Summary
The transcriptional control of CD1d-restricted NKT cell development has remained elusive. We report that PLZF (promyelocytic leukemia zinc finger; Zbtb16), a member of the BTB/POZ-ZF family of transcription factors which includes the CD4 lineage-specific c-Krox (Th-POK, Zbtb7b), is exquisitely specific to CD1d-restricted NKT cells and human MR1-specific MAIT cells. PLZF was induced immediately after positive selection of NKT cell precursors and PLZF-deficient NKT cells failed to undergo the intrathymic expansion and effector differentiation that characterize their lineage. Instead, they preserved a naïve phenotype and were directed to lymph nodes. Conversely, transgenic expression of PLZF induced CD4 thymocytes to acquire effector differentiation and migrate to non-lymphoid tissues. We suggest that PLZF is a transcriptional signature of NKT cells that directs their innate-like effector differentiation during thymic development.
Introduction
The development of thymic αβ cell lineages relies on signaling processes that are initiated upon TCR engagement by MHC or MHC-like ligands at the CD4+CD8+ double positive (DP) stage (reviewed in (He and Kappes, 2006; Singer and Bosselut, 2004)). Although overlapping with their positive selection, the lineage commitment of thymocytes can be separated and manipulated based on the expression of signature components of transcriptional networks involved in lineage decisions. For example, the antagonistic expression and function of c-Krox (Th-POK, Zbtb7b) and Runx3 define the CD4 and CD8 lineages, respectively (He et al., 2005; Setoguchi et al., 2008; Sun et al., 2005; Wildt et al., 2007). A fraction of MHC class-II restricted CD4 T cells further differentiates into CD25+ regulatory T cells characterized by the transcription factor Foxp3 (Fontenot et al., 2003; Hori et al., 2003).
Little is known, however, about the transcriptional regulation of unconventional αβ T cell lineages such as NKT cells, MAIT cells or CD8αα intraepithelial lymphocytes (reviewed in (Bendelac et al., 2007; Matsuda and Gapin, 2005; Shires et al., 2001; Treiner and Lantz, 2006)). There is evidence suggesting that, as for CD25+ regulatory T cells (Hsieh et al., 2004), their TCRs engage in higher avidity interactions with self ligands than conventional T cells, resulting in the acquisition of a permanently activated effector or memory phenotype. However, it remains unknown whether their resulting transcriptional profile is a mere combination of those already described for conventional naïve T cells and their various memory and effector products or whether essential signature transcription factors are induced, transiently or permanently, during the thymic development of these lineages (Matsuda et al., 2006; Shires et al., 2001).
NKT cells are a well-characterized conserved population of T cells that mainly express CD1d-restricted αβ TCRs made of an invariant Vα14-Jα18 chain in mouse (Vα24-Jα18 in human) combined with variable Vβ8, Vβ7 and Vβ2 chains (Vβ11 in human) (reviewed in (Bendelac et al., 2007; Godfrey and Berzins, 2007)). These canonical semi-invariant TCRs recognize conserved self and foreign glycosphingolipids such as iGb3 and microbial alpha-glycuronosylceramides or alpha-glycuronosyldiacylglycerols, respectively. Essential to their innate-like function in the mammalian immune system are the massive expansion that rare NKT cell precursors undergo in the thymus upon interaction with CD1d ligands and their subsequent acquisition of an effector phenotype along with receptors of the NK lineage. The effector functions are characterized by the ability to explosively release cytokines such as IL-4 and IFN-γ in tissue locations where NKT cells preferentially migrate, such as the liver sinusoids (Geissmann et al., 2005). The cellular stages of NKT cell development have been extensively characterized (reviewed in (Bendelac et al., 2007; Godfrey and Berzins, 2007; Matsuda and Gapin, 2005)). Rare DP precursors expressing a canonical semi-invariant TCR interact with CD1d ligands expressed by cortical thymocytes and, in the process, engage hemophilic interactions between SLAM family receptors which provide essential signals through the adaptor SAP, the Src kinase Fyn and downstream NFκB. The signaled DP reach a CD24high CD4+ CD8low CD69high stage, termed ‘stage 0’, similar to the transitional CD4 stage attained by positively selected conventional CD4 and CD8 T cells. NKT precursors then mature by downregulating CD24 and CD8 to reach a mature CD4+ stage with a naïve CD44low phenotype (‘stage 1’) similar to conventional CD4 cells. Unlike conventional CD4 cells, which egress the thymus as naïve cells, stage 1 NKT cells remain in the thymus and immediately engage in multiple rounds of cell division, activating low basal transcription of Th2 followed by Th1 cytokines. This expansion is accompanied by the induction of CD44, a marker of antigen experienced T cells, and of the β chain of IL-15 receptor, which is induced by the transcription factor T-bet (‘stage 2’). Upon egress from the thymus, these stage 2 NKT cells further differentiate to stage 3 cells that express a panoply of NK lineage receptors, including NK1.1, NKG2D, CD94 and inhibitory Ly49 isotypes. In mouse, a population of NKT thymocytes fail to migrate and persist as resident stage 3 cells in the thymus. Peripheral NKT cells preferentially reside in the liver, continuously crawling along the sinusoidal endothelial cells (Geissmann et al., 2005), but they are also present in the spleen, bone marrow, lung and gut. Their frequency among mature T cells in comparatively low in lymph nodes, however, due to the lack of expression of the homing receptors CD62L and CCR7. The homeostatic signals regulating their prolonged lifespan are mediated by IL-15, as is the case for other memory and effector T cells and NK cells.
Studies of mice lacking various transcription factors expressed by other lymphocyte subsets have revealed some intrinsic (and usually incomplete) defects in NKT cell development. Such factors include Ets-1 family members (Lacorazza et al., 2002; Walunas et al., 2000), Runx proteins (Egawa et al., 2005), Gata-3 (Kim et al., 2006) and T-bet (Matsuda et al., 2006), but their absence results in multiple defects affecting T cells, NK cells and NKT cells (Egawa et al., 2007; Intlekofer et al., 2005; Lacorazza et al., 2002; Pai et al., 2003; Samson et al., 2003; Taniuchi et al., 2002; Townsend et al., 2004). NFκB factors are important to complete the NKT cell intrathymic expansion phase (Sivakumar et al., 2003; Stanic et al., 2004) but are also involved in multiple other lymphocyte lineages and differentiation programs (Hayden and Ghosh, 2008). It has remained a challenge, therefore, to determine whether some transcription factors might be specifically expressed in NKT cells and contribute to their development in a unique manner.
Here, we identify PLZF (promyelocytic leukemia zinc finger; Zbtb16) as an early transcriptional regulator of NKT cell development. PLZF is a member of the BTB/POZZF family of transcription factors which includes Th-POK (Zbtb7b) and LRF (Zbtb7a), factors that have been demonstrated to be critical for the CD4 T cell and B cell fates, respectively, as well as BCL-6 which is essential for germinal center B cell differentiation (reviewed in (Bilic and Ellmeier, 2007; Kelly and Daniel, 2006)). PLZF was highly expressed all along NKT cell development from the earliest step marking the positive selection of double positive thymocyte precursors to the terminally differentiated stage of NKT cell development in peripheral tissues. With the notable exception of the cousin lineage of MR1-restricted innate-like effector MAIT cells (Treiner and Lantz, 2006), which expressed high levels of PLZF as well, PLZF was low or absent from conventional naïve, effector or memory CD4 and CD8 T cells, as well as from other unconventional effector populations such as intraepithelial γδ and αβ lymphocytes. The absence of PLZF abrogated both the expansion and the effector differentiation of NKT cells resulting in their reversal to a naïve phenotype and their tissue redistribution to the lymph nodes rather than the liver. Conversely, ectopic expression in the CD4 lineage converted CD4 thymocytes into effector cells that preferentially migrated to non-lymphoid tissues.
Results
Selective expression of PLZF in CD1d-restricted NKT cells
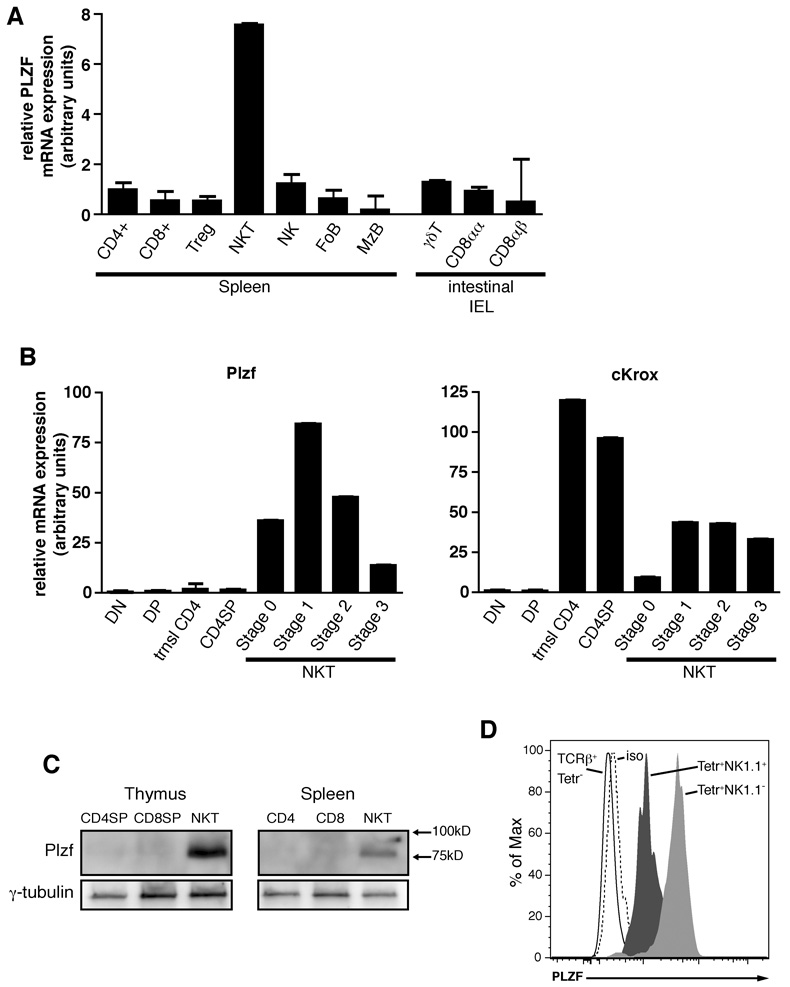
To identify novel transcription factors involved in NKT cell development, we performed a microarray screen on sorted NKT cells from C57BL/6 mice. Numerous probe sets mapping to Zbtb16 showed conspicuous over-expression in comparison to naive and activated conventional CD4+ and CD8+ T cells (data not shown). Similar results can be observed in microarray experiments published online (http://www.immgen.org). Quantitative PCR (qPCR) confirmed that PLZF transcripts were highly expressed in splenic NKT cells purified with specific CD1d-αGalCer tetramers and were low or absent in other splenic populations, including CD4 and CD8 T cells, CD25+ regulatory T cells, NK cells and follicular and marginal zone B cells (Fig. 1A). Strikingly, PLZF was low also in unconventional T cell lineages that constitutively express an effector phenotype. These lineages included intestinal intraepithelial lymphocytes (iIEL) expressing the αβ TCR along with CD8αα homodimers (CD8αα) or CD8αβ heterodimers (CD8αβ), or expressing the γδ TCR (Fig. 1A). Likewise, PLZF transcripts were low or absent in conventional CD44high (memory) CD4 and CD8 spleen cells or in Th1 and Th2 cells derived from splenic CD4 cells stimulated with anti-CD3 along with cytokine and anticytokine cocktails (data not shown; http://www.immgen.org).
Figure 1. PLZF Expression Is Specific to NKT Cells.
(A) Purified subsets of splenocytes and intestinal intraepithelial lymphocytes as indicated were assessed for PLZF mRNA expression by qRT-PCR. Data shown as mean +/− SD or triplicate PCR values.
(B) PLZF and c-Krox mRNA expression in thymic subsets and thymic NKT developmental stages as indicated. DN, CD4−8− double negative; DP, CD4+8+ double positive; trnsl CD4, transitional CD4+8lowCD24highTCRβhigh; CD4SP, CD4+8− single positive; NKT stage 0, CD24highCD69high; NKT stage 1, CD24lowCD44lowNK1.1−; NKT stage 2, CD24lowCD44highNK1.1−; NKT stage 3, CD24lowCD44highNK1.1+. Data shown as mean +/− SD of triplicate PCR analysis and representative of 2–3 independent experiments.
(C) PLZF western blot analysis of sorted CD1d-αGalCer+ NKT cells and conventional CD4 and CD8 cells (1x106 cells per lane) in the thymus and spleen, as indicated. Anti γ-tubulin was used as a loading control. Data are representative of 3 experiments
(D) Intracellular flow cytometry with anti-PLZF mAb D-9 of thymic NKT stage 1 + 2 cells (Tetr+NK1.1−, grey shaded), stage 3 (Tetr+NK1.1+; black shaded), and conventional T cells (TCRb+Tetr−), as indicated. The isotype control is the dashed histogram. Data are representative of 4 experiments.
To determine the stage of NKT cell development where PLZF is induced, we purified the different NKT stages from the mouse thymus using CD1d-αGalCer tetramers. All CD24low stages of NKT cell development from the CD44low NK1.1− naïve (stage 1) to CD44high NK1.1− effector (stage 2) to the terminally differentiated CD44high NK1.1+ (stage 3) expressed high levels of PLZF (Fig. 1B). Stage 3 cells in the thymus as in the spleen consistently expressed lower levels than thymic stages 1 and 2. The earliest NKT precursors (so-called stage 0) can be identified as CD24high tetramer+ cells which are uniformly CD69high and correspond to a mixture of CD4lowCD8low (DPlow) and CD4+CD8low cells that immediately follow positive selection (Benlagha et al., 2005). These cells, which are present at a low frequency of approximately 300 cells per thymus, were MACS-enriched based on CD1d-αGalCer tetramer staining from large pools of mice and further purified by flow cytometry sorting. As shown in Fig. 1B, high expression of PLZF transcript was already detected at this early stage. In comparison, sorted CD4+ CD8low thymocytes, the so-called transitional CD4 cells that represent post-selection CD69high precursors of both CD4 and CD8 lineage cells, exhibited low or undetectable transcript, similar to DN, DP and mature CD4 SP thymocytes (Fig. 1B). Thymic CD25 regulatory T cells and γδ T cells also expressed low PLZF levels (data not shown).
Interestingly, we found that the CD4 lineage-determining factor c-Krox (Th-POK, Zbtb7b), which belongs to the same family of BTB/POZ-ZF as PLZF, was also expressed at all stages of NKT cell development, starting immediately after positive selection in stage 0 cells (Fig. 1B). This expression mimics the pattern observed for conventional CD4 T cells and is consistent with the CD4+ phenotype of stage 0 and stage 1 NKT precursors.
To determine protein expression, we performed western blotting on sorted cell populations using an anti-PLZF polyclonal antibody specific for the C-terminal region of the protein. A protein with the expected 80–90KD molecular weight was easily detected in thymic NKT cells and, at weaker level, in splenic NKT cells as well, whereas conventional CD4 and CD8 cells had undetectable levels (Figure 1C). At the single cell level, intracellular flow cytometry with the mAb D-9 demonstrated expression in all CD1d-αGalCer tetramer-positive thymocytes, with high levels in NK1.1-negative NKT thymocytes (stage 1 and 2) and intermediate levels in NK1.1-positive cells (stage 3). In contrast, PLZF was undetectable in control single-positive thymocytes (Fig. 1D). Altogether, these results demonstrate the highly specific expression of PLZF in NKT cells and its early induction during thymic development concomitant with positive selection events.
Selective defect of NKT cell development in PLZF-deficient mice
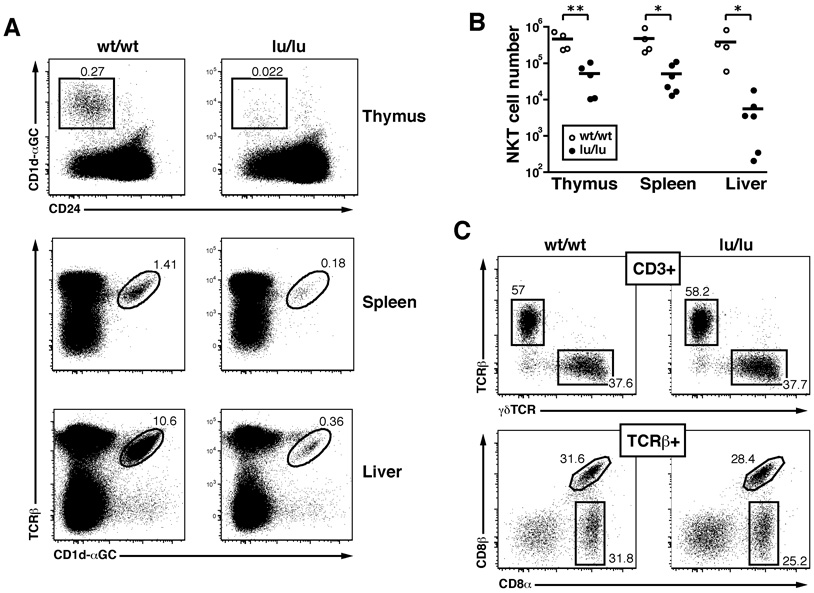
The Luxoid mutation arose spontaneously in a C3H/He line and was identified based on skeletal abnormalities and male sterility (Green, 1961). Luxoid represents a single base pair C-to-T transition which converts Arginine 234 to a nonsense codon, resulting in a protein that encompasses the BTB/POZ domain but lacks the other domains including the DNA binding region with all nine zinc fingers (Buaas et al., 2004). The Luxoid mice used in this study were backcrossed >30 times into C57BL/6. Heterozygous Luxoid mice were intercrossed to generate homozygous mutants (designated Zbtb16lu/lu) and examine their lymphoid compartments. Fig. 2A and B show that Zbtb16lu/lu littermates exhibited a drastic 9–70 fold reduction of NKT cells stained by specific CD1d-αGalCer tetramers in the thymus, spleen and liver in 4–8 week old mice. Other lymphocyte populations in thymus, spleen and peripheral lymph nodes, including conventional naïve and memory CD4 and CD8 T cells, B cells, NK cells, CD11c+MHC II+ and CD11b+ dendritic cells and macrophages, were unaffected (supplementary Fig. 1 and data not shown). Notably, unconventional T cell subsets including thymic γδ T cells (not shown) and all three intestinal intraepithelial effector lineages (TCR γδ, CD8αα TCRαβ, CD8αβ TCRαβ) were preserved (Fig. 2C)
Figure 2. Luxoid Mice Have a Selective Defect in NKT Cell Development.
(A) The frequency of NKT cells in the thymus, spleen, and liver of Luxoid (Zbtb16lu/lu) mice compared to their wt/wt littermate controls. NKT cells were gated (as shown) as CD24lowCD1d-αGC+ in the thymus and CD1d-αGC+TCRβ+ (or CD1d-αGC+B220− in some experiments) in the spleen and liver. Data are representative of 6 pairs of mice.
(B) Summary of NKT cell absolute numbers in individual thymuses, spleens, and livers of wt/wt and lu/lu littermates. * p<=0.05, ** p<=0.01.
(C) Unconventional intestinal IELs in Luxoid mice and their wt littermates. Upper row, frequency of αβ and γδ T cells among CD3+ gated IELs; lower row, frequency of CD8αα and CD8αβ T cells among TCRβ+ gated IELs. Absolute numbers of IELs were similar in wt and lu/lu mice. Data are representative of 5 littermate pairs in two independent experiments.
Intrinsic defect of NKT cell precursors revealed by mixed bone marrow chimeras
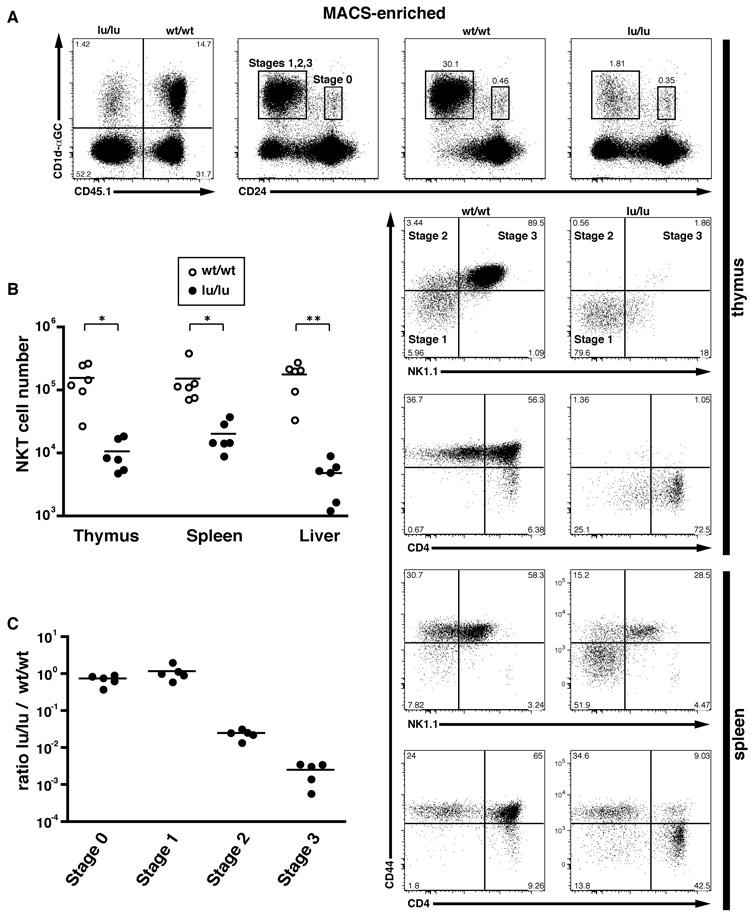
To determine whether the NKT cell defects were intrinsic to the NKT cell lineage or related to defects in their environment, B6.Jα18-deficient or B6 mice were lethally irradiated and reconstituted with a 1:1 or 1:3 mixture of CD45 allotype-marked wt and lu/lu bone marrow cells. While the wt and the lu/lu bone marrows showed a similar potency for generating myeloid and lymphoid lineages in these competitive chimeras, the lu/lu NKT cells were selectively decreased 7.5–37 fold on average in the thymus, spleen and liver compared to wt (Fig. 3A–B). Lu/lu cells, however, did not show impaired generation of naïve or memory CD4 and CD8 T cells (Supplementary Fig. 1A). Upon short term culture with anti-CD3 and polarizing cocktails of cytokines and anti-cytokine antibodies, the splenic CD4 T cells derived from the wt and the lu/lu compartments of these mixed chimeras expanded and polarized equivalently, further confirming that PLZF does not play a major role in conventional T cell expansion or differentiation into Th1 or Th2 effector subsets (Supplementary Fig. 1B). These findings establish therefore the intrinsic and highly specific nature of the developmental defect of NKT cell precursors in mice lacking PLZF.
Figure 3. Mixed Bone Marrow Chimeras Reveal a Cell-intrinsic NKT Cell Defect.
(A) CD45.2 lu/lu and CD45.1 wt/wt bone marrows were mixed at a 1:1 ratio and transferred into lethally irradiated Jα18−/− hosts. 6–8 weeks later, reconstituted animals were analyzed by flow cytometry. Individual thymus and spleen from the chimeras were MACS-enriched using CD1d-αGC tetramers prior to FACS analysis of NKT developmental subsets. Upper row, leftmost panel shows the relative proportion of CD45.2 (lu/lu) and CD45.1 (wt) NKT lineage cells in the thymus; right panels show the proportion of CD24high (stage 0) and CD24low (stage1, 2 and 3) cells in the total population (2nd panel from left) and in gated CD45.1 (wt) (3rd panel) and CD45.2 (lu/lu) (4th panel) CD1d-αGC+ cells. Wt and lu/lu CD24low NKT cells are further analyzed based on CD44 and NK1.1 expression, or CD44 and CD4 expression, as indicated for the thymus and spleen (bottom 2–5 rows). Data are representative of 6 individual chimeras.
(B) Summary of CD1d-αGC+ cell numbers in the wt and lu/lu compartments of individual mixed chimeras. Data are normalized based on the relative level of reconstitution by CD45.1 and CD45.2 bone marrows (e.g. NKT cell number in the wt CD45.1 compartment is divided by the CD45.1/(CD45.1+CD45.2) ratio measured for total lymphocytes. On average, the wt and lu/lu bone marrows contributed equally to the total lymphocyte population.
(C) Summary of the ratios of wt/wt over lu/lu CD1d-αGC+ cells at different stages of thymic development. Data are normalized based on the relative level of CD45.1 and CD45.2 reconstitution.
Early stage of the NKT cell development defect
To precisely and quantitatively determine the stages of NKT cell development affected by the absence of PLZF, we examined the thymus of the mixed bone marrow chimeras after NKT cell enrichment using MACS and CD1d-αGalCer tetramers (Fig. 3). CD45.1 wt and CD45.2 lu/lu cells were analyzed for their expression of CD24 to enumerate the earliest identifiable CD24high ‘stage 0’ cells that are the immediate CD69high product of positive selection, and for the expression of CD44 and NK1.1 to follow the progression from stage 1 to stage 2 to stage 3 among the CD24low cells. Stage 0 and stage 1 were clearly quantitatively preserved in the absence of PLZF in these competitive chimeras. In contrast, stage 2 and stage 3 cells were reduced to 2–3% of wild type, indicating that the developmental block occurred precisely prior to the massive intrathymic expansion and memory effector formation that characterizes NKT cell development (Fig. 3A and C). Thus, the residual population found in the lu/lu compartment was predominantly composed of stage 1 CD44low cells. Interestingly, a minor population amounting to 15–20% of these stage 1 thymocytes expressed low levels of NK1.1, suggesting that some reduced level of NK differentiation might procede in the absence of cell expansion and memory effector formation. In the spleen, a majority of lu/lu NKT cells expressed the CD44low naïve phenotype, suggesting that the thymic stage 1 cells were exported and survived in peripheral tissues. In addition, a population of CD44high NK1.1+ stage 3-like cells could be identified, suggesting that a minor fraction of NKT lineage cells were able to terminally differentiate in the absence of PLZF. Interestingly, these splenic stage 3-like cells exhibited a significant increase in the proportion of CD4− (DN) cells (Fig. 3A, bottom row), suggesting a role of PLZF in the maintenance of CD4 expression in the NKT lineage.
PLZF-deficient NKT cells are redistributed to peripheral lymph nodes and spleen
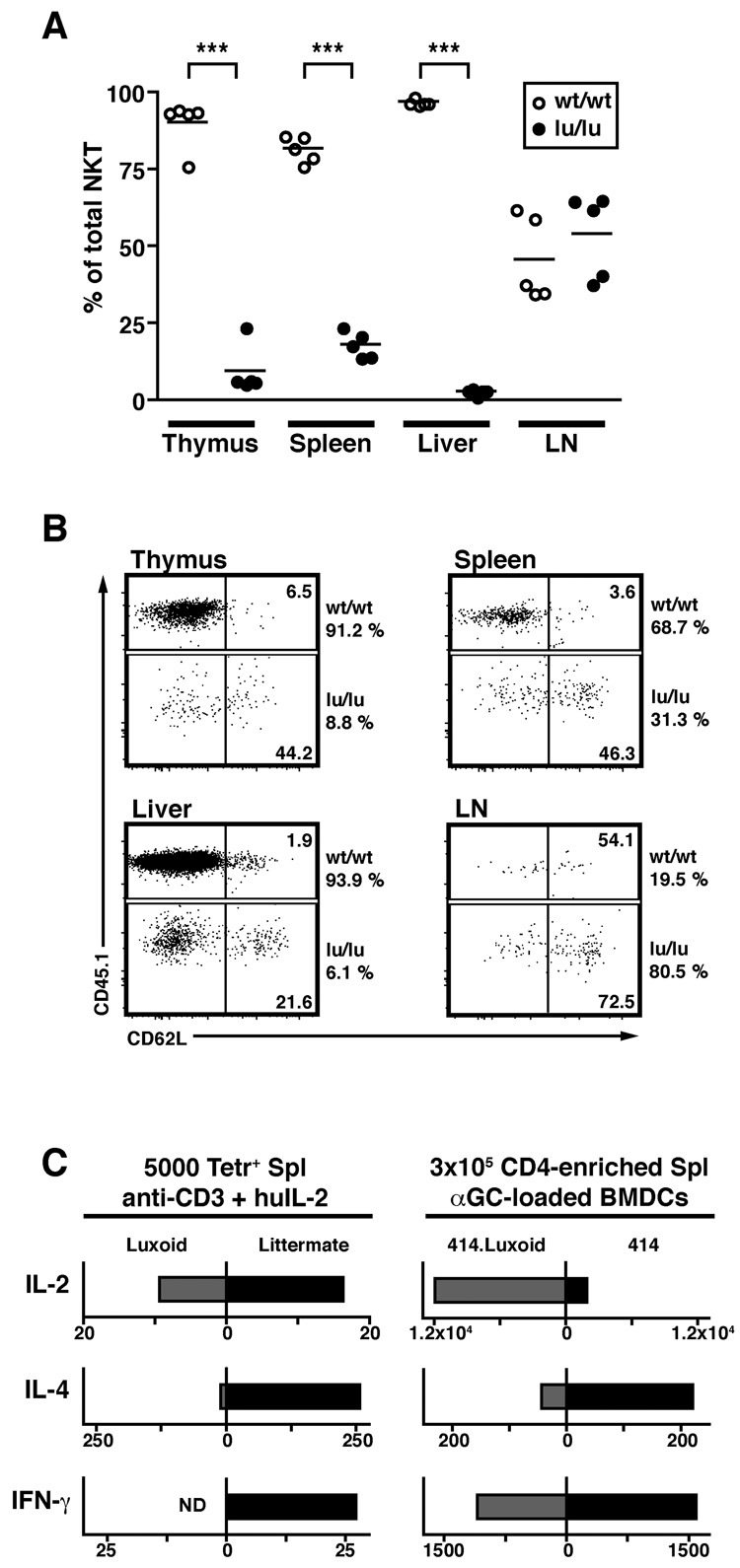
Although PLZF-deficient NKT cells were massively reduced in peripheral tissue, it was important to quantitatively determine whether the ‘naïve’ arrested stage 1 cells identified in the thymus could be exported and survive in the periphery. Strikingly, not only were such PLZF-deficient NKT cells found in the periphery, as shown in Fig. 3A, but, in some tissues where NKT cells are normally poorly represented, namely the peripheral lymph nodes, lu/lu NKT cells were present at the same or higher frequency than their wt counterparts (Fig. 4). This surprising result could be explained by the finding that, unlike the wt, they expressed high levels of CD62L, the homing receptor for high endothelial venules in lymph nodes and likely, therefore, were being redistributed towards lymph nodes. These findings demonstrate that the bulk of PLZF-deficient NKT cells maintain the naïve state that characterize stage 1 cells and survive in that stage without engaging into the subsequent expansion and effector formation leading to the terminal NKT cell differentiation.
Figure 4. PLZF-deficient NKT Cells Revert to a Naïve Phenotype.
(A) Summary of the percentages of wt/wt and lu/lu cells among CD1d-αGalCer+ cells in various tissues of individual mixed bone marrow chimeras. The percentages are normalized relative to the level of reconstitution of the total lymphocyte population by wt/wt and lu/lu cells, as assessed by CD45.1 and CD45.2 staining. For example, the ratio of CD45.1 CD1d-αGalCer+ cells over the total CD45.1 lymphocyte population is divided by (ratio of CD45.1+CD1d-αGalCer+ cells over the total CD45.1 lymphocyte population + ratio of CD45.2+CD1d-αGalCer+ cells over the total CD45.2 lymphocyte population) and multiplied by 100. *** p<=0.0001.
(B) Expression of CD62L by wt/wt and lu/lu CD1d-αGalCer+ cells in various tissues of mixed bone marrow chimeras. Data are representative of 5 chimeras.
(C) Cytokine secretion by Luxoid NKT cells. Left, cytokines released after stimulation of 5000 CD1d-αGalCer tetramer-sorted splenocytes (pooled from luxoid or wt littermates) with anti-CD3 + human IL-2; grey bars: lu/lu cells, black bars: +/+ littermate cells. A major increase in the IL-2/IL-4 ratio was also observed in a separate experiment comparing tetramer-sorted lu/lu vs wt splenocytes stimulated with αGalCer-loaded DC. Right, stimulation of 3x105 CD19- and CD8-depleted Vα14-Jα18 transgenic ‘414’ splenocytes with αGalCer-pulsed bone-marrow derived dendritic cells; grey bars: lu/lu 414 cells, black bars: +/+ 414 littermate cells. Supernatants were recovered after 2 days and values are expressed in pg/ml. Data are from an individual luxoid/littermate pair.
Attempts to evaluate the functional properties of PLZF-deficient NKT cells are complicated by the rarity of these cells. We FACS-purified the CD1d-αGalCer tetramer-positive cells arrested at stage 1 from the spleen of Luxoid mice and stimulated them with anti-CD3 + human IL2 or enriched these cells from Vα14 transgenic ‘414’ mice (Griewank et al., 2007) made PLZF-deficient by crossing to Luxoid mutants and stimulated them with α-GalCer-pulsed BMDC. In both experiments, a conspicuous defect of IL-4 production, the characteristic cytokine of NKT cells, was revealed by comparison with PLZF+/+ littermates (Fig. 4C). IL-2 was preserved or enhanced whereas IFN-γ was diminished or absent. Thus, in the absence of PLZF, NKT cells reverted to a stage resembling conventional CD4 T cells from which they normally diverge during thymic development.
Transgenic expression of PLZF in CD4 T cells
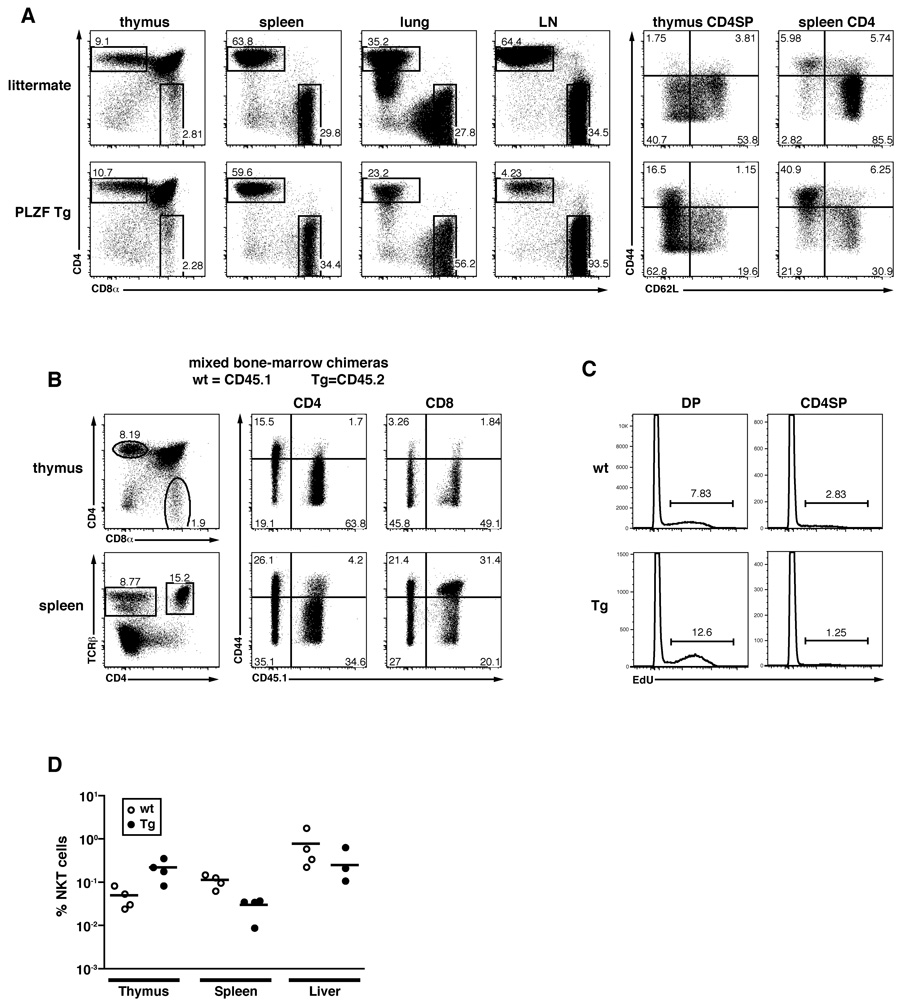
To test whether ectopic expression of PLZF would be sufficient to impart some aspects of the NKT cell differentiation program to conventional T cells, we transgenically expressed PLZF under control of the CD4 promoter. This promoter construct carries the intronic silencer that suppresses expression in CD8 cells and is exclusively active in double positive thymocytes and in CD4 lineage T cells (Sawada et al., 1994). Two independent lines, 1797 and 1960, were studied and showed similar results. PLZF-transgenic mice displayed relatively normal numbers of CD4 cells in the thymus, spleen, liver and lung, but exhibited a >10 fold reduction in the blood and lymph nodes where the remaining T cells were predominantly CD8 lineage cells (Fig. 5A and data not shown). This striking tissue redistribution was associated in part with the acquisition of a CD44highCD62Llow phenotype by a large cohort of CD4 cells in the thymus, spleen, liver and lung. Thus, the frequency of CD44highCD62Llow cells among CD4 cells was increased 5–10 times in transgenic CD4 single positive thymocytes, splenocytes lung and liver lymphocytes (Fig. 5A and data not shown). Markers of late NKT cell lineage differentiation such as CD122, NK1.1, NKG2D, however, were not expressed (data not shown). In contrast, despite their transient exposure to PLZF at the DP thymic stage, CD8 T cells did not exhibit apparent changes in frequency or phenotype in PLZF transgenic mice.
Figure 5. Transgenic Expression of PLZF converts CD4 thymocytes into effector cells.
(A) Left, CD4/CD8 FACS dot plots of lymphocytes from various organs of CD4 promoter-PLZF transgenic and wild type littermates (CD1d-αGC Tetramer-positive cells are gated out). Right, CD44/CD62L expression by CD4 cells of indicated organs gated as shown on the left panels. Data representative of two independent transgenic lines 1797 and 1960.
(B) PLZF transgenic CD4 cells in competitive chimeras. Wild-type (CD45.1+) and PLZF Tg (CD45.2+, line 1797) bone marrow cells were mixed 1:1 to reconstitute lethally irradiated wild type recipients. Thymus and spleen cells were stained as indicated (left panels) to gate CD4 and CD8 T cells for analysis of CD44 and CD45.1 (middle and right panels) Chimeras were analyzed 6.5 weeks after transfer. Data are representative of four individual chimeras.
(C) Cell cycle analysis 3 hours after in vivo injection of 100µg of 5-ethynyl-2'-deoxyuridine (EdU) into mixed bone marrow chimeras. Edu incorporation was detected by intracellular flow cytometry for DP (left panels) and CD4 single positive (right panels) thymocytes of wt (top panels) and PLZF-Tg (bottom panels) origin, as indicated. Similar values were obtained for splenic CD4 cells (not shown). Data are representative of three individual chimeras and two independent experiments.
(D) Frequency and tissue distribution of NKT cells in PLZF-transgenic mice. Data are from the same mixed bone marrow chimeras as in Fig. 5B and represent percentages of NKT cells among wild-type (CD45.1+) and PLZF Tg (CD45.2+) lymphocytes.
To test whether the changes associated with PLZF expression in CD4 cells were cell-intrinsic and to rule out the possibility that they might reflect homeostatic expansion following some CD4 lineage attrition, 1:1 mixed bone marrow (wt+PLZF Tg) radiation chimeras were generated. Fig. 5B clearly demonstrates that, even in the presence of competing wild-type T cells, the PLZF transgenic CD4 cells expressed the same effector phenotype as observed in transgenic mice. Further, wild type and PLZF transgenic progenitors generated equivalent numbers of CD4 T cells in the thymus and in the spleen. Again, the CD8 cells appeared unaltered. Altogether, the data establish that ectopic expression of PLZF in CD4 lineage cells is sufficient for their cell-intrinsic conversion into effector-type cells.
The PLZF-transgenic CD4 cells did not appear to be actively cycling, as shown by the very low level of EdU incorporation in thymic and splenic CD4 cells after a short pulse in vivo (Fig. 5C and data not shown). As a control, both wild-type and PLZF Tg DP thymocytes incorporated EdU as expected.
Finally, contrasting with the drastic consequences of ectopic expression of PLZF in CD4 cells, the overexpression of PLZF by PLZF-transgenic NKT cells did not seem to induce consistent changes across the range of tissues examined, including thymus, spleen and liver (Fig. 5D).
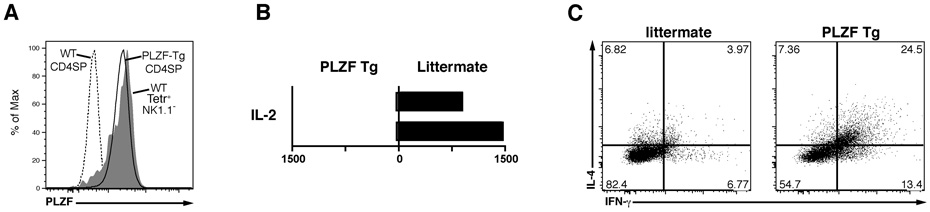
The level of PLZF protein expressed in transgenic CD4 cells (line 1797) was close to that normally expressed by stage 1 and stage 2 NKT thymocytes of wild-type mice (Fig. 6A), making these mice ideal to examine whether PLZF expression also changed the cytokine expression profile. Transgenic CD4 cells stimulated with anti-CD3 + human IL-2 produced 25–50 times less IL-2 than their wild-type littermate counterparts (Fig. 6B). Intracellular flow cytometry further revealed a 4–6 fold increased frequency of IL-4/IFN-γ double producer cells in PLZF-transgenic CD4 cells compared with wild-type (Fig. 6C). Such high frequency of Th1/Th2 double producers is a distinctive feature of the NKT cell differentiation program (Bendelac et al., 2007). These changes are the mirror image of those observed in NKT lineage cells developing in the absence of PLZF.
Figure 6. Cytokine profile of PLZF Transgenic CD4 T cells.
(A) Intracellular flow cytometry with anti-PLZF mAb D-9 of thymic PLZF-transgenic (line 1797) CD4 SP cells compared with wt CD4 SP and with wt NKT stage 1 + 2 cells thymocytes (Tetr+NK1.1−), as indicated.
(B) Decreased IL-2 secretion by PLZF-transgenic CD4 T cells. CD4 cells were MACS-enriched from spleens of PLZF-transgenic and littermate and stimulated with anti-CD3 + human IL-2 for two days. Values in pg/ml for 2 pairs of wt and Tg littermates, as indicated.
(C) Intracellular flow cytometry for IL-4 and IFN-γ in littermate and PLZF transgenic mice. Splenocytes were prepared and stimulated as in A, with the addition of brefeldin A for the last six hours of stimulation. Three pairs of wt and Tg littermates were analyzed. The percentages of dual IL-4/ IFN-γ producers were 24.5, 17.3, 26.1 in transgenics vs 3.97, 4.92, 7.34 in wt.
MR1-restricted MAIT cells express high levels of PLZF
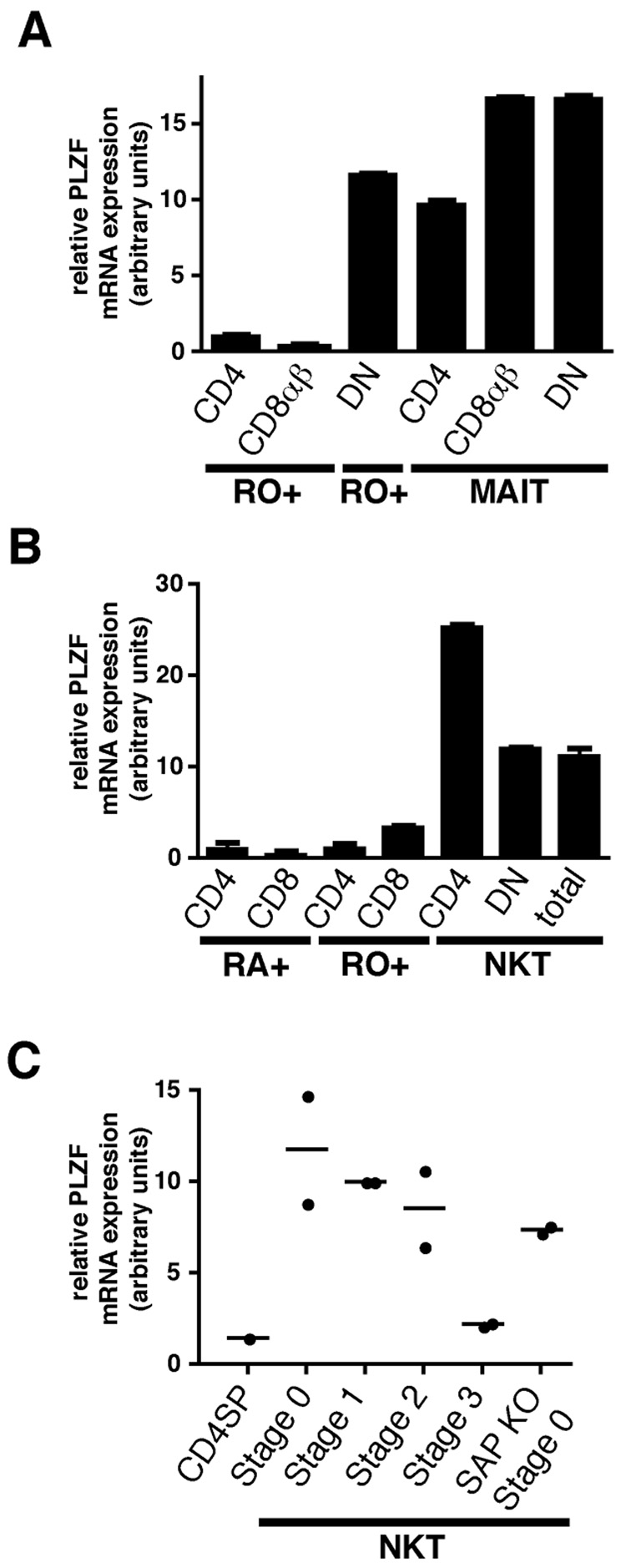
The other well characterized natural memory/effector lineage that is selected by a non-classical MHC class I-like ligand expressed on bone marrow derived cells in the thymus is the mucosal associated invariant T (MAIT) cell. MAIT cells express semi-invariant αβ TCRs with a canonical Vα19-Jα33 in mouse or Vα7.2-Jα33 in human that recognize MR1 and accumulate in the intestinal lamina propria where they can release Th1 and Th2 cytokines (reviewed in (Treiner and Lantz, 2006)). Although MAIT cells are very rare and difficult to purify in mouse, they are abundantly represented in human peripheral blood where they can be enriched using mAbs specific for Vα7.2 and CD45RO. Fig. 7A shows that, relative to RO+ (memory) CD4 or CD8 T cells, human MAIT cells expressed very high levels of PLZF, whether they belonged to the CD4, CD8αβ or DN (defined as CD4−CD8β−) subset of MAIT cells. In addition, high levels of PLZF were also detected in the DN RO+ TCRαβ population, an unusual PBL subset that includes some Vα24 NKT cells, MAIT cells, as well as many other NKT-like cells expressing diverse TCRs (Porcelli et al., 1993). As expected, human CD4 and DN Vα24 NKT cells, the homologs of mouse Vα14 NKT cells, also expressed high levels of PLZF compared to conventional RA+ naïve or RO+ memory CD4 and CD8 T cells (Fig. 7B). Altogether, the results demonstrate that expression of PLZF is a highly restricted characteristic of NKT cells and its cousin lineage of MAIT cells in mouse and human.
Figure 7. PLZF Expression in Human NKT and MAIT Cells and in SAP-deficient NKT thymocytes.
(A) Vα7.2+CD45RO+CD161+ MAIT cells were purified from fresh human PBL according to their expression of CD4 or CD8β. The DN subset of MAIT cells is defined as CD4− CD8β−. Control Vα7.2−RO+ memory cells were purified according to expression of CD4 and CD8β. The Vα7.2−RO+CD4−CD8β− (DN RO+) population is enriched in CD1d-restricted unconventional T cells.
(B) NKT cells were MACS-enriched from fresh human PBL using CD1d-αGC tetramer and FACS-sorted into total NKT cells or their CD4 and DN subsets. The NKT-depleted MACS fraction was used to FACS purify naïve (RA+) and memory (RO+) CD4 and CD8 conventional T cells.
(C) PLZF expression by SAP-deficient ‘stage 0’ NKT thymocytes. 2000 CD1d-αGC+thymocytes were sorted at different stages of NKT development for wild-type mice and at the arrested ‘stage 0’ for SAP-deficient mice. PLZF mRNA expression was measured by quantitative qRT-PCR. Data summarize two independent experiments. Control CD4 single positive (CDSP) are shown as negative control.
PLZF expression in SAP-deficient NKT cells
The adaptor SAP and its associated tyrosine kinase Fyn control early development of the NKT lineage by transducing signals emanating from homophilic interactions between SLAM family receptors such as SLAM and Ly108 expressed at the surface of cortical thymocytes (Griewank et al., 2007). SAP-deficient NKT lineage cells are arrested at stage 0, suggesting the possibility that SAP signaling may be involved in the induction of PLZF. Quantitative RT-PCR experiments, however, suggested that this may not be the case, as PLZF was already upregulated in SAP-deficient ‘stage 0’ NKT thymocytes (Fig. 7C).
Discussion
NKT cells are part of a relatively large group of T and B lymphocytes called ‘natural memory’ or ‘innate-like’ lymphocytes that are produced in the thymus or bone marrow in a constitutive memory or effector state and migrate to various tissues where they perform specialized functions of host defense against microbial aggression and tumors (Bendelac et al., 2001). Innate-like lymphocytes express germline-encoded TCRs and BCRs that often exhibit specific reactivity against microbial structures as well as basal autoreactivity to various self components. Prominent among innate-like T lymphocytes, mucosal γδ T cell subsets, CD1d-restricted liver-tropic NKT cells and MR1-restricted lamina propria MAIT cells have been well characterized. The epithelial layer of the small intestine also harbors large quantities of tissue resident effector T cells, which include γδ T cells, CD8αα TCRαβ lymphocytes that recognize non-classical β2-microglobulin associated MHC-like ligands, and conventional MHC class I-restricted CD8αβ TCRαβ lymphocytes.
Transgenic experiments have demonstrated that the germline-encoded TCRs and BCRs of innate-like lymphocytes faithfully drive the peculiar effector differentiation and migratory properties of their lineage of origin during development in the thymus or bone marrow (reviewed in (Bendelac et al., 2001)). However, the transcriptional networks resulting from these interactions and governing these unique lineages have not been elucidated. Various transcription factors have been implicated, but so far they lack exquisite lineage specificity and instead appear to control more generic aspects of development, memory/effector differentiation and homeostasis that are shared across various T and NK cell subsets. These include Ets-1 family proteins, Runx proteins, Gata-3, T-bet and NFκB (Egawa et al., 2005; Kim et al., 2006; Lacorazza et al., 2002; Matsuda et al., 2006; Stanic et al., 2004; Walunas et al., 2000). Thus, it remains unclear whether signature transcription factors (such as c-Krox and Foxp3 expressed in the CD4 or regulatory T cell lineages, respectively) may drive these unique unconventional lineages or whether a combination of factors shared across different lineages and functional programs ultimately compose their transcriptional network.
This report provides the first demonstration of a transcription factor that is specifically expressed at high levels in CD1d-restricted invariant NKT cells and is essential for their development. Notably, PLZF was low or absent in most of the other T cell populations of αβ and γδ T cells expressing similar effector and tissue resident properties, including all populations of intestinal intraepithelial lymphocytes, in conventional memory and effector CD4 and CD8 T cells generated in vivo or in vitro, and in NK cells. Consistent with these findings, PLZF did not appear to be required for the development of any of these populations.
Our survey of the other unconventional lineages identified high expression of PLZF in only one of them, the human MR1-restricted invariant MAIT cells, which are most closely related to NKT cells because of their usage of a semi-invariant canonical αβ TCR and their recognition of the non-classical MHC class I like ligand MR1 on hemopoietic cell types in the thymus and periphery (Treiner and Lantz, 2006). MAIT cells also express cytokine secretion properties and phenotypic markers of the memory/effector and NK lineage, including CD161 in humans where they have been best characterized. However, due to the rarity of MAIT cells in the mouse, particularly in the thymus where developmental stages have not been characterized, it remains to be clarified whether PLZF is expressed in mouse MAIT cells and whether it is necessary for their development.
Strikingly, PLZF was found to be induced very early in NKT cell development, as it was already expressed at intermediate levels in CD24high CD69high CD1d-αGalCer+ thymocytes, a population that includes DP and CD4 SP thymocytes that have just undergone positive selection by CD1d ligands expressed in the cortex. The stimulation of positively selected NKT precursors differs from that of conventional MHC-restricted thymocytes by at least two criteria. The ligands involved in NKT cell development have weak agonist properties as suggested by the low level stimulation of NKT hybridomas cocultured with cortical thymocytes in vitro (Bendelac, 1995). One ligand that can fully stimulate NKT cells is the endogenous lipid iGb3 (Zhou et al., 2004), although other ligands remain to be characterized (Porubsky et al., 2007). In addition, hemophilic interactions between SLAM family members expressed by cortical thymocytes during TCR/ligand recognition provide signaling through the adaptor SAP and Fyn that is essential to NKT cell development but does not appear to be involved in conventional T cell development (Griewank et al., 2007). It is tempting to speculate that some components of the SAP/Fyn signaling pathway may be responsible for PLZF induction. However, we have found that PLZF was still induced in signaled NKT thymocytes in the absence of SAP. Furthermore, studies in patients with the XLP syndrome associated with SAP deficiency suggest that the development of MAIT cells is independent of SAP signaling (O.L., unpublished results). Thus, the signal(s) leading to the early induction of PLZF remain to be identified.
PLZF induction in early NKT precursors paralleled that of c-Krox, a lineage determinating factor for the CD4 lineage. The regulation of CD4 and CD8 expression by developing NKT cells remains enigmatic. CD4 is unnecessary for the development or function of NKT cells (Bendelac et al., 1994), yet it is expressed on a large fraction of mature NKT cells, whereas CD8β is not expressed. By analogy with conventional CD4 cells, it is hypothesized that strong and persistent signaling by endogenous NKT ligands results in c-Krox expression and CD4 lineage commitment. Later, as developing NKT cells undergo cell division and acquire the effector phenotype (stage 2), a fraction of the CD4 cells downregulate CD4 to become DN, a phenomenon that has remained unexplained. Our unpublished results show that these DN nevertheless maintain c-Krox expression. It is also intriguing that PLZF-deficient NKT cells were relatively enriched in DN compared with CD4 cells, suggesting that PLZF may directly or indirectly regulate the impact of c-Krox on CD4 gene regulation.
Although the massive reduction of NKT cell numbers observed in the absence of PLZF might have suggested developmental arrest and death, detailed examination of the thymic precursors and of the few residual CD1d tetramer-positive cells in peripheral tissues suggested a more intriguing alternative explanation. In fact, NKT precursors were simply prevented from expanding and acquiring the memory effector differentiation in the thymus, whereas the minor population of their naïve precursors was fully preserved. Thus, whereas PLZF-deficient NKT cells were nearly absent in the tissues where effector NKT cells are normally found, such as the liver, the minor population of naïve CD44low NKT cells could still be detected in the periphery, particularly in the peripheral lymph nodes due to their high expression of the homing receptor CD62L which characterizes naïve cells. In mixed bone marrow chimeras, PLZF-deficient NKT cells were as frequent as their wild type counterparts in the lymph nodes, whereas they were outnumbered 37 to 1 in the liver. Furthermore, these arrested cells exhibited a cytokine profile reminiscent of naïve CD4 cells, with high IL-2 and low IL-4 and IFN-γ. These results suggest therefore that, while PLZF is required for the expansion and effector differentiation phase of NKT thymocyte development, its absence is nevertheless compatible with the survival and export of the earlier CD44low stage cells that resemble conventional CD4 SP cells. This conclusion is intriguing because it might be expected that, due to their autoreactive potential, such CD44low cells should become activated by self antigen anyway. However, stimulation by naturally expressed endogenous ligands is weak, since NKT hybridomas exposed to CD1d expressing cells secrete only very low levels of cytokines compared to their response to the addition of full agonists. In addition, it is possible that the levels of endogenous NKT ligands are downregulated in the periphery of healthy animals, as suggested by the observation that cortical thymocytes are among the cell types most consistently capable of stimulating NKT cell hybridomas (Park et al., 1998). Thus, as NKT cell precursors move away from the thymic cortex into the medulla and then migrate to the periphery, their stimulation by self ligands may decrease. Alternatively, chronic low level stimulation in the periphery might generate the minor population of stage 3 NKT cells found in PLZF-deficient mice. A similar set of questions has been raised about the autoreactivity of CD25+ regulatory T cells since it is not readily apparent, except after transfer of a regulatory T cell TCR into cells that are subsequently injected into a lymphopenic environment (Hsieh et al., 2004). In conclusion, these findings raise the possibility that, in the healthy state, the weak autoreactivity of NKT cells is more apparent in the thymus where it serves signaling functions for lineage specification than in the periphery.
It is notable that ectopic expression of PLZF using the CD4 promoter (with the intronic silencer) changed the naïve status of conventional CD4 lineage cells and converted them into effector-type cells with migratory and cytokine properties reminiscent of stage 2 NKT cells, including the double IL-4/IFN-γ production. Although the extent of this conversion, in particular the influence of TCR specificity, remains to be fully investigated, it appeared to be broad-based and cell-intrinsic, as evidenced by the equivalent representation of PLZF-deficient and wild type CD4 cells in 1:1 mixed bone marrow chimeras and by the scarcity of naïve recirculating CD4 cells found in blood or lymph nodes of PLZF-deficient mice. Furthermore, preliminary analysis suggested a normal polyclonal Vβ repertoire similar to wild type. Strikingly, the conversion may occur in the thymus itself, as only few CD4 thymocytes exhibited the naïve phenotype that characterizes their wild-type counterparts. CD8 T cells, however, failed to convert despite the transient expression of PLZF at the DP stage, suggesting a requirement for persistent PLZF expression or, alternatively, that the CD8 lineage may be refractory to PLZF. Altogether, loss and gain of function experiments establish the central role of PLZF in directing the effector differentiation program of NKT cells. However, PLZF alone was not sufficient to drive NKT cell differentiation all the way to the terminal NK-like stage 3, indicating that additional factors controlling this later stage remain to be discovered.
PLZF is a member of the BTB/POZ-ZF family of transcriptional regulators sharing a highly conserved BTB/POZ dimerization domain at the N-terminus, which can participate in homo- and hetero-dimerization, and a variable number of zinc-finger domains responsible for DNA binding at the C-terminus. This family includes c-Krox, which is induced upon TCR signaling in the thymus to direct the CD4 lineage (He et al., 2005; He et al., 2008; Sun et al., 2005; Wildt et al., 2007); LRF which represses T cell instructive signals produced by Notch in the bone marrow to preserve B cell progenitors (Maeda et al., 2007); and Bcl-6, MAZR, BAZF and ROG which also contribute to various aspects of lymphocyte development and function as well as to malignant hemopoiesis (reviewed in (Bilic and Ellmeier, 2007; Kelly and Daniel, 2006)). These factors are characterized by their transcriptional repression through recruitment of a repressor complex consisting of N-CoR, SMRT, sin3A/b, and histone deacetylases. PLZF is involved in the pro-oncogenic translocation (11;17)(q23;q21) with the retinoic acid receptor α (RARα) locus. The translocation generates a PLZF-RARα fusion protein consisting of the dimerization domains of PLZF and the DNA binding domains of RARα, resulting in the development of acute promyelocytic leukemia (APL) as a result of myeloid precursors failing to terminally differentiate (Alcalay et al., 2003; Grignani et al., 1998; Lin et al., 1998). PLZF expression has been found in some hematopoietic cells such as myeloid progenitors, but there is no report of hemopoietic defects in mice lacking PLZF. Instead, lack of PLZF in the Luxoid mouse or in a targeted knockout resulted in the progressive loss of spermatogonial stem cells (Buaas et al., 2004; Costoya et al., 2004), suggesting a role of PLZF in regulating cellular proliferation and differentiation.
Given the highly specific expression and function of PLZF in NKT cell development, further studies of the mechanisms underlying its induction and function will be warranted to understand the molecular events that specify this peculiar lineage.
Experimental Procedures
Mice
Luxoid (B6.C3-Zbtb16Lu/J) mice were obtained from The Jackson Laboratory and have been described elsewhere (Buaas et al., 2004). The Luxoid mutation spontaneously arose on the C3H/He inbred strain and was backcrossed to C57BL/10Gn for 69 generations and subsequently to C57BL/6J for over 35 generations at The Jackson Laboratory. C57BL/6 (CD45.2) and B6.SJL-Ptprca Pep3b/BoyJ (CD45.1) were from The Jackson Laboratory. B6.Jα18−/− mice have been described elsewhere (Cui et al., 1997) and the B6.SAP−/− mice were maintained in our laboratory (Griewank et al., 2007). For the generation of PLZF transgenic mice, PLZF was PCR amplified from cDNA prepared from FACS sorted C57BL/6 thymic NKT cells and cloned into a plasmid containing the minimal CD4 promoter/enhancer and the intronic silencer (Sawada et al., 1994). The linearized construct was injected into fertilized C57BL/6 oocytes, and the injected oocytes were implanted into pseudopregnant CD-1 (VAF+) outbred female mice. Transgenic mice were screened using PCR (forward primer: 5′-TGTGCCAGGGTCGGAGACAATAACGG-3′; reverse primer: 5′-TCCTGGCTGTCCACCATGATGACCAC-3′). Two founder lines #1797 and #1960 were studied. All mice were raised in a specific pathogen-free environment at the University of Chicago and experiments performed in accordance with the guidelines of the Institutional Animal Care and Use Committee.
Quantitative Real-time PCR
Total RNA was isolated from 20,000–100,000 FACS-sorted C57BL/6J primary cells using a combination of Trizol (Invitrogen) and RNeasy mini kit (Qiagen, Valencia, CA, USA) and was reverse-transcribed with random primers using AffinityScript qPCR cDNA Synthesis Kit (Stratagene). Some populations were prepared using a mixture of random primers and oligo-d(T). For rare ‘stage 0’ NKT thymocytes, pooled thymuses were first MACS-enriched using CD1d-αGC tetramer and anti-APC beads with autoMACS (Miltenyi Biotech) and subsequently sorted directly into lysis buffer. From these lysates, cDNA was prepared using the Cells-to-DNA II kit (Ambion) and random primers. Transcripts for murine PLZF (Zbtb16) were quantified using primers spanning intron 4 (Forward primer:5'-cccagttctcaaaggaggatg-3', Reverse primer: 5'-ttcccacacagcagacagaag-3'). Transcripts for murine c-Krox (Zbtb7b) were quantified using primers spanning intron 2 (Forward primer:5'- actggtgagaagccctttgc-3', Reverse primer: 5'- ttccgcatgtggatcttcag-3'). Transcripts were normalized to 18s ribosomal RNA (Forward primer: 5'-ttgactcaacacgggaaacc-3', Reverse primer: 5'-acccacggaatcgagaaaga-3').
For human NKT cell preparations, PBL were MACS-enriched with CD1d-αGC tetramers using magnetic beads and FACS sorted into total NKT cells or their CD4 and DN subsets. Total RNA from approximately 5x104 cells was prepared using a combination of Trizol (Invitrogen) and RNeasy mini kit (Qiagen), and reverse transcribed with Superscript III (Invitrogen) using oligo-d(T). For human MAIT cell preparations, PBL were stained and FACS sorted as Vα7.2+CD161+CD45RO+CD3+pan γδ−. MAIT subsets were further purified using CD4 and CD8β. Control memory T cell fractions were defined as CD3+γδ−Va7.2−CD45RO+ and further fractionated into CD4+CD8β−, CD4−CD8β+ or CD4−CD8β− (DN, including CD8αα). RNA from 200,000 cells was purified and reverse transcribed using a combination of random primers and oligo-d(T).
Transcripts for human PLZF were quantified using primers spanning intron 3 (Forward primer: 5'-tagtttgcggctgagaatgc-3', Reverse primer: 5'-accgcactgatcacagacaaag-3') and normalized to HPRT (Forward primer: 5'-gaaagggtgtttattcctcatgg-3', Reverse primer: 5'-ctcccatctccttcatcacatc-3').
All primers were generated using the Ensembl database and Primer3. Quantitative PCR was performed on a Stratagene Mx3005p using Brilliant SYBR Green qPCR Master Mix (Stratagene).
SDS-PAGE and Western Blotting
Thymuses and spleens from C57BL/6J mice were pooled and MACS-enriched for NKT cells using CD1d-αGC tetramers prior to FACS sorting using a combination of CD1d-αGC tetramer, anti-TCRβ, and anti-B220. Approximately 1x106 sorted cells were pelleted and frozen at −80C until use. Cell pellets were lysed on ice in TNE lysis buffer containing protease inhibitors, and separated by SDS-PAGE on a 10% mini-gel. The gel was transferred to PVDF membrane and blotted using a rabbit polyclonal antibody specific to the C-terminal region of human PLZF (ab39354; Abcam) at a concentration of 0.4mg/ml. Goat anti-rabbit-HRP secondary antibody was applied at a concentration of 0.08mg/ml (111-036-003; Jackson ImmunoResearch) and HRP was detected using ChemiGlow West reagent (Alpha Innotech). For loading control, a γ-tubulin specific antibody (T6557; Sigma) was used at a dilution of 1:10,000 dilution of ascites.
Flow Cytometry
CD1d-αGalCer tetramers were prepared as previously described (Benlagha et al., 2000). Fluorochrome labeled monoclonal antibodies (clone indicated in parentheses) against mouse B220 (RA3-6B2), CD1d (1B1), CD3e (17A2), CD4 (GK1.5 or L3T4), CD8α (53-6.7), CD8β (53-5.8), CD11b (M170), CD11c (HL3), CD21/35 (7G6), CD23 (B3B4), CD24 (M1/69), CD25 (PC61), CD45.1 (A20), CD45.2 (104), CD44 (IM7), CD62L (MEL-14), CD69 (H1.2F3), CD86 (GL-1), γδTCR (GL3), IA/IE (2G9), IL-4 (11B11), IFNγ (XMG1.2), NK1.1 (PK136), and TCRβ (H57-597), were purchased from eBioscience, BD Biosciences, or BioLegend. Fluorochrome-labeled monoclonal antibodies against human CD3 (UCHT1), CD4 (RPA-T4 or OKT4), CD8α (RPA-T8), CD8β (2ST8.5H7), CD45RA (HI100), CD45RO (IM247 or UCHL1), CD161 (DX12), pan γδ (Immu 510), Vα7.2 (3C10 (Martin et al, in preparation)), and Vα24 (C15), were purchased from eBioscience, BD Biosciences, and Immunotech.
For NKT cell MACS enrichment, cells were labeled with APC-conjugated CD1d-αGalCer tetramers, bound to anti-APC magnetic beads, and enriched on an autoMACS cell separator (Miltenyi Biotech) as described (Benlagha et al., 2005). Samples were analyzed on an LSRII (Becton Dickinson), or sorted on a FACSAria (Becton Dickinson) or MoFlo (Dako Cytomation), with doublet exclusion and DAPI staining of dead cells in most experiments. For intracellular flow cytometry of PLZF, thymocytes were first depleted of immature cells using anti-CD24 and autoMACS before staining for surface markers. Cells were then washed and blocked with goat serum (Southern Biotech), washed again and fixed using the permeabilization and fixation buffer “Foxp3 Staining Buffer Set” from eBioscience. Intracellular PLZF was detected using 4 µg/ml mouse monoclonal antibody D-9 (Santa Cruz) and FITC conjugated rat anti-mouse (eBioscience). Data was analyzed using FlowJo (Tree Star).
Cytokine Analysis
Cells were cultured on microplates coated with 10µg/ml anti-CD3 (clone 145-2C11) in the presence of 10 U/ml human IL-2 or were stimulated by irradiated αGC-loaded bone-marrow derived dendritic cells. Supernatants were stained for cytokines using Mouse Grp I Cytokine 23-Plex Panel (Biorad) and assayed using a Bioplex instrument. For intracellular flow cytometry, brefeldin A was added for the final 6 hours of culture and cells were permeabilized and stained for IL-4 and IFN-γ production by intracellular flow cytometry using BD BioSciences Cytofix/Cytoperm kit.
EdU Assay for Proliferation
Mice were injected iv with 1 mg of EdU (5-ethynyl-2'-deoxyuridine, Invitrogen) 3 hours before analysis. Thymocytes were processed and stained for flow cytometry according to the manufacturer’s instructions.
Generation of Mixed Bone Marrow Chimeras
6–8 week-old B6.Jα18−/− or B6 mice (CD45.2) were subjected to 900–950 Rads irradiation using a gamma cell 40 irradiator with a cesium source. 3–6 hours later, irradiated mice were injected intravenously with 2–10x106 bone marrow cells obtained from femurs of B6.Zbtb16lu/lu or B6.CD4p-PLZF transgenics (CD45.2) and B6.CD45.1 and mixed at 1:1 or 3:1 ratios. Mice were analyzed 6–9 weeks after irradiation.
Statistical Analysis
Stastical analysis was performed in Prism (Graphpad Software) using the unpaired t-test for Luxoid mice and the paired t-test for the mixed bone marrow chimeras.
Supplementary Material
Acknowledgements
We thank members of the Bendelac and Lantz laboratories for help and advice, Bana Jabri, Eric Bertolino and Harinder Singh for discussions; Ryan Duggan, David Leclerc and Mike Olson for expert assistance with cell sorting; the University of Chicago Transgenic Mouse Facility and Animal Resource Center. This work was supported by NIH RO1 AI038339 to AB; the University of Chicago Digestive Diseases Research Core Center (P30 DK42086). AB is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alcalay M, Meani N, Gelmetti V, Fantozzi A, Fagioli M, Orleth A, Riganelli D, Sebastiani C, Cappelli E, Casciari C, et al. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J Clin Invest. 2003;112:1751–1761. doi: 10.1172/JCI17595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A. Positive selection of mouse NK1+ T cells by CD1-expressing cortical thymocytes. J Exp Med. 1995;182:2091–2096. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nat Rev Immunol. 2001;1:177–186. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Killeen N, Littman D, Schwartz RH. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–1778. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Benlagha K, Wei DG, Veiga J, Teyton L, Bendelac A. Characterization of the early stages in thymic NKT cell development. Journal of Experimental Medicine. 2005;202:485–492. doi: 10.1084/jem.20050456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilic I, Ellmeier W. The role of BTB domain-containing zinc finger proteins in T cell development and function. Immunol Lett. 2007;108:1–9. doi: 10.1016/j.imlet.2006.09.007. [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet. 2004;36:647–652. doi: 10.1038/ng1366. [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet. 2004;36:653–659. doi: 10.1038/ng1367. [DOI] [PubMed] [Google Scholar]
- Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Valpha14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. [DOI] [PubMed] [Google Scholar]
- Egawa T, Eberl G, Taniuchi I, Benlagha K, Geissmann F, Hennighausen L, Bendelac A, Littman DR. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenot JD, Gavin MA, Rudensky AY. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–336. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- Geissmann F, Cameron TO, Sidobre S, Manlongat N, Kronenberg M, Briskin MJ, Dustin ML, Littman DR. Intravascular immune surveillance by CXCR6+ NKT cells patrolling liver sinusoids. PLoS Biol. 2005;3:e113. doi: 10.1371/journal.pbio.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey DI, Berzins SP. Control points in NKT-cell development. Nat Rev Immunol. 2007;7:505–515. doi: 10.1038/nri2116. [DOI] [PubMed] [Google Scholar]
- Green MC. The position of luxoid in linkage group II of the mouse. J Hered. 1961;52:297–300. doi: 10.1093/oxfordjournals.jhered.a107103. [DOI] [PubMed] [Google Scholar]
- Griewank K, Borowski C, Rietdijk S, Wang N, Julien A, Wei DG, Mamchak AA, Terhorst C, Bendelac A. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Shared principles in NF-kappaB signaling. Cell. 2008;132:344–362. doi: 10.1016/j.cell.2008.01.020. [DOI] [PubMed] [Google Scholar]
- He X, He X, Dave VP, Zhang Y, Hua X, Nicolas E, Xu W, Roe BA, Kappes DJ. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. [DOI] [PubMed] [Google Scholar]
- He X, Kappes DJ. CD4/CD8 lineage commitment: light at the end of the tunnel? Curr Opin Immunol. 2006;18:135–142. doi: 10.1016/j.coi.2006.02.003. [DOI] [PubMed] [Google Scholar]
- He X, Park K, Wang H, He X, Zhang Y, Hua X, Li Y, Kappes DJ. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- Hsieh CS, Liang Y, Tyznik AJ, Self SG, Liggitt D, Rudensky AY. Recognition of the peripheral self by naturally arising CD25+ CD4+ T cell receptors. Immunity. 2004;21:267–277. doi: 10.1016/j.immuni.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Intlekofer AM, Takemoto N, Wherry EJ, Longworth SA, Northrup JT, Palanivel VR, Mullen A, Gasink CR, Kaech SM, Miller JD, et al. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM. POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol. 2006;16:578–587. doi: 10.1016/j.tcb.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Kim PJ, Pai SY, Brigl M, Besra GS, Gumperz J, Ho IC. GATA-3 regulates the development and function of invariant NKT cells. J Immunol. 2006;177:6650–6659. doi: 10.4049/jimmunol.177.10.6650. [DOI] [PubMed] [Google Scholar]
- Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, Cordon-Cardo C, Mao S, Pandolfi PP, Nimer SD. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr., Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Maeda T, Merghoub T, Hobbs RM, Dong L, Maeda M, Zakrzewski J, van den Brink MR, Zelent A, Shigematsu H, Akashi K, et al. Regulation of B versus T lymphoid lineage fate decision by the proto-oncogene LRF. Science. 2007;316:860–866. doi: 10.1126/science.1140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda JL, Gapin L. Developmental program of mouse Valpha14i NKT cells. Curr Opin Immunol. 2005;17:122–130. doi: 10.1016/j.coi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Matsuda JL, Zhang Q, Ndonye R, Richardson SK, Howell AR, Gapin L. T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting C, Leiden JM, Glimcher LH, Ho IC. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- Park S-H, Roark JH, Bendelac A. Tissue specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–3134. [PubMed] [Google Scholar]
- Porcelli S, Yockey CE, Brenner MB, Balk SP. Analysis of T cell antigen receptor (TCR) expression by human peripheral blood CD4-8- a/b T cells demonstrates preferential use of several Vb genes and an invariant TCR a chain. J Exp Med. 1993;178:1–16. doi: 10.1084/jem.178.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porubsky S, Speak AO, Luckow B, Cerundolo V, Platt FM, Grone HJ. Normal development and function of invariant natural killer T cells in mice with isoglobotrihexosylceramide (iGb3) deficiency. Proc Natl Acad Sci U S A. 2007;104:5977–5982. doi: 10.1073/pnas.0611139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson SI, Richard O, Tavian M, Ranson T, Vosshenrich CA, Colucci F, Buer J, Grosveld F, Godin I, Di Santo JP. GATA-3 promotes maturation, IFN-gamma production, and liver-specific homing of NK cells. Immunity. 2003;19:701–711. doi: 10.1016/s1074-7613(03)00294-2. [DOI] [PubMed] [Google Scholar]
- Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- Shires J, Theodoridis E, Hayday AC. Biological insights into TCRgammadelta+ and TCRalphabeta+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE) Immunity. 2001;15:419–434. doi: 10.1016/s1074-7613(01)00192-3. [DOI] [PubMed] [Google Scholar]
- Singer A, Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv Immunol. 2004;83:91–131. doi: 10.1016/S0065-2776(04)83003-7. [DOI] [PubMed] [Google Scholar]
- Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanic AK, Bezbradica JS, Park JJ, Matsuki N, Mora AL, Van Kaer L, Boothby MR, Joyce S. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes. J Immunol. 2004;172:2265–2273. doi: 10.4049/jimmunol.172.4.2265. [DOI] [PubMed] [Google Scholar]
- Sun G, Liu X, Mercado P, Jenkinson SR, Kypriotou M, Feigenbaum L, Galera P, Bosselut R. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. [DOI] [PubMed] [Google Scholar]
- Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- Treiner E, Lantz O. CD1d- and MR1-restricted invariant T cells: of mice and men. Curr Opin Immunol. 2006;18:519–526. doi: 10.1016/j.coi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Walunas TL, Wang B, Wang CR, Leiden JM. Cutting edge: the Ets1 transcription factor is required for the development of NK T cells in mice. J Immunol. 2000;164:2857–2860. doi: 10.4049/jimmunol.164.6.2857. [DOI] [PubMed] [Google Scholar]
- Wildt KF, Sun G, Grueter B, Fischer M, Zamisch M, Ehlers M, Bosselut R. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J Immunol. 2007;179:4405–4414. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]
- Zhou D, Mattner J, Cantu C, 3rd, Schrantz N, Yin N, Gao Y, Sagiv Y, Hudspeth K, Wu YP, Yamashita T, et al. Lysosomal glycosphingolipid recognition by NKT cells. Science. 2004;306:1786–1789. doi: 10.1126/science.1103440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.