Abstract
Regulatory cis-acting variants account for a large proportion of gene expression variability in populations. Cis-acting differences can be specifically measured by comparing relative levels of allelic transcripts within a sample. Allelic expression (AE) mapping for cis-regulatory variant discovery has been hindered by the requirements of having informative or heterozygous single nucleotide polymorphisms (SNPs) within genes in order to assign the allelic origin of each transcript. In this study we have developed an approach to systematically screen for heritable cis-variants in common human haplotypes across >1000 genes. In order to achieve the highest level of information per haplotype studied, we carried out allelic expression measurements by using both intronic and exonic SNPs in primary transcripts. We used a novel RNA pooling strategy in immortalized lymphoblastoid cell lines (LCLs) and primary human osteoblast cell lines (HObs) to allow for high-throughput AE. Screening hits from RNA pools were further validated by performing allelic expression mapping in individual samples. Our results indicate that >10% of expressed genes in human LCLs show genotype-linked AE. In addition, we have validated cis-acting variants in over 20 genes linked with common disease susceptibility in recent genome-wide studies. More generally, our results indicate that RNA pooling coupled with AE read-out by second generation sequencing or by other methods provides a high-throughput tool for cataloging the impact of common noncoding variants in the human genome.
High-density genome-wide association studies (GWAS) are generating unbiased lists of common variants altering complex disease risk. The disease-associated variants are localized in blocks of linkage disequilibrium (LD) that overlap known genes. Relatively few of the replicated GWAS hits to date, or variants in LD blocks, are known to involve protein-coding variation (The Wellcome Trust Case Control Consortium 2007; Barrett et al. 2008; Bouatia-Naji et al. 2008; Hom et al. 2008; Willer et al. 2008); therefore, relevant causal polymorphisms may reside in regulatory regions and affect gene expression. The effects of polymorphisms in these cis-regulatory sequences are apparent through studies of gene expression variation within and between human populations (Morley et al. 2004; Cheung et al. 2005; Dixon et al. 2007; Goring et al. 2007; Stranger et al. 2007; Kwan et al. 2008; Schadt et al. 2008). Functional single nucleotide polymorphisms (SNPs) affecting gene expression are not easily identifiable as they may reside in nonconserved sequences surrounding a gene (Burton et al. 2007). Identification of cis-acting SNPs has the potential of not only providing insight on how noncoding variants alter disease risk but may also provide further understanding of how regulatory elements control gene expression.
Reported frequencies of common human cis-variants underlying differences in gene expression at the transcript level in blood-derived immortalized or primary cells varies from 2% to over 6% (Goring et al. 2007; Stranger et al. 2007). Additional layers of regulatory variation involved in differential expression of isoforms, such as alternatively spliced or terminated transcripts, were shown to influence ∼1% of measured genes (Kwan et al. 2008).
Cis-regulatory differences can be alternatively measured by allelic expression (AE). AE controls for environmental and trans-acting influences by measuring the relative expression of alleles within a sample rather than between samples (Pastinen and Hudson 2004). The AE patterns can be further mapped to cis-acting variants, provided that genotypes from these samples can be accurately phased (Pastinen et al. 2005; Serre et al. 2008). This approach yields direct as opposed to inferred relationships between polymorphisms and cis-regulatory differences. AE also detects epigenetic effects such as imprinting or random monoallelic expression (Gimelbrant et al. 2007; Pollard et al. 2008). To date, most allelic expression studies have used coding variants as marker SNPs to distinguish between the two alleles (Yan et al. 2002; Bray et al. 2003; Lo et al. 2003; Pant et al. 2006; Marioni et al. 2008; Pollard et al. 2008). This makes sampling inefficient since there are few informative SNPs per gene and this has limited the widespread use of allelic expression as a general tool to find heritable regulatory variation.
In this study, we use both unspliced primary transcripts as well as mRNAs as targets. This allows the use of SNPs both in coding and intronic regions to assess AE, which maximizes the information content per gene. Linkage disequilibrium (LD) between the causal variants and intragenic SNPs can lead to AE biased toward one allele. Secondly, we measured allelic expression in pooled DNA and RNA, which allows for the enrichment of heritable variation associated with a haplotype of interest (such as a disease-linked haplotype). Pooling of the samples is an efficient way to detect allelic expression based on differences in allele frequencies between the pools, which can then be subsequently validated and mapped in individual samples.
Results
Overall study
Although our aim was to develop a pipeline for screening and mapping allelic expression for genes that have been associated to complex traits in recent GWAS (n = 118) (Fig. 1), this method was also used for genes found in the ENCODE regions (n = 108), disease candidate genes (n = 262), and genes nearby previously identified GWAS loci or randomly chosen genes (n = 616) as detailed in Supplemental Table 1. For our primary discovery panel, we used RNA and DNA from Caucasian lymphoblastoid cells (CEU-LCLs) from the Human HapMap project (The International HapMap Consortium 2007). To demonstrate the utility of the approach in less well-characterized cell panels and assess tissue specificity of allelic expression, we also used RNA and DNA pools derived from human bone cells (osteoblasts, or HObs) of Swedish origin (Grundberg et al. 2008). All assays were performed using normalized sequencing (Ge et al. 2005). To validate a subset of the genes showing allelic expression, we measured AE in individual samples (as opposed to a pooled sample), which allowed us to map AE to haplotypes (Pastinen et al. 2005). In addition, we demonstrated the utility of second generation sequencing technologies by applying an allele-counting method on a 454 Life Sciences (Roche) GS-FLX sequencer.
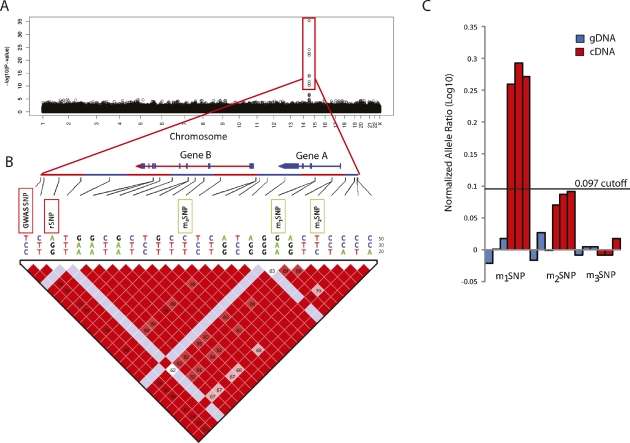
Figure 1.
(A) Hypothetical Manhattan plot for a disease GWAS, demonstrating a strong association to SNPs on chromosome 14, highlighted in red block. (B) Genomic region demonstrating an LD block spanning the GWAS SNPs. Marker SNPs (m1SNP, m2SNP, m3SNP) in genes (A and B), found in strong LD with the GWAS SNP, are used to assay for AE (rSNP, regulatory SNP). (C) Normalized allele ratio (log10) graph demonstrating different scenarios. Strong allelic expression is found for Gene A using m1SNP that tests for the same haplotypes found for the GWAS SNP. Marker m2SNP shows borderline AE for the same gene (Gene A) and tests for a different pattern of haplotypes. No AE is found for Gene B using m3SNP as the gDNA, and cDNA ratios are not significantly different from each other.
Pooling accuracy and cut-offs
In pilot experiments, we assessed the reproducibility of our pooling method for both gDNA and cDNA (RNA) pools. Due to our sample size and technical limitations in normalized Sanger sequencing for rare SNPs, we restricted the screening to common SNPs (minor allele frequency (MAF) ≥ 0.1). The accuracy of the cDNA pooling was indirectly assessed by testing the same SNP in two independently derived cDNA pools from the same panel of individuals (cell culture, RNA isolation, and pooling were all independent). Reproducible (r = 0.83) measurements of AE differences in independent pools (Supplemental Fig. 1) required consistency across triplicate measurements in individual pools (i.e., log mean normalized allele difference/standard deviation > 1.5) as well as greater than 1.25-fold difference between estimated allele frequencies in DNA and cDNA pools (i.e., log10 allele ratio Δ > 0.097), which were subsequently used as cut-offs.
Allelic expression screening in pooled LCLs samples by normalized Sanger sequencing
We selected a total of 2532 SNPs in 1103 genes for screening in the LCL pools (Supplemental Table 2), of which 829 genes were successfully amplified by RT–PCR. After removing 210 SNPs that failed quality control either due to background sequence or failed sequencing in one of the RNA pools, 757 genes (1745 SNPs) were successfully assayed for pooled AE. We identified 388 SNPs in 258 genes showing pooled AE differences above the threshold (Supplemental Table 3). Thirty-four percent (89/258) of genes had at least two SNPs showing a significant difference in AE. Furthermore, of the AE SNPs occurring in the same transcript and in moderate LD (r2 > 0.6) with one another, or if the same SNP was independently tested more than once, the same haplotypes were predicted to be overexpressed in all (47/47) cases. For the remaining 42 genes with multiple hits, incomplete or unknown LD between independent SNPs precluded the assessment of consistency. We further compared our results to the 56 genes found to be significantly AE in two recent studies (Pastinen et al. 2005; Serre et al. 2008). We note that 26 genes were screened for AE and of these, 17/26 genes (65%) were found to be AE. The sensitivity increased to 81% (17/21) (Supplemental Table 11), if two or more SNPs were assayed for the same gene in the pooled screening. Examples of genes showing positive allelic expression in our pooled assays and with previous evidence of cis-regulatory variation include CHI3L2, SERPINB10, and BTN3A2 (Pastinen et al. 2004; Cheung et al. 2005; Serre et al. 2008).
Comparison between the use of intronic and exonic SNPs
The head-to-head comparison of the performance of intronic vs. exonic SNPs in allelic expression is complicated by the fact that one would need to rely on SNPs that have perfect proxies (i.e., belong to the same haplotype) in exons and introns. For this reason, we have analyzed pairs of SNPs that are in high LD (r2 > 0.8) and mapped to an exon and an intron of the same transcript (Supplemental Table 15). We observe concordant data for 69% (34/49) of exonic and intronic pairs (r2 > 0.8). Sixteen percent (8/49) have AE by intronic but not by exonic SNP, while the reverse was true for 15% (7/49) of the data. Furthermore, the use of both intronic and exonic SNPs has led us to the identification of 157 more genes that show AE than achievable if we had only used exonic SNPs. Only 101 genes would have been identified had intronic SNPs been excluded and if we had assayed other available exonic SNPs (r2 > 0.8 with original intronic hit). In fact, only 23% of our hits are due to coding SNPs. A slight enrichment of exonic SNPs is seen among the AE hits as compared with intronic SNPs (20.2% vs. 14.2%). This data is consistent with our earlier observations (Pastinen et al. 2005) showing that the increased informativity of transcripts when measuring intronic SNPs comes with some cost on assay success rates.
Validation of LCL pooling data by allelic expression mapping
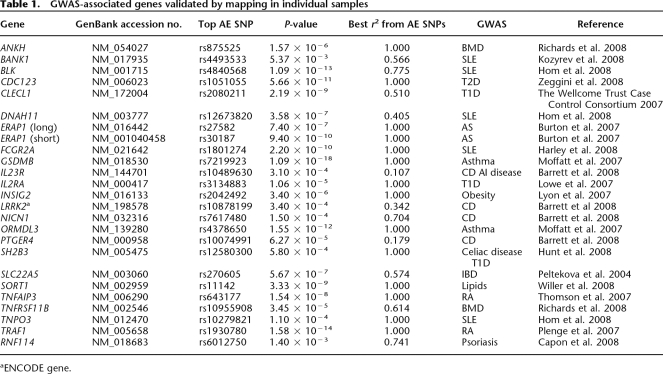
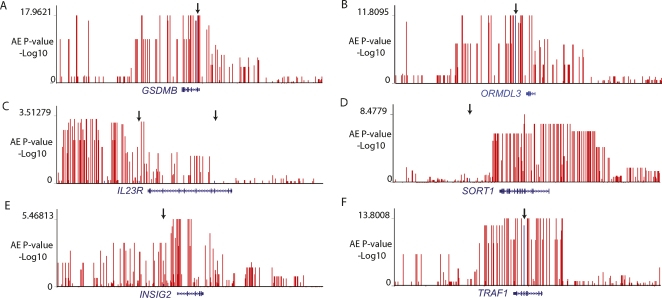
Allelic expression mapping in phased individual samples is a direct approach for validation of heritable cis-effects (Pastinen et al. 2005; Serre et al. 2008), and this was used for following up on the screening hits found in the LCL pools. Altogether, we were able to validate significant AE for 72% of the SNPs (135/188 SNPs) or 64% (63/99 genes) of the genes subjected to allelic expression mapping in individual samples (Table 1; Supplemental Tables 4, 5, 8). In most cases, multiple intragenic SNPs were used to derive individual AE calls as previously described (Pastinen et al. 2005) (see Supplemental Methods; Supplemental Figs. 2, 3). The linkage disequilibrium between screening hits and top AE mapping hits (±100 kb of gene) was strong: average r2 = 0.77 (95% CI 0.69–0.85). Regulatory haplotype mapping data from the validation experiments are shown for genes implicated in complex traits identified by GWAS (TRAF1, INSIG2, IL23R, SORT1, GSDMB, ORMDL3) in Figure 2.
Table 1.
GWAS-associated genes validated by mapping in individual samples
aENCODE gene.
Figure 2.
Allelic imbalance association mapping results for GWAS genes. Vertical red lines correspond to –log10(P-value) of the AE assay for each SNP tested. The arrow points to the GWAS SNPs, indicated by the vertical blue lines. (A) GSDMB (asthma); (B) ORMDL3 (asthma); (C) IL23R (Crohn’s disease); (D) SORT1 (Dyslipidemia); (E) INSIG2 (Obesity); (F) TRAF1 (Rheumatoid arthritis).
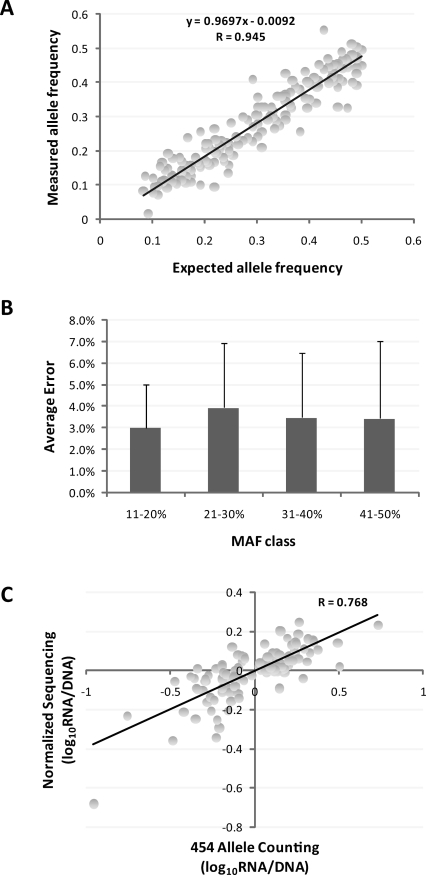
Figure 3.
(A) Comparison of expected and observed allele count (n = 278) in DNA pools derived from 55 CEU LCLs. (B) Average error with standard deviation (error bars) of allele frequencies derived from 454 counting as compared with known frequencies in the DNA pool. (C) Correlation of estimated fold difference (log10) in allele frequencies between cDNA and gDNA pools based on normalized sequencing (y-axis) and 454 allele counting (x-axis).
AE genes found in the random ENCODE regions
We designed pooled AE assays for 108 of the 146 genes found in the random ENCODE regions in order to get an unbiased estimate of the prevalence of cis-regulatory variants in human genes expressed in LCLs (Supplemental Table 1). There were no common SNPs reported for 5% (7/146) of the genes and we could not design successful assays targeting the 5′ haplotype for 21% (31/146) of the genes; therefore, these genes were not tested for AE. Sixty-five genes were successfully measured in the LCLs, and 19 showed screening evidence of AE. Of these, nine were validated by individual AE-mapping (Supplemental Table 4). This set (9/65) of validated LCL-expressed genes yields an estimate of 14% prevalence of common cis-regulatory haplotypes for human genes.
Coupling RNA pooling with digital allele counting on 454 GS-FLX
The second-generation sequencers, such as the 454 GS-FLX, allow single molecule amplification and allele counting, which can be used to measure allele frequency differences between nucleic acid pools of hundreds of targets. Therefore, we carried out a proof-of-principle experiment by using an allele-counting method in 300 previously assessed SNPs on a 454 GS-FLX sequencer. The allele-counting method allowed us to directly estimate the gDNA-pooling accuracy, since the exact expected allele frequencies of the gDNA pool for the HapMap CEU samples could be easily calculated (Fig. 3A). On average, a 3% error rate across MAFs ranging from 0.1 to 0.5 was found (Fig. 3B). The Pearson (r) correlation of allele difference estimation between the DNA and RNA pools and between normalized sequencing with consistent replicates and allele counting on the 454 GS-FLX was 0.77 (P < 2.2 × 10−16) (Fig. 3C; Supplemental Table 9).
Pooled allelic expression in human osteoblasts
In order to investigate the prevalence of AE in primary human cells, we tested a panel of 55 HObs. A total of 1460 SNPs in 685 genes were screened in the HOb pools (Supplemental Table 2). Of these, 498 genes were successfully amplified by RT–PCR, suggesting that 27% (187/685) of these genes are not expressed in the HObs. After removing 138 SNPs that failed quality control, 445 genes were successfully assayed for AE (Supplemental Table 6). We identified 276 SNPs in 180 genes (40%) with evidence of AE, including 66 (37%) genes that had at least two SNPs showing a significant difference in allelic expression. AE mapping is less efficient in unrelated HObs as compared with LCL trios due to less efficient phasing; therefore, the validation testing was restricted to a subset of AE SNPs (n = 11 loci) in individual heterozygotes from our panel. Of the 10 new loci (STAT4 has been previously validated) (Sigurdsson et al. 2008) subjected to validation in individual samples, three loci reached significance individually in this test, six demonstrated overexpression of the expected allele, and one showed equal distribution of AE-calls. The total number of AE calls in HObs toward expected vs. opposite direction in this experiment was 41 vs. 6 (binomial test P = 8.9 × 10−8; Supplemental Table 7) providing strong evidence that allele biases detected in the primary cell pools are due to cis-acting differences in expression.
Allelic expression comparison between LCLs and HObs
An analysis of the 685 genes screened in both the HObs and the LCLs revealed that 103 (15%) were only expressed in the LCLs, 84 (12%) were only expressed in the HObs, 414 (60%) were expressed in both, and 84 (12%) were not expressed in either cell type. Of the 586 genes that were successfully assessed for AE in both cell lines, significant AE was observed in 290 (49%) of the 586 genes, of which 28% are shared in both the HObs and the LCLs, 40% are LCL specific, and 32% of the genes are HOb specific (Supplemental Table 1). Approximately 70% of tissue-restricted AE occurs in genes where expression is detected in both tissues. In 36/46 (78%) genes where the same SNP yielded a positive screening hit, the AE was observed in the same direction between the tissues.
Gene network and pathway analysis for tissue-specific AE genes
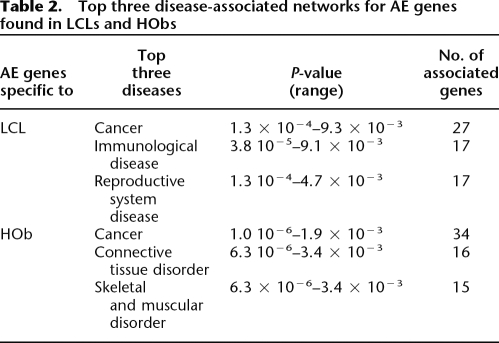
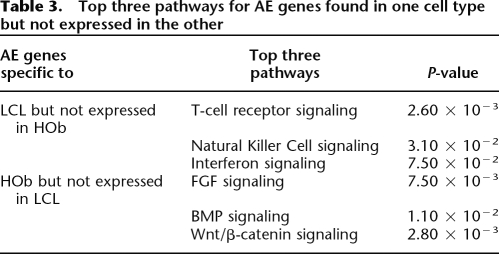
In order to determine the biological involvement of tissue-specific AE genes and any potential enrichment of biologically relevant pathways, we performed a gene ontology analysis using the ingenuity pathways analysis (IPA) system (Supplemental Table 12). AE specific to LCLs enriches for genes associated with cancer, immunological, and reproductive-related gene networks. AE specific to HObs showed enrichment of cancer related networks but are different from the LCLs by showing enrichment of gene networks involved in connective tissue and musculoskeletal disease (Table 2; Supplemental Tables 13, 14). Comparison of AE genes that are expressed in only one tissue but not in the other indicates that the LCL-only expressed genes are involved in t-cell receptor, natural killer cell, and interferon signaling pathways, while the HOb-only expressed genes are enriched in the FGF-, BMP-, and Wnt/β-catenin signaling pathways (Table 3).
Table 2.
Top three disease-associated networks for AE genes found in LCLs and HObs
Table 3.
Top three pathways for AE genes found in one cell type but not expressed in the other
Comparison between the normalized Sanger sequencing and the allele counting on the 454 GS-FLX
The mean and standard deviations from the known vs. measured gDNA allele frequencies were used to define cut-offs for calling AE from the 454 GS-FLX data (Supplemental Table 10). For both the LCLs and the HObs, converging results were observed in 71% of cases. In the nonconvergent cases, 11% were due to AE detection by the 454 and not by normalized Sanger sequencing, whereas 16% were due to AE detection by the normalized Sanger sequencing and not by the 454. In the remaining 2% of cases, the independent methods suggested overexpression of opposite alleles. We note that 454 GS-FLX allele counting in pools was carried out without replicates, so the “replicate consistency” rule applied on the normalized Sanger sequencing could not be used, which may have introduced a higher degree of sampling variability in the 454 data.
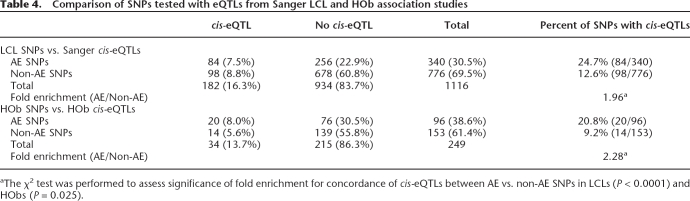
Comparison of allelic expression in LCLs and HObs to cis-eQTL data
For the CEU LCLs, we used the publicly available expression data (Stranger et al. 2007) to compare AE in RNA pools with cis-eQTL mapping. Of the 340 AE hits amenable to direct comparison, 25% of SNPs (84/340) had converging evidence for cis-regulatory variation (nominal P-value < 0.05 for eQTL association) (Table 4) and the same direction of effect was observed in 85% of convergent hits (Supplemental Table 3). In contrast, when we compared the non-AE screening SNPs, 13% of SNPs (98/776) had associated cis-eQTLs. Overall, there is a significant enrichment of cis-eSNPs with AE screening hits compared to non-AE hits (χ2 = 25.2, P < 10−5).
Table 4.
Comparison of SNPs tested with eQTLs from Sanger LCL and HOb association studies
aThe χ2 test was performed to assess significance of fold enrichment for concordance of cis-eQTLs between AE vs. non-AE SNPs in LCLs (P < 0.0001) and HObs (P = 0.025).
The AE SNPs were also compared with LCL cis-eQTLs identified using the Affymetrix exon array platform at both the gene and exonic level (GSE9372, Supplemental Table 3), and the results show similar levels of enrichment of cis-eQTLs for the AE SNPs compared with non-AE SNPs (χ2 = 25.0, P < 10−5).
For the HObs, we compared each of the AE SNPs with eQTLs identified from a panel of 98 HObs using the Illumina Ref-8 and Hap550K BeadArrays to obtain gene expression and genotype information, respectively (E. Grundberg, T. Kwan, B. Ge, K.C.L. Lam, V. Koka, H. Mallmin, O. Nilsson, and T. Pastinen, unpubl.). Direct comparison was available for 96 SNPs showing AE hits, of which 21% of SNPs (20/96) showed overlapping cis-eQTL (Supplemental Table 6). For the HOb SNPs that were not among AE hits, only 9% of SNPs (14/153) showed significant cis-eQTLs from the HOb data set (significance for eQTL overlap with hits vs. non-hits, χ2 = 5.1, P = 0.024).
Discussion
In this study, we present an approach to systematically screen for heritable cis-variants in hundreds of genes using cultured, immortalized lymphoblastoid cell lines (LCLs) or primary human osteoblast cell lines (HObs). Our two-stage approach measuring AE in primary transcripts allowed identification of cis-acting regulatory haplotypes in over 60 loci, most of which have not been observed in earlier AE studies using measurements of coding SNPs in individual samples (Pant et al. 2006; Gimelbrant et al. 2007; Bjornsson et al. 2008; Serre et al. 2008). Our results in the primary cell panel show that the method can be extended to more complex samples. We were able to validate 64% (63/99) of the AE genes by performing mapping in individual samples, and the success rate was higher for genes that were assayed with at least two SNPs (75%) compared with genes with only one SNP (48%). This was also apparent when we compared our data with previously identified AE genes (Pastinen et al. 2005; Serre et al. 2008) where our ability to detect AE increased from 65% to 81% when at least two SNPs were assayed per gene.
Our comparison between the use of intronic SNPs vs. exonic SNPs revealed that in 69% of cases there was no difference in the calls for these SNPs. However, the increase in the number of AE genes (and haplotypes) that we were able to identify using intronic SNPs clearly demonstrates that they are a valuable resource.
A previous study using DNA pooling coupled with low-throughput pyrosequencing (Lavebratt et al. 2004) showed a higher median error for allele frequency measurements in similarly sized DNA pools as compared with our results on high-throughput allele counting by 454 (4.9% vs. 2.9%). The same investigators suggested that increased pool size reduces error, particularly for common alleles. We speculate that accuracy of allele counting on the 454, coupled with increased RNA and DNA pool sizes, could improve the sensitivity and specificity of our approach. In addition, RNA sequencing directly from unamplified cDNA (Marioni et al. 2008) with deep sequencing of nuclear RNA pools (to enrich for primary transcripts) could rapidly provide a genome-wide catalog of cis-regulatory haplotypes across multiple cell types.
In comparison to cis-eSNP mapping, we observe a significant overlap with AE screening hits, but most variants (including validated screening hits) detected by one approach are not observed by the other. Similarly, cis-eSNP mapping on different platforms (Stranger et al. 2007; Kwan et al. 2008) targeting different parts of processed transcripts yield numerous associations not detected by one approach. In part, the apparent lack of overlap could be attributed to issues of statistical power and technical limitations of each method, but it is likely that a proportion of “approach-specific” variants are linked to true differences in processing of primary transcripts as well as confounding trans-acting effects (Wang et al. 2008). The detected and validated allelic expression for genes in the ENCODE random regions indicate that 14% of primary transcripts show genotype-dependent AE, which is likely an underestimate since we did not exhaustively tag common haplotypes overlapping the gene. Furthermore, our screening approach does not detect AE caused by variants unlinked to the primary transcript, which could account for up to one-third of cis-variants in Caucasian populations based on eQTL data (Emilsson et al. 2008). Our validation approach, mapping of allelic expression (Pastinen et al. 2005), allows detection of distal cis-variants as well and could be applied directly if haplotypes of interest do not overlap the transcript.
Tissue specificity of allelic expression was recently highlighted in mice (Campbell et al. 2008). Our AE screening data in LCLs and primary osteoblasts also implies that even for genes expressed in both tissues the same haplotypes (the osteoblast cohort is of Northern European origin similar to the origin of CEU LCLs) may exert different effects in ∼50% of cases. Furthermore, the overlap of AE hits (28%) between LCLs and HObs is almost identical to the eSNP overlap (30%) reported recently for complex human tissues (Schadt et al. 2008). Based on these results it is apparent that the expansion of the number of cell types and tissues used in population genomic studies is critical for cataloging noncoding functional variation.
We have also observed an enrichment of AE in gene networks related with immunological disease in the LCLs and with musculoskeletal disease when HOb-specific AE hits are considered. These data, as well as previous studies, suggest benefits from analyzing cis-regulatory variation in appropriate disease tissues (Emilsson et al. 2008; Johnson et al. 2008). However, screening for AE in tissue samples is still challenging because of sample heterogeneity and sample degradation (Li et al. 2004; Hamilton et al. 2006).
The follow-up of a subset of our AE screening hits by mapping cis-regulatory haplotypes in the cell panels have already yielded converging links between functional variants in LCLs or HObs with complex disease risk (Richards et al. 2008; Sigurdsson et al. 2008). In addition, we report 24 GWAS implied disease loci with cis-regulatory haplotypes falling into three categories where the disease-associated haplotype and regulatory haplotype are: (1) indistinguishable (e.g., TRAF1, ORMDL3, GSDMB, BLK, and TNFRSF11B), (2) partially overlapping, i.e., where the same alleles contribute to both the phenotype and the functional effect, but the top reported association does not coincide (e.g., INSIG2, IL23R, or IL2RA), (3) clearly distinct (e.g., TNFAIP3). In some cases, links between cis-eQTLs and the disease haplotype were made earlier, such as ORMDL3 and asthma (Moffatt et al. 2007). Our data confirms the cis-effect exerted by the disease-associated haplotype (Fig. 2) on ORMDL3, but uncovers a second strong functional effect of the same haplotype in cis-regulation of the GSDMB gene (Fig. 2). In the case of the GSDMB gene, the asthma-associated haplotype accounts for >90% of the allelic expression seen in CEU LCLs, and such strong size effects could be exploited in fine-mapping efforts. An example from the second category is provided by IL23R, where two regions of the gene have been associated with Crohn’s disease risk (Barrett et al. 2008). We observe a functional effect for the haplotypes overlapping the 5′ disease-associated region, and the functional variants could assist in dissecting this locus further by conditional or regression-based association analysis. However, we acknowledge that connecting the dots between allelic expression and disease association requires extensive functional validation, but the allelic expression association provides plausible hypotheses for the characterization of common disease haplotypes.
In summary, we have shown the widespread association of common haplotypes with the allelic expression of primary transcripts using a new RNA pooling approach. Similarities and differences between our and traditional approaches for the identification of regulatory variants underscore the importance of multiple methods in assessing gene expression. Coupled with recent data linking common SNPs to interindividual variation in chromatin structure (Kerkel et al. 2008; Maynard et al. 2008), this demonstrates the vast functional potential of noncoding variants and indicates the need for various approaches for population genomic dissection of common SNPs.
Methods
Cell culture
Lymphoblastic Cell Lines (LCLs): HapMap CEU immortalized LCLs from 60 unrelated individuals (The International HapMap Consortium 2005) were obtained from the Coriell Cell Repositories and were cultured as previously described (Pastinen et al. 2004, 2005; Serre et al. 2008). Five of the 60 LCLs were excluded due to consistent slow growth (GM12056, GM12236, GM12716, GM12717, and GM12875).
Human osteoblast cell lines (HObs): Human trabecular bone from the proximal femoral shaft was collected from 55 unrelated donors undergoing total hip replacement at the Uppsala University Hospital, Sweden. The primary cell culture was carried out as previously described (Grundberg et al. 2008).
RNA isolation/cDNA synthesis/DNA isolation
Nucleic acids were extracted from the cells and quality control was carried out applying previously described methods (Grundberg et al. 2008; Pollard et al. 2008). For pooled analysis of intronic SNPs, we enriched the heteronuclear RNA in the cDNA preparation by using gene-specific reverse-transcription targeting intronic sequences downstream of the SNP of interest. Gene-specific priming was carried out in pools of 200–400 SNPs in DNase I-treated total RNA samples using 0.1 uM concentration of each primer. For the validation of genes by allelic expression mapping in individual cell lines, we used standard random hexamer-based cDNA synthesis, thereby eliminating the potential biases introduced by gene-specific priming.
Gene and marker SNP selection
The genes and SNPs tested are listed in Supplemental Table 2. For GWAS hit regions, the SNPs were selected based on maximal LD with the reported disease-associated SNPs. Additional SNPs tagging other haplotypes in these genes were also selected to increase informativity of the validation test by allelic expression mapping in individual samples. For genes found in the ENCODE random regions, the SNPs were targeted to the 5′ haplotype blocks observed in CEU HapMap LCLs. SNPs in disease candidate genes and other genes without a priori information of phenotype associations were chosen to cover the most common haplotype blocks (in HapMap CEU).
Pooling of samples and allelic expression measurements by conventional, normalized sequencing
For our primary discovery panel, two different pools, one cDNA and one gDNA, were made from the 55 LCL samples. For the initial pilot experiments, two cDNA LCL pools were constructed. For each of these pools, the cell culture, the RNA isolation, and the pooling were all performed independently of each other. The concentration of each DNA and RNA sample was measured on three independent occasions using a standard spectrophotometer and a Nanodrop ND-1000 (NanoDrop Technologies) to minimize sampling error. Each marker SNP was amplified by PCR using dilutions from pooled stock solutions as a template (10 ng/10 uL PCR reaction). PCRs and RT–PCRs performed in three to six replicates were verified by agarose gel electrophoresis and sequenced using ABI Big Dye chemistry and capillary electrophoresis on an ABI 3730 sequencer (Applied Biosystems). Allelic expression levels for each SNP were assessed following a previously published method (Ge et al. 2005). Allelic expression ratios for the RNA samples were calculated using the normalized heterozygote ratios from the genomic DNA originating from the same samples. The same methods were used for the 55 HOb samples.
Allele-counting by high-throughput parallel sequencing
Fusion primers including 5′ extensions providing binding sites for the 454 Life Sciences Technology A and B primers were used. Similar to the normalized sequencing procedure described above, all SNPs were amplified by PCR using the pooled stock solution of cDNA and gDNA (LCLs or HObs) and were performed in triplicate. However, after amplification, each triplicate was pooled together and electrophoresed on an agarose gel for verification. Normalized quantities of 303 different PCR products were pooled to create RT–PCR and PCR product libraries. Both libraries were purified with a Montage PCR Centrifugal Filter Device from Millipore, and quantified using Quant-iT PicoGreen assay (Invitrogen). Dilutions of both libraries were then amplified by emulsion PCR using the GS emPCR Kit II (Roche) and subsequently sequenced in one direction on a GS-FLX instrument using two medium regions of a 70 × 75 GSPicoTiterPlate, as previously described (Margulies et al. 2005). The sequencing runs produced a total of 127,745 and 152,017 high-quality reads for the gDNA and cDNA SNP libraries, respectively. Mapping and alignment to the reference sequences was conducted by the Genome Sequencer FLX software version 1.1.02 (454 Life Sciences). The reads were filtered using the default FLX System software filters and primer sequences were trimmed from the ends of reads. The sequences were mapped to the target regions of the human reference genome (NCBI build 36.1) and pairwise aligned to the reference sequence using the GS Reference Mapper. Non-uniquely mapped reads were discarded. Multiple sequence alignments (MSAs) were constructed from the 454 pairwise alignments. For known SNPs, the frequency of each base in the specified column of the MSA was recorded.
Mapping of allelic expression associated SNPs in individual samples
Genes reported in GWAS were prioritized for validation. Similar to the normalized sequencing procedure described above, all intragenic SNPs used for validation were amplified by PCR, but using individual LCL samples that were known to be heterozygous for the marker SNP. Multiple SNPs were assayed for each gene in order to increase our power of detection and decrease our sampling variability. A list of the SNPs used for each locus is provided in Supplemental Table 8. Allelic expression level mapping was carried out essentially as previously described (Ge et al. 2005; Pastinen et al. 2005) with minor modifications (see Supplemental Methods for details). The population distribution of allelic expression in phased chromosomes was tested for association using a two-sided Fisher’s exact test as previously described (Pastinen et al. 2005, Serre et al. 2008). Allelic expression association mapping results for validated hits are shown in Supplemental Table 5.
Comparison of AE and non-AE SNPs against Sanger and HOb cis-eQTLs
For all SNPs assayed for AE against a given gene, we examined whether there was a significant cis-eQTL from the association analysis of the publicly available LCL CEU expression data against the same SNP (Stranger et al. 2007). For each gene tested, we retrieved the corresponding gene expression data from the Sanger data set. Genotypes for the CEU population were obtained from the HapMap database (release 23a). The whole-genome expression data and genotype data for our panel of 98 human osteoblast samples were obtained using the Illumina Ref-8 and Hap550K platforms, respectively. Using each gene–SNP combination, we performed a linear regression analysis on the expression/genotype data, implemented in the PLINK software package. Raw P-values were obtained from the regression using the standard asymptotic t-statistic, as well as the direction of effect (overexpressed allele). The cis-eQTLs were considered significant when the nominal P < 0.05.
Gene network and pathway analysis
The AE genes from human LCLs and HObs were uploaded in the Ingenuity Pathway Analysis (IPA) system (Ingenuity Systems, www.ingenuity.com). Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge and then overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Networks and pathways of these “Focus Genes” were then algorithmically generated, based on their connectivity. The IPA system identified the biological functions that were most significant to each data set. Fisher’s exact test was used to calculate a P-value determining the probability that each biological function assigned to the data set is due to chance only.
Canonical pathways analysis identified the pathways from the IPA library of canonical pathways that were most significant to the data set. The significance of the association between the data set and the canonical pathway was measured in two ways: (1) a ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway; (2) a Fisher’s exact test was used to calculate a P-value determining the probability that the association between the genes in the data set and the canonical pathway is explained by chance alone.
Acknowledgments
We thank the Genome Quebec and Genome Canada for funding. T.P. holds a Canada Research Chair (Tier 2).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.084798.108.
References
- Barrett J.C., Hansoul S., Nicolae D.L., Cho J.H., Duerr R.H., Rioux J.D., Brant S.R., Silverberg M.S., Taylor K.D., Barmada M.M., et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat. Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjornsson H.T., Albert T.J., Ladd-Acosta C.M., Green R.D., Rongione M.A., Middle C.M., Irizarry R.A., Broman K.W., Feinberg A.P. SNP-specific array-based allele-specific expression analysis. Genome Res. 2008;18:771–779. doi: 10.1101/gr.073254.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouatia-Naji N., Rocheleau G., Van Lommel L., Lemaire K., Schuit F., Cavalcanti-Proenca C., Marchand M., Hartikainen A.L., Sovio U., De Graeve F., et al. A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science. 2008;320:1085–1088. doi: 10.1126/science.1156849. [DOI] [PubMed] [Google Scholar]
- Bray N.J., Buckland P.R., Owen M.J., O'Donovan M.C. Cis-acting variation in the expression of a high proportion of genes in human brain. Hum. Genet. 2003;113:149–153. doi: 10.1007/s00439-003-0956-y. [DOI] [PubMed] [Google Scholar]
- Burton P.R., Clayton D.G., Cardon L.R., Craddock N., Deloukas P., Duncanson A., Kwiatkowski D.P., McCarthy M.I., Ouwehand W.H., Samani N.J., et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat. Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.D., Kirby A., Nemesh J., Daly M.J., Hirschhorn J.N. A survey of allelic imbalance in F1 mice. Genome Res. 2008;18:555–563. doi: 10.1101/gr.068692.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capon F., Bijlmakers M.J., Wolf N., Quaranta M., Huffmeier U., Allen M., Timms K., Abkevich V., Gutin A., Smith R., et al. Identification of ZNF313/RNF114 as a novel psoriasis susceptibility gene. Hum. Mol. Genet. 2008;17:1938–1945. doi: 10.1093/hmg/ddn091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon A.L., Liang L., Moffatt M.F., Chen W., Heath S., Wong K.C., Taylor J., Burnett E., Gut I., Farrall M., et al. A genome-wide association study of global gene expression. Nat. Genet. 2007;39:1202–1207. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- Emilsson V., Thorleifsson G., Zhang B., Leonardson A.S., Zink F., Zhu J., Carlson S., Helgason A., Walters G.B., Gunnarsdottir S., et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Ge B., Gurd S., Gaudin T., Dore C., Lepage P., Harmsen E., Hudson T.J., Pastinen T. Survey of allelic expression using EST mining. Genome Res. 2005;15:1584–1591. doi: 10.1101/gr.4023805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant A., Hutchinson J.N., Thompson B.R., Chess A. Widespread monoallelic expression on human autosomes. Science. 2007;318:1136–1140. doi: 10.1126/science.1148910. [DOI] [PubMed] [Google Scholar]
- Goring H.H., Curran J.E., Johnson M.P., Dyer T.D., Charlesworth J., Cole S.A., Jowett J.B., Abraham L.J., Rainwater D.L., Comuzzie A.G., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Grundberg E., Brandstrom H., Lam K.C., Gurd S., Ge B., Harmsen E., Kindmark A., Ljunggren O., Mallmin H., Nilsson O., et al. Systematic assessment of the human osteoblast transcriptome in resting and induced primary cells. Physiol. Genomics. 2008;33:301–311. doi: 10.1152/physiolgenomics.00028.2008. [DOI] [PubMed] [Google Scholar]
- Hamilton G., Allsop J.M., Patel N., Forton D.M., Thomas H.C., O’Sullivan C.P., Hajnal J.V., Taylor-Robinson S.D. Variations due to analysis technique in intracellular pH measurements in simulated and in vivo 31P MR spectra of the human brain. J. Magn. Reson. Imaging. 2006;23:459–464. doi: 10.1002/jmri.20524. [DOI] [PubMed] [Google Scholar]
- Harley J.B., Alarcon-Riquelme M.E., Criswell L.A., Jacob C.O., Kimberly R.P., Moser K.L., Tsao B.P., Vyse T.J., Langefeld C.D., Nath S.K., et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat. Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom G., Graham R.R., Modrek B., Taylor K.E., Ortmann W., Garnier S., Lee A.T., Chung S.A., Ferreira R.C., Pant P.V., et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N. Engl. J. Med. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- Hunt K.A., Zhernakova A., Turner G., Heap G.A., Franke L., Bruinenberg M., Romanos J., Dinesen L.C., Ryan A.W., Panesar D., et al. Newly identified genetic risk variants for celiac disease related to the immune response. Nat. Genet. 2008;40:395–402. doi: 10.1038/ng.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.D., Zhang Y., Papp A.C., Pinsonneault J.K., Lim J.E., Saffen D., Dai Z., Wang D., Sadee W. Polymorphisms affecting gene transcription and mRNA processing in pharmacogenetic candidate genes: detection through allelic expression imbalance in human target tissues. Pharmacogenet. Genomics. 2008;18:781–791. doi: 10.1097/FPC.0b013e3283050107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkel K., Spadola A., Yuan E., Kosek J., Jiang L., Hod E., Li K., Murty V.V., Schupf N., Vilain E., et al. Genomic surveys by methylation-sensitive SNP analysis identify sequence-dependent allele-specific DNA methylation. Nat. Genet. 2008;40:904–908. doi: 10.1038/ng.174. [DOI] [PubMed] [Google Scholar]
- Kozyrev S.V., Abelson A.K., Wojcik J., Zaghlool A., Linga Reddy M.V., Sanchez E., Gunnarsson I., Svenungsson E., Sturfelt G., Jonsen A., et al. Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat. Genet. 2008;40:211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- Kwan T., Benovoy D., Dias C., Gurd S., Provencher C., Beaulieu P., Hudson T.J., Sladek R., Majewski J. Genome-wide analysis of transcript isoform variation in humans. Nat. Genet. 2008;40:225–231. doi: 10.1038/ng.2007.57. [DOI] [PubMed] [Google Scholar]
- Lavebratt C., Sengul S., Jansson M., Schalling M. Pyrosequencing-based SNP allele frequency estimation in DNA pools. Hum. Mutat. 2004;23:92–97. doi: 10.1002/humu.10292. [DOI] [PubMed] [Google Scholar]
- Li J.Z., Vawter M.P., Walsh D.M., Tomita H., Evans S.J., Choudary P.V., Lopez J.F., Avelar A., Shokoohi V., Chung T., et al. Systematic changes in gene expression in postmortem human brains associated with tissue pH and terminal medical conditions. Hum. Mol. Genet. 2004;13:609–616. doi: 10.1093/hmg/ddh065. [DOI] [PubMed] [Google Scholar]
- Lo H.S., Wang Z., Hu Y., Yang H.H., Gere S., Buetow K.H., Lee M.P. Allelic variation in gene expression is common in the human genome. Genome Res. 2003;13:1855–1862. doi: 10.1101/gr.1006603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe C.E., Cooper J.D., Brusko T., Walker N.M., Smyth D.J., Bailey R., Bourget K., Plagnol V., Field S., Atkinson M., et al. Large-scale genetic fine mapping and genotype-phenotype associations implicate polymorphism in the IL2RA region in type 1 diabetes. Nat. Genet. 2007;39:1074–1082. doi: 10.1038/ng2102. [DOI] [PubMed] [Google Scholar]
- Lyon H.N., Emilsson V., Hinney A., Heid I.M., Lasky-Su J., Zhu X., Thorleifsson G., Gunnarsdottir S., Walters G.B., Thorsteinsdottir U., et al. The association of a SNP upstream of INSIG2 with body mass index is reproduced in several but not all cohorts. PLoS Genet. 2007;3:e61. doi: 10.1371/journal.pgen.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., Egholm M., Altman W.E., Attiya S., Bader J.S., Bemben L.A., Berka J., Braverman M.S., Chen Y.J., Chen Z., et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni J., Mason C., Mane S., Stephens M., Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard N.D., Chen J., Stuart R.K., Fan J.B., Ren B. Genome-wide mapping of allele-specific protein-DNA interactions in human cells. Nat. Methods. 2008;5:307–309. doi: 10.1038/nmeth.1194. [DOI] [PubMed] [Google Scholar]
- Moffatt M.F., Kabesch M., Liang L., Dixon A.L., Strachan D., Heath S., Depner M., von Berg A., Bufe A., Rietschel E., et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant P.V., Tao H., Beilharz E.J., Ballinger D.G., Cox D.R., Frazer K.A. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–339. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastinen T., Hudson T.J. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- Pastinen T., Sladek R., Gurd S., Sammak A., Ge B., Lepage P., Lavergne K., Villeneuve A., Gaudin T., Brandstrom H., et al. A survey of genetic and epigenetic variation affecting human gene expression. Physiol. Genomics. 2004;16:184–193. doi: 10.1152/physiolgenomics.00163.2003. [DOI] [PubMed] [Google Scholar]
- Pastinen T., Ge B., Gurd S., Gaudin T., Dore C., Lemire M., Lepage P., Harmsen E., Hudson T.J. Mapping common regulatory variants to human haplotypes. Hum. Mol. Genet. 2005;14:3963–3971. doi: 10.1093/hmg/ddi420. [DOI] [PubMed] [Google Scholar]
- Peltekova V.D., Wintle R.F., Rubin L.A., Amos C.I., Huang Q., Gu X., Newman B., Van Oene M., Cescon D., Greenberg G., et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat. Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- Plenge R.M., Seielstad M., Padyukov L., Lee A.T., Remmers E.F., Ding B., Liew A., Khalili H., Chandrasekaran A., Davies L.R., et al. TRAF1-C5 as a risk locus for rheumatoid arthritis–a genomewide study. N. Engl. J. Med. 2007;357:1199–1209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard K.S., Serre D., Wang X., Tao H., Grundberg E., Hudson T.J., Clark A.G., Frazer K. A genome-wide approach to identifying novel-imprinted genes. Hum. Genet. 2008;122:625–634. doi: 10.1007/s00439-007-0440-1. [DOI] [PubMed] [Google Scholar]
- Richards J.B., Rivadeneira F., Inouye M., Pastinen T.M., Soranzo N., Wilson S.G., Andrew T., Falchi M., Gwilliam R., Ahmadi K.R., et al. Bone mineral density, osteoporosis, and osteoporotic fractures: A genome-wide association study. Lancet. 2008;371:1505–1512. doi: 10.1016/S0140-6736(08)60599-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E., Molony C., Chudin E., Hao K., Yang X., Lum P.Y., Kasarskis A., Zhang B., Wang S., Suver C., et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre D., Gurd S., Ge B., Sladek R., Sinnett D., Harmsen E., Bibikova M., Chudin E., Barker D.L., Dickinson T., et al. Differential allelic expression in the human genome: A robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdsson S., Nordmark G., Garnier S., Grundberg E., Kwan T., Nilsson O., Eloranta M.L., Gunnarsson I., Svenungsson E., Sturfelt G., et al. A risk haplotype of STAT4 for systemic lupus erythematosus is over-expressed, correlates with anti-dsDNA and shows additive effects with two risk alleles of IRF5. Hum. Mol. Genet. 2008;17:2868–2876. doi: 10.1093/hmg/ddn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranger B.E., Nica A.C., Forrest M.S., Dimas A., Bird C.P., Beazley C., Ingle C.E., Dunning M., Flicek P., Koller D., et al. Population genomics of human gene expression. Nat. Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson W., Barton A., Ke X., Eyre S., Hinks A., Bowes J., Donn R., Symmons D., Hider S., Bruce I.N., et al. Rheumatoid arthritis association at 6q23. Nat. Genet. 2007;39:1431–1433. doi: 10.1038/ng.2007.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.Y., Fu Y., McPeek M.S., Lu X., Nuzhdin S., Xu A., Lu J., Wu M.L., Wu C.I. Complex genetic interactions underlying expression differences between Drosophila races: Analysis of chromosome substitutions. Proc. Natl. Acad. Sci. 2008;105:6362–6367. doi: 10.1073/pnas.0711774105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Wellcome Trust Case Control Consortium Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Yuan W., Velculescu V.E., Vogelstein B., Kinzler K.W. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- Zeggini E., Scott L.J., Saxena R., Voight B.F., Marchini J.L., Hu T., de Bakker P.I., Abecasis G.R., Almgren P., Andersen G., et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]