Abstract
Subtelomeres are concentrations of interchromosomal segmental duplications capped by telomeric repeats at the ends of chromosomes. The nature of the segments shared by different sets of human subtelomeres reflects their high rate of recent interchromosomal exchange. Here, we characterize the rearrangements incurred by the 15q subtelomere after it arose from a chromosome fission event in the common ancestor of great apes. We used FISH, sequencing of genomic clones, and PCR to map the breakpoint of this fission and track the fate of flanking sequence in human, chimpanzee, gorilla, orangutan, and macaque genomes. The ancestral locus, a cluster of olfactory receptor (OR) genes, lies internally on macaque chromosome 7. Sequence originating from this fission site is split between the terminus of 15q and the pericentromere of 14q in the great apes. Numerous structural rearrangements, including interstitial deletions and transfers of material to or from other subtelomeres, occurred subsequent to the fission, such that each species has a unique 15q structure and unique collection of ORs derived from the fission locus. The most striking rearrangement involved transfer of at least 200 kb from the fission-site region to the end of chromosome 4q, where much still resides in chimpanzee and gorilla, but not in human. This gross structural difference places the subtelomeric defect underlying facioscapulohumeral muscular dystrophy (FSHD) much closer to the telomere in human 4q than in the hybrid 4q–15q subtelomere of chimpanzee.
Subtelomeres are composed of interchromosomally duplicated sequences situated near the ends of chromosomes just proximal of telomeric repeats (Mefford and Trask 2002). Roughly half of the subtelomeric sequence in the human genome has moved or changed copy number since human and chimpanzee diverged (Linardopoulou et al. 2005). Due to this recent change, most subtelomeric duplications show variation in chromosomal location and copy number in the human population. Assays of the content of subtelomeres in other primates by fluorescence in situ hybridization (FISH) have revealed significant differences among species (Kingsley et al. 1997; Trask et al. 1998; Martin et al. 2002; van Geel et al. 2002b; Linardopoulou et al. 2005). However, no study has yet traced the evolution of a given subtelomere in primates by comparative analyses at the sequence level.
The evolution of subtelomeres is “reticulate” (Jackson et al. 2005; Huson and Bryant 2006). Recurrent shuffling of material among ends makes it difficult to distinguish structures that are identical by descent from the shared presence (or absence) of particular sequences. Gene conversion-like transfers between duplicated segments can obfuscate the timing of the original duplication. In order to reconstruct the history of a given subtelomere in the face of these complications, it is best to have, as reference, the sequence of an ancestral locus prior to its participation in subtelomeric rearrangements.
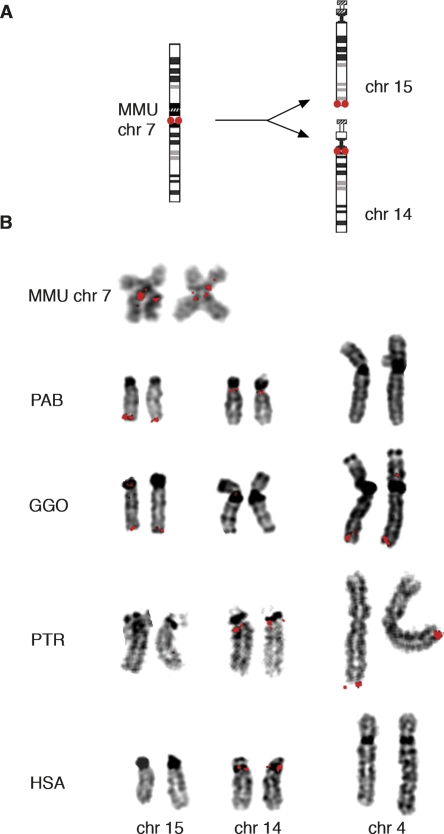
Here, we characterize the ancestral source of sequence that became the subtelomere of the long arm of chromosome 15 (15qter) in great apes due to a chromosome fission event. With the ancestral locus in hand, we reconstruct the changes incurred by this sequence after it was exposed to the recombination processes prevailing at chromosome ends. Previous studies had shown that human chromosomes 14 and 15 were generated by chromosome fission in the ancestor of great apes (Fig. 1A) (Wienberg et al. 1992; Murphy et al. 2001a; Ventura et al. 2003). Macaque chromosome 7 represents the ancestral state, as more distantly related organisms have the macaque-like configuration (Murphy et al. 2001b). Human chromosome 15 derives mostly from the short arm of this ancestral chromosome, and chromosome 14 from its long arm. The ancestral centromere was extinguished, and both fission products acquired new centromeres, such that the regions orthologous to human chromosomes 15 and 14 were arranged in a tail-to-head configuration in the ancestral state (Fig. 1A).
Figure 1.
Localization of the fission site within macaque BAC CH250-246c20 by FISH. (A) Ideograms showing cytogenetic manifestation of the fission of ancestral chromosome, represented by rhesus macaque chromosome 7, which created great ape chromosomes 15 and 14, here represented by the orangutan homologs (adapted from http://www.biologia.uniba.it/primates/). The red dots identify the location of the fission site region prior to the fission, and at 15qter and in 14q near the centromere immediately post-fission. This post-fission arrangement is grossly preserved in orangutan. (B) Chromosome locations of macaque BAC CH250-246c20 detected by FISH (red) pinpoint the fission site within its sequence and reveal post-fission rearrangements of the region in the hominids. The BAC hybridizes to rhesus macaque chromosome 7 (MMU), but produces two separate sets of signals in the chromosomes of all great apes except human. Chromosomes orthologous to human chromosomes 15, 14, and 4 are shown for orangutan (PAB), gorilla (GGO), chimpanzee (PTR), and human (HSA). FISH signals were not observed consistently with CH250-246c20 at any other chromosome locations in these species. See the text for explanations of the lack of signal from this BAC on the gorilla homolog of chromosome 14 and human and chimpanzee orthologs of chromosome 15. The double signals seen on some macaque chromatids, including those shown here, suggest that some of this BACs sequence might be duplicated around the centromere of macaque 7. MMU chr 7 refers to the chromosome number in the macaque karyotype. We use the chromosome numbering of the human karyotype for homologous chromosomes in gorilla, orangutan, and chimpanzee here and throughout this study.
We were particularly interested in this chromosomal fission, because it gave rise to a new subtelomere at 15qter. (The opposite side of the fission site became situated in the pericentromere of the long arm of chromosome 14, due to the subsequent addition of new centromeric and acrocentric sequences to the other chromosome end generated by the fission.) We have constructed detailed comparative maps of the orthologous 15qter subtelomeres in the great apes as well as the prefission state as it now exists in macaque. These analyses allow us to pinpoint the fission within an ancestral cluster of olfactory receptor (OR) genes, which is preserved at an internal location on macaque chromosome 7. Using the macaque sequence as reference, we can reconstruct many of the gross structural rearrangements, including loss of OR genes, that affected this region after the fission event situated it at the 15q terminus. These changes cumulatively result in species-specific 15q structures and sets of intact 15q OR genes.
This study also resolves the intertwined histories of primate 4q and 15q subtelomeres. In previous studies, we found multiple human subtelomeric copies of a sequence containing three OR genes (referred to as block 3 or f7501) (Trask et al. 1998; Linardopoulou et al. 2005). This sequence is single copy in other primate genomes. It maps to 15qter in orangutan, but to the q-arm terminus of the chimpanzee and gorilla homologs of human chromosome 4 (numbered chromosome 3 in the chimpanzee and gorilla karyotypes, but referred to hereafter as 4qter for clarity) (Trask et al. 1998; Linardopoulou et al. 2001, 2005; Mefford et al. 2001). The sequence is present at varied sets of locations in humans, but the sets consistently include 15q and exclude 4q. These results were difficult to reconcile with the ([human, chimpanzee] gorilla) trichotomy established from phylogenetic relationships of sequence variants at most genomic loci (Ruvolo 1997; Chen and Li 2001; Patterson et al. 2006), but suggested that block 3 originated at 15qter and subsequently transferred to 4qter prior to chimpanzee–gorilla speciation. Here, we show that this sequence derived from the internal fission site, then became situated at 15qter, and then was duplicatively transferred to 4q as part of a much larger segment.
The 4q subtelomere harbors the site of a molecular defect causing facioscapulohumeral dystrophy (FSHD) (Wijmenga et al. 1992). For still unknown reasons, this defect, a contraction of a tandemly repeated array of D4Z4 units, causes FSHD only when it occurs in the genomic and epigenetic context possessed by a subset of 4qter alleles (Lemmers et al. 2002, 2007). The significant features of this pathological context have not yet been defined (van der Maarel and Frants 2005; Lemmers et al. 2007). Therefore, the structural differences between human and chimpanzee 4q reported here might be relevant to the etiology of FSHD.
Results and Discussion
Building comparative maps of the fission site and its subtelomeric descendents
The human genome assembly (Build 36.1) has complete sequence coverage of one 15q allele spanning from chromosome 15-specific sequence, through ∼115 kb of sequence duplicated at various other chromosome ends, and terminating in telomere repeats (Fig. 2). The duplicated sequence can be divided into five segments (called blocks 1–5) distinguished by their different distributions among multiple chromosome ends in the human genome assembly (Linardopoulou et al. 2005). At least one other chromosome in the assembly shows a break in homology with respect to 15q sequence at each block boundary.
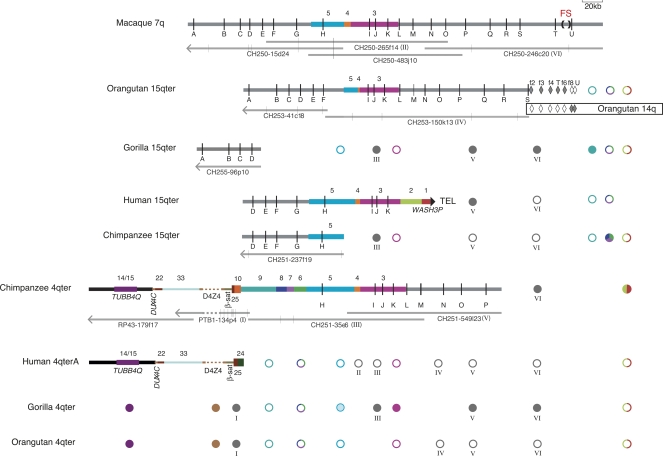
Figure 2.
Sequence content of the fission site region of macaque chromosome 7 and chromosomes 15q and 4q subtelomeres in primates, as compiled from sequenced BACs and targeted FISH and PCR assays. The annotated thick horizontal lines indicate sequence contigs derived from non-human BACs or the human genome assembly. The large filled triangle at the right end of human 15q indicates it as the only one of these sequence contigs to reach the terminal telomere-repeat arrays. Vertical black lines represent OR genes identified by letters A–U (see Supplemental Table 6 for official nomenclature for these genes). The fission site (FS), indicated by (), lies between ORs T and U. Colored bars represent sequence blocks known to be subtelomerically duplicated in the human genome (Linardopoulou et al. 2005); blocks 5, 4, and 3 derive from the fission site region. Horizontal gray bars indicate additional sequences orthologous to the rhesus macaque fission-site region. Orange-brown and gray dashes indicate unsequenced D4Z4 arrays in contigs and BAC PTB1-134p4, respectively. Filled or open diamonds indicate PCR assays on flow-sorted orangutan chromosomes 15 or 14 that were positive or negative, respectively (Supplemental Table 3). The spans of the constituent sequenced BACs are indicated by the thin gray lines under each non-human contig; vertical gray lines on these tracks indicate gaps in sequence contigs; left-pointing arrowheads indicate that the BAC sequence extends toward the centromere from portion shown. BACs used for FISH are indicated by roman numerals I–VI in parentheses. Circles summarize the results of FISH assays for sequence whose presence is not already evident from the sequence contigs; filled or open circles indicate the presence or absence, respectively, of a cumulative signal-intensity score at the cytogenetic location exceeding 25% of the maximum cumulative score observed in the experiment (Trask et al. 1998 for block 3; Supplemental Table 2). Note that the presence of a FISH signal does not necessarily mean that the entire probe sequence resides at that location. Gray circles labeled with roman numerals I–VI represent FISH results with BACs constituting the contigs. Colored circles correspond to block-specific FISH probes; multicolored circles indicate that the FISH probe spanned more than one block. Circles for gorilla 4q represent a compilation of FISH results (Supplemental Table 2) and our PCR assays for blocks 9, 8, 7, 6, and 5 sequences in 4q-derived, D4Z4-containing gorilla BACs identified by Bodega et al. (2007) (Supplemental Table 3). The block-5 circle is partially shaded to indicate that gorilla 4q is FISH negative for this sequence, but these gorilla 4q BACs are PCR positive only for the most distal block 5 assays. Note that FISH assays detect the presence/absence, but not the order, of sequences in a given subtelomere. Wherever possible, FISH-result symbols are aligned with the corresponding probe’s sequence in at least one contig; the FISH symbols for blocks 1/2, 9, and 6/7/8 are shown at the far right to indicate that these probe sequences, when detected by FISH, most likely lie telomeric of the sequence in the contigs. The sequence contigs are drawn to scale and aligned to facilitate comparison of their contents: macaque 7q, orangutan 15q, and human 15q contigs are aligned at OR gene I; chimpanzee 15q and 4q are aligned to human 15q at gene H; gorilla 15q is aligned to human 15q at gene D; human 4q is aligned to chimpanzee 4q at TUBB4Q. The diagrams of sequence contigs are truncated at arrowheads just proximal of gene A to exclude a complex region of tandem repeats that proved difficult to align across species, and the diagram for PTB1-134p4, is truncated at an arrowhead to indicate the extent of curated sequence from this BAC.
Although genome assemblies for chimpanzee, orangutan, and rhesus macaque exist, the regions orthologous to the fission site are not well represented and contain gaps and errors (data not shown). Therefore, we constructed BAC-based contigs of sequence orthologous to the 15q subtelomere in these non-human primates as well as gorilla. We screened BAC libraries constructed from each species’ genome using human 15q subtelomeric sequences (distal chromosome 15-specific and block-3 sequences) (Supplemental Table 1). We chose eight clones from these screens for sequencing based on restriction enzyme fingerprinting, PCR assays of sequence content, and chromosomal location determined by FISH (data not shown) in order to provide the most sequence coverage of 15q-orthologous regions in each species. We successfully assembled sequence contigs for portions of the 15qter subtelomere in the genomes of chimpanzee, gorilla, and orangutan, as well as the macaque sequence resembling the prefission state (Fig. 2). By searching public sequence databases, we identified two already sequenced macaque BACs that extend the macaque contig. Our 420-kb contig of macaque sequence crosses the fission site (Figs. 2 and 3 diagram the most relevant 405 kb). It spans 21 OR genes, named A through U here for simplicity. This contig shares over 150 kb with human 15q sequence. We also assembled a contig of the chimpanzee 4qter subtelomere that spans sequences orthologous to the human 4qter and 15qter subtelomeres (Fig. 2). In total, we sequenced eight non-human BACs and incorporated four previously sequenced BAC clones into contigs. Each BAC was sequenced to a read depth of ∼11; PCR was used to order and orient the resulting sequence contigs and close gaps. We assembled contigs of overlapping BACs meeting our criteria for being derived from the same chromosomal location, allowing for allelic variation (methods are detailed in Supplementary Text). A total of 39 gaps remain in our sequence contigs after filling in gaps with sequence available from overlapping BACs (Fig. 2).
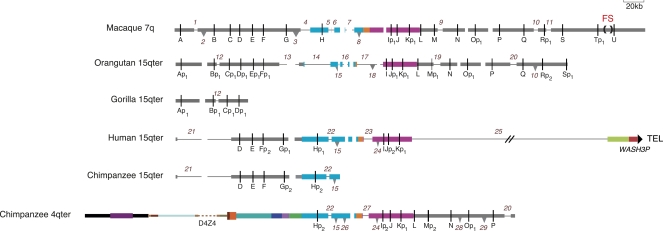
Figure 3.
Comparative map of the fission site region and its subtelomeric derivatives drawn to show all post-fission deletions and insertions involving >2.8 kb and OR pseudogenizing events. Thin black lines in the annotated sequence contigs indicate deletion of a sequence that is present in one or more other contigs. Inverted triangles indicate the presence of a sequence that is absent in one or more other contigs; the base of each triangle corresponds to the size of the inserted sequence. The direction of these changes (i.e., deletion vs. insertion) was inferred using parsimony in cases where more than one species shares an alternative structure and/or by examining the state of repeats at the breakpoints (Supplemental Table 4). These changes are numbered from left to right and down the figure; a change shared by more than one species carries the same number. In order to maximally align homologous sequences in the diagram, spaces are placed in contigs lacking each insertion at the position where an insertion is present in one or more other contigs. These spacers are not numbered. Rearrangement number 10 is shown as both insertion and deletion (INDEL), because its direction cannot be inferred from the available sequence. OR pseudogenes are indicated by “p,” and subscript numbers indicate independent or shared deactivating mutations. Not shown is the state of three gorilla OR genes, Ip3, J, and Kp1, which was determined by sequencing PCR products amplified from genomic DNA (Mefford et al. 2001, accession nos. EU685271–EU685273); FISH localizes these single-copy genes to gorilla 4q (Trask et al. 1998). The gorilla genome lacks gene H (Supplemental Table 3). The dashed turquoise bar in the orangutan 15q contig corresponds to a sequencing gap in insertion number 15. See the legend for Figure 2 for an explanation of other features.
We performed additional targeted FISH and PCR experiments to confirm and supplement our comparative sequence analyses, since only the human 15q sequence extends into the telomere repeat arrays. FISH was used to locate regions in primate chromosomes having homology with sequenced BACs and known human subtelomerically duplicated segments (Supplemental Table 2). PCR was used to assay primate genomic DNAs for the presence of OR genes flanking the fission site and various subtelomerically duplicated sequences (Supplemental Table 3). Figure 2 shows the content information drawn from these assays for 15q and 4q termini not already evident from the available sequence.
Pinpointing the fission site within the macaque sequence
We located the fission site within macaque BAC CH250-246c20 by FISH: This BAC produces signal midway along macaque chromosome 7, but its signal is split between 15qter and the 14q pericentromere in orangutan (Fig. 1B).
The following observations more precisely locate the fission site to an 8-kb interval between genes T and U in the macaque genome. First, our contig of orangutan 15qter sequence extends to gene S. Second, we find gene U’s best matching human ortholog and homology with ∼10 kb of flanking nonrepetitive sequence from the end of the macaque contig in the still fragmentary genome assembly of human 14q pericentromeric sequence. Third, PCR assays of flow-sorted orangutan chromosomes 15 and 14 localize the fission site to an 8-kb region between genes T and U: Products of expected size were amplified from flow-sorted orangutan chromosome 15 with the first five of seven PCR primer pairs designed from macaque sequence between genes S and U and from orangutan chromosome 14 with the last two (Fig. 2; Supplemental Table 3). This 8-kb region is not present in the orangutan genome assembly, and interspersed repeats occupy over 84% of this region in the macaque sequence, precluding further delineation of the breakpoint.
Structural changes in 15q subsequent to the fission event
Our 15q and 4q sequences and FISH analyses reveal multiple major changes to the OR cluster after it was split to form the 15q terminus (Figs. 1B, 2, 3). The 15q side of the fission site in macaque contains 20 OR genes (A–T), which serve as landmarks to describe these changes. The segment encompassing genes H–K corresponds to a sequence that duplicated in whole or in part to various human subtelomeres, subdividing the region into blocks 5, 4, and 3 (Linardopoulou et al. 2005). The orangutan 15qter subtelomere is grossly similar to the macaque sequence, but differs from it by 20 insertion/deletion events, each involving >2.8 kb (our arbitrary cutoff) and a total of >147 kb within the ∼300 kb compared (Fig. 3). Several interstitial deletions specific to the orangutan 15q sequence together remove genes G and H and much of block 5. This sequence appears to be lost from the orangutan genome by FISH (Linardopoulou et al. 2005) (Supplemental Table 2), genomic PCR assays (Supplemental Table 3), and searches of the orangutan assembly and trace sequence archives.
The 15q subtelomere has also undergone numerous gross changes along the human, chimpanzee, and gorilla lineages (Figs. 2, 3). A single interstitial deletion removed genes A–C in the ancestor of human and chimpanzee; the breakpoints of this deletion (number 21 in Fig. 3) are identical in human and chimpanzee. We find no vestiges of these genes in either genome by searching the archives of sequence traces. The human 15qter subtelomere is also missing distal genes L–T. Our PCR assays and trace-archive searches fail to detect these genes in the human genome, a result consistent with absence of FISH signal of macaque BAC 250-246c20, which spans genes N–U, on human 15q or any location other than the 14q pericentromere (Fig. 1B). On human 15qter, genes L–T are supplanted by WASH3P, a member of a recently identified family of subtelomeric genes (Linardopoulou et al. 2007). Complete and partial forms of WASH are duplicated at multiple chromosome ends in primates; the intact form spans blocks 1 and 2, and the partial form includes only block 1 (Fig. 2) (Linardopoulou et al. 2007). WASH sequence is not detected by FISH at the 15q subtelomere in gorilla, chimpanzee, or orangutan (Fig 2; Supplemental Table 2) (Linardopoulou et al. 2007), suggesting that this gene was appended to 15qter in the human lineage.
Although the chimpanzee 15qter sequence in BAC CH251-237f19 extends only into block 5, FISH analyses indicate that chimpanzee 15q has lost even more distal sequence than has human 15q. It lacks a detectable signal from block 3 (Trask et al. 1998) as well as from the region spanning genes N–U cloned in BAC CH250-246c20 (Fig. 1B), but appears by FISH to have acquired some other subtelomerically duplicated sequences known from the human genome (Fig. 2; Supplemental Table 2).
Our gorilla 15qter sequence consists of a single 184-kb BAC orthologous to the portion of the macaque contig farthest from the fission site. Only the most distal 63 kb subjected to our comparative analyses are depicted in Figures 2 and 3. Unlike human and chimpanzee 15qter, gorilla 15qter retains genes A–C. FISH and PCR show other differences in the unsequenced portion of gorilla 15q (Fig. 2). By FISH, gorilla 15q lacks the bulk of block 5 and block 3, yet it retains at least some part of the sequence in clone CH250-246c20 (Fig. 1B) and has acquired other subtelomerically duplicated sequences known from the human genome (block 9) (Fig. 2). PCR assays of the gorilla genome for genes N–U are negative for all but one gene (Q) (Supplemental Table 3). This inconsistency between FISH and PCR results might be explained by changes in gorilla sequence at primer sites (yielding false-negative PCRs). However, genes L–T also have no matches in the gorilla trace sequence archives. Therefore, the bulk of CH250-246c20 might indeed be lost from the gorilla genome, with local duplication of part of 246c20’s sequence accounting for the relatively robust FISH signal at gorilla 15q and 4q.
In addition to these gross structural alterations, the sequence has incurred numerous smaller deletion, insertion, or retrotransposition changes during the divergence of the higher primates. The numbers in Figure 3 identify changes >2.8 kb in size (see below for further analyses of these changes); smaller changes are reflected in Figure 3 by the slightly different spacing between the OR genes in the different species.
As for the chromosome-14 side of the fission site, the CH250-246c20 BAC hybridizes to the 14q pericentromere in human and chimpanzee as it does in orangutan (Fig. 1B). No 246c20 signal is seen on 14 in gorilla, likely due to deletion accompanying a gorilla-specific pericentromeric rearrangement (Yunis and Prakash 1982). The sequence assemblies of the pericentromeric regions of primate chromosomes, including human 14, are currently inadequate to track the fate of sequence from this side of the fission site in more detail.
Transfer of a large portion of the fission-site region to the 4q subtelomere
The fission-spanning BAC CH250-246c20 hybridizes to the 4qter subtelomere in chimpanzee and gorilla, but not human or orangutan (Fig. 1B). Thus, a portion of the CH250-246c20 sequence was transferred from an ancestral chromosome 15 to chromosome 4, presumably along with adjoining sequence that includes the previously mapped block 3 (Trask et al. 1998). Assuming that this transfer was a singular event, it must have occurred after the fission and orangutan speciation, but prior to gorilla–chimpanzee speciation. This scenario predicates the subsequent loss of fission site-derived sequence from 4q along the human lineage.
Our 542-kb contig of the chimpanzee 4q subtelomere provides a more detailed picture of this transfer and subsequent events. The chimpanzee 4q subtelomere is a hybrid structure, orthologous in the proximal part to the human 4q subtelomere and in the distal part to the human 15q subtelomere (Fig. 2). As in human 4q, the proximal chimpanzee 4q subtelomere contains the D4Z4 array, a beta-satellite array, and block 25. However, unlike in human 4q, the terminal 190 kb of the chimpanzee 4q contig derives from the fission-site region and spans from block 5 to gene P. We identified gene Q in chimpanzee genomic DNA by PCR (Supplemental Table 3). The more distal genes R, S, and T are likely missing entirely from the chimpanzee genome, as they are also not found by PCR or by searching the chimpanzee trace-sequence archives or genome assembly.
In chimpanzee 4q, block 5 is joined to ∼68 kb of sequence that is not found on human 4q or 15q, but is duplicated to varying extents on multiple human subtelomeres (Linardopoulou et al. 2005). Past recombination events have broken this region so as to define blocks 6–10 (Linardopoulou et al. 2005). Notably, precisely the same block-6-to-block-5 join seen in chimpanzee 4q is present in the human genome as part of multiple large subtelomeric duplications, albeit not on human 4q. Thus, this junction was formed prior to human–chimpanzee divergence.
Gorilla 4q is also a hybrid of human 4q and 15q sequences, but one that is structurally different from that of chimpanzee 4q. Gorilla 4q BACs identified by Bodega et al. (2007) localize by FISH to gorilla 4q and contain the D4Z4 repeat (data not shown; Bodega et al. 2007). They are PCR positive for part of block 5, block 4, and the proximal portion of block 3, but PCR negative for blocks 6, 7, 8, or 9 sequences and much of block 5 (Supplemental Table 3). This internal deletion is supported by insignificant FISH signals for these blocks on gorilla 4qter (Fig. 2; Supplemental Table 2).
No sequence from the fission site is detectable by FISH on orangutan chromosome 4q, but this orangutan subtelomere does contain other features of the 4q subtelomere, including the D4Z4 array (Fig. 2; Supplemental Table 2).
A parsimonious explanation for all of these observations is that block 5 and accompanying distal material from the fission site duplicatively transferred from 15qter to 4qter, either directly or indirectly, some time after the fission, but before human–chimpanzee–gorilla divergence. The 15q-to-4q duplication likely encompassed block 5 (still present on both chimpanzee 15q and 4q) as well as fission-site material distal of block 5, because gorilla 15q and 4q both retain some vestige of sequence cloned in CH250-246c20 and CH251-549l23 (the most distal clone in the chimpanzee 4q contig) (Fig. 2; Supplemental Table 2). The sequence group 10-9-8-7-6 might have resided on the ancestral 4qter before this transfer, or 10-9-8-7-6 might have been picked up as the fission-site sequence made the transit from 15q to 4q. In this transfer, block 5 joined block 6 and displaced sequence distal of block 6 on the recipient chromosome. Duplications encompassing this novel 6–5 join distributed later to other chromosomal ends in humans and possibly other species. Chimpanzee 4q retains the basic hybrid structure of 4q and 15q, but everything distal of block 25 was lost from human 4q and replaced with yet another subtelomerically duplicated sequence (block 24).
Post-speciation sequence exchange among paralogs
The sequence identities of human and chimpanzee copies of block 5 support the scenario involving duplication prior to human–chimpanzee divergence, but indicate an even more complicated history. If the duplication was followed by speciation and independent accrual of mutations, we would expect to find the human 15q and chimpanzee 15q copies to show higher identity to each other than either does to the copy found on chimpanzee 4q. Instead, the two chimpanzee copies are the most closely related pair of this trio. By comparing ∼19.1 kb of hand-curated, well-aligned block-5 sequences, we find that the chimpanzee 4q and 15q copies are only 1.43% diverged (Jukes-Cantor adjusted). They also share 43 derived mutations, including a 4-bp deletion that disrupts the ORF of gene H, not seen in the macaque or human 15q copies. In contrast, the human 15q and chimpanzee 15q copies are 1.65% diverged and share 22 derived mutations not seen in chimpanzee 4q; the human 15q and chimpanzee 4q copies are 1.64% diverged and share 13 derived mutations not seen in chimpanzee 15q. Collectively, these data indicate that the copies of block 5 on chimpanzee 15q and human 15q are no longer true orthologs. None of the other sequenced block-5 paralogs in the human genome is more closely related than any other to the chimpanzee 15q or 4q copies. In fact, the human block-5 paralogs are all very closely related to each other (>98.89% identity), indicating that they are the products of post-speciation duplication or ectopic exchange.
One explanation for the incongruity we observe between sequence phylogeny and the duplication timing suggested by the shared presence of particular subtelomeric structures in primates (see above) invokes at least one post-speciation gene-conversion event between block-5 paralogs in chimpanzee. A “copy-paste” exchange event could have made the chimpanzee 15q and 4q copies more similar to each other than expected for copies generated by a duplication preceding human–chimpanzee speciation (e.g., 1.61%, the human–chimpanzee sequence divergence measured for nearby 15q unique sequence). This explanation is plausible given the prevalent signs of past gene conversion-like exchanges in human subtelomeric sequences (Linardopoulou et al. 2005). Indeed, the program GeneConv identifies a putative gene conversion between the chimpanzee 15q and 4q sequences with high statistical support (Bonferonni-corrected KA P value < 0.002). An alternative possibility is that the derivative products of the 15q-to-4q translocation were segregating in the shared ancestor of human, chimpanzee, and gorilla, and then differentially sorted among the three lineages. The true history of block-5 sequences may prove more complicated when the sequences of all paralogs detected by FISH on other human and chimpanzee chromosomes (Supplemental Table 2) are determined.
Relationship of human 4q structural variants to the chimpanzee 4q subtelomere
Human 4qter alleles can be subdivided into two major groups, A and B, on the basis of sequence-content differences immediately distal of the D4Z4 array (Lemmers et al. 2002). Deletions of the D4Z4 array give rise to FSHD only when they occur on the 4qA form (Lemmers et al. 2002, 2004, 2007). Our chimpanzee 4q sequence surrounding the D4Z4 array confirms the proposal made by van Geel et al. (2002a) that the human 4qA allele is ancestral to the 4qB allele. The human 4qA allele and chimpanzee 4q share a divergent terminal D4Z4 repeat known as pLAM (van Deutekom et al. 1993) (data not shown) as well as a stretch of beta satellite distal of the D4Z4 array (Fig. 2). 4qB alleles lack these features (Lemmers et al. 2002; van Geel et al. 2002a) (data not shown). Our chimpanzee 4q sequence contains a gap in the D4Z4 array due to BAC-insert instability and difficulties assembling sequence of this tandem array. However, the available sequence transitioning from the D4Z4 array into sequence centromeric and telomeric of it suggests that the array lies in the same orientation in chimpanzee and human 4q. Like the human 4q subtelomere, the chimpanzee 4q subtelomere also contains a proximal partial inverted D4Z4 repeat, DUX4C (Fig. 2). As noted above, the similarities between human 4qA and chimpanzee 4q end ∼11 kb distal of the D4Z4 array; chimpanzee 4qter terminates with at least 260 kb of sequence that has not been detected on any characterized human 4qter allele.
Mechanistic insights
The sequence at the fission site gives potential clues about the mechanism by which the ancestral chromosome split here to form two chromosomes. The break occurred within an 8-kb region of the macaque genome now densely packed with repetitive elements, all but perhaps one of which (an L1PA5) was present in the region before the fission. The L1PA5 is 6.6% diverged from its consensus; all of the rest are >12% diverged from their consensus and thus predate the macaque–human split. The sequence delineating the fission site has features that might have made it susceptible to break to form two chromosomes, such as inverted repeats and AT-richness. It is 66.8% AT, similar to other genomic regions predisposed to breakage (Zlotorynski et al. 2003; Zhang and Freudenreich 2007). We also discovered two sets of overlapping inverted repeats within LINEs in the 8-kb fission region. The arms of inverted repeat pairs are 378 and 355 bp, 72.4% and 73.3% identical, and separated by 701 and 41 bp, respectively. While inverted-repeat structures like this are not unique to the fission site, they can be associated with chromosome breakage, as homologous sequences can undergo intrastrand base pairing, forming extruded hairpin structures that can block replication fork progression (Lobachev et al. 2007).
All subsequent significant structural rearrangements of the fission site region appear to have been mediated by nonhomologous end joining (NHEJ) of double-strand breaks (DSBs) or retrotransposition. Comparison of the available orthologous sequences identifies 29 structural changes involving >2.8 kb of sequence. We could infer the direction of change (e.g., insertion vs. deletion) by using parsimony in cases where more than one species shares an alternative structure and/or by examining the state of repeats at the breakpoints. The fission site region has incurred a total of 19 deletions (including the translocation-mediated deletion joining block 3 to block 2 in human 15q), nine insertions, and one change for which the direction could not be inferred (thus, an INDEL) (Fig. 3; Supplemental Table 4). The nine insertions include eight LINE retrotransposition events and one retrotransposition to create a SUZ12 processed pseudogene. Rearrangements mediated by NHEJ can be distinguished from those mediated by nonallelic homologous recombination by comparing the derived and original sequences at each rearrangement breakpoint (Linardopoulou et al. 2005). All 19 deletions and the aforementioned INDEL bear the molecular signatures of NHEJ (Supplemental Table 4). Of the 38 double-strand breaks healed by NHEJ to form deletions, 68% occurred in repetitive elements, a fraction not significantly different from the repeat occupancy of the regions analyzed (55%).
Fates of OR genes from the fission locus
Each primate genome analyzed retains a unique collection of intact OR genes derived from the fission site (Fig. 3). The 21 genes in the macaque cluster are >13% diverged from each other (synonymous changes expressed as a percentage of possible synonymous changes, i.e., dS), which greatly exceeds the average dS of macaque and human sequences elsewhere in the genome (∼7%) (Chen and Li 2001). This divergence and the presence of a clear ortholog for each of the 21 macaque genes in at least one other higher primate’s sequence indicate that all members of this gene set existed prior to the fission event. The topology of the phylogenetic tree of all orthologous ORs from the fission site region now residing in sequenced portions of 15q or 4q (Supplemental Fig. 1) confirms that only the second chimpanzee copy of gene H arose by duplication subsequent to the fission. Orthologs for all other genes form gene-specific clades whose topologies roughly recapitulate the species tree. Note that this tree excludes recent duplicates of genes H, I, J, and K on other human chromosome ends (Mefford et al. 2001; Linardopoulou et al. 2005); the present analyses reveal that the initial source for this dispersal was 15q.
Only macaque has retained the full complement of 21 genes from the ancestral fission site region. Each of the other species has lost at least one of 14 of these genes by deletion. If additional genes were present in the common ancestor, they have been lost from all genomes studied. In addition, 16 of the 21 genes have acquired more subtle mutations, rendering them pseudogenes in subsets of the species (Fig. 3; Supplemental Fig. 1). Only 16 of the 21 macaque genes encode an intact ORF, and other primates analyzed retain yet smaller fractions of intact ORFs from the original set. Fourteen of the ORs are pseudogenes in multiple species, and in all but two of these cases, the orthologous pseudogenes were created independently (Fig. 3). A total of 31 independent inactivating mutations (including deletions) are needed to account for all of the missing or defective ORs in the sequence contigs represented in Figure 3. As a result of these many changes, each species has retained a unique collection of intact OR genes from the original set of 21.
Concluding remarks
Our in-depth analysis of the region orthologous to the chromosome 15q subtelomere in five primate species revealed numerous large structural changes as well as many subtle mutations affecting the functionality of OR genes. We had the unusual opportunity to trace changes in this sequence that arose after it was relocated from a relatively tranquil internal chromosome location to a recombination-prone terminal position. Despite the reticulate evolution of subtelomeres, it is possible to order some events when equipped with the orthologous sequences now residing at 15q and 4q in great apes and the internal locus in macaque. Only genes K and O appear to have been pseudogenes before the fission, as all orthologs of each gene share the same debilitating mutation. Once the same OR cluster was positioned near a chromosome end, it incurred numerous deletions, insertions, and translocations, such that the 15q fission locus is grossly altered in human, chimpanzee, and gorilla. We show here that all of the major changes incurred by the locus were mediated by NHEJ or retrotransposition. Such structural changes could contribute to phenotypic diversity of closely related species whose genomes have high sequence identity overall. While the OR genes at 15qter are probably not critical for survival, each species possesses a unique repertoire of 15q ORs, perhaps altering olfactory abilities. We have identified OR genes in other primates that have been lost from the human genome, and vice versa. In addition, it is possible that the fission event and subsequent structural changes have divorced these OR genes from their original regulatory context, which might have altered the relative expression of the intact genes, as expression of some OR genes is known to require interaction with an enhancer that lies up to 2 Mb away (Serizawa et al. 2003; Fuss et al. 2007). However, we showed previously that gene I is transcribed from multiple human subtelomeres (Linardopoulou et al. 2001), and Zhang et al. (2007) report that OR genes D, E, F, G, and U are expressed in human olfactory epithelium (genes H–K were not included on their microarray used to assay expression in this tissue). Determining the phenotypic effect of the changes to this set of OR genes will require access to high-quality mRNA from primate olfactory epithelium and development of robust methods to identify ligands for particular ORs.
Second to the fission event itself, the most significant and largest structural change to the fission locus sequence has resulted in grossly different structures of human 4qter and chimpanzee 4qter. The chromosome 4q subtelomere is of particular interest because it contains the locus responsible for FSHD. FISH studies have examined the chromosome 4q subtelomere in other species (Clark et al. 1996; Winokur et al. 1996; van Geel et al. 2002b), but our study is the first to analyze the genomic structure of this locus in non-human primates at the sequence level. We have characterized one chimpanzee allele of 4qter, which is structurally similar to the human 4qA allele to a point just distal of the D4Z4 array. Beyond that point, we find >200 kb of sequence, at least 190 kb of which is derived from 15qter, on 4qter in the chimpanzee (and likely gorilla) genome, but not human. Future studies will be needed to show whether this gross difference in genomic context and/or proximity to the potential epigenetic silencing effects of the telomere significantly influence the relative expression of distal 4q genes in human and chimpanzee.
Methods
BAC libraries constructed from rhesus macaque, orangutan, gorilla, and chimpanzee DNA were screened for clones containing sequence orthologous to chromosome-specific and duplicated sequence from human 15qter. Cytogenetic locations of BACs and cloned subtelomeric duplication segments were determined by FISH as described (Trask et al. 1998; Linardopoulou et al. 2005). BACs derived from the fission site were sequenced and assembled to produce “comparative-grade” finished sequence. PCR assays of selected subtelomeric BACs and primate genomic DNA were performed as in Linardopoulou et al. (2005). Orangutan chromosomes 14 and 15 were flow-sorted and subjected to PCR assays as described (Mefford et al. 2001). OR gene phylogenetics, nomenclature, and subfamily assignment were determined following standard methods. Methodological details for all of these procedures are provided in Supplemental Text.
Acknowledgments
We thank Elena Linardopoulou for advice, Baoli Zhu and Mikhail Nefedov for construction of BAC libraries, and Yoko Kuroki and Asao Fujiyama for chimpanzee BACs. This work was supported by NIH grants RO1-GM057070 and RO1-DC004209 (B.J.T.), T32-HG00035 (M.K.R.), and U01 HG002523 (P.d.J.). The following individuals were key contributors within the NISC Comparative Sequencing Program: Robert Blakesley, Gerard Bouffard, Shelise Brooks, Jyoti Gupta, Jacquelyn Idol, Jennifer McDowell, Baishali Maskeri, Jennifer Vogt, and Alice Young.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. AC188481, AC183330, AC150715, AC149242, AC148620, AC148535, AC173434, AC186245, AC205763, AC150448, AC183669, AC197422.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083170.108.
References
- Bodega B., Cardone M.F., Muller S., Neusser M., Orzan F., Rossi E., Battaglioli E., Marozzi A., Riva P., Rocchi M., et al. Evolutionary genomic remodelling of the human 4q subtelomere (4q35.2) BMC Evol. Biol. 2007;7:39. doi: 10.1186/1471-2148-7-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.C., Li W.H. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 2001;68:444–456. doi: 10.1086/318206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L.N., Koehler U., Ward D.C., Wienberg J., Hewitt J.E. Analysis of the organisation and localisation of the FSHD-associated tandem array in primates: Implications for the origin and evolution of the 3.3 kb repeat family. Chromosoma. 1996;105:180–189. doi: 10.1007/BF02509499. [DOI] [PubMed] [Google Scholar]
- Fuss S.H., Omura M., Mombaerts P. Local and cis effects of the H element on expression of odorant receptor genes in mouse. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- Huson D.H., Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jackson M.S., Oliver K., Loveland J., Humphray S., Dunham I., Rocchi M., Viggiano L., Park J.P., Hurles M.E., Santibanez-Koref M. Evidence for widespread reticulate evolution within human duplicons. Am. J. Hum. Genet. 2005;77:824–840. doi: 10.1086/497704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley K., Wirth J., van der Maarel S., Freier S., Ropers H.H., Haaf T. Complex FISH probes for the subtelomeric regions of all human chromosomes: Comparative hybridization of CEPH YACs to chromosomes of the Old World monkey Presbytis cristata and great apes. Cytogenet. Cell Genet. 1997;78:12–19. doi: 10.1159/000134616. [DOI] [PubMed] [Google Scholar]
- Lemmers R.J., de Kievit P., Sandkuijl L., Padberg G.W., van Ommen G.J., Frants R.R., van der Maarel S.M. Facioscapulohumeral muscular dystrophy is uniquely associated with one of the two variants of the 4q subtelomere. Nat. Genet. 2002;32:235–236. doi: 10.1038/ng999. [DOI] [PubMed] [Google Scholar]
- Lemmers R.J., Wohlgemuth M., Frants R.R., Padberg G.W., Morava E., van der Maarel S.M. Contractions of D4Z4 on 4qB subtelomeres do not cause facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2004;75:1124–1130. doi: 10.1086/426035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers R.J., Wohlgemuth M., van der Gaag K.J., van der Vliet P.J., van Teijlingen C.M., de Knijff P., Padberg G.W., Frants R.R., van der Maarel S.M. Specific sequence variations within the 4q35 region are associated with facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2007;81:884–894. doi: 10.1086/521986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E., Mefford H.C., Nguyen O., Friedman C., van den Engh G., Farwell D.G., Coltrera M., Trask B.J. Transcriptional activity of multiple copies of a subtelomerically located olfactory receptor gene that is polymorphic in number and location. Hum. Mol. Genet. 2001;10:2373–2383. doi: 10.1093/hmg/10.21.2373. [DOI] [PubMed] [Google Scholar]
- Linardopoulou E.V., Williams E.M., Fan Y., Friedman C., Young J.M., Trask B.J. Human subtelomeres are hot spots of interchromosomal recombination and segmental duplication. Nature. 2005;437:94–100. doi: 10.1038/nature04029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulou E.V., Parghi S.S., Friedman C., Osborn G.E., Parkhurst S.M., Trask B.J. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 2007;3:e237. doi: 10.1371/journal.pgen.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev K.S., Rattray A., Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: Causes and consequences in eukaryotic cells. Front. Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- Martin C.L., Wong A., Gross A., Chung J., Fantes J.A., Ledbetter D.H. The evolutionary origin of human subtelomeric homologies–or where the ends begin. Am. J. Hum. Genet. 2002;70:972–984. doi: 10.1086/339768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H.C., Trask B.J. The complex structure and dynamic evolution of human subtelomeres. Nat. Rev. Genet. 2002;3:91–102. doi: 10.1038/nrg727. [DOI] [PubMed] [Google Scholar]
- Mefford H.C., Linardopoulou E., Coil D., van den Engh G., Trask B.J. Comparative sequencing of a multicopy subtelomeric region containing olfactory receptor genes reveals multiple interactions between non-homologous chromosomes. Hum. Mol. Genet. 2001;10:2363–2372. doi: 10.1093/hmg/10.21.2363. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Page J.E., Smith C., Desrosiers R.C., O'Brien S.J. A radiation hybrid mapping panel for the rhesus macaque. J. Hered. 2001a;92:516–519. doi: 10.1093/jhered/92.6.516. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Stanyon R., O'Brien S.J. Evolution of mammalian genome organization inferred from comparative gene mapping. Genome Biol. 2001b;2:REVIEWS0005. doi: 10.2286/gb-2001-2-6-reviews0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Richter D.J., Gnerre S., Lander E.S., Reich D. Genetic evidence for complex speciation of humans and chimpanzees. Nature. 2006;441:1103–1108. doi: 10.1038/nature04789. [DOI] [PubMed] [Google Scholar]
- Ruvolo M. Molecular phylogeny of the hominoids: Inferences from multiple independent DNA sequence data sets. Mol. Biol. Evol. 1997;14:248–265. doi: 10.1093/oxfordjournals.molbev.a025761. [DOI] [PubMed] [Google Scholar]
- Serizawa S., Miyamichi K., Nakatani H., Suzuki M., Saito M., Yoshihara Y., Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Trask B.J., Friedman C., Martin-Gallardo A., Rowen L., Akinbami C., Blankenship J., Collins C., Giorgi D., Iadonato S., Johnson F., et al. Members of the olfactory receptor gene family are contained in large blocks of DNA duplicated polymorphically near the ends of human chromosomes. Hum. Mol. Genet. 1998;7:13–26. doi: 10.1093/hmg/7.1.13. [DOI] [PubMed] [Google Scholar]
- van der Maarel S.M., Frants R.R. The D4Z4 repeat-mediated pathogenesis of facioscapulohumeral muscular dystrophy. Am. J. Hum. Genet. 2005;76:375–386. doi: 10.1086/428361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deutekom J.C., Wijmenga C., van Tienhoven E.A., Gruter A.M., Hewitt J.E., Padberg G.W., van Ommen G.J., Hofker M.H., Frants R.R. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum. Mol. Genet. 1993;2:2037–2042. doi: 10.1093/hmg/2.12.2037. [DOI] [PubMed] [Google Scholar]
- van Geel M., Dickson M.C., Beck A.F., Bolland D.J., Frants R.R., van der Maarel S.M., de Jong P.J., Hewitt J.E. Genomic analysis of human chromosome 10q and 4q telomeres suggests a common origin. Genomics. 2002a;79:210–217. doi: 10.1006/geno.2002.6690. [DOI] [PubMed] [Google Scholar]
- van Geel M., Eichler E.E., Beck A.F., Shan Z., Haaf T., van der Maarel S.M., Frants R.R., de Jong P.J. A cascade of complex subtelomeric duplications during the evolution of the hominoid and Old World monkey genomes. Am. J. Hum. Genet. 2002b;70:269–278. doi: 10.1086/338307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M., Mudge J.M., Palumbo V., Burn S., Blennow E., Pierluigi M., Giorda R., Zuffardi O., Archidiacono N., Jackson M.S. Neocentromeres in 15q24-26 map to duplicons which flanked an ancestral centromere in 15q25. Genome Res. 2003;13:2059–2068. doi: 10.1101/gr.1155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienberg J., Stanyon R., Jauch A., Cremer T. Homologies in human and Macaca fuscata chromosomes revealed by in situ suppression hybridization with human chromosome specific DNA libraries. Chromosoma. 1992;101:265–270. doi: 10.1007/BF00346004. [DOI] [PubMed] [Google Scholar]
- Wijmenga C., Hewitt J.E., Sandkuijl L.A., Clark L.N., Wright T.J., Dauwerse H.G., Gruter A.M., Hofker M.H., Moerer P., Williamson R., et al. Chromosome 4q DNA rearrangements associated with facioscapulohumeral muscular dystrophy. Nat. Genet. 1992;2:26–30. doi: 10.1038/ng0992-26. [DOI] [PubMed] [Google Scholar]
- Winokur S.T., Bengtsson U., Vargas J.C., Wasmuth J.J., Altherr M.R., Weiffenbach B., Jacobsen S.J. The evolutionary distribution and structural organization of the homeobox-containing repeat D4Z4 indicates a functional role for the ancestral copy in the FSHD region. Hum. Mol. Genet. 1996;5:1567–1575. doi: 10.1093/hmg/5.10.1567. [DOI] [PubMed] [Google Scholar]
- Yunis J.J., Prakash O. The origin of man: A chromosomal pictorial legacy. Science. 1982;215:1525–1530. doi: 10.1126/science.7063861. [DOI] [PubMed] [Google Scholar]
- Zhang H., Freudenreich C.H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007;27:367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., De la Cruz O., Pinto J.M., Nicolae D., Firestein S., Gilad Y. Characterizing the expression of the human olfactory receptor gene family using a novel DNA microarray. Genome Biol. 2007;8:R86. doi: 10.1186/gb-2007-8-5-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotorynski E., Rahat A., Skaug J., Ben-Porat N., Ozeri E., Hershberg R., Levi A., Scherer S.W., Margalit H., Kerem B. Molecular basis for expression of common and rare fragile sites. Mol. Cell. Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]