Abstract
Small RNAs regulate the genome by guiding transcriptional and post-transcriptional silencing machinery to specific target sequences, including genes and transposable elements (TEs). Although miniature inverted-repeat transposable elements (MITEs) are closely associated with euchromatic genes, the broader functional impact of these short TE insertions in genes is largely unknown. We identified 22 families of MITEs in the Solanaceae (MiS1–MiS22) and found abundant MiS insertions in Solanaceae genomic DNA and expressed sequence tags (EST). Several Solanaceae MITEs generate genome changes that potentially affect gene function and regulation, most notably, a MiS insertion that provides a functionally indispensable alternative exon in the tobacco mosaic virus N resistance gene. We show that MITEs generate small RNAs that are primarily 24 nt in length, as detected by Northern blot hybridization and by sequencing small RNAs of Solanum demissum, Nicotiana glutinosa, and Nicotiana benthamiana. Additionally, we show that stable RNAi lines silencing DICER-LIKE3 (DCL3) in tobacco and RNA-dependent RNA polymerase 2 (RDR2) in potato cause a reduction in 24-nt MITE siRNAs, suggesting that, as in Arabidopsis, TE-derived siRNA biogenesis is DCL3 and RDR2 dependent. We provide evidence that DICER-LIKE4 (DCL4) may also play a role in MITE siRNA generation in the Solanaceae.
Small interfering RNA (siRNA) and microRNA (miRNA) play fundamental roles in eukaryotic gene regulation, antiviral defense, genome organization, and maintenance of genome integrity. These small RNAs are processed from RNA precursors by type III RNase enzymes, referred to as DICER or DICER-LIKE proteins (DCL), and incorporated into RNA induced silencing complexes (RISC) to mediate post-transcriptional gene silencing (PTGS) by translational arrest or mRNA cleavage, or into RNA-induced transcriptional silencing complexes (RITS) to mediate transcriptional gene silencing (TGS) by repressive DNA and histone modifications (Baulcombe 2006; Li and Ding 2006; Ding and Voinnet 2007; Henderson and Jacobsen 2007; Slotkin and Martienssen 2007). In Arabidopsis, small RNA biogenesis enzymes are encoded by gene families in which the individual members have specialized functions in parallel pathways. DCL1 and hyponastic leaves 1 (HYL1) produce miRNAs that have a major role in developmental regulation (Carrington and Ambros 2003; Chen 2005; Poethig et al. 2006). DCL4, DCL2, and DCL3 act hierarchically in response to RNA virus infection by generating viral siRNAs (Ding and Voinnet 2007). DCL4, RNA-dependent RNA polymerase 6 (RDR6), and suppressor of gene silencing 3 (SGS3) are involved in trans-acting siRNA (ta-siRNA) accumulation and regulate juvenile–adult transition (Poethig et al. 2006). DCL2, DCL1, RDR6, and Nuclear RNA polymerase D 1A (NRPD1A) act on natural antisense transcripts and produce nat-siRNA in response to abiotic stress (Borsani et al. 2005). More recently, RDR2, NRPD1A, and SNF2, in conjunction with DCL4, have been implicated in non-cell-autonomous silencing (Dunoyer et al. 2007; Smith et al. 2007).
Small RNA pathways also regulate the activity of transposable elements—both DNA transposons and retrotransposons—via a class of transposable element (TE)-derived 24-nt siRNAs that guide transcriptional gene silencing (TGS), generated by a pathway involving DCL3, RDR2, NRPD1A, and ARGONAUTE 4 (AGO4) (Henderson and Jacobsen 2007). Most TEs are concentrated in highly repetitive regions near centromeres and telomeres, where siRNA-dependent chromatin modifications establish vast stretches of heterochromatin that dominate the genomic makeup of many organisms (Bennetzen and Kellogg 1997). However, isolated examples have also uncovered the co-option of TE siRNAs in controlling the expression of genes bearing TE insertions. For example, the expression of FLC and FWA, which govern flowering time, is regulated by siRNA-guided TGS targeting of a Mutator-family TE and directly repeated SINE elements inserted in the FLC intron (Liu et al. 2004) and FWA promoter (Chan et al. 2004), respectively. It is thought that TE mobility may provide the evolutionary capacity to alter patterns of gene regulation via the insertion of TEs within “gene islands.”
A particular class of truncated DNA transposons, miniature inverted-repeat transposable elements (MITEs), may play a role in the co-option of TE-derived siRNAs in gene regulation. MITEs, which are generally <600 bp, lack open reading frames, and thus their mobility depends on the activity in trans of transposases encoded by cognate full-length autonomous transposons (Jiang et al. 2003, 2004b). The full-length DNA transposons are hypothesized to be the evolutionary progenitors of MITEs, based on sequence conservation between miniature inverted-repeat transposable elements (MITEs) and autonomous transposon terminal inverted repeats (TIRs) and target site duplications (TSDs). MITEs have amplified to a high copy number in several plant genomes, as in rice, which contains more than 90,000 MITEs grouped into approximately 100 different families (Feschotte et al. 2003; Jiang et al. 2004b; Juretic et al. 2004). In the rice and Arabidopsis genomes, the majority of MITEs are inserted in the euchromatin arms of chromosomes (Feng et al. 2002; Santiago et al. 2002; Wright et al. 2003), rather than in the heterochromatin. This observed close physical association between MITEs and plant genes (Wessler 1998; Mao et al. 2000) has provoked the hypothesis that MITEs play a role in gene regulation, as predicted of “controlling elements” (McClintock 1951). However, prior to the recent discovery of small RNA pathways, a plausible explanation for mechanisms of MITE-based gene regulation, and for functional roles of TEs in general, has been incomplete. The recent proposal that several families of miRNAs in humans, Arabidopsis, and rice are derived from MITEs (Piriyapongsa and Jordan 2007, 2008; Piriyapongsa et al. 2007) gives further, tantalizing evidence that MITEs may serve a major evolutionary role in co-opting TE silencing pathways for gene regulation, not only in plants, but also across the eukaryotic spectrum. However, investigation of biogenesis of small RNAs derived from MITEs has not been thoroughly studied in plants, and the roles of MITEs in general, particularly when associated with plant genes, remain largely unexplored.
Research in the Solanaceae family, which comprises more than 3000 species including the important crop plants tomato (Solanum lycopersicum), potato (Solanum tuberosum), eggplant (Solanum melongena), pepper (Capsicum annuum), and tobacco (Nicotiana tabacum), has produced comparative genomic resources—including the sequencing of multiple alleles of several resistance gene loci (Chang et al. 2002; Song et al. 2003; Schornack et al. 2004; Huang et al. 2005; Kuang et al. 2005) that facilitate identification and analysis of TEs. Using comparative sequence analysis and the related empty site (RESite) method (Le et al. 2000), we examined homeologous R-gene regions from different haplotypes of the same or closely related Solanaceae species to identify 22 MITE families. Using Northern blot hybridization analysis, we show that all MITE families tested generate small RNAs in Solanaceae species, and we identified specific MITE-derived siRNAs by small RNA sequencing. We determined that MITE small RNA are generated by a pathway similar to that required for TE-derived siRNA biogenesis in Arabidopsis by silencing each of the four DICER-LIKE (DCL) homologs in N. tabacum and RDR2 in S. tuberosum, demonstrating that silencing of DCL3 and RDR2 causes a reduction in MITE-derived small RNAs. However, in contrast to Arabidopsis, our findings suggest a role for DCL4 in MITE small RNA biosynthesis in the Solanaceae. These results reveal the presence of MITE-derived small RNAs in the Solanaceae and provide a foundation for a systematic and in-depth analysis of the roles of MITEs in small RNA-mediated gene regulation and evolution in Solanaceae.
Results
Identification and analysis of MITE families in Solanaceae
Identification and classification of MITEs
We identified 22 families of MITEs in Solanaceae (MiS1–MiS22) within six syntenic tomato, potato, and tobacco R-gene regions based on shared structural similarities and the analysis of related empty sites (RESite) (Le et al. 2000). Fifteen of these MiS families are novel, while seven have been described previously (Table 1). Based on their similarity to the characteristic features of known MITEs, including TIR and TSD sequence composition, MiS2–MiS4, MiS6–MiS11, MiS12–MiS17, and MiS18–MiS19 were grouped into hAT-like, Mutator-like, Stowaway-like, and Tourist-like superfamilies, respectively (Table 1). MiS1, MiS5, and MiS20–22 share no similarity with any known MITE superfamily. Eight MiS families have members that exceed the length limitations (<600 bp) that arbitrarily define MITEs, but retain MITE properties, including high copy number, lack of an encoded transposase, association with genes, and conserved structures (TIRs, TSDs); hence, we identified these families as MiS and included them in our analysis. We extended our Solanaceae MITE sequence data set using the representative member of each MiS family as a query in BLASTN searches of Solanaceae sequences in the GenBank nonredundant database and retrieved MiS sequences with E-values < e-5, the minimal stringency necessary to detect a 30-nt stretch of identical sequence within an 80-nt MITE. The full-length sequence of the original MiS data set retrieved from GenBank is provided in Supplemental Data S1.1. Consistent with previous reports on MITE association with genes, we found several MITE insertions located near genes and within introns, exons, 5′- and 3′-untranslated regions (UTRs), and promoters. The location of 198 members of MiS1–22 and their positions within a gene or location near a gene is provided in Supplemental Table S1.1. Notably, Solanaceae MITEs are located upstream and downstream of genes, within promoters, 5′- and 3′-untranslated regions, introns, and exons.
Table 1.
Structural features of MITE families in Solanaceae
(TSD) Target site duplication; (TIR) terminal inverted repeat.
Estimation of genome-wide copy number
MITEs often exist in high copy number, and we hypothesized that Solanaceae genomes would contain abundant MiS sequences. To assess the abundance and amplification of MiS, we used the sequence of a representative MiS from each family MiS(X)-1 (Supplemental Data S1.1) as queries in BLASTN searches of tomato and potato BAC-end (http://www.tigr.org/tdb/sol/sol_ma_blast.shtml) and tobacco genomic DNA (NCBI Nicotiana tabacum gss) databases and extrapolated the number of hits retrieved to estimate genome-wide copy numbers as described in Supplemental Table S1.2. An extended data set of N. tabacum MiS1, MiS5, and MiS17 sequences is provided in Supplemental Data S1.2. Several MiS families from the hAT-like (MiS2, MiS4), Stowaway-like (MiS12, MiS13, MiS16, MiS17), and Tourist-like superfamilies (MiS18, MiS19) are represented in all three species, ranging from an estimated 100 to several thousand copies, which suggests an origin that predates the divergence of these species. In contrast, the estimated copy numbers of the MiS1 family are much higher in tobacco (>1000) than in tomato and potato (<60), while most Mutator-like MiS (MiS6–MiS11) have higher estimated copy numbers in tomato and potato (approximately 400–3200) than in tobacco (0–32), suggesting that amplifications have occurred after the divergence of Solanum and Nicotiana. The most abundant family was MiS22, which retrieved 2000 and 1874 hits, respectively, in the tomato and potato BAC-end sequence databases, extrapolating to an estimated 3516 and 9802 copies in the respective genomes. While we expect errors in our extrapolation, owing to incomplete genome sequence data sets for all three species and the bias of the MITE query sequences to identify more MiS hits in their cognate genomes as compared to diverged genomes, our results nonetheless indicate that many MITE families have amplified to high copy numbers in Solanaceae genomes, a property consistent with other genomes.
Structural, functional, and evolutionary implications of MITE insertions
In addition to their characteristic TIR and TSD sequence composition, and high copy number, MITEs have features with evolutionary and functional implications. For example, MITEs can capture and move gene sequences to new locations; MITE genetic and structural properties implicate cognate autonomous elements as both progenitors and activators of MITE transposition, and MITEs are often associated with genes. While analyzing MiS insertions, we discovered three families of elements—MiS5, MiS2, and MiS1—that are of particular interest because features of some family members provide insight into their evolutionary and functional properties.
Tandem MITE complex
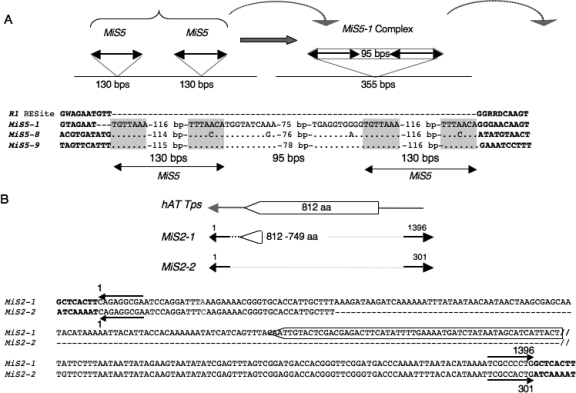
Analysis of MiS5-1, a 355-bp insertion in an R1 resistance gene homolog (R1b-11, AC149266, 1364–1718 bp) in the Solanum demissum Chromosome 5 R1-gene cluster, revealed that it is a complex comprising two tandem, nearly identical 130-bp MiS5 elements (with identical TIRs and two mismatches overall) separated by 95 bp of intervening sequence (Fig. 1A). Additional MiS5 complexes were identified in S. demissum and other Solanum species (Supplemental Data S1.1). For example, MiS5-8, a 355-bp complex (S. lycopersicum, AF411809, 126,063–126,418 bp), and MiS5-9, a 357-bp complex (S. tuberosum, TA45582_4113, 250–606 bp), share 83% and 91% overall identity with MiS5-1, respectively, but have unrelated sequences flanking each insertion. Analyses of 51 RESites for MiS5-1 identified among S. demissum R1 homologs lacking MiS5-1 suggest that MiS5 does not generate a TSD but rather a 3-bp target-site deletion. We found no insertions or deletions within the 51 RESites, while the R1b-11 homolog containing the complex MiS5-1 insertion showed a 3-bp deletion at the target site (Fig. 1A).
Figure 1.
Structures of MiS5 and MiS2. (A) Several MiS5 members are found as complex with an intervening sequence. A model is presented in which tandem insertions of the same MiS5 element generate a MiS complex that captures and transposes intervening sequence. (Bold black lines flanked by arrowheads, TIRs) MiS. The sequences of the MiS5-1 RESite in S. demissum (R1; AC149265.1) and three MiS5 complexes, MiS5-1 in S. demissum (AC149266, 1364–1718), MiS5-8 in S. lycopersicum (AF411809, 126,063–126,418), and MiS5-9 in S. tuberosum (TA45582_4113, 250–606), are shown. (Dots) Nucleotide identity; (dashes in R1) the RESite lacking MiS5 insertion; (dashes in MiS5-1) a 3-bp deletion at its insertion site; (gray) TIRs; (bold) flanking sequence. (B) MiS2-1 (1396 bp) and MiS2-2 (301 bp) are hAT-type elements in GenBank accessions AC149265, 50,964–52,375, and AC149488, 68,757–69,073, respectively. (Open arrowhead) The MiS2-1 sequence is predicted to encode a truncated product with similarity to the C terminus (749–812 amino acids) of candidate S. demissum hAT transposase (AAT39314). Conserved sequences between MiS2-1 and MiS2-2 are shown aligned. (Horizontal arrows above and below the aligned sequences) The 9-bp MiS2 TIRs. (Bold) The 8-bp TSDs generated by the MiS2 insertions. The MiS2-1 sequence enclosed in the open arrow is predicted to encode amino acids 794–812 of the candidate S. demissum hAT transposase.
Numerous MiS5 (∼130 bp) sequences were also found in Solanaceae BAC-end, GenBank nonredundant databases, and EST databases (Supplemental Tables S1 and S2.1). Retention of nucleotide identity in the intervening sequences of distinct MiS5 complexes indicates that tandem MITEs have the capacity to be excised and transpose as a complex, thereby providing a mechanism that “captures” and carries intervening sequence to a new genomic location.
Autonomous transposon progenitors of MITEs
MITEs are activated by and hypothesized to be derived from full-length autonomous DNA transposons. Consistent with this hypothesis, we identified two highly homologous MITEs in S. demissum, MiS2-1 (AC149265, 50,964–52,375) and MiS2-2 (AC149488, 68,757–69,073), which share ∼85% overall nucleotide identity and contain nearly identical hAT superfamily TIR sequences. MiS2-1 is 1396 bp long and contains a 192-bp sequence homologous to a putative hAT transposase (AAT39314, 749–812 amino acids), while MiS2-2 is only 301 bp long and has no predicted transposase coding sequence (Fig. 1B; see Supplemental Data S1.1 for sequences of MiS2-1 and MiS2-2). It appears that MiS2-1 and MiS2-2 are different derivatives of a progenitor hAT superfamily transposon. We also identified a candidate progenitor of the Mutator-like MiS10 family, an autonomous Mutator-like TE in S. demissum (AC149302, 44,107–50,193), which has a characteristic 9-bp A/T-rich TSD and ∼350-bp MiS10-like TIRs (Supplemental Data S1.1) and encodes a predicted MuDR-like transposase.
A MITE serves as an alternative exon
MiS1-1, a 186-bp insertion, was identified in intron 3 of the tobacco mosaic virus (TMV) resistance gene, N (U15605), by sequence comparison between the N gene of Nicotiana glutinosa and the NH gene (A535010) of N. tabacum, a candidate N ortholog, with a predicted product sharing 88% amino acid identity with N (Fig. 2A; Stange et al. 2004). The MiS1 inserted sequence was used as a BLASTN query of GenBank, leading to the discovery of a second sequence, 87 bp in length, in the third intron of the N gene, as well as a third MiS1 sequence, 193 bp in length, located 387 bp upstream of the Nicotiana sylvestris RAN-1 gene (AY563050). These three elements exhibit 80%–85% nucleotide identity (data not shown). They share the 6-bp terminal inverted-repeat (IR) sequences but are flanked by unrelated sequences, indicating that the elements are members of a novel MITE family, which we have named MiS1 with members MiS1-1, -2, and -3, corresponding to the 186-, 87-, and 193-bp elements, respectively (Table 1; Supplemental Data S1.1).
Figure 2.
MiS1 is expressed as an alternative exon and as a 3′-UTR. (A) The structure of the N gene (U15605) and a candidate N ortholog, NH (A506320), showing (boxes) exons and (angled lines) introns. The full-length Ns transcript includes (gray boxes) exons 1–5. The alternatively spliced NL transcript includes exons 1–5 and the (AE; black box) alternative exon and is predicted to encode a truncated product by an AE-encoded frameshift. Intron 3 has two MITE insertions (MiS1-1 and MiS1-2). The entire AE is located within MiS1-1. The insertion of MiS1-1 in N is shown relative to the RESite in NH; with the AE highlighted in black. (Boxes) MiS1 TIRs; (numbers) indicate nucleotide positions in base pairs. (Bold, underlined lettering) A sequence complementary to 21 nt of MiS1 N. glutinosa siRNA, sNg_317-22-1 (5′-TCCTCTTTCTCTGCAATATTGT-3′), located within the alternative exon of MiS1-1. (Underline) Putative polyadenylation termination sequences. (B) MiS1 elements inserted in opposite orientations in different genes. (Boxes with arrowheads) The coding regions of two N. benthamiana transcript assemblies, TA10655_4100 (http://plantta.tigr.org/cgi-bin/plantta_report.pl?ta=TA10655_4100) and TC10011 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/tc_report.pl?tc=TC10011&species=n_benthamiana). They are predicted to encode a hypothetical protein and WRKY transcription factor 22, respectively (based on BLASTX searches of GenBank) (see Supplemental Table S2.1). (Gray and black lines with arrowheads showing orientation) A MiS1 element is inserted in the 3′-UTR of each transcript in complementary orientations. The sequences of the MiS1 insertions are shown, indicating the putative dsRNA formed between TA_10655_4100 and TC10011. (Boxes) MiS1 TIRs. Forty-six bases of TA10655_4100 are omitted. (Bold, underlined lettering in TC10011) A sequence complementary to N. glutinosa MiS1 siRNA, sNg_1919_19_1 (5′-TAAGACCCCTTTATTTATA-3′) (Table 2). (C) A predicted (MFOLD) hairpin structure of the reverse complement of the MiS9 sequence within S. tuberosum EST DN938187 (405–108 nt) is shown (Supplemental Table S2.1). DN93187 shares homology with Solanum ESTs whose predicted products exhibit strong similarity to the C-terminal portion of Arabidopsis DCL2 (Supplemental Table S2.1). (Bold, underlined lettering in DN93187) A sequence complementary to S. demissum MiS9 siRNA, sSd_2627_24_1 (5′-TAAGACCCCTTTATTTATA-3′) (Table 2).
N is transcribed into two mRNA isoforms (NS and NL), and expression of NL containing a 70-bp alternative exon is required for full resistance to TMV. Approximately 8 h following TMV infection, the relative abundance of the alternatively spliced NL isoform increases (Whitham et al. 1994; Dinesh-Kumar and Baker 2000). MiS1-1 provides the 70-bp alternative exon for the NL mRNA isoform, and the alternative splice sites are located within the MITE sequence (Fig. 2A). The MiS1 element is not found in other closely related N homologs in GenBank (AF211528, AY438027, AY438028, AY535010, AC151803, AJ300266, AC154033), and the deduced amino acid sequence encoded by the N alternative exon has no significant similarity to any known protein. Based on BLASTN searches of the tobacco genomic sequence database (NCBI Nicotiana tabacum gss), the MiS1 family is present in high copy numbers in the tobacco genome (Supplemental Table S1.2). We conclude that the N alternative exon was generated through the insertion of MiS1-1 into intron 3 of the N gene and provides an example of a MITE insertion contributing coding sequence to a functional alternative-splicing product.
MITEs are transcribed
Our original MiS data set revealed the close association of members of each MITE family with genes. We hypothesized that, in addition to the example of MiS1-1 expression as the alternative exon of N, other Solanacae MITEs would be found in association with genes as read-through transcripts or transcripts arising from MiS insertion into a nearby promoter. To assess expression of MiS, we performed BLASTN searches in tomato, potato, tobacco, and Nicotiana benthamiana EST Transcript Assembly (TA) databases (http://tigrblast.tigr.org/euk-blast/plantta_blast.cgi; Childs et al. 2007) using a representative MiS sequence (MiSX-1) from each family (Supplemental Data S1.1) as a query. We identified approximately 666 ESTs containing MiS insertions with E-values of <e-5 and found that each MiS family is represented in at least one of the three EST databases (Supplemental Fig. S1; Supplemental Tables S1.2 and S2.1). Generally, there was a positive correlation between the number of hits detected in EST databases and the estimated copy number in genomic DNA (Supplemental Fig. S1; Supplemental Table S1.2), for example, high copy number MiS families, such as MiS3, MiS17, and MiS18 in tobacco, and MiS13, MiS17, and MiS22 in tomato and potato, also retrieved a high number of EST hits. As revealed by analysis of genomic sequence, MiS22 yielded the most hits in EST databases, being present in 101 sequences (0.05% of the total) in the potato EST database.
In 104 of the 666 TAs containing a Solanaceae MITE-related sequence, there was sufficient information to resolve the position of the insertion within the predicted gene. Of these, 46 of the insertions were located in the 3′-UTR, 11 in the 5′-UTR, one serving as an alternative exon located in an intron (in the N gene, discussed above), and 46 in coding regions, disrupting ORFs (Supplemental Table S2.1). Some insertions of a MITE family were observed in both “+” and “−” (complementary) orientations. Interestingly, five of the 10 MiS1 elements found in the EST/TA databases were inserted in the 3′-UTR of the predicted corresponding transcripts. The 3′-UTRs of these transcripts are terminated within the MiS1 sequences, before the TIR, regardless of the orientation of the insertion, raising the possibility that MiS1 family members may encode polyadenylation signals for transcript termination and poly(A) addition. We identified candidate polyadenylation signal sequences, near upstream elements (NUE), far upstream elements (FUE), and cleavage elements (CE), FUE.1 and NUE.3 (AATAAA) in 93% and CE.1 (TAATTA) (Shen et al. 2008) in 11% of the 76 MiS1 tobacco sequences surveyed, whereas the FUE.1/NUE.3 and CE.1 signal sequences were identified in only 33% and 7% of the 1007 surveyed MiS17 tobacco sequences. A sequence alignment of two of N. benthamiana transcript assemblies containing MiS1 elements in complementary orientations in their 3′-UTRs is shown in Figure 2B and described further below.
Detection and cloning of MiS small RNA
Recently, transposon-derived small RNA has been shown to play an important role in silencing retrotransposon and Mutator-type elements via TGS and PTGS mechanisms (Xie et al. 2004; Slotkin et al. 2005; Sridhar et al. 2007). Our discovery that Solanaceae genomes carry candidate autonomous transposon progenitors and high copy numbers of MITEs led us to hypothesize that MiS small RNAs may play a role in transcriptional silencing of MITEs as described for transposon regulation in Arabidopsis and other species. Our finding that Solanacae MITEs are transcribed and co-transcribed with adjacent genes, presumably by RNA polymerase II (Pol II), and may form predicted double-stranded RNA (dsRNA) or single-stranded hairpin precursors of small RNAs, suggested that post-transcriptional silencing may also play a role in MITE regulation. A putative dsRNA structure formed from “complementary insertions” of MiS1 is depicted in Figure 2B. MiS1 family members were found in sense and antisense orientations in the 3′-UTRs of two N. benthamiana EST transcript assemblies, TA10655_4100 (http://plantta.tigr.org/cgi-bin/plantta_report.pl?ta=TA10655_4100) and TC10011 (http://compbio.dfci.harvard.edu/tgi/cgi-bin/tgi/tc_report.pl?tc=TC10011&species=n_benthamiana) (Fig. 2B; Supplemental Table S2.1), which are predicted to encode a hypothetical protein and a product homologous to WRKY transcription factor 22, respectively. If transcripts bearing these complementary MiS sequences are coexpressed, we hypothesize that stretches of dsRNA form through pairing of 3′-UTR MiS1 sequences and may functionally mimic natural antisense transcripts precursors of regulatory small RNAs (Borsani et al. 2005).
An example of a hairpin-like structure formed from an MiS9-containing transcript (DN938187) with a long TIR (∼100 nt or more) is shown in Figure 2C.
MiS small RNA detection
As a first step to test our hypothesis that Solanaceae MITEs generate small regulatory RNAs, we performed Northern blot hybridization analysis using probes for specific MiS families to analyze the low-molecular-weight RNA fractions isolated from Nicotiana benthamiana, N. tabacum SR1, N. tabacum SR1:NN, N. glutinosa, S. demissum, S. tuberosum Kennebec, S. lycopersicum, Oryza sativa, and Arabidopsis thaliana (Fig. 3; probes described in Supplemental Table S3.1). Each MiS probe detected small RNAs in several Solanaceae species, with 24-nt RNA constituting the predominant size class (Fig. 3A–E).
Figure 3.
MiS small RNAs detected in Solanum and Nicotiana species by Northern blot hybridization. Forty micrograms of small RNA-enriched from total RNA isolated from Solanaceae species was size separated using a 20% acrylamide gel, blotted, and hybridized with MiS-derived probes (see Supplemental Table S3.1 for a description of probes). (A) Hybridization probe for (upper panel) Solanum-MiS17 probe; (lower panel) Nicotiana-MiS17; (B) Nicotiana-MiS1 probe; (C) Solanum-MiS15 probe; (D) Solanum-MiS5 probe; (E) Solanum-MiS8 probe; (F) Os-Ditto probe; (G) MIR165. An EtBr stain is shown for each gene as a sample loading control. (At) A. thaliana, Col-0; (Os) O. sativa Nipponbare; (Nb) N. benthamiana; (NtS) N. tabacum, SR1; (NtTG34) N. tabacum, SR1:NN; (Ng) N. glutinosa; (StKE) S. tuberosum, Kennebec; (StPI) S. tuberosum, Pentland Ivory; (Sd) S. demissum; (Sb) S. bulbocastanum; (StL) S. tuberosum, Katahdin leaves; (StF) S. tuberosum, Katahdin flowers; (SlL) S. lycopersicum VF36 leaves; (SlF) S. lycopersicum, VF36 flowers.
The MiS probes detected small RNAs most strongly in their species of origin and in closely related species. For example, the MiS17 probe, amplified from a pool of S. demissum and S. tuberosum genomic DNA, only detected a strong signal in Solanum species (Fig. 3A, top panel), while an MiS17 probe amplified from a pool of N. benthamiana, N. tabacum, and N. glutinosa genomic DNA only detected a strong signal in Nicotiana species (Fig. 3A, middle panel). Similarly, the MiS1 probe, amplified from N. glutinosa genomic DNA, displayed a strong signal in N. glutinosa and in N. benthamiana, but a much weaker signal in N. tabacum and in Solanum species (Fig. 3B), while the MiS18, MiS5, and MiS8 probes, amplified from S. demissum genomic DNA, only displayed strong signals in Solanum species (Fig. 3C–E, respectively). None of the MiS probes detected small RNA in Arabidopsis or rice. As a control, a probe made from the rice Tourist-superfamily MITE Ditto displayed a much stronger hybridization signal in rice than in other species, and a probe for the conserved miRNA MIR165 (Supplemental Table S3.2; Reinhart et al. 2002) displayed a strong signal in all species (Fig. 3, F and G, respectively).
MiS small RNA sequence
We hypothesized that the ∼24-nt MiS-related small RNAs detected in Northern blots were generated by a NRPD1A–RDR2–DCL3 pathway, similar to that previously described for biogenesis of 24-nt repeat-associated siRNA for transcriptional silencing of transposons in Arabidopsis. Based on our predictions of paired transcripts and hairpin structures, we postulated that small RNA pathways associated with post-transcriptional gene silencing could also produce MiS small RNAs.
The genomic origin, mechanism of biogenesis, and potential function of small RNAs can be inferred from their sequence, length, and 5′-terminal base composition. To test our hypotheses on the origin, biogenesis, and potential function of MITE small RNAs detected by Northern blot hybridization analyses, we cloned and sequenced small RNAs from S. demissum, N. benthamiana, and N. glutinosa to obtain a total of 4421, 1408, and 1806 small RNA reads of good quality, respectively (Supplemental Data S2.1, S2.2, and S2.3). Sequenced S. demissum small RNAs between 15 and 25 nt account for 69.2% of the total S. demissum data set; those below 15 nt comprise the second largest group and account for 19.8%, and small RNAs between 26 and 40 nt account for 9.9% (Fig. 4A). 5′-Terminal nucleotide composition analysis showed an enrichment for 5′ A and T residues in the 15- to 25-nt size class of small RNAs, with this enrichment diminishing in the larger and smaller size classes (Fig. 4B).
Figure 4.
Characteristics of cloned small RNAs. (A) Length distribution of small RNAs of the total S. demissum data set (4421 smRNAs). (Black bars) Indicate the percentage of small RNAs in each size class among the total data set. (B) Proportion of the total S. demissum small RNAs within each of the four size classes with A or T or G or C as the 5′-terminal nucleotide. Within each size class, (gray bar) the portion of small RNAs with a 5′-terminal A or T; (white bars) the portion with a 5′-terminal G or C. (C) Size distribution and 5′-terminal nucleotide composition of miRNAs ranging from 15 to 27 nt in length. (Black bar) The total number of cloned miRNA in each size class. The number of miRNAs with a 5′ (dark gray bar) A, (gray bar) T, (light gray bar) G, or (white bar) C residue. (D) Size distribution and 5′-terminal nucleotide composition of MiS small RNAs ranging from 15 to 31 nt in length. The 5′ nucleotide composition is indicated as in C.
We attempted to establish the identity of each nonredundant small RNA between 15 and 40 nt. To identify known miRNAs in this data set, we performed BLASTN searches of the miRbase (http://microrna.sanger.ac.uk/sequences/index.shtml), which identified 38 unique candidate miRNA sequences corresponding to 74 reads within the S. demissum small RNA data set and 30 unique candidate miRNA sequences within the combined N. benthamiana and N. glutinosa data set (Supplemental Table S2.2; Griffiths-Jones 2004; Griffiths-Jones et al. 2006). Analysis of candidate miRNA sequences confirmed the preference of a 5′-terminal T residue for 35 of 38 S. demissum and 27 of 30 Nicotiana candidate miRNAs described for AGO1-associated miRNAs (the S. demissum results are summarized in Fig. 4C; Supplemental Table 2.2; Pilcher et al. 2007; Mi et al. 2008). Candidate miRNA targets were identified in Solanaceae EST/TA databases for 14 of the 38 unique S. demissum miRNAs and for eight of the 30 unique Nicotiana miRNAs (Supplemental Table S2.2). The Nicotiana miR403 target was validated by 5′-RACE analysis (Supplemental Fig. S2.3).
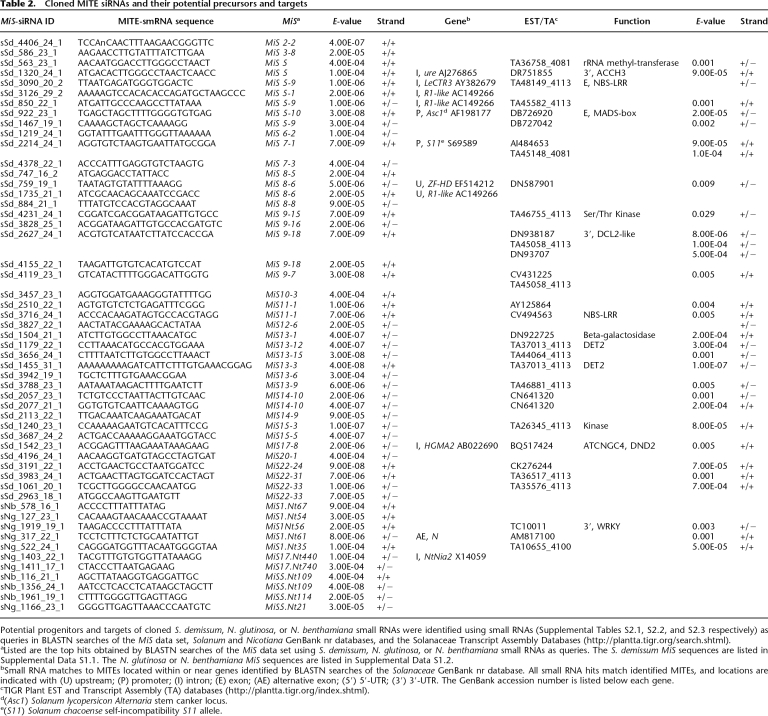
To identify MiS small RNAs, we used small RNAs as queries in BLASTN searches of Solanaceae MITE sequences. Notably, 43 nonredundant MiS small RNA sequences from S. demissum and 11 nonredundant MiS small RNA sequences from N. benthamiana and N. glutinosa were identified (Table 2). These small RNAs matched members of the MiS1–MiS3, MiS5–MiS15, MiS17, MiS20, and MiS22 families, and some were found in both plus and minus orientations (Table 2). Consistent with the size classes detected in the small RNA blots, the size distribution of cloned MiS small RNA peaked at 24 nt. The majority of other MiS small RNAs were 21 nt, 22 nt, or 23 nt in length (Fig. 4D). In contrast to cloned candidate miRNAs, a preference for a 5′-terminal A residue was observed for MiS small RNAs (Fig. 4D). Based on recent findings showing AGO-mediated sorting of small RNAs based on 5′-terminal nucleotide composition, we hypothesize that the 24-nt MiS small RNAs with a 5′-terminal adenosine may be recruited by AGO4 and involved in siRNA-mediated DNA methylation (RdDM) and transcriptional silencing, whereas AGO2 may recruit 21-nt MiS smRNAs with 5′ A residues (Mi et al. 2008). BLASTN searches of Solanaceae nr GenBank databases identified potential MITE progenitors and/or targets of MiS smRNA-mediated transcriptional silencing within gene promoters (Asc1 and S11, the Alternaria stem canker locus and the Solanum chacoense self-incompatibility S11 allele, respectively), introns (LeCTR3, ure, R1-like), or upstream regions of genes (R1-like) (Table 2). BLASTN searches against the TIGR Plant Transcript Assembly database identified candidate hits with E-values < 0.01 for 25 of the 43 MiS small RNAs (Table 2). Some MiS small RNAs retrieved multiple hits. One MiS1-1 small RNA from N. glutinosa, sNg_317_22_1 (Table 2), matched (+/−) a sequence within the 70-bp MiS1-1-derived alternative exon of the N gene (Fig. 2A; Table 2). The characteristic sequence, length, and terminal base composition of S. demissum, N. benthamiana, and N. glutinosa small RNAs support the hypothesis that the 24-nt MiS small RNAs are generated from MITEs by an NRPD1A–RDR2–DCL3 pathway. The finding that adenosine is the 5′-terminal nucleotide of most 24-nt MiS small RNAs supports the hypothesis that these RNAs interact with AGO4 for siRNA-dependent DNA methylation and transcriptional silencing. DCL2- and DCL4-dependent biogenesis pathways may also generate 21–22-nt MITE small RNAs that mediate post-transcriptional regulation of MITE expression.
Table 2.
Cloned MITE siRNAs and their potential precursors and targets
Potential progenitors and targets of cloned S. demissum, N. glutinosa, or N. benthamiana small RNAs were identified using small RNAs (Supplemental Tables S2.1, S2.2, and S2.3 respectively) as queries in BLASTN searches of the MiS data set, Solanum and Nicotiana GenBank nr databases, and the Solanaceae Transcript Assembly Databases (http://plantta.tigr.org/search.shtml).
aListed are the top hits obtained by BLASTN searches of the MiS data set using S. demissum, N. glutinosa, or N. benthamiana small RNAs as queries. The S. demissum MiS sequences are listed in Supplemental Data S1.1. The N. glutinosa or N. benthamiana MiS sequences are listed in Supplemental Data S1.2.
bSmall RNA matches to MITEs located within or near genes identified by BLASTN searches of the Solanaceae GenBank nr database. All small RNA hits match identified MITEs, and locations are indicated with (U) upstream; (P) promoter; (I) intron; (E) exon; (AE) alternative exon; (5′) 5′-UTR; (3′) 3′-UTR. The GenBank accession number is listed below each gene.
cTIGR Plant EST and Transcript Assembly (TA) databases (http://plantta.tigr.org/index.shtml).
d(Asc1) Solanum lycopersicon Alternaria stem canker locus.
e(S11) Solanum chacoense self-incompatibility S11 allele.
Genetic requirements for MITE small RNA biogenesis
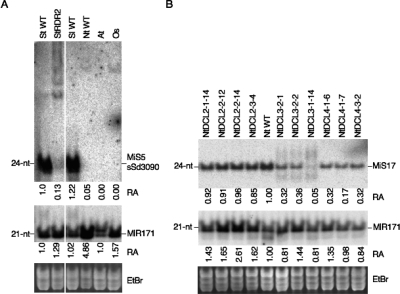
To test the hypothesis that MiS 21–24-nt small RNAs are products of DCL2-, DCL3-, or DCL4-dependent siRNA pathways, we investigated the genetic requirements for their biogenesis. We generated RNAi lines targeting candidate orthologs of DCL2, DCL3, and DCL4 in N. tabacum, and an RNAi line targeting the RDR2 ortholog of S. tuberosum. Supplemental Tables S4.1, S4.2, and S4.3 and Supplemental Figures S2.1 and S2.2 provide details on vector construction and characterization of N. tabacum RNAi lines. Characterization of transgenic NtDCL RNAi lines demonstrated that N. tabacum orthologs of Arabidopsis DCL2-4 loci were nonredundantly silenced (Supplemental Fig. S2.2). Low-molecular-weight RNA fractions were isolated from mature leaf tissue of tobacco NtDCL and potato RDR2 RNAi lines and wild-type (WT) controls, and blotted and hybridized with MiS5-1 small RNA probe, sSd_3090_20, or the MiS17 probe (Supplemental Table S3.2), to assess MiS small RNA accumulation in the RNAi lines.
Accumulation of 24-nt MiS5-1-related small RNA, detected with the sSd_3090_20_2 (Table 2) hybridization probe, was significantly reduced, with a relative abundance of 0.13 in the potato RDR2 RNAi line compared to wild-type (WT) levels. This reduction was accompanied by the appearance of several longer RNA species of ∼60 to 100 nt (Fig. 5A). The sSd_3090_20 probe detected MiS5 small RNA in WT potato and tomato, but not in tobacco, Arabidopsis, or rice, which may reflect either the low copy number of MiS5 in the tobacco genome or divergence of Solanum and Nicotiana MiS5 sequences (Supplemental Fig. S1; Supplemental Table S.1). In contrast, the MIR171 probe detected conserved MIR171 at similar levels in the potato RDR2 RNAi line and wild-type potato (Fig. 5A).
Figure 5.
RNA-dependent RNA polymerase (RDR2) and DICER-LIKE (DCL) are involved in the biogenesis of MiS small RNAs. Small RNA-enriched fractions isolated from wild type and RNAi lines were prepared and analyzed by Northern blot hybridization as described in Figure 3. Blots were hybridized with MiS and miRNA probes (see Supplemental Table S3.2 for a description of probes). (A) Detection of MiS5 (Sd3090 probe) small RNA and MIR171 in (St WT) wild-type S. tuberosum Katahdin; (St rdr2) S. tuberosum Katahdin RDR2 RNAi line; (Sl WT) wild-type S. lycopersicum VF36; (Nt WT) wild-type N. tabacum SR1; (At) wild-type Arabidopsis Col; and (Os) wild-type Oryza sativa Nipponbare. (B) Detection of MiS17 plus strand small RNA and MIR171 in wild-type tobacco SR1 and N. tabacum RNAi lines targeting (DCL2) DCL2-1, DCL2-2, and DCL2-3; (DCL3) DCL3-2 and DCL3-2; and (DCL4) DCL4-1 and DCL4-3 (for construction and characteristics of tobacco RNAi lines, see Supplemental Figs. S2.1 and S2.2; Supplemental Tables S4.1–S4.4). (A,B, bottom panels) The ethidium bromide (EtBr) staining of gels as loading controls. The numbers below the hybridization panels in A and B indicate the relative abundance (RA) of siRNA detected by Northern blot hybridization in each sample compared to StWT in A and NtWT in B. Relative siRNA abundance has been normalized (WT = 1.0) according to EtBr staining of gels in A and B.
In tobacco, four DCL2 RNAi lines—NtDCL2-1-14, NtDCL-2-12, NtDCL-2-14, and NtDCL2-3-4—showed a slight increase in MIR171 accumulation but no detectable change in MiS17 small RNA accumulation compared to wild type (Fig. 5B). In contrast, NtDLC3 and NtDCL4 lines showed a threefold to 20-fold reduction in MiS17 small RNA, but no significant change in the accumulation of MIR171 (Fig. 5B) compared to wild type. Intriguingly, shorter small RNAs of 21 to 22 nt as well as longer small RNAs of ∼30 nt and greater appear in the NtDCl3 lines, but not in the NtDCL4 lines.
These data indicate that MiS small RNA biogenesis involves RDR2, DCL3, and possibly DCL4. In Arabidopsis, RDR2 and DCL3 operate in a pathway generating ∼24-nt siRNAs that guide TGS; however, a role for DCL4 in generating endogenous 24-nt small RNA is unexpected and has not been observed in Arabidopsis.
Discussion
Solanaceae MITES contribute to evolution of gene complexity
MITEs are closely associated with genes (Wessler 1998; Mao et al. 2000), but the functional impact of these genic insertions has not been clearly established. Our investigation revealed a novel mechanism by which MITEs may assert gene regulation—via small RNA pathways. However, we also identified several MITEs that have the capacity to alter gene structure, thus playing the traditionally appreciated role of TEs in the evolution of new genes and transcriptome diversity. In future studies of MITE function, it will be important to account for the variety of mechanisms, both structural and small RNA-mediated, by which MITEs have an impact on gene regulation and genome evolution.
Our analysis of MiS demonstrates that MITEs can create new gene structures in several ways. We found that the N alternative exon, which generates the NL mRNA isoform, is encoded by MiS1-1. It is possible that MiS1 contributes to regulation of N expression at the transcriptional and/or post-transcriptional level. TMV infection might induce transient modification of TGS mechanisms that lead to changes in N transcription. Virus infection could also cause changes in post-transcriptional regulation of N expression by temporarily inhibiting PTGS and MiS1 siRNA-guided cleavage of the NL mRNA. The premature translation termination codon induced by the MiS1 alternative exon could render NL mRNA a target of nonsense-mediated decay, a post-transcriptional surveillance pathway. The predicted product of NL mRNA, a truncated TIR-NB protein (Ntr), may play a functional role in TMV resistance and participate in TIR domain-dedent recognition and oligomerization of N (Mestre and Baulcombe 2006; Burch-Smith et al. 2007). Additionally, we found evidence that MiS1 may provide a polyadenylation signal when located in the 3′-UTR of Nicotiana transcripts, indicating that MiS1 may have a general capacity to generate novel 3′-truncated polyadenylated transcripts. Furthermore, as with other TEs, insertion of MiS in exons can introduce premature stop codons, and insertion and subsequent excision can introduce short insertions and deletions, which contribute to the diversity of R genes in particular (Kuang et al. 2005). We also found that MiS5-1 has the capacity to capture and move host sequence (Fig. 1A), similar to Pack-MULEs reported in Arabidopsis, rice, and maize (Jiang et al. 2004a). All of these mechanisms are expected to contribute to the complexity of the genome.
A model for MITE small RNAs in gene regulation
Our identification of MiS small RNAs and multiple genes bearing MiS insertions, which could serve as both origins of small RNA and targets for small RNA-mediated silencing, suggests that MiS silencing pathways may be co-opted for regulation of genes bearing MiS, similar to the regulation imposed by small RNA-mediated regulation via the SINE-like direct repeats in the FWA promoter (Chan et al. 2004). This hypothesis can be extended by proposing that the amplification and diversification of MiS and other mobile elements may contribute to evolution of networks of coordinately regulated genes via insertion and subsequent selection of homologous elements in multiple genes, thus providing target sites for co-regulation by RISC or RITS complexes loaded with target-specific MiS and other TE small RNAs. Indeed, two studies have implicated MITEs as negative transcription regulators that may be targets of TGS: investigation of the LAT59 gene-defined Stowaway-Le1, which is located in the promoter, as a negative regulator of expression (Twell et al. 1991; Bureau and Wessler 1994), and inhibition of DNA methylation of the rice ubiquitin2 promoter, harboring Kiddo and MDM1, resulted in an increase in the ubiquitin2 transcript (Yang et al. 2005). Arabidopsis small RNAs are sorted into distinct AGO complexes, and this sorting has important functional implications. AGO1 preferentially recruits small RNAs with a 5′-terminal uridine, and AGO2 and AGO4 recruit small RNAs with a 5′-terminal adenosine (Mi et al. 2008). Consistent with these findings, we determined that the 5′-terminal residue of the majority of our candidate miRNAs is uridine. The 5′-terminal residue of the majority of MiS small RNAs was determined to be adenosine, suggesting that MiS smRNAs interact with AGO4 as described for repeat-associated siRNAs (rasiRNAs) involved in RNA-directed DNA methylation, histone modification, and transcriptional gene silencing of transposons and other repeats in Arabidopsis. AGO2 may recruit MiS 21-nt RNAs with 5′-terminal A residues.
Secondary siRNA generated from PTGS may also play a role in these hypothetical MiS regulatory networks. The identification of multiple conserved MiS sequences in ESTs databases, and their potential to form siRNA precursors from dsRNA via sense–antisense pairing or hairpin structures from RNA transcribed from inverted repeats suggests that MiS may have the capacity to generate small RNAs similar to nat-siRNAs or NRPD1A-independent siRNAs derived from single-stranded hairpin precursors. These siRNAs potentially target gene transcripts bearing MiS insertions for PTGS. The identification of Nicotiana MiS1 small RNAs matching paired 3′-UTRs containing complementary MiS1 insertions (Fig. 2B; Table 2) suggests a potential role for MiS1 small RNAs in post-transcriptional gene silencing. The presence of a transcribed MiS9 insertion in the 3′-UTR of tomato and potato ESTs/TAs encoding putative DCL2 products (Supplemental Table S2.1) and identification of MiS9 small RNA with perfect matches to the predicted MiS9 ssRNA hairpin also supports the hypothesized role of MITE-derived siRNAs in post-transcriptional silencing. In this regard, it is interesting to note that deep sequencing of tomato small RNAs revealed that TAPIR fold-back elements produce small RNAs in a miRNA-like pattern (Moxon et al. 2008). In the cases of pseudogenes with MiS insertions, PTGS may provide the ability to generate secondary siRNAs that regulate functional homologs.
The biogenesis of MITE siRNA
Using RNAi knockdown lines, we showed that RDR2, DCL3, and possibly DCL4 contribute to MiS small RNA accumulation in different ways. As expected, knockdown of RDR2 and DCL3 significantly reduced 24-nt MiS small RNA accumulation and, in addition, resulted in the appearance of novel small RNA species. In the StRDR2 RNAi line, several larger MiS5 RNAs, some with a length of ∼100 nt, were observed. In the NtDCL3-1 and NtDCL3-2 RNAi lines, novel size classes of ∼21–22-nt novel small RNAs and >30 nt were detected. These species were not observed in the NtDCL4, NtDCL2, or StRDR2 RNAi lines.
Our results support the hypothesis that biogenesis of the majority of MiS small RNA involves an NRPD1A/RDR2/DCL3/AGO4-dependent core pathway involved in transcriptional silencing and DNA methylation in Arabidopsis (Henderson and Jacobsen 2007). The smaller 21–22-nt RNAs detected in the NtDCL3 RNAi lines could result from DCL4 and DCL2 cleavage of accumulated MiS dsRNA precursor. The absence of these small RNAs in potato RDR2 RNAi lines indicates that their biogenesis may depend on RDR2. Processing of RDR2 products by DCL4 has been observed in plants expressing ssRNA hairpin structures from transgenes for induced silencing (Henderson and Jacobsen 2007; Smith et al. 2007). Nevertheless, a role for DCL4 in MiS small RNA accumulation is interesting and awaits further investigation.
Understanding the interactions of different silencing pathway components in production of TE-derived small RNAs will be resolved with genetic analysis that incorporates lines carrying mutations in multiple silencing RNA pathway loci. These lines will be required in experimental systems to investigate the potential functional roles of Solanaceae small RNAs. Given the high copy number of gene-associated transposons in Solanaceae, their potential to generate both RNA polymerase IV-dependent and -independent substrates for small interfering RNAs, and the significant functional redundancies documented at multiple steps in the silencing pathway (Henderson et al. 2006; Smith et al. 2007; Zhang et al. 2007), it will be important to systematically account for the different mechanisms of TE-derived small RNAs and their potential functional roles in gene silencing.
Methods
MITE analyses
Transposable elements were investigated at six resistance gene loci: the R1 locus on Chromosome 5 of S. demissum (Kuang et al. 2005), the major late blight resistance cluster (MLB) on Chromosome 11 of S. demissum (H. Kuang and B. Baker, unpubl.), the N locus in tobacco (Whitham et al. 1994), the Bs4 locus in tomato (Schornack et al. 2004), the Prf/Pto locus in tomato (Chang et al. 2002), and the RB locus on Chromosome 8 of Solanum bulbocastanum (Song et al. 2003). Sequences of two or three haplotypes of R1, Bs4, N, Prf/Pto, and RB were obtained from GenBank accessions (R1: AC135288, AC139840, AC142505, AC144791, AC145120, AC146506, AC149265, AC149266, AC149267, AC149287, AC149288, AC149290, AC149291, AC149301, AC149487, AC150162, AC151803, AC151815, AC151957, AC154033; Bs4: AY438027, AY438028; N: U15605, A535010 (NH); Prf/Pto: AF220602, AF220603; RB: AY303170, AY303171).
MITEs were detected using the related empty site (RESite) method, which confirms and delimits a MITE sequence by comparing its flanking sequence with a homologous region (Le et al. 2000). Sequences from syntenic regions or homologs of R-genes were aligned using ClustalX (Thompson et al. 1994). All insertions >60 bp were investigated for transposable element structural features such as TIRs and TSDs. To obtain additional MITEs, the sequences of all identified MITEs were used to perform BLASTN queries (E < e−5) in GenBank and in the TIGR Plant Transcript Assembly databases of tomato, potato, tobacco, pepper, N. benthamiana, and Petunia hybrida (http://tigrblast.tigr.org/euk-blast/plantta_blast.cgi) (Childs et al. 2007). Copy numbers were estimated by performing BLASTN searches with a representative member of each MiS family (Supplemental Data S1.1) in potato and tomato bacterial artificial chromosome (BAC) clone end and tobacco genomic sequence (http://www.tigr.org/tdb/sol/sol_ma_blast.shtml; http://www.tigr.org/tdb/sol/sol_ma_blast.shtml1; NCBI Nicotiana tabacum gss; Supplemental Table S1.2). MITEs may generally be considered to be shorter than 600 bp; however, we identified MITE families that included longer members, which we classified with their shorter counterparts based on the following criteria: We define MITEs with significantly similar internal sequences (<e−5 for BLASTN) to be members of the same MITE family, and MITEs with no significant similarity to be members of different MITE families, and further define MITE families with related TIR sequences as superfamilies. Each MITE was given a unique, two-number designation MiS(X)-X, with the first number identifying the family and the second identifying the individual member. All MITEs identified previously in Solanaceae species were included in this study for further analysis (Oosumi et al. 1995; Pozueta-Romero et al. 1996; Rebatchouk and Narita 1997; Mao et al. 2001; Schenke et al. 2003; Guyot et al. 2005).
Small RNA cloning and analyses
Total RNA (600 μg) was resolved by electrophoresis on a denaturing 15% polyacrylamide gel. A gel slice containing RNAs from the range of ∼10–60 nt was excised, and RNA was eluted by incubating in 0.3 M NaCl overnight at 4°C and ethanol-precipitated. Eluted RNAs were ligated to RNA/DNA chimeric adaptors (5′-ATCGTAGGCACCUGAAA-3′ and 5′-pUUUCTGTA GGCACCATCAATT-3′). The final ligation product was resolved through denaturing polyacrylamide gel electrophoresis, gel-eluted, ethanol-precipitated, and reverse-transcribed using an antisense reverse primer and superscript III RT (Invitrogen). The resulting cDNA was PCR-amplified using sense and antisense primers, and the products were resolved on a native 15% polyacrylamide gel, eluted, and BanI-digested. Digestion products were ligated to the TOPO-TA cloning vector (Invitrogen), and transformed plasmid DNA was purified and sequenced from clones using standard sequencing methods.
A Perl program was written to extract small RNA sequence from the raw ABI files by recognizing the adaptor sequences with a two-edit distance (insertion, deletion, and mismatch, with each count as one edit distance). All small RNAs between 1 and 100 nt in length were collected in FASTA format and numbered numerically from 1 to 4421. For example, the identifier sSd_1915_24 refers to S. demissum small RNA number 1915 and is 24 nt in length. Statistics on the small RNA size and end composition were compiled from the total small RNA data set contained in the FASTA file. A nonredundant small RNA FASTA formatted file was generated by removing redundant small RNA sequences and appending a read number to the small RNA identifier (Supplemental Data S2.1). MITE small RNAs (47 reads) and miRNAs (74 reads) were identified by BLASTN searches against a database of all MiS sequences (MiS1-22) and against the miR database http://microrna.sanger.ac.uk/sequences/index.shtml (Griffiths-Jones 2004; Griffiths-Jones et al. 2006), using a cut-off E-value of le-3. Small RNAs derived from rRNA and ribosomal genes (812 reads), tRNA (25 reads), and non-MiS transposable elements (18) were identified by BLASTN search against the TIGR Plant Repeat database (Ouyang and Buell 2004) and NCBI nr nucleotide database. Small RNAs that remained unidentified by the above searches (3323 reads) were placed into an unknown small RNA file. Candidate targets were predicted for all small RNAs by BLASTN against TIGR Plant Transcript Assembly databases (http://tigrblast.tigr.org/euk-blast/plantta_blast.cgi).
Small RNA detection
To detect MiS small RNA accumulation in various Solanaceae species, RNA was extracted from young leaf tissue of O. sativa Nipponbare, A. thaliana Columbia, N. benthamiana, N. tabacum SR1, N. tabacum SR1:NN (TG34), N. glutinosa, S. demissum, S. tuberosum Kennebec and Katahdin, S. lycopersicum VF36, and from floral tissues of S. demissum and S. lycopersicum plants, using TRIzol reagent (Sigma-Aldrich) as described previously (Chellappan et al. 2005), and blots carrying 40 μg of total RNA per lane were hybridized with random-primed DNA probes using amplified MITE sequences as templates (Supplemental Table S3.1). Hybridization and subsequent washes were performed as described (Chellappan et al. 2005).
To determine the genetic requirements for MiS small RNA accumulation, low-molecular-weight RNA was extracted from the mature leaf tissues of O. sativa, A. thaliana, S. tuberosum Katahdin, S. lycopersicum VF36, N. tabacum SR1, and from the RNAi knockdown lines in S. tuberosum Katahdin and N. tabacum SR1, as described previously (Diaz-Pendon et al. 2007), and blots carrying 10 μg per lane of low-molecular-weight enriched RNA were hybridized with end-labeled DNA oligonucleotides (Supplemental Table S3.2) to detect sSd3090_20_1 (MiS5), MIR171, and MiS17 small RNAs. Hybridization and subsequent washes were done as described (Diaz-Pendon et al. 2007). Blots were exposed to phosphor storage screens and scanned using a PhosphorImager (Molecular Dynamics). The quantification of band density was determined by Quantity One software (Bio-Rad, version 4.6.5) using the Volume Rectangle Tool.
Generation of transgenic DCL and RDR2 RNAi lines
To generate tobacco RNAi lines targeting the different DCL genes, Arabidopsis DCL2, DCL3, and DCL4 protein sequences were used as queries for tBLASTN searches against the NCBI Nicotiana tabacum gss database. All Nicotiana sequence hits were collected and categorized as one of the DCL candidate homologs according to their BLASTX scores against the Arabidopsis reference protein (Supplemental Table S4.2). Nicotiana DNA fragments corresponding to each DCL homolog were amplified using the primers described in Supplemental Table S4.1 and cloned into a pENTR/D/TOPO vector (Invitrogen). For each DCL homolog, unique constructs were generated to target different regions of the DCL gene; these constructs are distinguished by a numerical appendix, for example, NtDCL2-1, NtDCL2-2, and NtDCL2-3 (Supplemental Fig. S2.1; Supplemental Table S4.2). The DNA fragments (Supplemental Table S4.3) were introduced into a pHELLSGATE8 vector by LR reaction (Invitrogen) to produce a hairpin construct, sequenced, and transferred to Agrobacterium strain LBA4404 for transformation of N. tabacum SR1. Different lines generated with the same construct are distinguished by a line number, for example, Nt DCL2-1-14 (see Supplemental Fig. S2.2). Transgenic T0 generation plants were screened for silencing of DCL transcript expression by RT-PCR (Supplemental Fig. S2.2; Supplemental Table S4.4), and positive transformants were selected for small RNA analyses. A 413-bp cDNA fragment corresponding to the N. benthamiana RDR2 cDNA sequence (AY722009) was amplified from potato cv. Katahdin cDNA. The primers used for amplification included 5′-CACCCGCCAGAAGGTCTGTGTATC-3′ (RC13-forward) and 5′-TCACAGTGATCGACAACAGATTGC-3′ (RC14-reverse). Construct design and transformation procedures were performed essentially as described by Bhaskar et al. (2008). Transgenic potato (cv. Katahdin) plants silenced for RDR2, St RDR2, were verified by RT-PCR.
Acknowledgments
We are indebted to Aaron Friedman for his many contributions to the project including in-depth critical discussions and management of project resources, and for skillful editing of this manuscript. We are grateful to David Hantz and Julie Calfas for excellent care of greenhouse plants. This work was supported by the National Science Foundation (NSF) Plant Genome Research Program, DBI-0218166, and the United States Department of Agriculture, CRIS 5335-22000-006-00D.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.078196.108.
References
- Baulcombe D.C. Short silencing RNA: The dark matter of genetics? Cold Spring Harb. Symp. Quant. Biol. 2006;71:13–20. doi: 10.1101/sqb.2006.71.052. [DOI] [PubMed] [Google Scholar]
- Bennetzen J.L., Kellogg E.A. Do plants have a one-way ticket to genomic obesity? Plant Cell. 1997;9:1509–1514. doi: 10.1105/tpc.9.9.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaskar P.B., Raasch J.A., Kramer L.C., Neumann P., Wielgus S.M., Austin-Phillips S., Jiang J. Sgt1, but not Rar1, is essential for the RB-mediated broad-spectrum resistance to potato late blight. BMC Plant Biol. 2008;8:8. doi: 10.1186/1471-2229-8-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsani O., Zhu J., Verslues P.E., Sunkar R., Zhu J.K. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith T.M., Schiff M., Caplan J.L., Tsao J., Czymmek K., Dinesh-Kumar S.P. A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol. 2007;5:e68. doi: 10.1371/journal.pbio.0050068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau T.E., Wessler S.R. Stowaway: A new family of inverted repeat elements associated with the genes of both monocotyledonous and dicotyledonous plants. Plant Cell. 1994;6:907–916. doi: 10.1105/tpc.6.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington J.C., Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chan S.W., Zilberman D., Xie Z., Johansen L.K., Carrington J.C., Jacobsen S.E. RNA silencing genes control de novo DNA methylation. Science. 2004;303:1336. doi: 10.1126/science.1095989. [DOI] [PubMed] [Google Scholar]
- Chang J.H., Tai Y.S., Bernal A.J., Lavelle D.T., Staskawicz B.J., Michelmore R.W. Functional analyses of the Pto resistance gene family in tomato and the identification of a minor resistance determinant in a susceptible haplotype. Mol. Plant Microbe Interact. 2002;15:281–291. doi: 10.1094/MPMI.2002.15.3.281. [DOI] [PubMed] [Google Scholar]
- Chellappan P., Vanitharani R., Fauquet C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. 2005;102:10381–10386. doi: 10.1073/pnas.0504439102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. MicroRNA biogenesis and function in plants. FEBS Lett. 2005;579:5923–5931. doi: 10.1016/j.febslet.2005.07.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs K.L., Hamilton J.P., Zhu W., Ly E., Cheung F., Wu H., Rabinowicz P.D., Town C.D., Buell C.R., Chan A.P. The TIGR Plant Transcript Assemblies database. Nucleic Acids Res. 2007;35:D846–D851. doi: 10.1093/nar/gkl785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Pendon J.A., Li F., Li W.X., Ding S.W. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinesh-Kumar S.P., Baker B.J. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. 2000;97:1908–1913. doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.W., Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunoyer P., Himber C., Ruiz-Ferrer V., Alioua A., Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- Feng Q., Zhang Y., Hao P., Wang S., Fu G., Huang Y., Li Y., Zhu J., Liu Y., Hu X., et al. Sequence and analysis of rice chromosome 4. Nature. 2002;420:316–320. doi: 10.1038/nature01183. [DOI] [PubMed] [Google Scholar]
- Feschotte C., Swamy L., Wessler S.R. Genome-wide analysis of mariner-like transposable elements in rice reveals complex relationships with stowaway miniature inverted repeat transposable elements (MITEs) Genetics. 2003;163:747–758. doi: 10.1093/genetics/163.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyot R., Cheng X., Su Y., Cheng Z., Schlagenhauf E., Keller B., Ling H.Q. Complex organization and evolution of the tomato pericentromeric region at the FER gene locus. Plant Physiol. 2005;138:1205–1215. doi: 10.1104/pp.104.058099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson I.R., Jacobsen S.E. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Henderson I.R., Zhang X., Lu C., Johnson L., Meyers B.C., Green P.J., Jacobsen S.E. Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 2006;38:721–725. doi: 10.1038/ng1804. [DOI] [PubMed] [Google Scholar]
- Huang S., van der Vossen E.A., Kuang H., Vleeshouwers V.G., Zhang N., Borm T.J., van Eck H.J., Baker B., Jacobsen E., Visser R.G. Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 2005;42:251–261. doi: 10.1111/j.1365-313X.2005.02365.x. [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Hirochika H., Eddy S.R., McCouch S.R., Wessler S.R. An active DNA transposon family in rice. Nature. 2003;421:163–167. doi: 10.1038/nature01214. [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004a;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- Jiang N., Feschotte C., Zhang X., Wessler S.R. Using rice to understand the origin and amplification of miniature inverted repeat transposable elements (MITEs) Curr. Opin. Plant Biol. 2004b;7:115–119. doi: 10.1016/j.pbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Juretic N., Bureau T.E., Bruskiewich R.M. Transposable element annotation of the rice genome. Bioinformatics. 2004;20:155–160. doi: 10.1093/bioinformatics/bth019. [DOI] [PubMed] [Google Scholar]
- Kuang H., Wei F., Marano M.R., Wirtz U., Wang X., Liu J., Shum W.P., Zaborsky J., Tallon L.J., Rensink W., et al. The R1 resistance gene cluster contains three groups of independently evolving, type I R1 homologues and shows substantial structural variation among haplotypes of Solanum demissum. Plant J. 2005;44:37–51. doi: 10.1111/j.1365-313X.2005.02506.x. [DOI] [PubMed] [Google Scholar]
- Le Q.H., Wright S., Yu Z., Bureau T. Transposon diversity in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2000;97:7376–7381. doi: 10.1073/pnas.97.13.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Ding S.W. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu. Rev. Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., He Y., Amasino R., Chen X. siRNAs targeting an intronic transposon in the regulation of natural flowering behavior in Arabidopsis. Genes & Dev. 2004;18:2873–2878. doi: 10.1101/gad.1217304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Wood T.C., Yu Y., Budiman M.A., Tomkins J., Woo S., Sasinowski M., Presting G., Frisch D., Goff S., et al. Rice transposable elements: A survey of 73,000 sequence-tagged-connectors. Genome Res. 2000;10:982–990. doi: 10.1101/gr.10.7.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao L., Begum D., Goff S.A., Wing R.A. Sequence and analysis of the tomato JOINTLESS locus. Plant Physiol. 2001;126:1331–1340. doi: 10.1104/pp.126.3.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Mestre P., Baulcombe D.C. Elicitor-mediated oligomerization of the tobacco N disease resistance protein. Plant Cell. 2006;18:491–501. doi: 10.1105/tpc.105.037234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S., Cai T., Hu Y., Chen Y., Hodges E., Ni F., Wu L., Li S., Zhou H., Long C., et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moxon S., Jing R., Szittya G., Schwach F., Rusholme Pilcher R.L., Moulton V., Dalmay T. Deep sequencing of tomato short RNAs identifies microRNAs targeting genes involved in fruit ripening. Genome Res. 2008;18:1602–1609. doi: 10.1101/gr.080127.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosumi T., Garlick B., Belknap W.R. Identification and characterization of putative transposable DNA elements in solanaceous plants and Caenorhabditis elegans. Proc. Natl. Acad. Sci. 1995;92:8886–8890. doi: 10.1073/pnas.92.19.8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S., Buell C.R. The TIGR Plant Repeat Databases: A collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 2004;32:D360–D363. doi: 10.1093/nar/gkh099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilcher R.L., Moxon S., Pakseresht N., Moulton V., Manning K., Seymour G., Dalmay T. Identification of novel small RNAs in tomato (Solanum lycopersicum) Planta. 2007;226:709–717. doi: 10.1007/s00425-007-0518-y. [DOI] [PubMed] [Google Scholar]
- Piriyapongsa J., Jordan I.K. A family of human microRNA genes from miniature inverted-repeat transposable elements. PLoS One. 2007;2:e203. doi: 10.1371/journal.pone.0000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J., Jordan I.K. Dual coding of siRNAs and miRNAs by plant transposable elements. RNA. 2008;14:814–821. doi: 10.1261/rna.916708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J., Marino-Ramirez L., Jordan I.K. Origin and evolution of human microRNAs from transposable elements. Genetics. 2007;176:1323–1337. doi: 10.1534/genetics.107.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poethig R.S., Peragine A., Yoshikawa M., Hunter C., Willmann M., Wu G. The function of RNAi in plant development. Cold Spring Harb. Symp. Quant. Biol. 2006;71:165–170. doi: 10.1101/sqb.2006.71.030. [DOI] [PubMed] [Google Scholar]
- Pozueta-Romero J., Houlne G., Schantz R. Nonautonomous inverted repeat alien transposable elements are associated with genes of both monocotyledonous and dicotyledonous plants. Gene. 1996;171:147–153. doi: 10.1016/0378-1119(96)00007-8. [DOI] [PubMed] [Google Scholar]
- Rebatchouk D., Narita J.O. Foldback transposable elements in plants. Plant Mol. Biol. 1997;34:831–835. doi: 10.1023/a:1005855008823. [DOI] [PubMed] [Google Scholar]
- Reinhart B.J., Weinstein E.G., Rhoades M.W., Bartel B., Bartel D.P. MicroRNAs in plants. Genes & Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago N., Herraiz C., Goni J.R., Messeguer X., Casacuberta J.M. Genome-wide analysis of the Emigrant family of MITEs of Arabidopsis thaliana. Mol. Biol. Evol. 2002;19:2285–2293. doi: 10.1093/oxfordjournals.molbev.a004052. [DOI] [PubMed] [Google Scholar]
- Schenke D., Sasabe M., Toyoda K., Inagaki Y.S., Shiraishi T., Ichinose Y. Genomic structure of the NtPDR1 gene, harboring the two miniature inverted-repeat transposable elements, NtToya1 and NtStowaway101. Genes Genet. Syst. 2003;78:409–418. doi: 10.1266/ggs.78.409. [DOI] [PubMed] [Google Scholar]
- Schornack S., Ballvora A., Gurlebeck D., Peart J., Baulcombe D., Ganal M., Baker B., Bonas U., Lahaye T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004;37:46–60. doi: 10.1046/j.1365-313x.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- Shen Y., Ji G., Haas B.J., Wu X., Zheng J., Reese G.J., Li Q.Q. Genome level analysis of rice mRNA 3′-end processing signals and alternative polyadenylation. Nucleic Acids Res. 2008;36:3150–3161. doi: 10.1093/nar/gkn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin R.K., Martienssen R. Transposable elements and the epigenetic regulation of the genome. Nat. Rev. Genet. 2007;8:272–285. doi: 10.1038/nrg2072. [DOI] [PubMed] [Google Scholar]
- Slotkin R.K., Freeling M., Lisch D. Heritable transposon silencing initiated by a naturally occurring transposon inverted duplication. Nat. Genet. 2005;37:641–644. doi: 10.1038/ng1576. [DOI] [PubMed] [Google Scholar]
- Smith L.M., Pontes O., Searle I., Yelina N., Yousafzai F.K., Herr A.J., Pikaard C.S., Baulcombe D.C. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Bradeen J.M., Naess S.K., Raasch J.A., Wielgus S.M., Haberlach G.T., Liu J., Kuang H., Austin-Phillips S., Buell C.R., et al. Gene RB cloned from Solanum bulbocastanum confers broad spectrum resistance to potato late blight. Proc. Natl. Acad. Sci. 2003;100:9128–9133. doi: 10.1073/pnas.1533501100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar V.V., Kapoor A., Zhang K., Zhu J., Zhou T., Hasegawa P.M., Bressan R.A., Zhu J.K. Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature. 2007;447:735–738. doi: 10.1038/nature05864. [DOI] [PubMed] [Google Scholar]
- Stange C., Matus J.T., Elorza A., Arce-Johnson P. Identification and characterization of a novel tobacco mosaic virus resistance N gene homologue in Nicotiana tabacum plants. Funct. Plant Biol. 2004;31:149–158. doi: 10.1071/FP03160. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twell D., Yamaguchi J., Wing R.A., Ushiba J., Mccormick S. Promoter analysis of genes that are coordinately expressed during pollen development reveals pollen-specific enhancer sequences and shared regulatory elements. Genes & Dev. 1991;5:496–507. doi: 10.1101/gad.5.3.496. [DOI] [PubMed] [Google Scholar]
- Wessler S.R. Transposable elements and the evolution of gene expression. Symp. Soc. Exp. Biol. 1998;51:115–122. [PubMed] [Google Scholar]
- Whitham S., Dinesh-Kumar S.P., Choi D., Hehl R., Corr C., Baker B. The product of the tobacco mosaic virus resistance gene N: Similarity to toll and the interleukin-1 receptor. Cell. 1994;78:1101–1115. doi: 10.1016/0092-8674(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Wright S.I., Agrawal N., Bureau T.E. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 2003;13:1897–1903. doi: 10.1101/gr.1281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G., Lee Y.H., Jiang Y., Shi X., Kertbundit S., Hall T.C. A two-edged role for the transposable element Kiddo in the rice ubiquitin2 promoter. Plant Cell. 2005;17:1559–1568. doi: 10.1105/tpc.104.030528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Henderson I.R., Lu C., Green P.J., Jacobsen S.E. Role of RNA polymerase IV in plant small RNA metabolism. Proc. Natl. Acad. Sci. 2007;104:4536–4541. doi: 10.1073/pnas.0611456104. [DOI] [PMC free article] [PubMed] [Google Scholar]