Abstract
MicroRNAs (miRNAs) are small endogenous RNAs that pair to sites in mRNAs to direct post-transcriptional repression. Many sites that match the miRNA seed (nucleotides 2–7), particularly those in 3′ untranslated regions (3′UTRs), are preferentially conserved. Here, we overhauled our tool for finding preferential conservation of sequence motifs and applied it to the analysis of human 3′UTRs, increasing by nearly threefold the detected number of preferentially conserved miRNA target sites. The new tool more efficiently incorporates new genomes and more completely controls for background conservation by accounting for mutational biases, dinucleotide conservation rates, and the conservation rates of individual UTRs. The improved background model enabled preferential conservation of a new site type, the “offset 6mer,” to be detected. In total, >45,000 miRNA target sites within human 3′UTRs are conserved above background levels, and >60% of human protein-coding genes have been under selective pressure to maintain pairing to miRNAs. Mammalian-specific miRNAs have far fewer conserved targets than do the more broadly conserved miRNAs, even when considering only more recently emerged targets. Although pairing to the 3′ end of miRNAs can compensate for seed mismatches, this class of sites constitutes less than 2% of all preferentially conserved sites detected. The new tool enables statistically powerful analysis of individual miRNA target sites, with the probability of preferentially conserved targeting (PCT) correlating with experimental measurements of repression. Our expanded set of target predictions (including conserved 3′-compensatory sites), are available at the TargetScan website, which displays the PCT for each site and each predicted target.
MicroRNAs are ∼22-nucleotide (nt) endogenous RNAs that derive from distinctive hairpin precursors in plants and animals (Bartel 2004). After incorporation into a silencing complex, which contains at its core an Argonaute protein, an miRNA can pair to an mRNA and thereby specify the post-transcriptional repression of that protein-coding message, either by transcript destabilization, translational repression, or both. MicroRNAs constitute one of the more abundant classes of gene-regulatory molecules in animals, with hundreds of distinct miRNAs confidently identified in both human and mouse (Landgraf et al. 2007). A central goal for understanding the functions of all these small regulatory RNAs has been to determine which messages are targeted for repression.
The search for biological targets of metazoan miRNAs has benefited greatly from the comparative analysis of orthologous mRNAs. Targets of miRNAs can be predicted above the background of false-positive predictions by requiring conserved Watson–Crick pairing to the 5′ region of the miRNA, known as the miRNA seed (Lewis et al. 2003). Because so many messages have preferentially preserved their pairing to miRNA seeds, targets can be predicted simply by searching for conserved 6–8mer matches to miRNA seed region (Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005). Four types of seed-matched sites are known to be selectively conserved (Lewis et al. 2005): the 6mer site, which perfectly matches the 6-nt miRNA seed, the 7mer-m8 site, which comprises the seed match supplemented by a Watson–Crick match to miRNA nucleotide 8, the 7mer-A1 site, which comprises the seed match supplemented by an A across from miRNA nucleotide 1, and the 8mer site, which comprises the seed match supplemented by both the m8 and the A1 (Fig. 1A). Supporting the validity of seed-matched target predictions, cellular messages that either decrease following miRNA addition or increase following miRNA disruption preferentially contain seed matches (Lim et al. 2005; Giraldez et al. 2006; Rodriguez et al. 2007), with the following hierarchy of site efficacy: 8mer > 7mer-m8 > 7mer-A1 > 6mer (Grimson et al. 2007; Nielsen et al. 2007). The same is true when examining protein levels (Baek et al. 2008; Selbach et al. 2008).
Figure 1.
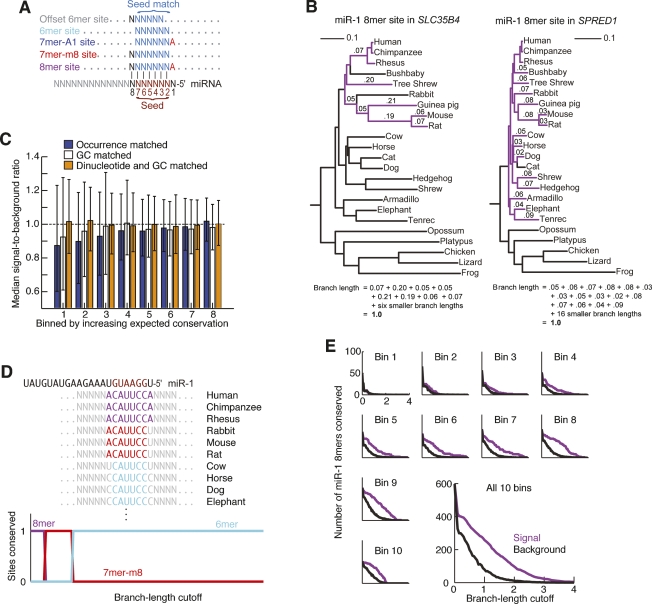
Method for detecting preferential conservation of miRNA sites. (A) Sites matching the miRNA seed region. All four canonical sites (colored) share six contiguous Watson–Crick matches to the miRNA seed (nucleotides 2–7); the offset 6mer contains six contiguous matches to nucleotides 3–8. (B) Phylogenetic conservation of individual sites. Each panel represents a miR-1 8mer site conserved to a branch length of 1.0. The 3′UTR of SLC35B4 falls into the fourth-most poorly conserved bin, which has a phylogeny with relatively long branch lengths (left), whereas the 3′UTR of SPRED1 falls into the most well-conserved bin, which has a phylogeny with shorter branch lengths (right). Branch segments connecting the species with sites are colored purple, with numbers indicating the lengths of the longer segments. The lengths of the segments connecting the species are summed to yield the branch-length score (equation). (C) Performance of controls matched for number of occurrences in human 3′UTRs, GC content, or expected conservation as calculated by a first-order Markov model. For each possible RNA 7mer, conservation rates (signal) and average conservation rates for 50 control 7mers (background) were calculated for all 3′UTRs at a branch-length cutoff of 1.0. Error bars indicate 25th and 75th percentiles. Because most 7mer motifs are not selectively maintained, well-performing controls should yield median signal-to-background ratios near 1.0, with low variability for every bin. (D) Nested site conservation. Because the seed-matched sites are interrelated, we subtract conserved instances of extended sites from those of shorter sites. This hypothetical site is an 8mer match to miR-1 in a human 3′UTR that is more broadly conserved as a 7mer-m8 than as an 8mer site, and as a 6mer than as a 7mer-m8 site. As a result of nested subtraction, our method considers this site an 8mer at low branch-length cutoffs but not a 6mer or 7mer. At moderate branch-length cutoffs, it switches to a conserved 7mer-m8 site, and at high branch-length cutoffs it switches to a conserved 6mer site but not a 7mer or 8mer. (E) Signal and background for the miR-1 8mer site. For each UTR bin 1 through 10, with bin 1 having the least conserved UTRs and bin 10 the most conserved, the number of miR-1 sites conserved at the indicated branch-length cutoff is plotted with estimated background (small plots). Results for all 10 bins were then combined to represent the aggregate signal and background for this site (large plot).
In addition to its utility for predicting the identities of the regulatory targets, comparative sequence analysis has provided fundamental insights regarding features of mRNA sites required for effective miRNA recognition. For example, a systematic analysis of matches to 7-nt segments spanning the length of the miRNAs showed that only those matching the 5′ region of the miRNA are conserved more than expected by chance, thereby defining the seed region as the key determinant of miRNA specificity (Lewis et al. 2003). Additional analyses of preferential conservation uncovered the importance of non-Watson–Crick recognition of an A across from miRNA nucleotide 1 and of an A or U across from nucleotide 9 (Lewis et al. 2005; Nielsen et al. 2007). Comparative analyses revealed targeting in open reading frames (ORFs) (Lewis et al. 2005; Stark et al. 2007a) but also supported experimental findings that sites are more effective if they fall outside the path of the ribosome (Grimson et al. 2007). Comparative analyses supported the importance of other features of site context, including positioning within high local AU composition (Grimson et al. 2007; Nielsen et al. 2007), away from the centers of long UTRs (Gaidatzis et al. 2007; Grimson et al. 2007; Majoros and Ohler 2007), and near to nucleotides that can pair to miRNA nucleotides 13–16 (Grimson et al. 2007).
Comparative analysis has also revealed the wide scope of metazoan miRNA targeting, indicating that many genes of mammals, flies, and worms are miRNA targets (Brennecke et al. 2005; Krek et al. 2005; Lewis et al. 2005; Xie et al. 2005; Lall et al. 2006). For example, combining the 3′UTR conservation attributed to miRNA seed matching with that from ORFs indicates that over one third of human protein-coding genes have been under selective pressure to maintain pairing to miRNAs (Lewis et al. 2005). Moreover, the selective depletion of seed-matching sites in messages highly expressed in the same tissues as the miRNAs implies frequent nonconserved targeting (Farh et al. 2005; Stark et al. 2005).
Ever since the availability of whole-genome multiple alignments (Blanchette et al. 2004), sites have been considered conserved if they are retained at orthologous locations in every genome under consideration and considered “nonconserved” or poorly conserved if they are missing or have changed in one of the genomes. This binary approach has been very productive but becomes less suitable now that the alignments include more than a few genomes. Requiring conservation in every species of a 28-genome alignment would exclude sites that are under strong selective pressure to be conserved in many genomes yet are missing at the orthologous position in some genomes either because of lineage-specific loss, gain, or substitution, or because of imperfections in sequencing, assembly, or alignment. To capture more of the conserved sites, a quantitative approach has been developed that makes the reasonable assumption that aligned sites within orthologous genes have a single origin and measures the portion of the phylogenetic tree that retains each site by summing the branch length over which each site has been preserved (Kheradpour et al. 2007). Because it represents an estimate of the amount of evolutionary time over which a site has been conserved, this branch-length score yields a multivalued metric that accounts for phylogenetic relationships between the species studied (Kheradpour et al. 2007). The score is interpreted by selecting a branch-length cutoff that separates more conserved and less conserved sites. Sliding the branch-length cutoff from zero to the total length of all branches enables tuning of sensitivity and specificity. This method has been applied to the 12-genome alignments of flies to predict conserved miRNA sites with sensitivity substantially improved over the previous binary approach (Kheradpour et al. 2007; Ruby et al. 2007) but has yet to be applied to mammalian site conservation.
While whole-genome alignments have made it simple to detect the conservation of sites in orthologous locations of genes, it is much more difficult to distinguish those sites or motifs under selective pressure to be maintained from those conserved by chance. A general attempt to detect preferential conservation of any motif used a simple Z-score test but did not control for genomic location or sequence characteristics (Xie et al. 2005). Another approach, developed for detecting maintenance of miRNA sites, has been to generate cohort sets of miRNA-like sequences, then determine the number of conserved sites that match these control sequences and use this as the estimate of chance conservation (Lewis et al. 2003). When choosing these controls carefully so as to avoid sites underrepresented in mRNA sequences, this approach has been effective for evaluating sets of miRNA sites in aggregate (Lewis et al. 2003, 2005; Brennecke et al. 2005; Krek et al. 2005; Stark et al. 2005; Lall et al. 2006; Kheradpour et al. 2007; Ruby et al. 2007). As previously implemented, however, this approach breaks down when examining individual miRNA-site interactions because of a failure to account adequately for differing mutational biases, dinucleotide conservation rates, and local conservation rates.
Here we develop an improved method for quantitatively evaluating site conservation and apply it to the study of vertebrate miRNA targeting. The improved sensitivity uncovered classes of sites, including offset seed matches and 3′-compensatory sites, whose conservation previously had not been detected with confidence. Overall, we find three times as many preferentially conserved sites as detected previously, thereby increasing the known scope and density of conserved miRNA regulatory interactions.
Results and Discussion
Detection of seed match conservation with increased sensitivity and statistical power
When using a branch-length metric to evaluate motif conservation, the first step is to build a phylogenetic tree based on the genomic regions under investigation (Kheradpour et al. 2007), which in our case was 3′UTRs. One major innovation of our method was to build the phylogeny in a way that controlled for the conservation of individual UTRs. Because mutation, gene conversion, and crossover rates vary throughout the genome (Wolfe et al. 1989; Hwang and Green 2004; Kauppi et al. 2004), different UTRs have substantially different background conservation levels. Moreover, some vertebrate genomes have low coverage and are missing a substantial fraction of genes, also affecting the apparent background conservation. Most methods for detecting positive selection take into account local conservation rates (Yang and Bielawski 2000) (for example, Ks in Ka/Ks), but genome-scale methods for detecting purifying selection have thus far not accounted for this factor. In addition to differences in basal conservation rates, UTRs have sequence-dependent functions apart from miRNAs, which can influence conservation levels. A site falling within a UTR with high overall conservation is far less likely to be conserved due to miRNA targeting than is one falling within a rapidly evolving UTR (Lewis et al. 2005). Any method that treats all the UTRs the same greatly overestimates purifying selection of sites in well-conserved UTRs and underestimates purifying selection of sites in poorly conserved UTRs.
Starting with a 28-way vertebrate whole-genome alignment that included 18 placental mammals and 10 other vertebrates (Miller et al. 2007), we extracted the human 3′UTRs and homologous regions from the 22 non-fish genomes. The five fish genomes were excluded because they lacked a sufficient amount of aligned 3′UTR sequence. To help control for individual UTR conservation, 3′UTRs were separated by conservation rate into 10 equally sized bins, and a unique set of branch lengths based on 3′UTR sequence alignments was constructed for each bin (Fig. 1B; Supplemental Fig. 1). The conservation of a given sequence (e.g., an 8mer miR-1 site in a particular 3′UTR) was then assessed by summing the total branch length in the phylogenetic tree connecting the subset of species having the sequence perfectly aligned, using the tree representing the bin of the 3′UTR under investigation. This branch-length value had no units, with a value of 1.0 corresponding to the average conservation of a single nucleotide in similar UTRs, and thus resembled a non-normalized version of the branch-length score described by Kheradpour et al. (2007). In our analyses, however, a site in a more divergent UTR needed to be conserved in fewer orthologs to achieve the same branch-length value because the branch lengths in a phylogeny representing the more divergent UTRs were longer than those of one representing more conserved UTRs. For example, the 8mer miR-1 site found within in the human SLC35B4 UTR received the same value as the site within the SPRED1 UTR, even though it was present in fewer aligned genomes (Fig. 1B).
Because sequences can be conserved by chance or for many reasons other than functional miRNA targeting, the branch-length values were only interpretable when considered within the context of the estimated background conservation. Our method attempted to control for many factors that can affect the conservation level of a short sequence of length k (a k-mer), including GC content, dinucleotide content, the interrelation of miRNA seed-match types, genome alignment quality, and the local conservation rate. The combined effects of all of these factors on background conservation were estimated based on em-pirically observed conservation of k-mers as opposed to theoretical calculations. As done previously (Lewis et al. 2003, 2005; Brennecke et al. 2005; Krek et al. 2005; Stark et al. 2005; Lall et al. 2006; Kheradpour et al. 2007; Ruby et al. 2007), the expected fraction of sites conserved due to miRNA recognition was estimated using a cohort of k-mers with similar properties, which were presumed to be subject to the same evolutionary pressures except for the possible miRNA regulatory relationship. Because we did not allow conserved k-mers that were seed matches for miRNAs with any known conservation, and because the discovery rate of highly conserved vertebrate miRNAs has dropped dramatically in recent years, the control k-mers can be assumed devoid of conservation due to miRNA targeting. Three substantial improvements to the estimation of background conservation were introduced. First, we matched control k-mers using an expected conservation based on both the k-mer’s GC content and the expected conservation of its constituent dinucleotides, which enabled a more rigorous and accurate estimate of background conservation levels for individual miRNAs (Fig. 1C). Previous methods matched k-mers based on their abundance in human 3′UTRs, which is adequate when analyzing large groups of miRNAs, but this variable is poorly correlated to conservation for individual miRNAs (Fig. 1C) and can be affected by evolutionary avoidance of k-mers, a known property of miRNA seed matches (Farh et al. 2005; Stark et al. 2005). Second, we created mutually exclusive seed-match classes by subtracting the signal and the background of larger seed matches (e.g., 8mers) from the smaller seed matches that could be contained within them (e.g., 7mers, Fig. 1D). This protected against double-counting conservation while increasing sensitivity by more closely matching control k-mer sizes to the observed conservation (see Supplemental material for discussion). Third, the estimate of background conservation controlled for the conservation of individual UTRs, in that control cohorts were analyzed using the same 10 phylogenetic trees and the same 10 UTR data sets were employed for analysis of authentic sites. Without this improvement, different members of the control cohort had widely varying conservation. By reducing this variability, more precise background estimates were achieved, which enabled more sensitive detection of site conservation. Thus, we calculated 10 distributions of branch-length values for both signal and background using both the k-mer and its set of controls and then summed these distributions to compile the overall signal and background distributions for each k-mer (Fig. 1E; Supplemental Discussion). These three innovations all helped to control for the background conservation specific to individual seed-match sites, enabling statistically sound comparisons between the conservation of seed-match types, between seed matches to different miRNAs, and even between individual sites.
At least 44,000 sites are selectively maintained because of miRNA targeting
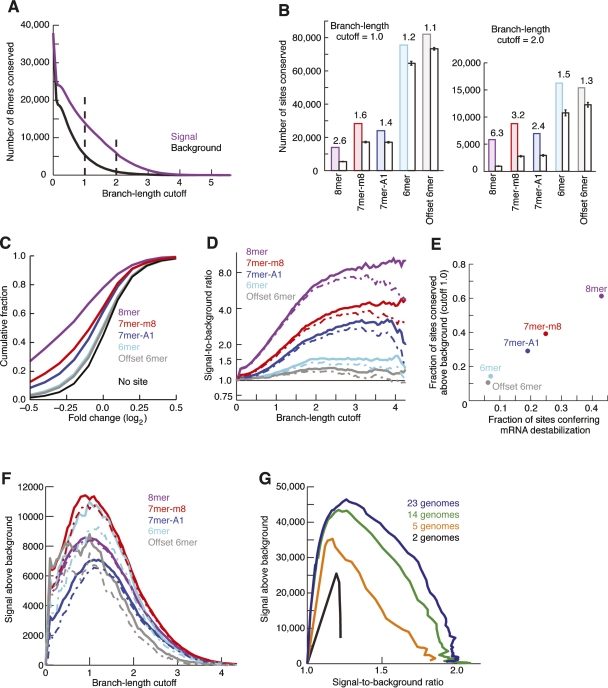
We first looked for excess conservation of seed matches for a set of highly conserved miRNAs that appear to have been present since the last common ancestor of all 23 vertebrate species under consideration, defined as those mammalian miRNAs also found in chicken, lizard, or fish, which fell into 87 families based on the identity of nucleotides 2–8 (Supplemental Table 1). For each k-mer, representing a single seed-match type for a particular miRNA, the distribution of branch-length values was compiled for sites present in human 3′UTRs. As the branch-length-value cutoff was increased from zero, the number of sites that matched control sequences decreased faster than did the number matching authentic miRNA seed matches (Fig. 2A). At any particular branch-length cutoff, if the number of conserved sites of a k-mer (the signal) was higher than that of control sequences (the background), the excess conservation was attributed to purifying selection. We use the term “background” instead of “noise” because the latter term may connote variance in the background estimate as opposed to the estimate itself. The number of sites conserved above background reflects the sensitivity of the analysis, whereas the ratio of signal to background reflects its specificity.
Figure 2.
Conservation of major seed-match types. (A) Conservation of 8mer sites for 87 broadly conserved miRNA families. High-sensitivity and high-specificity cutoffs are highlighted with broken lines at 1.0 and 2.0, respectively. (B) Conservation and background estimate for mutually exclusive site types at high sensitivity (left) and high specificity (right). The signal-to-background ratio is indicated above the pair of bars. Error bars indicate one standard deviation in the estimated background, based on subsampling of individual control k-mers. (C) Efficacy of offset 6mer sites. Microarray data monitoring mRNA destabilization following transfection of 11 miRNAs was analyzed as described previously (Grimson et al. 2007). Shown is the cumulative distribution of changes for transcripts containing exactly one offset 6mer site and no other canonical sites in their 3′UTR. For comparison, previously reported analyses of messages with single canonical sites are also shown (Grimson et al. 2007). (D) Signal-to-background ratio for indicated sites at increasing branch-length cutoff. Broken lines indicate 5% lower confidence limit (z-test). (E) Correlation of site conservation rate and experimental efficacy. Fraction of sites conserved above background was calculated as ([Signal – Background]/Signal) at a branch-length cutoff of 1.0. The minimal fraction of sites conferring destabilization was determined from the cumulative distributions (C), considering the maximal vertical displacement from the no-site distribution (correcting for the bumpiness of the distributions as described previously [Grimson et al. 2007]). (F) Estimates of signal above background for the major site types. Broken lines indicate 5% lower confidence limit (z-test). (G) Aggregate conservation above background for all major site types when using using subsets of genomes. To facilitate overlay of the plots, the X-axis is signal-to-background ratio rather than branch-length cutoff. The 14-genome subset represents the non-fish species originally available in the UCSC 17-way alignments. The five-genome subset contains human, mouse, rat, dog, and chicken, and the two-genome subset contains only human and mouse.
We first considered the three 7–8mer seed-match types (8mer, 7mer-m8, 7mer-A1), which correlate most strongly with targeting efficacy (Grimson et al. 2007; Nielsen et al. 2007) and are among the miRNA matches currently used to predict conserved targets of metazoan miRNAs (Fig. 2B; Brennecke et al. 2005; Grun et al. 2005; Krek et al. 2005; Lewis et al. 2005; Lall et al. 2006; Ruby et al. 2006, 2007; Gaidatzis et al. 2007). At a branch-length cutoff of 2.0, a large majority of these sites were in excess of the background (Fig. 2B, right). However, this high specificity came at a price, with many more sites detected above background at a less stringent cutoff of 1.0 (Fig. 2B).
Our more precise estimate of background conservation enabled robust detection of purifying selection for 6mer seed matches that were not part of the larger, 7–8mer seed-matched sites (Fig. 2B). Ten thousand sites were conserved above background—a high number when considering the marginal efficacy of these 6mer sites, as measured by monitoring mRNA destabilization or protein output after adding or disrupting miRNAs (Grimson et al. 2007; Nielsen et al. 2007; Baek et al. 2008; Selbach et al. 2008). Analysis of mRNA expression following ectopic addition of miRNAs into HeLa cells indicated that an offset 6mer matching miRNA positions 3–8 (Fig. 1A) mediated mRNA destabilization approaching that of the seed-matched 6mer, matching positions 2–7 (Fig. 2C), although the effects of the seed-match 6mer were still significantly stronger (P = 0.03, two-sided KS test). This marginal yet detectable activity prompted us to explore the possibility that these offset 6mer sites might also be selectively maintained. Our analysis, subtracting conservation due to 7- or 8-nt seed-matched sites as well as that attributed to matching seeds of related miRNAs, indicated that a small but detectable fraction of these offset 6mers were indeed selectively maintained (Fig. 2B). Because these 6mer sites are so abundant in 3′UTRs, this small fraction corresponded to thousands of sites under purifying selection, which have been missed by algorithms that search for only seed-matched sites.
The result for the offset 6mer raised the question of whether 6mer matches to nearby miRNA segments might also be selectively maintained. Analysis of matches to miRNA segments 1–6, 4–9, and 5–10, excluding those sites that also possessed seed matches, revealed no 6mer segments with appreciable signal above background (Supplemental Fig. 2). Parallel analyses of the mRNA expression data also failed to reveal 6mer sites with efficacy approaching that of the 6mer site corresponding to segment 3–8. We therefore focused on the selective conservation of the five types of sites that matched the seed region, one 8mer, two 7mers, and two 6mers, which we refer to as the 6mer and the offset 6mer (Fig. 1A).
When examined over a broad range of branch-length cutoffs, signal-to-background ratios plateaued at a branch-length cutoff of about 3 (Fig. 2D), which exceeded the maximal branch length of the more highly conserved UTR bins. Larger signal-to-background ratios implied higher fractions of seed matches under selection. For example, a signal-to-background ratio of 4.0 corresponds to 75% of matches being under purifying selection and thus presumably having conserved function. Regardless of the cutoff, the hierarchy of signal-to-background ratios remained constant, with 8mer > 7mer-m8 > 7mer-A1 > 6mer > offset 6mer. Moreover, the signal-to-background ratio of the five site types, which indicated the fraction of sites under selection, corresponded well with the minimal fraction of sites conferring transcript destabilization following microRNA transfection, indicating a striking correlation between the selective maintenance of site types and their efficacy (Fig. 2E).
When considering the number of selectively maintained sites, a moderate branch-length cutoff of 1.0 yielded the highest signal above background (Fig. 2F). Increasing cutoffs from 1.0 to 2.0 yielded a tradeoff between increased specificity (Fig. 2D) and decreased sensitivity (Fig. 2F). For the five individual site types, the number of selectively maintained sites showed little correlation with the signal-to-background ratio. For example, the signal-to-background ratio for the 6mer (1.2 at branch length 1.0) was far lower than that for the 8mer (2.6 at branch length 1.0), but signal above background for the 6mer (10,970 at branch length 1.0) was at least as high as that of the 8mer (8543 at branch length 1.0). Thus, 3′UTRs acquire and maintain marginally effective target sites in similar numbers as they do more highly effective sites. The 7mer-m8 sites appear most important in terms of the number of sites under selection (Fig. 2F), whereas 8mers are the most important in terms of the proportion of sequences under selection and, equivalently, the power for prediction of individual targets (Fig. 2D).
Summing together the signal and background estimates for the five site types at the most sensitive conservation cutoff (1.0) yielded 46,441 ± 2175 sites conserved above background (Fig. 2F), an average of 534 ± 25 per miRNA family (98 ± 2, 128 ± 7, 80 ± 8, 126 ± 22, 101 ± 14 for 8mer, 7mer-m8, 7mer-A1, 6mer, and offset 6mer sites, respectively). This number of sites was nearly three times higher than the most sensitive previous estimate, which had required perfect 6mer conservation in each of human, mouse, rat, and dog to detect 13,044 3′UTR sites conserved above background, or 210 sites conserved per miRNA family (Lewis et al. 2005). Several factors contributed to this large increase in the estimate of selectively maintained miRNA sites, including the improved methodology, larger and more accurate UTR and miRNA data sets, new genomes, and improved genome quality. To determine whether the principal factor was the newly available genomes, we performed the same analysis on subsets of genomes, keeping the UTR and miRNA data sets and methodology constant (Fig. 2G). The sensitivity was robust to the removal of a large number of genomes, suggesting that with current methods, the estimate of the number of selectively maintained sites will remain relatively constant with the addition of newly sequenced genomes.
Detection of selectively maintained sites with higher sensitivity implied that the number of conserved miRNA targets is far higher than previously estimated. Starting with all the sites detected at a given conservation cutoff and then randomly removing for each site type the number of sites corresponding to the predicted background in the relevant UTR bin yielded 9909 ± 302 genes targeted at a branch-length cutoff of 1.0. Using this method of sampling conserved sites, only 7% of genes had multiple conserved sites for the same miRNA family. Thus, for each miRNA family, the number of conserved targets (497 ± 49) approached the number of conserved sites (534 ± 25). Although more sites above background were predicted at the conservation cutoff of 1.0, the number of genes targeted reached a maximum of 10,739 ± 564 at a branch-length cutoff of 0.6, which corresponded to 57.8% ± 3.0% of the human RefSeq data set. This percentage is about twice that of the most sensitive previous estimate (Lewis et al. 2005). Again, the number of targets per miRNA family (438 ± 60) approached the number of sites conserved above background per miRNA family (462 ± 28). Nonetheless, 72% of the 10,739 targeted messages had sites to multiple miRNA families, with an average of 4.2 sites per targeted 3′UTR. Indeed, the observed twofold increase in targeted UTRs from a threefold increase in site detection meant that our analysis added many additional newly predicted sites to previously predicted targets, thereby increasing not only the number of predicted targets but also the density of predicted targeting.
Sites with seed bulges and mismatches are rarely under selection
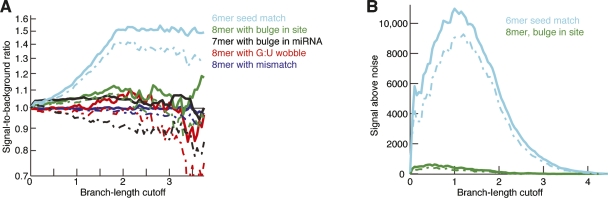
Having found a large and statistically significant number of conserved 6mer sites (Fig. 2F), despite their marginal efficacy (Fig. 2C), we investigated the possibility of selective conservation of imperfect seed matches, which also display severely compromised efficacy. Reasoning that if any mismatched sites were selectively maintained they would include those with the least disruptive mismatches, we focused on 8mer matches containing either a single mismatch, a G:U wobble, a bulged nucleotide within the site, or a bulged nucleotide within the miRNA. In contrast to the canonical seed-matched types, these imperfect sites displayed little enrichment of conservation (Fig. 3A). For all four mismatched classes, signal-to-background ratio hovered near 1.0, rarely exceeding 1.1 at any branch-length cutoff, indicating that the number of sites under selection was at most a small fraction of the total (Fig. 3A). The 8mer with a bulge in the site was the only class for which the 5% confidence limit on the ratio consistently exceeded 1.0. This class of sites appeared to have a few hundred sites conserved above background, a number 10 times less than that of even the weakest seedmatched class (Fig. 3B). We cannot exclude the possibility that a very small fraction of other mismatched sites might also have been selectively maintained. However, because of the low signal-to-background ratio and low 5% confidence estimate for the number of sites under selection, we conclude that seed-mismatched sites are hardly ever selectively maintained and that including a substantial number of such sites when predicting targets would greatly compromise prediction specificity. These conclusions are supported by recent proteomic experiments demonstrating poor efficacy of targets predicted by methods that allow sites with seed mismatches (Baek et al. 2008; Selbach et al. 2008).
Figure 3.
Occasional preferential conservation of imperfect sites. (A) Signal-to-background ratio for sites with the indicated single-nucleotide mismatches and bulges. A mismatch or G:U wobble must occur opposite miRNA seed nucleotides 2–7. A bulge in the site must occur between bases that pair to consecutive seed nucleotides 2–7, and a site creating a bulge must involve a 7mer match that skips one of the seed nucleotides 2–7. Results for the canonical 6mer site (Fig. 2D) are included for comparison. Broken lines indicate 5% lower confidence limit (z-test). (B) Weak signal above background for a class of imperfect sites found to have a significantly positive signal-to-background ratio. Results for the canonical 6mer site (Fig. 2F) are included for comparison. Broken lines indicate 5% lower confidence limit (z-test).
Selective maintenance of sites with a bulge in the site but not those with a bulge in the miRNA corresponds well with previous analyses of plant miRNA targeting (Mallory et al. 2004). This constraint observed in both plant and animal lineages can be explained by the idea that the Argonaute protein binds the miRNA backbone, preorganizing the miRNA seed region such that the Watson–Crick face is poised for pairing to the message (Bartel 2004). These contacts to the backbone in the seed region, presumably present before and after binding, would constrain the seed backbone, spacing each seed nucleotide such that a bulge in the miRNA would impose a gap in the site that would be difficult to span without disrupting adjacent pairs. In contrast, a bulged nucleotide in the site would be extruded into solvent and therefore more readily accommodated.
Pairing to the 3′ end of miRNAs displays small but measurable excess conservation
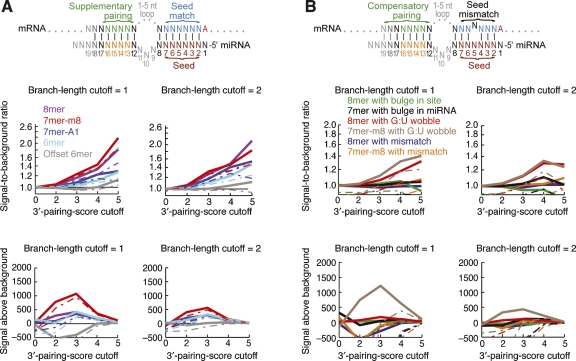
Although pairing to the 3′ region of the miRNA has long been thought to be consequential, evidence that such pairing enhances the efficacy of mammalian seed-matched sites has been obtained only recently (Grimson et al. 2007). Such sites in which 3′ pairing productively augments seed pairing are called “3′-supplementary sites.” Productive 3′ pairing optimally centers on miRNA nucleotides 13–16 and the UTR region directly opposite this miRNA segment (Fig. 4A, top). Like seed pairing, 3′ pairing appears relatively insensitive to predicted thermostability and instead quite sensitive to pairing geometry, preferring contiguous Watson–Crick pairs uninterrupted by bulges, mismatches, or G:U wobbles. These features are captured in a 3′-pairing score, which awards one point for each contiguous Watson–Crick pair matching miRNA nucleotides 13–16 and a half point for each contiguous pair extending the pairing in either direction. Pairing segments offset from the miRNA are then penalized by subtracting a half point for each nucleotide of offset beyond ±2 nucleotides from the register directly opposite the miRNA, and then sites are assigned the score of the highest scoring pairing segment (Grimson et al. 2007). For example, the site shown in Figure 4A (top), which has seven contiguous, well-positioned pairs would be assigned a score of 5.5. Sites with scores ≥3 display modestly increased efficacy and conservation (Grimson et al. 2007).
Figure 4.
Conserved pairing to the 3′ ends of miRNAs. (A) Preferential occurrence of pairing to the 3′ region of the miRNA, which can supplement canonical sites (diagrammed at top). Signal-to-background ratio (top two graphs) and signal above background (bottom two graphs) are plotted at conservation branch-length cutoffs of 1.0 (left) and 2.0 (right). Five percent confidence limits are shown as dashed lines. (B) Preferential occurrence of pairing to the 3′ region of the miRNA, which can compensate for mismatches or bulges in seed pairing (diagrammed at top). As in A, except that seed matches were replaced as indicated with sites containing a mismatched, G:U wobble, or bulged nucleotide.
We set out to determine for each site type the selective maintenance of 3′-supplementary pairing. At specified cutoffs for branch length and 3′-pairing score we determined the number of sites with supplementary 3′ pairing, estimating the background by repeating the analysis with a chimeric miRNA set created by swapping all possible 5′ and 3′ ends for miRNAs within our 87 miRNA families. For each site, the 3′ pairing score used was the maximum over all members of the miRNA family. For each of the four seed-matched types, and especially for the 7mer-m8 site, selective maintenance of 3′-supplementary pairing was confidently observed (Fig. 4A). As expected for a biological signal, specificity increased with greater conservation and with a greater 3′-pairing score. Sensitivity peaked at a pairing score cutoff of 3.0, indicating that as few as 3–4 well-positioned supplementary pairs were selectively maintained (Fig. 4A). However, even at this sensitive cutoff, only 2281 ± 537 seed-matched sites had preferentially conserved 3′ pairing. Assuming that sites with selectively maintained 3′ pairing were also drawn from the pool of ∼44,000 sites with selectively maintained matches to the seed region, we estimate that only 4.9% ± 1.1% of all preferentially conserved sites have preferentially conserved 3′ pairing. Nonetheless, for those rare sites with high 3′ pairing scores, consideration of supplemental pairing provided a useful boost to the overall signal-to-background ratio. For example, for the 49 8mer sites with 3′-pairing scores ≥5.0 and branch-length values ≥2.0, the aggregate signal-to-background ratio was estimated to be 13:1 (calculated as 6.3 × 2.1, using values from Figs. 2D and 4A, respectively), implying that the conservation of these individual sites was confidently attributed to miRNA targeting. For the remaining 95.1% of selectively maintained seed matches, which do not have preferential conservation of pairing to the 3′ end of miRNAs, the 3′ region of the miRNA might still interact with the message, but in a way that does not favor matches over mismatches and therefore does not add detectably to targeting specificity.
Selective maintenance of 3′-compensatory sites
Pairing to the 3′ portion of the miRNA can not only supplement a 7–8mer match, it can also compensate for a single-nucleotide bulge or mismatch in the seed region, as illustrated by the let-7 miRNA sites in lin-41 and the miR-196 site in HOXB8 (Vella et al. 2004; Yekta et al. 2004). Such sites are called “3′-compensatory sites” (Fig. 4B, top). Previous analyses of mRNA array datafailed to detect efficacy of 3′-compensatory sites, even when considering the principles that uncovered consequential 3′-supplementary pairing, which are embodied in the 3′-pairing score (Grimson et al. 2007). This failure is attributed to the idea that 3′-compensatory sites are exceedingly rare, presumably because the amount of pairing needed to compensate for seed mismatches is greater than that needed to supplement seed-matched sites, and as a result, a significant association is difficult to detect (Grimson et al. 2007). Supporting this idea, all experimentally validated examples of metazoan 3′-compensatory sites involve pairing that centers on miRNA nucleotides 13–17 and extends to at least 9 contiguous Watson–Crick pairs, which corresponds to a 3′-pairing score ≥6.5 (Vella et al. 2004; Yekta et al. 2004).
An analysis of preferential site conservation could be more sensitive than that of array data, both because site-conservation analysis captures sites mediating translational repression without detectable mRNA destabilization and because site-conservation analysis can simultaneously consider many more miRNAs. With this possibility in mind, we examined the prevalence of 3′-compensatory sites, applying the same methodology as used for 3′-supplementary sites. Sites with single-nucleotide mismatches or bulges had no enrichment in conserved or nonconserved 3′ pairing exceeding the 5% confidence threshold, regardless of the cutoffs (Fig. 4B). This result indicated that most of the bulged sites conserved above background (Fig. 3) are not conserved because they are 3′-compensatory sites. However, 7–8mer sites with a single G:U wobble had confidently enriched pairing at the relatively high 3′-pairing-score cutoff of 4.0. At a branch-length cutoff of 1.0, the number of sites within the 5% confidence interval numbered only 399, an average of only 4.5 per miRNA family. Thus, our results support previous assertions that mismatched seed sites with 3′-compensatory pairing are only rarely under selective pressure to be conserved (Brennecke et al. 2005; Lewis et al. 2005). Perhaps because such sites with extensive pairing to the 3′ portion of the miRNA possess much more informational complexity than do the 7–8mer perfect matches and therefore emerge much less frequently and are harder to maintain in evolution, these 3′-compensatory sites appear to be used only rarely for biological targeting, comprising 1.5% ± 0.7% of the conserved sites in mammals. The somewhat higher number of 3′-supplementary sites implies that 3′ pairing, when consequential, is principally a supplementary feature of canonical seed sites and more rarely plays a role in conferring activity to imperfect seed sites.
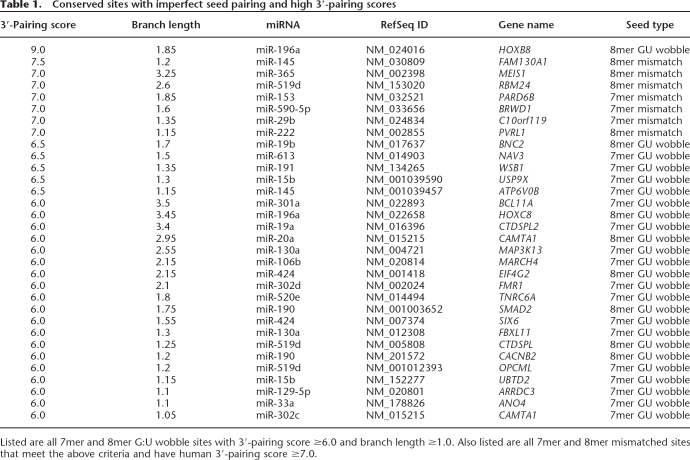
The paucity of 3′-compensatory sites poses special challenges for confidently detecting conserved biological sites above background. Our use of the 3′-pairing score and our observation that the G:U class of mismatches was more frequently compensated by conserved 3′ pairing both represented important inroads into meeting this challenge. The 25 conserved G:U sites with 3′-pairing scores ≥6 include the miR-196 site in the HOXB8 3′UTR, which has an off-scale 3′-pairing score of 9.0, and a similar miR-196 site in the HOXC8 3′UTR (Yekta et al. 2004). These 25 sites, together with the seven mismatched sites with scores ≥7, are listed in Table 1 and will be included in the next release of TargetScan predictions (targetscan.org). Bulged sites with high 3′-pairing scores (≥6) did not appear preferentially conserved and thus are not included in the list.
Table 1.
Conserved sites with imperfect seed pairing and high 3′-pairing scores
Listed are all 7mer and 8mer G:U wobble sites with 3′-pairing score ≥6.0 and branch length ≥1.0. Also listed are all 7mer and 8mer mismatched sites that meet the above criteria and have human 3′-pairing score ≥7.0.
Mammalian-specific miRNAs have few selectively maintained seed matches
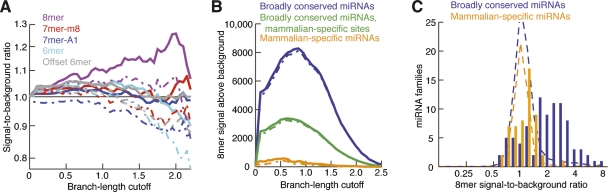
An early study found that sites matching broadly conserved vertebrate miRNAs were more likely to be maintained than those matching mammalian-specific miRNAs (Lewis et al. 2003). Since then, target-prediction specificity has been estimated using only those miRNAs conserved to fish or chicken (Krek et al. 2005; Lewis et al. 2005; Gaidatzis et al. 2007), raising the question of whether the more recently emerged miRNAs have acquired enough conserved targets to detect any conservation signal above background. To address this question, we assembled a set of 53 miRNAs that were present in diverse placental mammals but absent in all sequenced chicken, lizard, and fish genomes (Supplemental Table 2). Examining the placental mammal subset of the phylogeny, we found little preferential conservation for sites matching these mammalian-specific miRNAs (Fig. 5A). In contrast to sites matching broadly conserved miRNAs (Fig. 2D), the full 8mer was the only seed-match type with signal-to-background ratio consistently and confidently above 1.0 (Fig. 5A), and its ratio was no higher than that of offset 6mers matching broadly conserved miRNAs.
Figure 5.
Conservation of sites matching mammalian-specific miRNAs. (A) Signal-to-background ratio for sites matching 53 mammalian-specific miRNA families in 18 placental mammals, otherwise as in Figure 2D. (B) Signal above background for 8mer sites matching either broadly conserved or mammalian-specific miRNAs in 18 placental mammals, otherwise as in Figure 2F. Analysis of 8mer sites matching broadly conserved miRNAs considers either all sites (blue) or excludes those sites conserved beyond placental mammals (green). (C) Signal-to-background ratios for 8mer sites matching individual miRNAs in orthologous 3′UTRs of placental mammals at optimal sensitivity (branch-length cutoff of 0.85). For the broadly conserved miRNA set, conservation signal excludes sites conserved beyond placental mammals. Distributions expected if miRNA targeting conferred no preferential conservation were estimated using the average signal-to-background ratio of 8mer controls selected for each site, considering GC content and dinucleotide-based conservation (broken lines). Expectations differed between the two sets because of different miRNA numbers and different dinucleotide compositions.
The performance of the mammalian-only set also differed from that of the broadly conserved set when considering signal above background, with far fewer 8mers conserved above background (Fig. 5B). These differences could be due either to inherent differences in the miRNA sets, such as the level and breadth of expression, or to differential evolutionary time available for beneficial site emergence. To help differentiate between these possibilities, we screened matches to the broadly conserved miRNAs, removing all sites with seed matches conserved beyond mammals, thereby limiting the set of 8mer sites to those more likely to have arisen in mammals after the divergence of mammals and other vertebrates. This removed more than half of the conserved sites matching the broadly conserved miRNAs, showing that part of the reason for the higher number of sites is the much greater time available for beneficial site emergence. However, even when considering the more restricted set of sites, and after normalizing for the numbers of miRNAs in the two sets, the broadly conserved miRNAs had more than four times as many selectively maintained 8mer matches per miRNA than did the mammalian-specific miRNAs (Fig. 5B), suggesting that the level and breadth of miRNA expression are also important factors. Combining the signal above background for all site types, the differential appeared far greater, with the mammalian miRNAs averaging only 10.9 ± 3.0 selectively maintained sites.
To determine whether a few of the mammalian-only miRNAs might have conservation patterns resembling those of the broadly conserved set, we examined the conservation of 8mer seed matches corresponding to individual miRNAs. As expected, the signal-to-background ratios of the broadly conserved miRNAs fell mostly outside the control distribution (Fig. 5C). The observation that most fell outside the control distribution illustrated that the high signals observed for these broadly conserved miRNAs in aggregate also applied to most of them individually and could not be attributed to chance overlap of a few seed matches to a few highly conserved regulatory sequences. In contrast, the mammalian-specific miRNAs had a distribution only slightly shifted from that of the controls, with an excess of ∼5–10 miRNAs in the 1.5–2.5 signal-to-background range. We conclude that only a small subset of mammalian-specific miRNAs (including most prominently miR-487b, miR-127-3p, miR-379, and miR-543; Supplemental Table 2) have a measurable number of selectively maintained sites and caution that for the majority of mammalian-specific miRNAs, the observation that a site is conserved provides no evidence that it has biological function.
Estimating confidence for selective maintenance of individual sites
The widespread scope of conserved targeting brings to the fore the question of which of these many miRNA target interactions can be predicted with confidence. One raw indicator is the conservation branch length of the site—clearly, more highly conserved sites are more likely to be under selection, particularly when controlling for differential UTR conservation, as in our method. However, our branch-length values did not account for the type of site and its sequence features. For example, a branch length of 1.0 for an 8mer is far more compelling evidence for selective maintenance than a branch length of 3.0 for an offset 6mer (Fig. 2D). To control for site type and sequence features, we used our previously described controls to calculate a signal-to-background ratio (S/B) for each site at each branch length. For the purpose of evaluating individual sites, assessing controls at each branch length instead of at each branch-length cutoff is necessary to avoid crediting poorly conserved sites for having the same sequence as many highly conserved sites. We then converted this S/B to a probability of conserved targeting (PCT), which is approximately equal to (S/B − 1)/(S/B) (or near zero, for sites with S/B < 1). This score reflected the Bayesian estimate of the probability that a site is conserved due to selective maintenance of miRNA targeting rather than by chance or any other reason not pertinent to miRNA targeting, allowing for uncertainty in the S/B ratio. For predicting biologically conserved target sites, the score represented 1 – FDR, where FDR was our estimate of the false-discovery rate. This deceptively simple value incorporated knowledge of the conservation level of a particular site, the seed-match type, the number of selectively maintained seed matches for the particular miRNA, the background conservation level for the k-mer, and the UTR conservation context.
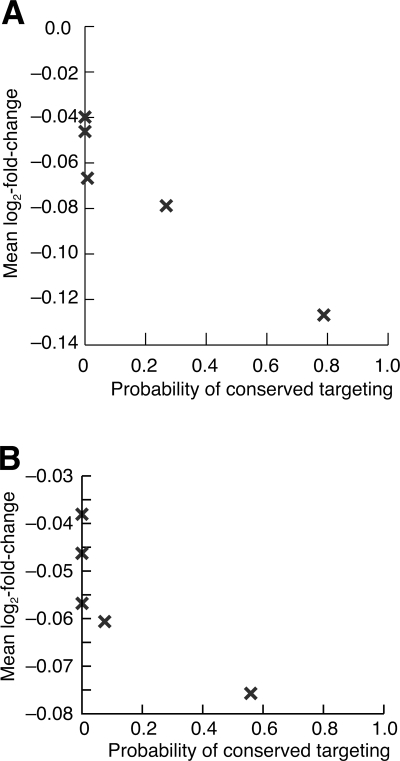
The PCT provides a useful criterion for assessing the biological relevance of predicted miRNA–target interactions. Selectively maintained sites are more likely to have detectable biological function, are more likely to have functions in experimental animals that are pertinent to humans, and tend to be more effective (Grimson et al. 2007; Nielsen et al. 2007). To illustrate the ability of this measure to predict site efficacy, we turned to experimental data examining mRNA destabilization after introducing miRNAs (Grimson et al. 2007). As expected, site PCT correlated with the mean level of mRNA destabilization (Fig. 6A). In addition, we tested a subset of miRNA sites that were previously considered nonconserved because they were not found in mouse, rat, or dog alignments (Lewis et al. 2005). Those sites newly recognized as selectively maintained were substantially more responsive to the transfected miRNAs than were those still lacking a signal for conserved targeting (Fig. 6B).
Figure 6.
Correlation of PCT with mRNA destabilization. (A) Destabilization of human messages with exactly one 7mer-m8 3′UTR site to a transfected miRNA (Grimson et al. 2007). Messages were grouped into six equal bins based on the site PCT. (B) Destabilization of human messages with exactly one 7mer-m8 3′UTR site to a transfected miRNA (Grimson et al. 2007), considering only those sites that were not conserved in either mouse, rat, or dog.
Having established a more sensitive and precise tool for evaluating site conservation, we overhauled the TargetScan web resource, separating conserved and highly conserved 3′UTR targets of each miRNA based on the PCT values of their 7–8mer sites, compiling an aggregate probability of conserved targeting (aPCT) for those targets with multiple sites for that miRNA, calculated as aPCT = 1 – (FDRsite1 × FDRsite2 × FDRsite3 . . .). To capture sites that were missing in the annotated human 3′UTRs but present in the mouse annotations, a mouse-centric version of the analyses was performed in parallel. This improved web resource, called TargetScan version 5.0, also lists the PCT for each 7–8mer site, with the option of sorting the predicted targets of each miRNA by their aPCT. Retained in TargetScan 5.0 are site context scores, which consider features such as the AU content in the vicinity of the site and the position of the site within the message, to predict the function and quantitative efficacy of each site (Grimson et al. 2007). Because the context scores are determined based on information completely orthogonal to site conservation, the PCT values and the context scores provide independent and complementary information useful for predicting the biological relevance and efficacy of each site.
The widespread scope of conserved targeting
One of the key results of applying our new methods for quantitatively evaluating site conservation to the study of miRNA targeting was the sheer number of preferentially conserved sites. When considering only the 6–8mer perfect 3′UTR matches to the seed regions of broadly conserved miRNAs, 46,441 ± 2175 sites were conserved above background, implying that 57.8% ± 3.0% of the human genes are conserved miRNA targets. Considering also imperfectly matched sites added another 625 ± 239 preferentially conserved sites to the total (Fig. 3B); 3′-compensatory sites added another 714 ± 315 (Fig. 4B), sites matching more narrowly conserved miRNAs added another 578 ± 158 (Fig. 5B), altogether modestly raising the estimated number of preferentially conserved 3′UTR sites to 48,360. Not considered are the human miRNAs that have escaped confident annotation. However, because these miRNAs have remained unannotated, in part because they are more narrowly conserved and have more restrictive expression domains, our results for the mammalian-specific miRNAs suggest that eventual consideration of these unannotated miRNAs will add only modestly to the current picture of conserved targeting. We suspect that a more significant increase will come by considering targeting in ORFs, which is thought to be widespread, although less than that in 3′UTRs (Lewis et al. 2005; Stark et al. 2007b). Further increases will come with improved UTR annotations and the consideration of alternative 3′UTRs. Anticipating these additional sites, we can say with confidence that over 60% of human protein-coding genes are conserved targets of miRNAs, with our best estimate of the actual fraction of human protein-coding genes under pressure to maintain pairing to miRNAs falling above this, at about two thirds of all protein-coding genes.
One surprise of our analysis was the substantial number of selectively maintained 6mer and offset 6mer sites, numbering 10,970 ± 1909 and 8803 ± 1276, respectively. Indeed, when excluding these sites and considering only the 7–8mer sites, our estimate of the number of conserved targets per broadly conserved miRNA dropped to 292 ± 18, which corresponded to 44.5 ± 3.4% of the RefSeq genes. With the exception of an offset 6mer site for let-7 miRNA in the human LIN28 3′UTR (Wu and Belasco 2005), 6mer sites typically have very poor efficacy when examined experimentally, as if the majority are inert, and nearly all the rest have very marginal influence on protein output (Fig. 2C; Grimson et al. 2007; Nielsen et al. 2007; Baek et al. 2008; Selbach et al. 2008). Yet 3′UTRs selectively maintain these 6mer target sites in similar numbers as they do 7 and 8mer sites. Of course, 6mers are much easier to access by mutation and then preserve from mutation, but what would be the selection pressure to preserve such minor effects? One conundrum in the field of miRNA-mediated gene regulation is why so many 7–8mer sites would be conserved so broadly in metazoan 3′UTRs, when each down-regulates protein output by very little—nearly always less than 50% and usually less than 30%, which would only very rarely produce observable phenotypic consequences (Baek et al. 2008). But the 7–8mer conundrum pales when considering the selective maintenance of 6mers, which appear to tweak gene expression so finely that the effects are difficult to detect at the molecular level, let alone the phenotypic level.
One way to reconcile the high number of preferentially conserved 6-nt sites with the modest efficacy of these sites is to propose that these conserved 6mers are not preferentially conserved based on their activity as 6-nt sites, but instead, represent the inactive (or less active) decay products of preferentially conserved 7–8mer sites. When considering this explanation, two scenarios could contribute the conserved 6mer signals. In one scenario, the extended site has degraded to one of the 6mers in the human lineage. For example, a 7mer site that functions in most animals but has decayed to an inactive 6mer in primates would have contributed to our count of conserved 6mers in human UTRs. In the second scenario, the extended site is conserved in the UTR of human and other species but exceeds the branch-length cutoff only when including additional species in which it has degraded to one of the 6mers. For example, if the 8mer site conserved in the SPRED1 UTR had degraded to an inactive 6mer in hedgehog, then the site would have contributed to our count of conserved 8mers at branch-length cutoff of 0.9, but not at 1.0; at a cutoff of 1.0 it would have contributed to our count of conserved 6mers. When performing the analyses so as to exclude these two scenarios, our estimate of the number of preferentially conserved sites per highly conserved miRNA family dropped 35%, to an average of 77 ± 22 for 6mer sites and 69 ± 15 for offset 6mer sites (Supplemental Discussion). Thus, some of the preferential conservation of 6-nt matches to the seed region can be explained by their relationship with conserved 7mers, but thousands of 6mers and offset 6mers are selectively maintained without the aid of 7mer conservation. Moreover, because these decreases in preferentially conserved 6-nt sites imply corresponding increases in preferentially conserved 7–8mer sites, consideration of these scenarios does not change our estimates of the total number of preferentially conserved sites and the total number of conserved mammalian targets.
Our observation that these 6mer sites have been selectively maintained so frequently implies the existence of widespread pressure for finely adjusted protein output with surprisingly narrow tolerances for optimal expression in different cell types. The observation that this selective pressure persists over such long branch lengths indicates that these optimal expression levels and narrow tolerances persist in the face of many other changes over surprisingly long evolutionary distances. These results become somewhat easier to reconcile when considering the possibility that 6mers might impart more robust repression in particular cells or conditions that have yet to be accessed experimentally. Hinting at this possibility is the more readily detected activity of 6mers matching miR-430 during the maternal-to-zygotic transition, a developmental stage at which the miR-430 family comprises most of the miRNA molecules in the animal (Giraldez et al. 2006). An alternative possibility is that the 6mer and offset 6mer impart some function other than repressing protein output. For example, if transient miRNA association played a widespread role in mRNA subcellular localization, then many 6mer sites could be conserved without imparting any repression. In either scenario—widespread and persistently narrow gene-expression tolerances or expanded miRNA function—the discovery that so many 6mers and offset 6mers are selectively maintained sets the stage for important future insights into miRNA biology.
Methods
Alignments and phylogeny
Sequences (3′UTR) were defined as the longest 3′UTR for each human RefSeq gene (Pruitt et al. 2005), resulting in 18,577 unique UTRs for 18,577 genes. Human UTR boundaries were used to acquire orthologous UTRs from the 28-genome vertebrate sequence alignments from the UCSC genome browser (Karolchik et al. 2008). The average phylogeny was determined using the branching structure provided by UCSC, and branch lengths were estimated by running DNAML (part of the PHYLIP package [Felsenstein 1989]) on the UTR alignments. To define UTR conservation bins, human UTRs were ranked using the median branch length over single bases. Then, DNAML was run on the sequences in each UTR bin, creating phylogenies with the same branching structure but modified branch lengths scaled for each UTR bin.
miRNA sequences
The broadly conserved miRNA set comprised all 79 families of miRBase entries (release 10.0) with both a human and zebrafish miRNA sharing the same seed sequence (Supplemental Table 1). Additionally, we included 8 human miRNA families with mouse miRBase entries and a conserved foldback in lizard, chicken, frog, or another fish genome, yielding a total of 87 miRNA families. To generate the mammalian-specific miRNA set, miRbase entries for human miRNAs conserved to mouse were manually inspected for aligned sequences from most of the sequenced mammals, perfect conservation of the seed sequence within placental mammals that had aligned sequence, no conservation in species other than placental mammals, and good predicted folding characteristics in most placental mammals. Seven of the remaining miRNAs whose seed matches had strong excess conservation (>2.0 S/B ratio) in species other than placental mammals were also removed (reasoning that this excess conservation was likely caused by an miRNA ortholog that escaped detection because of imperfect alignments or sequencing coverage), leaving 53 mammalian-specific miRNAs (Supplemental Table 2). A list of miRNAs left out of the mammalian-specific category for various reasons is found in Supplemental Table 3.
Background estimation
For each seed-match k-mer, possible control k-mers were first filtered to remove seed matches to other miRNAs, and to have the same length, number of G + C bases, and possible matches to PUF proteins (UGU[A/G]) as the seed match. In the case of 8mer and 7mer-A1 matches, the controls were also constrained to have an A in the 3′-most nucleotide. For each branch-length cutoff, the 50 control k-mers with the closest expected conservation rate to the seed-match k-mer were selected. Expected conservation was calculated using a first-order Markov model with parameters derived from the empirical dinucleotide conservation rate in 3′UTRs at a particular branch-length cutoff. In other words, control k-mers were picked to exactly match length, GC content, and PUF binding sites, and to closely match the expected conservation rate based on the dinucleotide content. The background estimate was the number of occurrences of the k-mer multiplied by the average fraction of sites conserved in control k-mers at the same branch length. As done for the signal, background was determined for each UTR bin separately, and then at each branch length, background from the 10 UTR bins was summed to generate the overall background distribution. At each branch length, confidence intervals were calculated using the Gaussian distribution with mean equal to the background estimate and standard deviation calculated using the background estimates given by individual control k-mers as opposed to the average over all 50.
Seed-match types
For the five major seed-match types (8mer, 7mer-m8, 7mer-A1, 6mer, offset 6mer), raw signal and background values were calculated without regard to whether a shorter seed match was nested within a longer one. Then, for each miRNA and for each branch-length cutoff, the signal for the 8mer match was subtracted from the signal of the corresponding 7mer-m8 match, and the 8mer background estimate was subtracted from the 7mer-m8 estimate. Similarly, 8mer signal and background were subtracted from signal and background of the other seed-match types, 7mer-m8 signal and background were subtracted from that of both 6mers and offset 6mers, and 7mer-A1 signal and background was subtracted from that of 6mers. In cases in which a particular seed match could be considered a 7mer-m8 match to one miRNA and a 7mer-A1 to another miRNA, it was considered only as a 7mer-m8 match to avoid double-counting. Likewise, seed matches ambiguous between 6mers and offset 6mers were considered as only 6mers. Seed matches with mismatches, G:U wobbles, or bulges were filtered so that they did not contain perfect seed matches to other miRNAs.
3′ pairing
Pairing scores (3′) were calculated as previously described (Grimson et al. 2007). For sites matching miRNA families comprising miRNAs with different 3′ sequences, the 3′-pairing score used was the maximum score for any member of the miRNA family. The branch length of a site at a particular 3′-pairing score was calculated for the set of species having both the seed match conserved and a 3′-pairing score at least that of the human score. Control 3′-pairing scores and conservation were estimated by swapping every miRNA seed and 3′-end family, and recalculating 3′-pairing scores for every combination. Background estimate and confidence intervals were calculated as before, except that there were 86 control 3′-end families for each swap instead of 50 control k-mers. For compensatory pairing sites in Table 1, orthologous sites were considered conserved if their 3′-pairing score was greater than 6.0, regardless of the human pairing score. This was done to capture well-conserved human sites that have recently extended already strong 3′ pairing.
Probability of conserved targeting
For each human seed-match site, the conservation signal and background were calculated using methods analogous to those described above. Rather than using a branch-length cutoff, we used a branch-length window with a size set to meet a minimum of 20 occurrences of the site in human UTRs and calculated the fraction of seed matches conserved within the branch-length window. In the few cases with less than 20 total occurrences of the seed match, all corresponding sites were assigned a PCT of 0. For the remaining sites, the PCT was defined as E[(S − B)/S] where B, the background estimate, is a constant, and S is a random variable. S is defined as max{Sobs/N, B}, where Sobs is the observed signal and N is the total number of occurrences, distributed around the observed total Nobs as Poisson(Nobs). Because the background estimate is based on a mean of many measurements, but the signal is based on a single observation, we fix the number of conserved occurrences (Sobs) and allow the total number of occurrences (N) to vary. Thus, the PCT, which ranges between 0 and 1, corresponds to a Bayesian estimate of the probability that a site conserved to a particular branch length is conserved due to miRNA targeting. For some miRNAs, we observed high variability of PCT values for sites with close branch-length values; therefore, we implemented a smoothing procedure when deriving PCT values reported at the TargetScan site (Supplemental Discussion).
Acknowledgments
We thank members of the Bartel and Burge laboratories for helpful discussions, and the Broad Institute of MIT and Harvard, the Human Genome Sequencing Center at the Baylor College of Medicine, the Genome Sequencing Center at Washington University, and the Department of Energy’s (DOEs) Joint Genome Institute for providing access to unpublished vertebrate genome sequences. We also thank George Bell, Katherine Gurdziel, Prathapan Thiru, Bingbing Yuan, and Fran Lewitter for implementing our improved methods at the TargetScan website. This work was supported by a Krell Institute/Department of Energy Computational Sciences Graduate Fellowship (to R.C.F.) and by grants from the NIH to C.B.B. and D.P.B.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.082701.108.
References
- Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP—phylogeny inference package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- Gaidatzis D., van Nimwegen E., Hausser J., Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Mishima Y., Rihel J., Grocock R.J., Van Dongen S., Inoue K., Enright A.J., Schier A.F. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Wang Y.L., Langenberger D., Gunsalus K.C., Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput. Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang D.G., Green P. Bayesian Markov chain Monte Carlo sequence analysis reveals varying neutral substitution patterns in mammalian evolution. Proc. Natl. Acad. Sci. 2004;101:13994–14001. doi: 10.1073/pnas.0404142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Kuhn R.M., Baertsch R., Barber G.P., Clawson H., Diekhans M., Giardine B., Harte R.A., Hinrichs A.S., Hsu F., et al. The UCSC Genome Browser Database: 2008 update. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L., Jeffreys A.J., Keeney S. Where the crossovers are: Recombination distributions in mammals. Nat. Rev. Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- Kheradpour P., Stark A., Roy S., Kellis M. Reliable prediction of regulator targets using 12 Drosophilagenomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lall S., Grun D., Krek A., Chen K., Wang Y.L., Dewey C.N., Sood P., Colombo T., Bray N., Macmenamin P., et al. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Majoros W.H., Ohler U. Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics. 2007;8:152. doi: 10.1186/1471-2164-8-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallory A.C., Reinhart B.J., Jones-Rhoades M.W., Tang G., Zamore P.D., Barton M.K., Bartel D.P. MicroRNA control of PHABULOSA in leaf development: Importance of pairing to the microRNA 5′ region. EMBO J. 2004;23:3356–3364. doi: 10.1038/sj.emboj.7600340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W., Rosenbloom K., Hardison R.C., Hou M., Taylor J., Raney B., Burhans R., King D.C., Baertsch R., Blankenberg D., et al. 28-way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen C.B., Shomron N., Sandberg R., Hornstein E., Kitzman J., Burge C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt K.D., Tatusova T., Maglott D.R. NCBI Reference Sequence (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez A., Vigorito E., Clare S., Warren M.V., Couttet P., Soond D.R., van Dongen S., Grocock R.J., Das P.P., Miska E.A., et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Stark A., Kheradpour P., Parts L., Brennecke J., Hodges E., Hannon G.J., Kellis M. Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res. 2007a;17:1865–1879. doi: 10.1101/gr.6593807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007b;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vella M.C., Choi E.Y., Lin S.Y., Reinert K., Slack F.J. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3′UTR. Genes & Dev. 2004;18:132–137. doi: 10.1101/gad.1165404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., Sharp P.M., Li W.H. Mutation rates differ among regions of the mammalian genome. Nature. 1989;337:283–285. doi: 10.1038/337283a0. [DOI] [PubMed] [Google Scholar]
- Wu L., Belasco J.G. Micro-RNA regulation of the mammalian lin-28 gene during neuronal differentiation of embryonal carcinoma cells. Mol. Cell. Biol. 2005;25:9198–9208. doi: 10.1128/MCB.25.21.9198-9208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Bielawski J.P. Statistical methods for detecting molecular adaptation. Trends Ecol. Evol. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]