Abstract
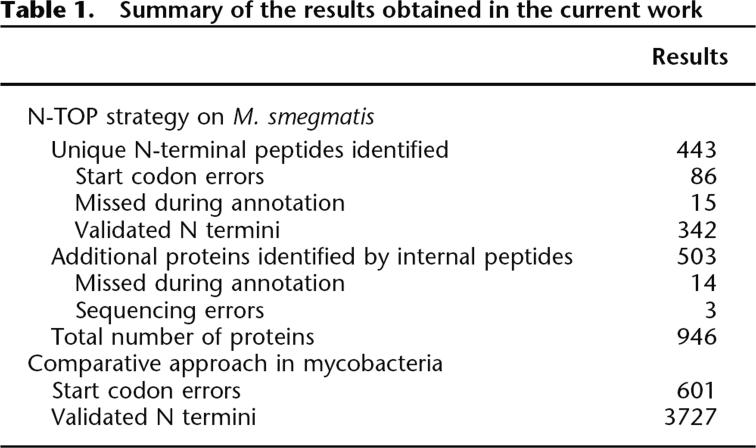
The progress in sequencing technologies irrigates biology with an ever-increasing number of genome sequences. In most cases, the gene repertoire is predicted in silico and conceptually translated into proteins. As recently highlighted, the predicted genes exhibit frequent errors, particularly in start codons, with a serious impact on subsequent biological studies. A new “ortho-proteogenomic” approach is presented here for the annotation refinement of multiple genomes at once. It combines comparative genomics with an original proteomic protocol that allows the characterization of both N-terminal and internal peptides in a single experiment. This strategy was applied to the Mycobacterium genus with Mycobacterium smegmatis as the reference, and identified 946 distinct proteins, including 443 characterized N termini. These experimental data allowed the correction of 19% of the characterized start codons, the identification of 29 proteins missed during the annotation process, and the curation, thanks to comparative genomics, of 4328 sequences of 16 other Mycobacterium proteomes.
The increasing availability of data from multiple genome sequencing projects provides biologists with an invaluable framework to integrate experimental results and design new experiments at different scales. However, several recent studies have highlighted the prevalence of gene prediction errors, even in the “simple” prokaryotic genomes. Genome sequencing itself represents a non-negligible source of errors (Weinstock 2000), but despite major advances, most inconsistencies result from in silico predictions (Galperin et al. 1998). Among these errors, the incorrect prediction of initiation codons in prokaryotic genomes is particularly widespread (Aivaliotis et al. 2007). For instance, error rates in start codon prediction vary from 10% to 44% in Halobacterium salinarum and Natromonas pharaonis (Aivaliotis et al. 2007), depending on the gene prediction program used. This reality is often underestimated or even ignored by biologists, even though the correct definition of genes is determinant for subsequent in silico and experimental studies. For example, by altering the definition of the coding sequence of a gene, an erroneous start codon can hamper the detection of regulatory motifs on the genome or even mask another gene in a compact genome (Salgado et al. 2000; Edwards et al. 2005). Moreover, the protein sequence itself can be either truncated or extended, leading to errors in bioinformatics protein characterization (function, localization, etc.) and, obviously, to major difficulties in protein expression experiments (Trivedi et al. 2004; Horie et al. 2007). The second highly prejudicial error encountered in prokaryotic genome annotation is under-prediction of small genes or genes exhibiting an unusual composition. The accumulation of erroneous information in genomic and protein databases will continue to grow since features are frequently transferred from annotated to unknown sequences (Doerks et al. 1998), which only amplifies the errors.
To break this vicious circle and to cope with the multiplication of prokaryotic genome data, including many projects aimed at exploring genetic diversity within a genus or a species by multiple-strain sequencing (Liolios et al. 2008), one cannot rely solely on manual curation. In this context, the proteogenomic approach, i.e., annotation refinement through proteomics, is promising and has already been used to investigate several bacterial genomes (Jaffe et al. 2004a, b; Wang et al. 2005; Gupta et al. 2007, 2008), revealing the expression of genes annotated as pseudogenes as well as some completely missed genes or some errors in start codon annotation. However, these high-throughput studies do not focus on the N-terminal identification of proteins, limiting the correction of gene boundaries. In contrast, other methods have aimed at the specific identification of N-terminal peptides from the digest of a protein extract (Gevaert et al. 2003; McDonald et al. 2005; McDonald and Beynon 2006), but these methods imply the loss of all internal peptides, which is a major drawback both for protein and proteome coverage.
Here, we report an original strategy coupling a new N-terminal-oriented proteomic (N-TOP) method with comparative genomics. The concept underlying this “ortho-proteogenomic” approach is to characterize large sets of N-terminal and internal peptides in a single run for a reference organism and to propagate the obtained experimental information to orthologs of closely related species. We have developed two original strategies: (1) a new straightforward mass spectrometry (MS)-based workflow relying on a single specific N-terminal labeling of the intact proteins, preserving internal peptides, and (2) a conservative comparative approach to curate the annotation of multiple closely related microbial genomes simultaneously. Mycobacterium was chosen as a first study example for this strategy since 17 complete genomes of Mycobacterium strains or species are available, including important pathogens such as M. tuberculosis, M. leprae, and M. ulcerans. Within the Mycobacterium genus, M. smegmatis is ideally suited to test our combined approach since it is a model species for experiments (a fast-growing and nonpathogenic species) that exhibits a large repertoire of genes. Our proteogenomic approach allowed the experimental identification of 946 proteins from M. smegmatis, revealing 29 new proteins missed during the annotation process. In the same experiment, 443 N-terminal peptides of the 946 proteins were characterized. These N termini sequences revealed an error rate of 19% in the prediction of the initiation codon. Comparative analysis applied to the sequences of the 16 other mycobacteria resulted in 4328 curated protein sequences. Besides the immediate value of these data to the whole scientific community working on Mycobacterium, the ortho-proteogenomic method presented here should initiate a new step in genome annotation and sequence database curation. The data used in this study are available at ftp://ftp-igbmc.u-strasbg.fr/pub/lecompte/Msmegmatis.
Results
TMPP labeling of protein N termini: Workflow establishment
N-succinimidyloxycarbonylmethyl)tris(2,4,6-trimethoxyphenyl) phosphonium bromide (TMPP-Ac-OSu) was selected as the labeling reagent in order to obtain a better ionization efficiency, simplified fragmentation, and retention times allowing a better separation of N-terminal peptides analyzed by liquid chromatography-coupled mass spectrometry (LC-MS/MS) after enzymatic digestion. A new workflow was developed and experimental conditions were setup for N-terminal protein labeling from a total biological extract with all the usual detergents, chaotropic agents, and reduction conditions used for protein extraction, including membrane proteins (Xiong et al. 2005). It was necessary to replace dithiothreitol (DTT) by tributylphosphine (TBP) in the labeling buffer since DTT hindered the TMPP reaction.
Derivatization conditions on model peptides have been described (Roth et al. 1998; Adamczyk et al. 1999; Huang et al. 1999; Sadagopan and Watson 2000; Czeszak et al. 2004; Chamot-Rooke et al. 2007), but they were totally inadequate for complex protein extracts in the presence of detergents. In our case, a large excess of TMPP must be used for complex extract labeling. Prior to LC-MS/MS analysis, all traces of TMPP must be removed, as TMPP retention time is close to that of TMPP-derivatized peptides and thus interferes with their MS detection. Several methods to remove the excess were tested, including different membrane filtration devices, but they induced dramatic peptide material losses and did not allow complete elimination of TMPP. Finally, a one-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D SDS-PAGE) step was shown to be ideal for removing excess reagent and had the additional advantages of being compatible with strong detergents and of reducing the complexity of protein extracts prior to LC-MS/MS.
Application of the workflow to model proteins
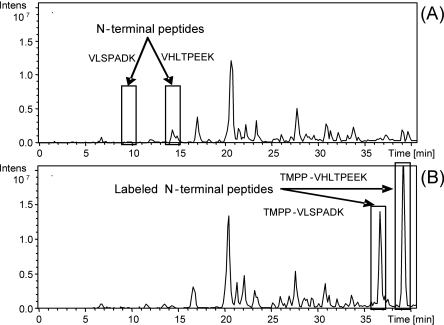
Purified hemoglobin alpha and beta chains solubilized in the labeling buffer used for biological samples (see labeling buffer in Methods) were employed as the model system. TMPP-derivatized and -nonderivatized hemoglobin chains were separated on 1D SDS-PAGE and digested in-gel, and the peptide extracts were analyzed by LC-MS/MS (see Supplemental Methods S1 for more details). Recognition of N-terminal peptides was based on the characteristic mass shift caused by the TMPP labeling (+572.18 Da). Figure 1 shows that the N-terminal derivatized peptides have an increased retention time due to the addition of the hydrophobic TMPP group, in contrast to internal peptides. This implies that the lysine side chain ε-amines were preserved fully intact by a careful control of pH at 8.2 during the labeling. For studies of very complex biological samples, this retention time shift allows a better LC-MS/MS analysis of TMPP-derivatized peptides since it shifts the elution times toward a less complex part of the chromatogram.
Figure 1.
Comparison of base peak chromatograms from LC-MS/MS of hemoglobin digests. (A) Without N-terminal protein labeling; (B) with N-terminal protein labeling.
Figure 1 also shows that the TMPP labeling significantly increases the ionization efficiency. The two peaks of N-terminal peptides that were minor in the native form become major after derivatization, whereas all internal peptides remain unchanged. So, TMPP derivatization introduces a permanent positive charge and a hydrophobic group resulting in an enhancement of the ionization efficiency. These results show that the established workflow provides efficient N-terminal peptide identification in a complex mixture of proteins.
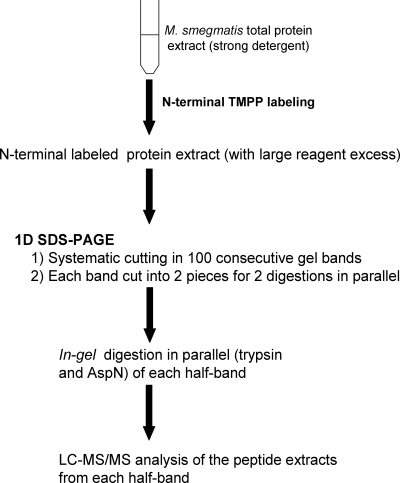
Application of the workflow to the proteome of M. smegmatis
The workflow established for the M. smegmatis proteome is summarized in Figure 2. After N-terminal labeling, the total protein extract was loaded on a 1D SDS-PAGE. The gel was systematically cut into 100 horizontal bands, and each band was divided into two equal gel slices for enzymatic digestion with trypsin and endoproteinase AspN. Finally, after in-gel digestion, the peptide extracts were analyzed by LC-MS/MS. Due to their high complexity, a 170-min LC gradient was optimized from 10% to 70% CH3CN with a slope of 0.35% per minute. N-terminal-labeled and internal native peptides were identified using Mascot MS/MS data searches against the complete genomic sequence rather than protein sequence databases, avoiding problems associated with computational predictions and annotations (Choudhary et al. 2001; Kuster et al. 2001; Oshiro et al. 2002; Jaffe et al. 2004a, b; Fermin et al. 2006; Gupta et al. 2007; Tanner et al. 2007).
Figure 2.
Analytical workflow.
The N-TOP strategy led to the identification of 443 unique N-terminal peptides (from 591 N-terminal sequences: 361 tryptic and 230 AspN sequences) and to a total of 946 validated proteins (Table 1). The two digestion modes appeared to be complementary but also allowed the cross-validation of the determined N termini in 148 cases (Supplemental Fig. S2). Trypsin digestion was the most efficient since it led to the identification of >80% of all identified start codons. All experimentally determined N-terminal sequences, internal sequences, protein lists, and N-terminal spectra are available at ftp://ftp-igbmc.u-strasbg.fr/pub/lecompte/Msmegmatis and the MGF peak lists are available at http://tranche.proteomecommons.org/ using the following hash:
Table 1.
Summary of the results obtained in the current work
 |
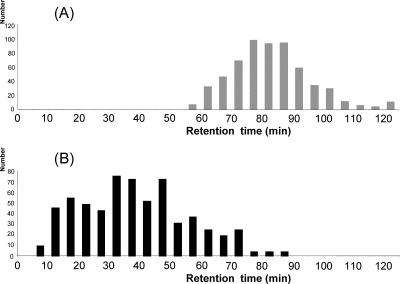
The improved ionization efficiency of the labeled peptides on model proteins could be generalized at the proteome level. The impact on reversed phase chromatography was also of major importance since the analysis of the chromatograms revealed two distinct elution zones: the labeled N-terminal peptides (Fig. 3A) and the native internal peptides (Fig. 3B). These two zones are partially overlapping because, despite derivatization, very hydrophilic-labeled peptides can be eluted before very hydrophobic native peptides. Nevertheless, this overall shift in chromatographic behavior prevented ionization suppression due to internal peptides and improved the characterization of labeled peptides.
Figure 3.
Comparison of the peptide retention times. (A) Retention time of the 591 N-terminal-labeled sequences. (B) Retention time of a random selection of 600 identified internal peptides.
In addition, the developed workflow had the advantage of not modifying internal peptides, allowing their simultaneous identification in a single classical LC-MS/MS analysis, even in the case of post-translational modifications. These internal peptides allowed the identification of 503 additional proteins and confirmed 92% of the protein identifications based on the N-terminal peptide.
Definition of rules for N-terminal peptide identification
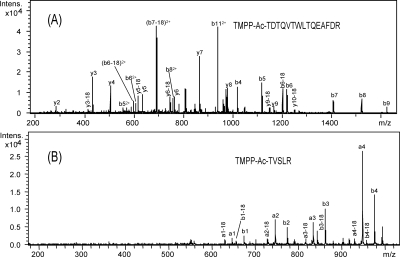
Protein identification was commonly performed for internal peptides (see Methods). However, for N-terminal peptide identification, the Mascot search parameter settings had to be carefully adapted, since the TMPP labeling has a strong influence on the N-terminal labeled peptide fragmentation behavior, especially for doubly charged precursors. Considering that peptide scores from classical search engines are based mainly on the fragmentation behavior of native fully tryptic peptides, each fragmentation spectrum of a potential derivatized N-terminal peptide (whose first amino acid of the sequence is coded by a start codon or by the first codon following a start codon, ATG, GTG, or TTG) was manually validated using several criteria. The first criterion was the retention time shift described above. The second criterion was the fragmentation pattern observed for a large set of derivatized peptides, which allowed us to improve previous observations on a few model peptides (Adamczyk et al. 1999; Sadagopan and Watson 2000, 2001) and to establish fragmentation “rules.” Generally, due to the permanent positive charge, N-terminal fragment ions (an and bn) are predominantly observed from enzymatic TMPP-derivatized peptides, with intensities depending on the peptide sequences (Fig. 4). As an example, the CID spectrum of a triply charged labeled peptide (Fig. 4A) displays a series of singly and doubly charged bn ions plus a few y-type ions. Fragmentation of doubly charged labeled peptides generates fewer peaks (mostly N-terminal fragments) than unlabeled ones, especially in the lower mass range (Fig. 4B). This is expected because only C-terminal fragments can be present below 630 m/z (the mass of the a-type fragment corresponding to the TMPP-labeled glycine). This particular pattern of fragmentation for doubly charged peptides is specific to the TMPP-labeled peptides. For these peptides, in spite of high-quality fragmentation spectra, Mascot ion scores are generally lower than for unlabeled peptides because few fragments remain in the lower mass range (Supplemental Table S5). In addition, the presence of very intense a-type ions is not common with ESI-Ion Trap mass spectrometers; they are observed with MALDI-TOF/TOF spectrometers, for example. So it was necessary to add these a-type fragments into the searched ESI-IT fragments used in Mascot searches to prevent false negative identifications. Finally, the last criteria taken into account were the length of the peptide and the identification of additional internal peptides. In the case of single-peptide assignments, all peptide sequences shorter than seven amino acids were excluded.
Figure 4.
CID spectra of N-terminal-labeled peptides. (A) Triply charged labeled peptide. (B) Doubly charged labeled peptide.
All N-terminal sequences collected in this study begin with, or begin immediately after, a start codon. Other labeled peptides (whose first amino acid of the sequence is not coded by a start codon or by the first codon following a start codon), for which N-terminal labeling was probably made after endogenous digestion of proteins (for example, signal peptide cleavage), were discarded at the initial step of the validation process (a total of 32 peptides), and no suggestions have been made on the position of any start codon not determined exactly. In this context, it was also interesting to observe the N-terminal methionine cleavages in the experimental sequences. Indeed, the final list of validated N-terminal sequences can be divided into two subsets: N-terminal sequences with and without methionine removal. The N-terminal methionine excision process occurs in all organisms and involves methionine aminopeptidase (MAP), whose activity has been reported to be linked mainly to the side chain size of the penultimate amino acid of the protein (Frottin et al. 2006). In our experimental data, the occurrence of methionine removals correlates well with the penultimate amino acids (Supplemental Table S4), in agreement with previously determined rules (Hirel et al. 1989; Link et al. 1997). This suggests that our start site data set does not contain sequences corresponding to endogenously digested proteins; otherwise, we would have observed random methionine cleavages. The manual validation has improved our ability to interpret TMPP-labeled peptide fragmentation spectra and allowed us to establish precise criteria and threshold scores that will facilitate the automation of the workflow in subsequent studies (Supplemental Table S5) and that allow estimation of the false discovery rate in our start site data set.
Comparison of the experimental proteome versus the predicted proteome
The experimentally determined sequences were compared with the predicted protein sequences (Table 1) from The Institute for Genomic Research (http://www.tigr.org/, released in 2005), revealing 86 errors (19%) in start codon prediction. 19% of the wrongly predicted sequences were too short, while 81% were too long. This error rate is in agreement with previous estimates from studies of other G+C-rich prokaryotes (Aivaliotis et al. 2007).
In addition, we detected three sequencing errors in the genome that had been reported previously (Deshayes et al. 2007), and we identified 29 proteins that were missed in the initial annotation. Start codons were identified for 15 of these proteins. M. smegmatis protein sequences with experimental start codon validation or correction and proteins missing in the annotation have been submitted to the Swiss-Prot database (http://expasy.org/sprot/) and are available at ftp://ftp-igbmc.u-strasbg.fr/pub/lecompte/Msmegmatis.
These results demonstrate that proteomics data can allow the identification of new proteins that would otherwise have been missed by classical computational annotation. They also confirm the utility of preserving internal peptides and clearly establish the need to work with genomic sequences rather than predicted proteins in order to identify new coding regions and start codons upstream of the predicted start sites.
Comparative genomic approach to correct mycobacteria sequences
We have capitalized on our large set of experimentally determined N-terminal sequences in order to correct predicted sequences in 16 other Mycobacterium genomes using a comparative genomic approach. Each reference sequence of M. smegmatis with an experimentally determined N terminus was aligned to its orthologs in mycobacteria, and the N-terminal region of the multiple alignment was analyzed (see example in Supplemental Fig. S3). A total of 4648 protein sequences were processed: 3727 N-terminal sequences (80% of the initial set) were validated and 601 (13%) were corrected. Since we adopted stringent parameters to avoid propagation errors, the remaining 320 sequences (7%) were not modified despite some discrepancy with the reference sequence of M. smegmatis. We thus obtained a conservative estimate of error rates in start prediction in Mycobacterium ranging from 9% to 21% (14% on average; see Supplemental Table S6). The validated and corrected sequences of mycobacteria are available at ftp://ftp-igbmc.u-strasbg.fr/pub/lecompte/Mycobacteria.
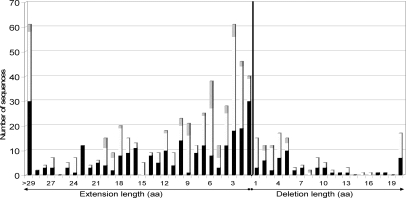
As observed in the comparison of the predicted proteome to experimental data, the large majority (84%) of detected errors are due to an erroneous 5′ extension of the gene (Fig. 5). Sixty percent of deletions are short (at most five amino acids), while 75% of artificial extensions are longer than five amino acids, with 61 encompassing more than 29 amino acids. Such major errors will seriously compromise subsequent in silico and/or experimental studies. Interestingly, minor initiation codons (GTG and TTG) are overrepresented in the corrected start codons (48% and 8%, respectively) compared with 35% and 4% reported for M. tuberculosis protein genes, for example (Cole et al. 1998). This suggests an overprediction of ATG as the translational start codon, either by gene prediction programs or during manual annotation.
Figure 5.
Distribution of false extension and deletion lengths in mycobacteria (including M. smegmatis). Bars are shaded proportionally to the number of each actual initiation codon after correction: (black) ATG, (white) GTG, (gray) TTG.
Discussion
Recently, several proteomic initiatives have highlighted some inherent limits of in silico gene predictions in genomes, especially concerning exon/intron boundaries and start codon predictions. Our “ortho-proteogenomic” study represents the first quantitative evaluation of start codon prediction in public prokaryotic genome annotations at a genus level. The prediction error rates from 9% to 21% are surprisingly high considering that we are working on well studied pathogenic bacteria, such as M. tuberculosis H37Rv, whose genome has been annotated twice (Cole et al. 1998; Camus et al. 2002) and has benefited from the input of numerous experimental studies. The relative homogeneity of error rates within the Mycobacterium genus clearly demonstrates that errors cannot be attributed to a particular in silico annotation pipeline. Although the quality of genome annotation can be improved by combining different methods during the reannotation process, gene prediction tools are still faced with intrinsic limits. In the case of M. smegmatis, new annotations have been released since we began this work. In the latest version of the protein sequence database downloaded in 2007 from http://www.uniprot.org/, some annotation errors have been corrected: The error rate decreased from 19% to 13% for start codon prediction, and 29 proteins were missed in the initial annotation versus four in the latest one. However, the comparison between the two annotation releases (Supplemental Table S7) reveals that some new errors have been introduced in the latest version, emphasizing the limits of the reannotation processes.
In this context, there is an urgent need to promote genome annotation refinement and protein database curation by large-scale proteomic analysis (the so-called proteogenomics approach). The N-TOP workflow established for the M. smegmatis proteome is an optimized process involving standard proteomic techniques coupled with a fast one-step specific N-terminal protein labeling of a complete proteome. It combines the advantages of the usual proteogenomic approach and of N-terminal-specific techniques since it allows the characterization of both internal and N-terminal peptides in the same experiment. In comparison with other complete proteome analyses, our method results in a similar level of proteome coverage (946 proteins identified). For example, in 2005, Wang and colleagues (Wang et al. 2005) used multidimensional chromatography and tandem mass spectrometry to analyze the M. smegmatis proteome under 25 different growth conditions and obtained 901 distinct proteins. In order to compare our method with another widespread proteomic approach, we performed a classical 2D-gel-based analysis of the M. smegmatis proteome (see Supplemental Methods S1) and identified 846 proteins with the same identification protocol. Thus, the N-TOP workflow provides satisfactory coverage and, in addition, offers the invaluable identification of a large set of N-terminal peptides. Indeed, we obtained only 173 N-terminal peptides with the 2D-gel-based analysis (data not shown) versus 443 with the N-TOP strategy. When comparing with other N-terminal specific approaches, the N-TOP strategy raised results similar to the largest published data set for prokaryotes (Aivaliotis et al. 2007) in a single experiment, using the extract corresponding to one culture condition and with a reduced number of LC-MS/MS analyses. Applying the same experimental workflow to extracts obtained under different growth conditions (rich media, different additives) and phase cultures would allow the identification of additional N-terminal peptides from proteins expressed specifically during these phases or in these media (Wang et al. 2005; Gupta et al. 2007) and would thus allow an increase of the overall proteome coverage. With an established and optimized protocol, the 4328-curated sequences obtained in this study can be produced with two person weeks of work using three instrument-weeks, and thus can constitute a reproducible part of a genomics pipeline.
An efficient and synergic integration of high-throughput experimental data with in silico predictions is a challenging task requiring standardization, automation, and portability. Both the proteomic and bioinformatic communities are becoming aware of this problem as attested by several recent initiatives, such as the development of standards for MS data representation (Orchard and Hermjakob 2008) or new software such as PepLine (Ferro et al. 2008) that allow easier mapping of MS/MS data on eukaryotic genomic sequences. The straightforward ortho-proteogenomic workflow presented here uses standard proteomic and bioinformatic techniques and thus can be applied routinely in any proteomic laboratory. In the case of the Mycobacterium genus, our conservative comparative approach, the “ortho” part of ortho-proteogenomic, was particularly fruitful since one experimentally determined N terminus allows us to correct/validate roughly 10 genes simultaneously. With 10 bacterial genuses totaling 172 complete genome sequences at the time of writing, our strategy represents a cost-effective and promising means to curate the huge amount of incoming genomic data. More generally, it would be interesting to extend our strategy of propagation to more distantly related organisms (within a class, for instance) by sampling the extreme representative species at the experimental level and integrating the data in the evolutionary context of ortholog alignment.
Methods
N-terminal derivatization of M. smegmatis protein extract
Unless otherwise specified, all chemicals were obtained from Sigma. A solution of 0.1 M of (N-succinimidyloxycarbonyl-methyl)tris(2,4,6-trimethoxyphenyl) phosphonium bromide (TMPP-Ac-OSu) in CH3CN:water (2:8, v/v) was added at a molar ratio of 200:1 to 500 μg of M. smegmatis protein extract (see Supplemental Methods S1 for bacterial strains, growth conditions, and protein preparation) solubilized in labeling buffer (50 mM Tris-HCl, 8 M urea, 2 M thiourea, pH 8.2, 1 mM phenylmethylsulfonyl fluoride, 1 mM EDTA, 5 mM TBP [Bio-Rad Laboratories], protease inhibitor mixture [Roche], 10% CH3CN, 1% SDS). After a quick mix, the reaction was maintained at room temperature for 1 h. Residual derivatizing reagent was quenched by adding a solution of 0.1 M hydroxylamine at room temperature for 1 h.
1D SDS-PAGE separation
M. smegmatis N-terminal labeled protein extract was finally supplemented with glycerol at a concentration of 10%. Proteins were then separated on a 5%–20% 1D SDS-PAGE (20 cm × 20 cm) on a PROTEAN II (Bio-Rad) apparatus at 5 mA for 10 min and 10 mA overnight. The gel was stained with Colloidal blue. The whole lane was systematically cut into 100 bands of ∼2 mm, which were processed for enzymatic digestion. Each band was cut into two pieces for further digestion with two different enzymes (trypsin and AspN) and mass spectrometry analysis with an Agilent 1100 Series capillary LC system (Agilent Technologies) coupled to an HCT Ultra ion trap (Bruker Daltonics) (see Supplemental Methods S1 for more details about in-gel digestion and mass spectrometry analysis).
M. smegmatis genome database construction
The complete genome sequence of M. smegmatis was downloaded from the TIGR (http://www.tigr.org/). Using an in-house script, the 6.98-Mbp sequence was fragmented into regular segments to generate a nucleic acid database, which was imported into a local Mascot server and translated into six reading frames on the fly.
Identification validation
The MS/MS data were analyzed using the Mascot 2. 2. 0. algorithm (Matrix Science) to search against the constructed M. smegmatis genome database. For protein identification from internal peptides, the searches were performed with carbamidomethylation of cysteines and oxidation of methionines specified as variable modifications. For the two digestion modes, trypsin and AspN_ambic, a maximum of one missed cleavage was tolerated; 0.5 Da error in MS and MS/MS search modes was tolerated. Proteins identified with at least three internal peptides with a Mascot ion score greater than 25 were validated. For the estimation of the protein identification false positive rate, a target-decoy database search was performed (for review, see Elias and Gygi 2007). In this approach, peptides are matched against a concatenated database composed of the target database and a decoy database consisting of sequence-reversed entries. Applying the same criteria, we estimated the false positive rate to be <1%.
For N-terminal peptide identification, the searches were performed on genome database subsets under the same conditions as for internal peptides, except that N-terminal modification with TMPP was setup in Mascot (+572.18 Da) as a variable modification, and semi-trypsin or semi-AspN_ambic were used as digestion enzymes. The fragmentation spectra of the putative labeled N-terminal peptides were manually inspected, taking into account the contribution of the N-terminal TMPP group (chromatographic retention time shift, particular fragmentation pathways, and charge states of the peptides).
Comparative genomic approach to correct mycobacteria sequences
We used 16 strains of the Mycobacterium genus having a complete genome sequence (see Supplemental Methods S1 for the list of strains of the Mycobacterium genus used). A nucleic acid database of the Mycobacterium genomic sequences extracted from GenBank was constructed, and a database of the corresponding protein sequences was retrieved from Uniprot. The 443 proteins of M. smegmatis with an experimentally determined N terminus were compared with this protein database using BLASTP (Altschul et al. 1997). For each protein, the detected homologs were included in a clustered multiple alignment of complete sequences constructed using the PipeAlign program suite (Plewniak et al. 2003). Sequences sharing at least 70% identity with the M. smegmatis reference sequence or belonging to the same cluster within the multiple alignments were selected for N-terminal comparison. If the first amino acids of the sequence to be validated were aligned with the first amino acids of the reference sequence, the sequence was assumed to be correct. Otherwise, the protein sequence was localized on its genomic sequence by a TBLASTN search, and the genomic sequence upstream of or downstream from the current start codon was searched for alternative initiation codons. The N terminus of the sequence was corrected if an initiation codon was found within ±9 bp (three amino acids) around the reference start codon. The complete comparative process was performed automatically by TCL/TK scripts, available on request.
Acknowledgments
We thank Raymond Ripp for his assistance during this work and Julie Thompson for a critical reading of the manuscript. This work was supported by institutional funds from INSERM, CNRS, and ULP, by “Protéomique et génie des protéines” (project no. PGP 04-013), ANR-05-BLAN-0407-02, and ANR no. 2007 PFTV 018 01 grants. The “Fondation pour la Recherche Médicale” is also acknowledged for the acquisition of a high-resolution mass spectrometer.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.081901.108.
References
- Adamczyk M., Gebler J.C., Wu J. Charge derivatization of peptides to simplify their sequencing with an ion trap mass spectrometer. Rapid Commun. Mass Spectrom. 1999;13:1413–1422. doi: 10.1002/(SICI)1097-0231(19990730)13:14<1413::AID-RCM657>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., et al. Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 2007;6:2195–2204. doi: 10.1021/pr0700347. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus J.C., Pryor M.J., Medigue C., Cole S.T. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- Chamot-Rooke J., van der Rest G., Dalleu A., Bay S., Lemoine J. The combination of electron capture dissociation and fixed charge derivatization increases sequence coverage for O-glycosylated and O-phosphorylated peptides. J. Am. Soc. Mass Spectrom. 2007;18:1405–1413. doi: 10.1016/j.jasms.2007.04.008. [DOI] [PubMed] [Google Scholar]
- Choudhary J.S., Blackstock W.P., Creasy D.M., Cottrell J.S. Interrogating the human genome using uninterpreted mass spectrometry data. Proteomics. 2001;1:651–667. doi: 10.1002/1615-9861(200104)1:5<651::AID-PROT651>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- Czeszak X., Morelle W., Ricart G., Tetaert D., Lemoine J. Localization of the O-glycosylated sites in peptides by fixed-charge derivatization with a phosphonium group. Anal. Chem. 2004;76:4320–4324. doi: 10.1021/ac049767q. [DOI] [PubMed] [Google Scholar]
- Deshayes C., Perrodou E., Gallien S., Euphrasie D., Schaeffer C., Van-Dorsselaer A., Poch O., Lecompte O., Reyrat J.M. Interrupted coding sequences in Mycobacterium smegmatis: Authentic mutations or sequencing errors? Genome Biol. 2007;8:R20. doi: 10.1186/gb-2007-8-2-r20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T., Bairoch A., Bork P. Protein annotation: Detective work for function prediction. Trends Genet. 1998;14:248–250. doi: 10.1016/s0168-9525(98)01486-3. [DOI] [PubMed] [Google Scholar]
- Edwards M.T., Rison S.C., Stoker N.G., Wernisch L. A universally applicable method of operon map prediction on minimally annotated genomes using conserved genomic context. Nucleic Acids Res. 2005;33:3253–3262. doi: 10.1093/nar/gki634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias J.E., Gygi S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Fermin D., Allen B.B., Blackwell T.W., Menon R., Adamski M., Xu Y., Ulintz P., Omenn G.S., States D.J. Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 2006;7:R35. doi: 10.1186/gb-2006-7-4-r35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M., Tardif M., Reguer E., Cahuzac R., Bruley C., Vermat T., Nugues E., Vigouroux M., Vandenbrouck Y., Garin J., et al. PepLine: A software pipeline for high-throughput direct mapping of tandem mass spectrometry data on genomic sequences. J. Proteome Res. 2008;7:1873–1883. doi: 10.1021/pr070415k. [DOI] [PubMed] [Google Scholar]
- Frottin F., Martinez A., Peynot P., Mitra S., Holz R.C., Giglione C., Meinnel T. The proteomics of N-terminal methionine cleavage. Mol. Cell. Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- Galperin M.Y., Walker D.R., Koonin E.V. Analogous enzymes: Independent inventions in enzyme evolution. Genome Res. 1998;8:779–790. doi: 10.1101/gr.8.8.779. [DOI] [PubMed] [Google Scholar]
- Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G.R., Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- Gupta N., Tanner S., Jaitly N., Adkins J.N., Lipton M., Edwards R., Romine M., Osterman A., Bafna V., Smith R.D., et al. Whole proteome analysis of post-translational modifications: Applications of mass-spectrometry for proteogenomic annotation. Genome Res. 2007;17:1362–1377. doi: 10.1101/gr.6427907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N., Benhamida J., Bhargava V., Goodman D., Kain E., Kerman I., Nguyen N., Ollikainen N., Rodriguez J., Wang J., et al. Comparative proteogenomics: Combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res. 2008;18:1133–1142. doi: 10.1101/gr.074344.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirel P.H., Schmitter M.J., Dessen P., Fayat G., Blanquet S. Extent of N-terminal methionine excision from Escherichia coli proteins is governed by the side-chain length of the penultimate amino acid. Proc. Natl. Acad. Sci. 1989;86:8247–8251. doi: 10.1073/pnas.86.21.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie M., Fukui K., Xie M., Kageyama Y., Hamada K., Sakihama Y., Sugimori K., Matsumoto K. The N-terminal region is important for the nuclease activity and thermostability of the flap endonuclease-1 from Sulfolobus tokodaii. Biosci. Biotechnol. Biochem. 2007;71:855–865. doi: 10.1271/bbb.60326. [DOI] [PubMed] [Google Scholar]
- Huang Z.H., Shen T., Wu J., Gage D.A., Watson J.T. Protein sequencing by matrix-assisted laser desorption ionization-postsource decay-mass spectrometry analysis of the N-Tris(2,4,6-trimethoxyphenyl)phosphine-acetylated tryptic digests. Anal. Biochem. 1999;268:305–317. doi: 10.1006/abio.1998.3085. [DOI] [PubMed] [Google Scholar]
- Jaffe J.D., Berg H.C., Church G.M. Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics. 2004a;4:59–77. doi: 10.1002/pmic.200300511. [DOI] [PubMed] [Google Scholar]
- Jaffe J.D., Stange-Thomann N., Smith C., DeCaprio D., Fisher S., Butler J., Calvo S., Elkins T., FitzGerald M.G., Hafez N., et al. The complete genome and proteome of Mycoplasma mobile. Genome Res. 2004b;14:1447–1461. doi: 10.1101/gr.2674004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster B., Mortensen P., Andersen J.S., Mann M. Mass spectrometry allows direct identification of proteins in large genomes. Proteomics. 2001;1:641–650. doi: 10.1002/1615-9861(200104)1:5<641::AID-PROT641>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Link A.J., Robison K., Church G.M. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- Liolios K., Mavromatis K., Tavernarakis N., Kyrpides N.C. The Genomes On Line Database (GOLD) in 2007: Status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2008;36:D475–D479. doi: 10.1093/nar/gkm884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L., Beynon R.J. Positional proteomics: Preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nat. Protocols. 2006;1:1790–1798. doi: 10.1038/nprot.2006.317. [DOI] [PubMed] [Google Scholar]
- McDonald L., Robertson D.H., Hurst J.L., Beynon R.J. Positional proteomics: Selective recovery and analysis of N-terminal proteolytic peptides. Nat. Methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- Orchard S., Hermjakob H. The HUPO proteomics standards initiative—easing communication and minimizing data loss in a changing world. Brief. Bioinform. 2008;9:166–173. doi: 10.1093/bib/bbm061. [DOI] [PubMed] [Google Scholar]
- Oshiro G., Wodicka L.M., Washburn M.P., Yates J.R., Lockhart D.J., Winzeler E.A. Parallel identification of new genes in Saccharomyces cerevisiae. Genome Res. 2002;12:1210–1220. doi: 10.1101/gr.226802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plewniak F., Bianchetti L., Brelivet Y., Carles A., Chalmel F., Lecompte O., Mochel T., Moulinier L., Muller A., Muller J., et al. PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res. 2003;31:3829–3832. doi: 10.1093/nar/gkg518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth K.D., Huang Z.H., Sadagopan N., Watson J.T. Charge derivatization of peptides for analysis by mass spectrometry. Mass Spectrom. Rev. 1998;17:255–274. doi: 10.1002/(SICI)1098-2787(1998)17:4<255::AID-MAS1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Sadagopan N., Watson J.T. Investigation of the tris(trimethoxyphenyl)phosphonium acetyl charged derivatives of peptides by electrospray ionization mass spectrometry and tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2000;11:107–119. doi: 10.1016/S1044-0305(99)00127-0. [DOI] [PubMed] [Google Scholar]
- Sadagopan N., Watson J.T. Mass spectrometric evidence for mechanisms of fragmentation of charge-derivatized peptides. J. Am. Soc. Mass Spectrom. 2001;12:399–409. doi: 10.1016/S1044-0305(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Salgado H., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J. Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner S., Shen Z., Ng J., Florea L., Guigo R., Briggs S.P., Bafna V. Improving gene annotation using peptide mass spectrometry. Genome Res. 2007;17:231–239. doi: 10.1101/gr.5646507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi O.A., Arora P., Sridharan V., Tickoo R., Mohanty D., Gokhale R.S. Enzymic activation and transfer of fatty acids as acyl-adenylates in mycobacteria. Nature. 2004;428:441–445. doi: 10.1038/nature02384. [DOI] [PubMed] [Google Scholar]
- Wang R., Prince J.T., Marcotte E.M. Mass spectrometry of the M. smegmatis proteome: Protein expression levels correlate with function, operons, and codon bias. Genome Res. 2005;15:1118–1126. doi: 10.1101/gr.3994105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock G.M. Genomics and bacterial pathogenesis. Emerg. Infect. Dis. 2000;6:496–504. doi: 10.3201/eid0605.000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y., Chalmers M.J., Gao F.P., Cross T.A., Marshall A.G. Identification of Mycobacterium tuberculosis H37Rv integral membrane proteins by one-dimensional gel electrophoresis and liquid chromatography electrospray ionization tandem mass spectrometry. J. Proteome Res. 2005;4:855–861. doi: 10.1021/pr0500049. [DOI] [PubMed] [Google Scholar]