Abstract
Insulators are DNA elements that prevent inappropriate interactions between the neighboring regions of the genome. They can be functionally classified as either enhancer blockers or domain barriers. CTCF (CCCTC-binding factor) is the only known major insulator-binding protein in the vertebrates and has been shown to bind many enhancer-blocking elements. However, it is not clear whether it plays a role in chromatin domain barriers between active and repressive domains. Here, we used ChIP-seq to map the genome-wide binding sites of CTCF in three cell types and identified significant binding of CTCF to the boundaries of repressive chromatin domains marked by H3K27me3. Although we find an extensive overlapping of CTCF-binding sites across the three cell types, its association with the domain boundaries is cell-type-specific. We further show that the nucleosomes flanking CTCF-binding sites are well positioned. Interestingly, we found a complementary pattern between the repressive H3K27me3 and the active H2AK5ac regions, which are separated by CTCF. Our data indicate that CTCF may play important roles in the barrier activity of insulators, and this study provides a resource for further investigation of the CTCF function in organizing chromatin in the human genome.
Insulators, which are DNA elements that prevent inappropriate interactions between the neighboring regions of the genome, can be functionally classified into enhancer blockers and barriers. The enhancer-blocking insulators prevent enhancers from interacting with unrelated genes, and the barrier insulators protect genes and regulatory regions from the adjacent heterochromatin or repressive domain-mediated effects, thus preventing position effects (Gerasimova and Corces 1996; Bell et al. 1999; Felsenfeld et al. 2004). Identified originally in Drosophila, insulators are known to bind proteins that mediate the insulator activity (Gerasimova and Corces 2001). While several such proteins have been identified in Drosophila, the only major insulator-binding protein identified in vertebrates is CTCF (CCCTC-binding factor) (Bell et al. 1999; Gerasimova and Corces 2001; West et al. 2002; Felsenfeld et al. 2004).
CTCF, a ubiquitously-expressed 11-zinc finger protein, is a critical transcription factor, which is involved in transcriptional activation and repression in addition to binding the chromatin insulators (Ohlsson et al. 2001; Gaszner and Felsenfeld 2006; Williams and Flavell 2008). It was originally identified as a repressor (Lobanenkov et al. 1990; Filippova et al. 1996) and later shown to be an activator of transcription (Vostrov and Quitschke 1997). Recently, it has been implicated in X chromosome inactivation (Filippova et al. 2005; Xu et al. 2007). The enhancer-blocking insulator activity of CTCF was first demonstrated at the HS4 insulator located at the 5′ end of the chicken beta-globin locus (Bell et al. 1999). The insulator function of CTCF has also been implicated in imprinting at the Igf2/H19 locus (Bell and Felsenfeld 2000; Hark et al. 2000; Kanduri et al. 2000; Fedoriw et al. 2004).
Recently, several genome-scale mapping experiments for CTCF-binding sites have been performed for a better understanding of the CTCF function. A study in mouse identified ∼200 CTCF-bound DNA fragments displaying enhancer-blocking activity (Mukhopadhyay et al. 2004). In a computational analysis of the human conserved noncoding elements, nearly 15,000 potential CTCF-binding sites were identified (Xie et al. 2007). A recent chromatin immunoprecipitation with microarray hybridization (ChIP-chip) study in human IMR90 cells identified 13,804 CTCF-binding regions (Kim et al. 2007). A cell-type invariance of CTCF binding was reported in this study by comparing the binding sites in IMR90 cells with that of the 232 sites identified in U937 cells (Kim et al. 2007).
In our earlier chromatin immunoprecipitation with massively parallel sequencing (ChIP-seq) studies, we had observed CTCF-binding sites flanking active domains with the region outside being histone H3K27 trimethylated (H3K27me3), a modification associated with the repressed regions of chromatin (Barski et al. 2007). Even though initial studies of chicken HS4 insulator suggested the importance of the CTCF-binding sites for its barrier activity, later dissection of this insulator showed that CTCF was not required for this activity (Recillas-Targa et al. 2002). While a few other studies in the recent past have suggested a barrier activity for CTCF (Cho et al. 2005; Filippova et al. 2005), there has been no direct evidence for this (Gaszner and Felsenfeld 2006). In order to examine whether CTCF is indeed involved in the barrier activity, it is important to delineate the relationship between CTCF-binding sites and the repressive and active domains of the genome.
In this study we investigated the potential role of CTCF in delimiting the repressive genomic domains. To identify CTCF-bound genomic sites at high resolution, we analyzed the ChIP-seq data from HeLa and Jurkat cells obtained in this study along with the ChIP-seq data from resting human CD4+ T cells (Barski et al. 2007) using the binding-site identification algorithm, SISSRs (site identification from short sequence reads) (Jothi et al. 2008). Our data revealed an extensive overlap of the CTCF-binding sites across the genome between the different cell types studied. A subset of the CTCF-binding sites was significantly associated with the boundaries of H3K27me3 domains, suggesting a possible repressive domain barrier function. Interestingly, the potential domain barrier activity of CTCF was cell-type-specific. We observed strong cell-type-specific phasing of nucleosomes at the CTCF-binding sites. We found that the histone H2AK5 acetylation (H2AK5ac) marked the active regions of the genome and was complementary to H3K27me3. CTCF binding in between these two domains further reinforces its potential role in the barrier insulator function.
Results
CTCF-binding sites overlap extensively between cell types
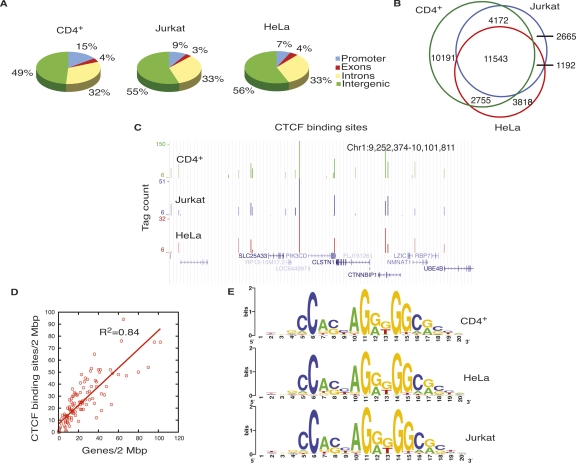
To identify the CTCF-bound genomic sites at high resolution, we analyzed ChIP-seq data from HeLa and Jurkat cells generated in this study, along with the ChIP-seq data from resting human CD4+ T cells (Barski et al. 2007) using SISSRs (Jothi et al. 2008). We identified 28,661, 19,308, and 19,572 CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells, respectively. Though a majority of CTCF-binding sites were located in the intergenic regions, many occupied other regions of the genome as well (Fig. 1A). Extensive overlap (40%–60%) of CTCF-binding sites was observed between the three cell types (Fig. 1B,C). We then compared the CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells to the binding sites in IMR90 cells, reported in another study (Kim et al. 2007). Over 56% of IMR90 CTCF sites overlapped with one or more of the CTCF sites in the three cell types considered in this study, confirming the general cell-type invariance of CTCF binding (Kim et al. 2007). Consistent with the previous observations (Kim et al. 2007), the gene-rich regions were enriched in CTCF-binding sites (Fig. 1D). Motif analysis (see Methods) on the identified CTCF sites revealed a consensus DNA-binding motif, which was identical in all of the three cell types studied (Fig. 1E). The consensus motif was very similar to the previously identified one (Kim et al. 2007). Over 90% of all CTCF sites identified in this study in HeLa cells, CD4+ T cells, and Jurkat cells contained the consensus motif. Though the identified motif appeared to be the major CTCF-binding sequence, a significant number of the sites lack the identified consensus sequence. This result is consistent with a recent study, which found that CTCF binds to genomic regions that lack the motif (Kim et al. 2007). Binding of CTCF to diverse sequences by using different combinations of its 11-zinc fingers has also been well documented (Filippova et al. 1996; Burcin et al. 1997; Ohlsson et al. 2001; Gaszner and Felsenfeld 2006; Filippova 2008).
Figure 1.
CTCF-binding sites overlap among different cell types. (A) Genome-wide distribution of CTCF-binding sites in CD4+ T cells, Jurkat cells, and HeLa cells. (B) CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells overlap extensively. (C) Overlap of CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells in an 850-kb region in chromosome 1, shown as custom tracks on the UCSC genome browser (Karolchik et al. 2008). (D) The densities of genes and CTCF-binding sites per 2 Mbp in chromosome 1. X-axis, number of genes/2 Mbp; y-axis, number of CTCF-binding sites/2 Mbp. (E) CTCF-binding motif in CD4+ T cells, HeLa cells, and Jurkat cells.
CTCF is enriched at the chromatin domain boundaries
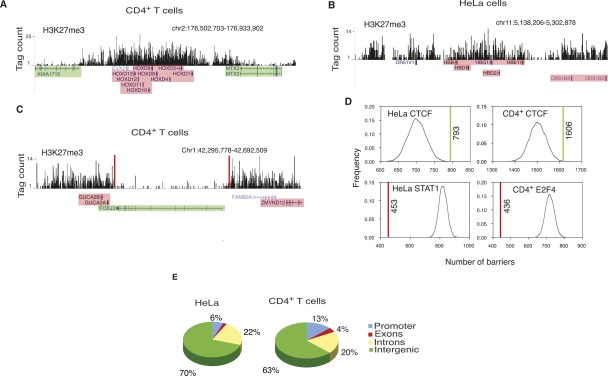
Since H3K27me3 marks the repressive regions of the genome, we decided to use H3K27me3 signals to identify the repressive chromatin domains. The genome-wide distribution of H3K27me3 in human CD4+ T cells (Barski et al. 2007) and HeLa cells was determined using ChIP-seq. As shown in Figure 2, A and B, H3K27me3 marked several repressed regions of the genome in both the cell types. To identify the repressive domains across the genome, we searched for contiguous stretches of H3K27me3-modified regions (see Methods for details). This analysis revealed 39,900 and 32,704 such domains in CD4+ T cells and HeLa cells, respectively. Two-thirds of the domains ranged in length between 5 kb and 25 kb in both the cell types (Supplemental Fig. S1).
Figure 2.
CTCF is enriched at the H3K27me3 domain boundaries. (A) Histone H3K27me3 pattern at the HOXD locus in CD4+ T cells; (B) beta-globin locus in HeLa cells shown as custom tracks on the UCSC genome browser (Karolchik et al. 2008). (Green) Expressed genes; (pink) silent genes. (C) A 400-kb region in chromosome 1 in CD4+ T cells. The barrier CTCF sites, shown as red bars, could be involved in maintaining the FOXJ3 gene free of H3K27me3, thus keeping it active, while the GUCA2A and GUCA2B genes that are within the H3K27me3 domain are silent. (Green) Expressed genes; (pink) silent genes. (D) In HeLa and CD4+ T cells, colocalization of the barrier CTCF sites with H3K27me3 domain boundaries (green lines) is higher compared with the colocalization between the randomly distributed sites and H3K27me3 domain boundaries (blue curve). The colocalization of the STAT1 and E2F4-binding sites (red line) with the H3K27me3 domain boundaries is lower than the colocalization between the randomly distributed sites and H3K27me3 domain boundaries (blue curve) in HeLa and CD4+ T cells. (E) Distribution of the barrier CTCF sites in HeLa and CD4+ T cells.
To identify the CTCF-bound genomic regions that may act as domain barriers, we searched for those CTCF-binding sites that occur near the H3K27me3 domain boundaries (edges of H3K27me3 domains; see Methods for details). As a conservative estimate, based on the enrichment of CTCF-binding sites near domain boundaries (Supplemental Fig. S2), we chose 1 kb as the maximal distance between the domain boundary and CTCF-binding site for it to be classified as a barrier. Since this 1-kb threshold could be too large for smaller domains, which could be ∼1–5 kb in length, we imposed an additional restriction, which requires that the distance between the H3K27me3 domain boundary and the CTCF-binding site be within 10% of the domain length (see Methods).
We identified 1606 and 793 CTCF-binding sites as barrier sites in CD4+ T cells and HeLa cells, respectively (Fig. 2C). The probability of this many CTCF sites colocalizing with the domain boundaries by chance is very low (P < 3 × 10−3 for CD4+ T cells and P < 10−4 for HeLa cells; see Methods for details). As a negative control, we tested the association of other unrelated proteins with the H3K27me3 domain boundaries. In CD4+ T cells, we mapped E2F4-binding sites by ChIP-seq (S. Cuddapah, R. Jothi, D.E. Schones, K. Cui, and K. Zhao, unpubl.) and identified 13,565 sites using SISSRs. We also identified 22,415 STAT1-binding sites in unstimulated HeLa S3 cells from the ChIP-seq data published previously (Robertson et al. 2007). Our analysis revealed that the total number of CTCF sites that occurred at the H3K27me3 domain boundaries in both the cell types (green lines) was higher than that of the randomly generated sites (blue curve) (Fig. 2D). Conversely, the occurrence of STAT1 and E2F4 sites (red lines) at the H3K27me3 domain boundaries was lower than that of the randomly generated sites (blue curve) (Fig. 2D). These results suggest a possible involvement of CTCF in the barrier function at the boundaries of repressive chromatin domains. Even though only 1578 domains in CD4+ T cells and 771 domains in HeLa cells were associated with CTCF binding, it should be noted that our criteria for classifying CTCF-binding sites as barrier sites is stringent, and that the actual number of CTCF barriers in the genome could be much more than what we have reported, as the CTCF sites away from the domain boundaries could also function as barriers, possibly through a looping mechanism. Most of the barriers occurred in the intergenic regions in both HeLa cells and CD4+ T cells (Fig. 2E). Motif analysis of the barrier CTCF sites revealed consensus DNA-binding motifs in both CD4+ T cells and HeLa cells, which were identical to the motif found for the “all CTCF” sites (Fig. 1E). We could not find any secondary motifs associated with the barrier CTCF sites.
CTCF binding at barriers are cell-type-specific
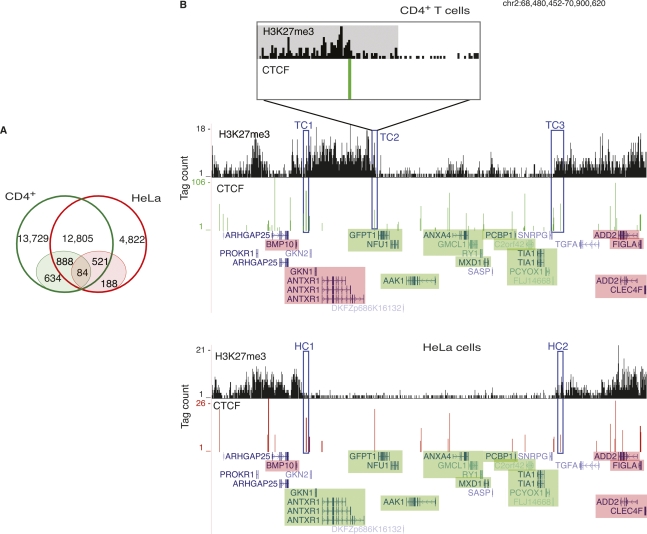
In spite of the significant overlap of CTCF-binding sites between the cell types (Fig. 1B), there was almost no overlap in the barrier CTCF sites between CD4+ T cells and HeLa cells, indicating that the CTCF barriers are highly cell-type-specific (Fig. 3A). We identified several genomic regions that bound CTCF in both cell types, but were barriers of H3K27me3 domains only in one cell type but not the other. Among 1409 such sites, 888 were barriers in CD4+ T cells and 521 were barriers in HeLa cells (Fig. 3A). For example, in HeLa cells (Fig. 3B, bottom) a 2.5-Mbp region in chromosome 2 contained a cluster of expressed genes with very low levels of H3K27me3 compared with the surrounding regions, where the genes are silent. The barrier CTCF sites in HeLa cells (HC1 and HC2) could be involved in keeping this locus free of H3K27me3. Despite occupying the same locus in CD4+ T cells, the CTCF site that might be functioning as a barrier of H3K27me3 in HeLa cells (HC1) clearly did not appear to perform the same function in CD4+ T cells (TC1) (Fig. 3B, top). The region downstream of this CTCF-binding site (TC1) was associated with H3K27me3 and the GKN1 and ANTXR1 genes were silent in CD4+ T cells, while these genes were active in HeLa cells. However, no CTCF sites were identified as barriers in this region in CD4+ T cells, which may be caused by a stringent definition of barrier CTCF sites. Since we observed an enrichment of CTCF-binding sites up to 5 kb into the H3K27me3 domain boundaries (Supplemental Fig. S2), we relaxed the barrier definition to include the CTCF sites within 10% of the domain or 5000 bp, whichever is smaller. This definition identified 3583 and 1089 barrier sites in CD4+ T cells and HeLa cells, respectively. Even under this relaxed definition, the CTCF-binding site TC2 (Fig. 3B, top) failed to qualify as a barrier site, as it was 5869 bp away from the domain boundary. However, a closer examination of the H3K27me3 patterns near TC2 (Fig. 3B, inset; H3K27me3 domain is shaded gray) revealed a steep decrease of H3K27me3 on one side of the CTCF site, suggesting that this CTCF site likely acts as a barrier. Therefore, we could have underestimated the number of potential barriers bound by CTCF.
Figure 3.
Barrier CTCF sites are cell-type-specific. (A) While over 50% and 74% of the CTCF-binding sites in CD4+ T cells and HeLa cells, respectively, overlap (open circles), only 5% and 11% of the barrier CTCF sites in the respective cell types overlap. The barrier CTCF sites are shaded. (B) A 2.5-Mbp region in chromosome 2 in CD4+ T cells (top) and HeLa cells (bottom). (Green) Expressed genes; (pink) silent genes. (HC1, HC2) Barrier CTCF sites in HeLa cells. A locus, which is occupied by CTCF in both CD4+ T cells and HeLa cells, could be functioning as a barrier in HeLa cells (HC1), but not in CD4+ T cells (TC1). (TC2, TC3) CTCF-binding sites, which could potentially be playing the role of domain barriers in CD4+ T cells. (Inset) An enlargement of the 30-kb region (TC2) in CD4+ T cells. The H3K27me3 domain is shaded gray.
CTCF demarcates active and repressive regions of the genome
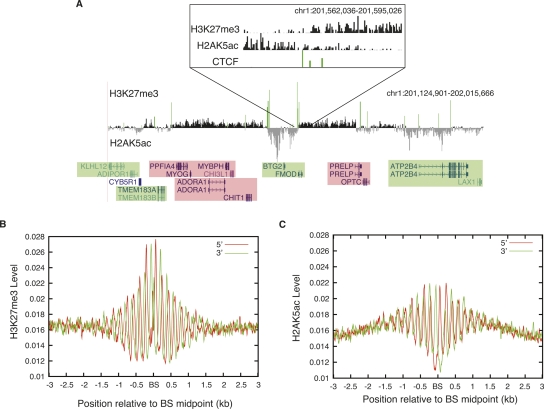
The acetylation of several histone residues is known to mark the active regions of the genome (Roh et al. 2005, 2006; Berger 2007; Li et al. 2007; Wang et al. 2008). In a recent study, we mapped the acetylation of 18 histone lysines in CD4+ T cells (Wang et al. 2008). Interestingly, we found that the domains enriched with H3K27me3 and acetylation existed adjacent to each other in several genomic loci. As shown in Figure 4A, these histone modifications were evidently complementary and they appeared to be separated by CTCF binding (green bars). The H2AK5 acetylation marked the active regions of the chromatin, with the genes residing within being expressed. As shown in Figure 4A (inset), increasing levels of H3K27me3 on one side of the CTCF-binding site and H2AK5ac on the other were observed, while the levels of these modifications at the CTCF-binding sites are low. The presence of both the modifications at the CTCF-binding sites (Fig. 4A, inset) could be caused by heterogeneity in cell populations. CTCF-binding sites mark the boundaries of the H3K27me3 and H2AK5ac domains, which strongly suggests a role for CTCF in separating the active and repressed regions of the genome (Fig. 4A, inset). In order to examine the distribution patterns of H3K27me3 and H2AK5ac surrounding CTCF-binding sites in greater detail, we aligned all of the nonpromoter (at least 5 kb away from an annotated TSS) CTCF-bound regions in CD4+ T cells and plotted the H3K27me3 and H2AK5ac profiles. We observed a striking phasing pattern of these signals in a 2-kb region surrounding the CTCF-binding sites (Fig. 4B,C).
Figure 4.
CTCF-binding sites separate active and repressive domains. (A) CTCF-binding sites separate the chromatin domains marked by H3K27me3 and H2AK5ac in a 900-kb region in chromosome 1. The H3K27me3 and H2AK5ac loci are complementary to each other and appear to be separated by CTCF-binding sites (green bars). The active genes are shaded green and the silent genes are shaded pink. (Inset) The transition between the H3K27me3 and H2AK5ac domains separated by CTCF sites. (B) The H3K27me3 profiles were plotted surrounding the nonpromoter (at least 5 kb away from the promoter) CTCF-binding sites in CD4+ T cells. (BS) CTCF-binding site. Red line indicates the reads from the sense strand and green line indicates reads from the antisense strand of DNA. (C) The H2AK5ac profiles were plotted surrounding the nonpromoter CTCF-binding sites in CD4+ T cells. (BS) CTCF-binding site.
The chromatin architecture at CTCF-binding sites is cell-type-specific
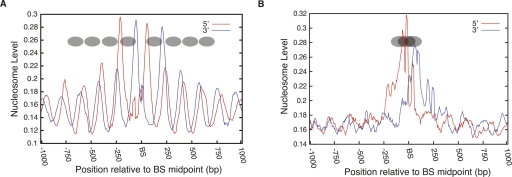
Since the chromatin used for analyzing H3K27me3 and H2AK5ac was mononucleosomal particles, the observed phasing pattern in Figure 4B,C suggests strong nucleosome positioning surrounding the CTCF-binding sites. To examine the nucleosome positioning directly, we analyzed the distribution of nucleosomes surrounding the CTCF-binding sites using the data from our recent study, which mapped the nucleosomes across the human genome in CD4+ T cells (Schones et al. 2008). This analysis indicated that CTCF bound to a linker region between two well-positioned nucleosomes, and the positioned nucleosomes extended on either side of the CTCF-binding site (Fig. 5A), which is consistent with earlier studies (Filippova et al. 2001; Kanduri et al. 2002;Fu et al. 2008). Though there was a high degree of overlap in CTCF-binding sites between CD4+ T cells and HeLa cells, about 26% of CTCF-binding sites were unique to HeLa cells. To determine whether the positioning of nucleosomes flanking the CTCF sites is cell-type invariant, we examined the CD4+ T cells nucleosome profiles at the CTCF-binding sites that were specific to HeLa cells (i.e., no binding detected in CD4+ T cells). The nucleosome profile at these HeLa-specific binding sites indicates that a nucleosome is occluding the CTCF-binding site and no other periodically positioned nucleosomes were present (Fig. 5B). The noisier nucleosome peak in Figure 5B, which occludes the CTCF-binding site, suggests overlapping of nucleosome positions. This could also explain the lack of other periodically positioned nucleosomes flanking the CTCF-binding site (Fig. 5B). These results suggest that the chromatin architecture at CTCF-binding sites is cell-type specific. We then compared the nucleosome phasing pattern between the CD4+ T cell-specific CTCF-binding sites and the sites shared by the CD4+ T cells and HeLa cells (Supplemental Fig. S3). The overall pattern of nucleosome phasing was similar between these two sets of CTCF-binding sites, although a significant nucleosome signal overlapping the CTCF sites was observed in the CD4+ T cells-only sites (Supplemental Fig. S3A). One possible explanation for this result is that CTCF binds to only one of the two alleles at these sites or binds only in a fraction of the CD4+ T cells.
Figure 5.
Nucleosomes are strongly positioned at the CTCF-binding sites and the positioning is cell-type-specific. (A) Nucleosomes at the CTCF-binding sites in CD4+ T cells are phased. CTCF binds to the linker region surrounded by well-positioned nucleosomes. The deduced nucleosomes are indicated as ovals. (BS) CTCF-binding site. (B) Nucleosome positions in CD4+ T cells at the HeLa cell-specific CTCF-binding sites. A nucleosome occludes the CTCF-binding site where no binding is detected in CD4+ T cells. The possible nucleosome positions on the CTCF-binding site are indicated as ovals.
Discussion
Insulators can be functionally divided into enhancer blockers, which prevent enhancers from activating unrelated genes, and domain barriers, which protect genomic regions from the adjacent heterochromatin or repressive domain-mediated effects (Felsenfeld et al. 2004). While CTCF has been suggested to possess barrier activity, no direct evidence exists thus far (Gaszner and Felsenfeld 2006). In this study, we addressed this question by identifying genome-wide H3K27me3-associated repressive chromatin domains and CTCF-binding sites in the primary CD4+ T cells, HeLa cells, and Jurkat cells. Interestingly, we find that the H2AK5ac-associated chromatin domains are located adjacent to the H3K27me3 domains and harbored expressed genes. We also find that CTCF-binding sites are significantly enriched at the boundaries between the H3K27me3 and H2AK5ac domains, indicating that CTCF may be involved in the chromatin barrier function.
Analysis of nucleosome positioning in the vicinity of nonpromoter CTCF-binding sites indicates that CTCF binds to the linker region between nucleosomes, and the nucleosomes surrounding the functional binding sites are well positioned. Furthermore, the CTCF sites that are bound in HeLa cells, but not bound in CD4+ T cells, have a nucleosome positioned right over the binding sites, which render them inaccessible (Fig. 5B). Periodic positioning of nucleosomes flanking the CTCF-binding sites has been observed earlier (Filippova et al. 2001; Kanduri et al. 2002; Fu et al. 2008). While an earlier study on the nucleosome positions at the H19 locus concluded that the positioning of the nucleosomes regulate CTCF interaction with its target site, but CTCF itself does not position nucleosomes (Kanduri et al. 2002), a recent study attributed chromatin remodeling function to CTCF (Fu et al. 2008). From our results, though it is clear that chromatin architecture plays a role in cell-type-specific CTCF/target interaction, it is not clear whether CTCF binds to positioned nucleosomal regions or whether the nucleosomes are positioned as a result of CTCF binding.
We find that CTCF can bind to the same locus in different cell types, but may function as barrier in one cell type but not in the other (Fig. 3B). This strongly suggests that CTCF binding alone may not be sufficient to mediate the barrier function of chromatin insulators and a secondary event may be required for specificity. This secondary event could be the binding of a secondary protein, as several proteins have been shown to interact with CTCF (Donohoe et al. 2007; Wallace and Felsenfeld 2007; Rubio et al. 2008; Stedman et al. 2008; Wendt et al. 2008). Besides being an insulator-binding protein, CTCF performs several functions, and it remains to be seen whether the varying functions of CTCF also depend on the interacting proteins. The association of CTCF with nucleophosmin and CHD8 has been suggested to be involved in its insulation function (Yusufzai et al. 2004; Wallace and Felsenfeld 2007). YY1 has been shown to be associated with CTCF in X chromosome inactivation (Donohoe et al. 2007). Taken together, these results point toward CTCF being a dynamic regulator of cellular functions whose specific roles could be defined by the factor(s) that associate with it.
The cohesin protein, which shares the consensus motif and colocalizes extensively with CTCF, has been suggested to function as a transcriptional insulator (Wendt et al. 2008). It would be interesting to investigate the role of cohesin in the barrier activity of CTCF. A conditional deletion of CTCF has found no evidence of spreading of the repressive histone modifications into the beta-globin 3′ HS1 locus (Splinter et al. 2006), suggesting an interaction of CTCF with another protein(s) that might play the role of barrier in the absence of CTCF. However, the regulation of chromatin domains by proteins independent of CTCF is also possible.
Thus, the function of CTCF appears to be regulated at least at two levels. The first level of regulation involves binding of CTCF to the target sites where the periodic nucleosome positioning precedes or succeeds binding of CTCF, both scenarios being possible, depending on the locus. The next level of regulation appears to involve the binding of interacting proteins, thus providing its functionality. Identification of the factors responsible for cell-type-specific chromatin remodeling, along with the identification of interacting proteins would be important to understand the mechanism of maintenance of chromatin architecture. The genome-wide CTCF-binding sites in multiple cell types and the cell-type-specific barrier CTCF sites identified in this study will be important resources not only for the understanding of the organization of the genome, but also for deciphering the cellular regulations that CTCF is involved in.
Methods
CD4+ T cell purification and cell culture
Human CD4+ T cells were purified as described (Barski et al. 2007). HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1 mM glutamine. Jurkat cells were maintained in RPMI supplemented with 10% fetal bovine serum and 1 mM glutamine.
ChIP-seq and gene expression analysis
For chromatin immunoprecipitation (ChIP) using a CTCF antibody (Upstate 07-729), the formaldehyde cross-linked cells were sonicated to obtain DNA fragments ranging in size from 150 to 200 bp. Cluster generation and sequencing were performed as described earlier (Barski et al. 2007) and sequence tags were mapped to the human genome using the Illumina Analysis Pipeline. MNase digested, mononucleosome-sized DNA fragments were used for H3K27me3 and H2AK5ac ChIPs (Barski et al. 2007; Wang et al. 2008). The gene expression profiles of CD4+ T cells (Schones et al. 2008) and HeLa cells were analyzed using the Affymetrix HG-U133 Plus 2.0 chip.
Binding-site identification
Sequenced short reads from the ChIP-seq experiments were processed using the SISSRs algorithm (Jothi et al. 2008) to identify genome-wide binding-site locations. Reads overlapping satellite repeat regions were eliminated from the SISSRs analysis. SISSRs V1.2 with “u” and “c” option was run with the following settings: average DNA fragment length = 200 bp; scanning window size w = 2; false discovery rate D = 10−3. More than 95% of the identified binding sites defined in this manner were within 400 bp in length. We identified a total of 28,661, 19,308, and 19,572 CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells, respectively. We also identified 13,565 E2F4-binding sites from ChIP-seq data from CD4+ T cells (S. Cuddapah, R. Jothi, D.E. Schones, K. Cui, and K. Zhao, unpubl.), and 22,415 STAT1 binding sites from ChIP-seq data for unstimulated HeLa cells (Robertson et al. 2007).
Genome-wide distribution of binding sites
Genome-wide distribution of CTCF-binding sites and barrier CTCF sites was determined with reference to RefSeq genes downloaded from UCSC genome browser (Karolchik et al. 2008). The 2-kb region centered on the transcription start site (TSS) was defined as the promoter.
Correlation between binding site and gene density
The entire genome was scanned using a 2-Mbp window, and the number of genes and CTCF-binding sites that fall within each window were recorded. Correlation between the gene density and CTCF-binding site density was assessed by fitting a linear regression.
Motif analysis
MEME (Bailey et al. 2006) with default parameters was used to identify statistically over-represented consensus motifs within the inferred binding sites. Over 90% of the CTCF-binding sites in CD4+ T cells, HeLa cells, and Jurkat cells contained the inferred consensus sequence.
Barrier site identification
The mapped H3K27me3 reads were first grouped into 1000-bp summary windows, following which “islands” of summary windows enriched with H3K27me3 tags were identified using an approach similar to that used in our earlier study (Barski et al. 2007). The islands identified in this manner are referred to as H3K27me3 domains. A total of 39,900 and 32,704 domains were identified in CD4+ T cells and HeLa cells, respectively. A CTCF-binding site, denoted by genomic coordinate x, is defined as a barrier site relative to a H3K27me3 domain d of length l, only if the distance between x and the domain boundary is at most the smaller of l/10 and 1000 bp. This definition yielded 1606 and 793 barrier CTCF sites in CD4+ T cells and HeLa cells, respectively. Using the same barrier site definition, we found that 436 E2F4-binding sites in CD4+ T cells, and 453 STAT1-binding sites in HeLa cells, qualify as barrier sites.
Data availability
The following data has been deposited in the NCBI GEO (under accession no. GSE12889): ChIP-seq raw seqeuence tags from CD4+ T cells (CTCF and H3K27me3), HeLa cells (CTCF and H3K27me3), and Jurkat cells (CTCF); H3K27me3 domains in CD4+ T cells and HeLa cells; barrier CTCF sites in CD4+ T cells and HeLa cells and HeLa cell gene expression (HG-U133 Plus 2.0 chip) data.
Statistical analysis
Barrier site identification
In order to assess the possibility that the identified barrier CTCF sites (1606 in CD4+ T cells and 793 in HeLa cells) colocalize with domain boundaries just by chance, we performed 10,000 trials of the following randomization experiment. In each trial, the observed CTCF sites were reassigned to random positions in the genome, and the number of reassigned CTCF sites classified as barrier sites was recorded. The P-value is then the fraction of times (over 10,000 trials) the number of CTCF sites classified as barrier sites in the random trial experiment is at least as much as the observed number of barrier CTCF sites. The smaller is the fraction (P-value), the higher the significance. The P-values were 4 × 10−3 and 3 × 10−4 for barrier CTCF sites in HeLa and CD4+ T cells, respectively. For both the negative control datasets (E2F4 and STAT1), the P-value was 1.
Acknowledgments
This work was supported by the Intramural Research Program of the National Heart, Lung, and Blood Institute, National Institutes of Health. The gene expression analysis using Affymetrix DNA microarrays was performed by NHLBI DNA microarray Core Facility. We thank Artem Barski, Andrew Smith, and L. Aravind for helpful comments and discussions.
Footnotes
[Supplemental material is available online at www.genome.org. The ChIP-seq and gene expression data from this study have been submitted to NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE12889.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.082800.108.
References
- Bailey T.L., Williams N., Misleh C., Li W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- Berger S.L. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Burcin M., Arnold R., Lutz M., Kaiser B., Runge D., Lottspeich F., Filippova G.N., Lobanenkov V.V., Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol. Cell. Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D.H., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Donohoe M.E., Zhang L.F., Xu N., Shi Y., Lee J.T. Identification of a Ctcf cofactor, Yy1, for the X chromosome binary switch. Mol. Cell. 2007;25:43–56. doi: 10.1016/j.molcel.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Fedoriw A.M., Stein P., Svoboda P., Schultz R.M., Bartolomei M.S. Transgenic RNAi reveals essential function for CTCF in H19gene imprinting. Science. 2004;303:238–240. doi: 10.1126/science.1090934. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G., Burgess-Beusse B., Farrell C., Gaszner M., Ghirlando R., Huang S., Jin C., Litt M., Magdinier F., Mutskov V., et al. Chromatin boundaries and chromatin domains. Cold Spring Harb. Symp. Quant. Biol. 2004;69:245–250. doi: 10.1101/sqb.2004.69.245. [DOI] [PubMed] [Google Scholar]
- Filippova G.N. Genetics and epigenetics of the multifunctional protein CTCF. Curr. Top. Dev. Biol. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Fagerlie S., Klenova E.M., Myers C., Dehner Y., Goodwin G., Neiman P.E., Collins S.J., Lobanenkov V.V. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 2001;28:335–343. doi: 10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- Filippova G.N., Cheng M.K., Moore J.M., Truong J.P., Hu Y.J., Nguyen D.K., Tsuchiya K.D., Disteche C.M. Boundaries between chromosomal domains of X inactivation and escape bind CTCF and lack CpG methylation during early development. Dev. Cell. 2005;8:31–42. doi: 10.1016/j.devcel.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Fu Y., Sinha M., Peterson C.L., Weng Z. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008;4:e1000138. doi: 10.1371/journal.pgen.1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaszner M., Felsenfeld G. Insulators: Exploiting transcriptional and epigenetic mechanisms. Nat. Rev. Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Corces V.G. Boundary and insulator elements in chromosomes. Curr. Opin. Genet. Dev. 1996;6:185–192. doi: 10.1016/s0959-437x(96)80049-9. [DOI] [PubMed] [Google Scholar]
- Gerasimova T.I., Corces V.G. Chromatin insulators and boundaries: Effects on transcription and nuclear organization. Annu. Rev. Genet. 2001;35:193–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- Jothi R., Cuddapah S., Barski A., Cui K., Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 2008;36:5221–5231. doi: 10.1093/nar/gkn488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C.F., Wolffe A., Ohlsson R., Lobanenkov V.V. Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 2000;10:853–856. doi: 10.1016/s0960-9822(00)00597-2. [DOI] [PubMed] [Google Scholar]
- Kanduri M., Kanduri C., Mariano P., Vostrov A.A., Quitschke W., Lobanenkov V., Ohlsson R. Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell. Biol. 2002;22:3339–3344. doi: 10.1128/MCB.22.10.3339-3344.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 Update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lobanenkov V.V., Nicolas R.H., Adler V.V., Paterson H., Klenova E.M., Polotskaja A.V., Goodwin G.H. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Mukhopadhyay R., Yu W., Whitehead J., Xu J., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., et al. The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 2004;14:1594–1602. doi: 10.1101/gr.2408304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlsson R., Renkawitz R., Lobanenkov V. CTCF is a uniquely versatile transcription regulator linked to epigenetics and disease. Trends Genet. 2001;17:520–527. doi: 10.1016/s0168-9525(01)02366-6. [DOI] [PubMed] [Google Scholar]
- Recillas-Targa F., Pikaart M.J., Burgess-Beusse B., Bell A.C., Litt M.D., West A.G., Gaszner M., Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes & Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh T.Y., Cuddapah S., Cui K., Zhao K. The genomic landscape of histone modifications in human T cells. Proc. Natl. Acad. Sci. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio E.D., Reiss D.J., Welcsh P.L., Disteche C.M., Filippova G.N., Baliga N.S., Aebersold R., Ranish J.A., Krumm A. Proc. Natl. Acad. Sci. Vol. 105. 2008. CTCF physically links cohesin to chromatin; pp. 8309–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the β-globin locus. Genes & Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008;27:654–666. doi: 10.1038/emboj.2008.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vostrov A.A., Quitschke W.W. The zinc finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 1997;272:33353–33359. doi: 10.1074/jbc.272.52.33353. [DOI] [PubMed] [Google Scholar]
- Wallace J.A., Felsenfeld G. We gather together: Insulators and genome organization. Curr. Opin. Genet. Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zang C., Rosenfeld J.A., Schones D.E., Barski A., Cuddapah S., Cui K., Roh T.Y., Peng W., Zhang M.Q., et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., et al. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008;451:796–801. doi: 10.1038/nature06634. [DOI] [PubMed] [Google Scholar]
- West A.G., Gaszner M., Felsenfeld G. Insulators: Many functions, many mechanisms. Genes & Dev. 2002;16:271–288. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- Williams A., Flavell R.A. The role of CTCF in regulating nuclear organization. J. Exp. Med. 2008;205:747–750. doi: 10.1084/jem.20080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Donohoe M.E., Silva S.S., Lee J.T. Evidence that homologous X-chromosome pairing requires transcription and Ctcf protein. Nat. Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- Yusufzai T.M., Tagami H., Nakatani Y., Felsenfeld G. CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell. 2004;13:291–298. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The following data has been deposited in the NCBI GEO (under accession no. GSE12889): ChIP-seq raw seqeuence tags from CD4+ T cells (CTCF and H3K27me3), HeLa cells (CTCF and H3K27me3), and Jurkat cells (CTCF); H3K27me3 domains in CD4+ T cells and HeLa cells; barrier CTCF sites in CD4+ T cells and HeLa cells and HeLa cell gene expression (HG-U133 Plus 2.0 chip) data.