge B., Sladek R., Sinnett D., Harmsen E., Bibikova M., Chudin E., Barker D.L., Dickinson T., et al. Differential allelic expression in the human genome: A robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen R., Fan J.B., Campbell D., Chang W., Chen J., Doucet D., Yeakley J., Bibikova M., Wickham Garcia E., McBride C., et al. High-throughput SNP genotyping on universal bead arrays. Mutat. Res. 2005;573:70–82. doi: 10.1016/j.mrfmmm.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Shridhar V., Bible K.C., Staub J., Avula R., Lee Y.K., Kalli K., Huang H., Hartmann L.C., Kaufmann S.H., Smith D.I. Loss of expression of a new member of the DNAJ protein family confers resistance to chemotherapeutic agents used in the treatment of ovarian cancer. Cancer Res. 2001;61:4258–4265. [PubMed] [Google Scholar]
- Smyth G.K. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Smyth G.K., Michaud J., Scott H.S. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K.H., Pena-Hernandez K.E., Davis J.W., Arthur G.L., Duff D.J., Shi H., Rahmatpanah F.B., Sjahputera O., Caldwell C.W. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67:2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- Vorwerk P., Wex H., Bessert C., Hohmann B., Schmidt U., Mittler U. Loss of imprinting of IGF-II gene in children with acute lymphoblastic leukemia. Leuk. Res. 2003;27:807–812. doi: 10.1016/s0145-2126(03)00014-6. [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- Willenbrock H., Juncker A.S., Schmiegelow K., Knudsen S., Ryder L.P. Prediction of immunophenotype, treatment response, and relapse in childhood acute lymphoblastic leukemia using DNA microarrays. Leukemia. 2004;18:1270–1277. doi: 10.1038/sj.leu.2403392. [DOI] [PubMed] [Google Scholar]
- Zhang B., Kirov S., Snoddy J. WebGestalt: An integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]