Abstract
Tissue-specific differentially methylated regions (tDMRs) have been identified and implicated for their indispensable involvement in mammalian development and tissue differentiation. In this report, a quantitative DNA methylation analysis was performed for 13 human orthologous regions of recently confirmed mouse tDMRs by using Sequenom Mass Array, by which bisulfite-treated fragments are quantitatively detected using time of flight masspectroscopy analysis. Eight regions were shown as tDMRs in various tissues from three independent individuals. Testis DNA samples from eight individuals were also analyzed for methylation. Interestingly, there is evidence that the DNA methylation level is divergent among individuals. DNA methylation levels of five testis-specific DMRs were significantly inversely correlated with the number of spermatocytes. However, a positive correlation was seen at tDMRs located near the TRIM38 and CASZ1 genes. Our results indicate that tDMRs are conserved between mouse and human and may have an important role in regulating tissue function, differentiation and aging.
Introduction
Methylation of cytosine in CpG dinucleotides is an inheritable modification of DNA in the mammalian genome that does not alter the nucleotide sequence [1; 2]. Generally, cytosine residues in the CpGs are methylated in the genome, especially within non-coding DNA, introns, repetitive sequences and potentially active transposable elements, resulting in long-term silencing [3; 4]. However, most CpG clusters, called CpG islands, are frequently found in the proximal promoter regions of many genes and are unmethylated during normal cell development. Nonetheless, recent genome-wide DNA methylation searches indicate that 4 to17% of CpG sites are different in methylation among tissues and developmental processes [5–8].
DNA methylation plays an important role in the diverse genomic process, such as in gene regulation, chromosomal stability, parental imprinting and X-inactivation [9]. It is also indicated that aberrant DNA methylation patterns or the epi-mutation of gene promoters impair normal transcription in either germ-line or somatic cells, causing abnormal development associated with various diseases, such as cancer [10–13]. Despite its importance and function originally postulated for this epigenetic modification, the function and characteristics of DNA methylation are still poorly understood, especially in individual human tissues. It is therefore important to analyze the tissue-specific DNA methylation of human individuals in detail.
In our previous studies we have found 5% or more of mouse promoter is differentially methylated among tissues [6] and seven out of 14 orthologous regions of mouse tDMRs also showed tDMR in humans [14]. Here we describe the quantitative DNA methylation analysis of the human genomic regions homologous to an additional 13 confirmed mouse tDMRs by sequenom MassArray analysis in order to investigate the characteristics of tDMRs in various tissues from three independent individuals. The human testis-specific DMRs are also further analyzed to see whether the percentage of DNA methylation in testis DNA is associated with the number of the spermatocytes in seminiferous tubule and the age of tissue specimens.
Materials and methods
Human samples
Normal tissue was obtained from organ donation from autopsy cases after receiving the informed, written consent of bereaved families or relatives at the Pathology Division, School of Medicine, Nihon University, Tokyo, Japan. The samples were immediately frozen and stored at −80°C. At the same time, a part of each sample was fixed and paraffin slides were prepared and stained by routine hematoxylin and eosin (H&E) staining to observe pathological features. Eight autopsy cases from males between 58 and 81 years old (average 66.5) were used in this study. All experiments were conducted in accordance with the guidelines approved by the Ethics Committee of Nihon University School of Medicine.
Mouse samples
Mouse samples were obtained from C57BL/6J (B6). Tissues (colon, liver, muscle, kidney, testis and brain) from 12-week-old male mice were purchased from The Jackson Laboratory. Mice were purchased at 5–6 weeks of age and maintained for three generations maximum in the animal facility at the Roswell Park Cancer Institute under IACUC approval. Mouse genomic DNAs were kindly gifted for this analysis.
Bisulfite treatment and PCR
The METHPRIMER program (http://www.urogene.org/methprimer/index1.html) [15] was used to design bisulfite PCR primers (Supplementary Table 1). All primers were purchased from Operon or Sigma Genosys. Genomic DNA from each sample was prepared using QuickGene-800 (FUJIFILM) and was subjected to bisulfite modification by using the EZ DNA Methylation kit (Zymo Research). The bisulfite-treated genomic DNA was amplified by Taq DNA polymerase (Roche) (2 minutes at 94°C followed by 40 cycles of 15 seconds at 94°C, 30 seconds at 50–60°C and 30 seconds at 72°C with a 4-minute final extension at 72°C). The PCR products obtained were subjected to gel electrophoresis analysis and then forwarded to Sequenom MassARRAY analysis. 100% methylated human DNA was purchased from Promega.
Sequence analysis
The databases used for sequence analysis, BLAT, can be found on the UCSC Genome Bioinformatics website (http://genome.brc.mcw.edu/). The mouse tDMR sequence is the sequence of a NotI-PvuII or NotI-PstI Restriction Landmark fragment [6]. The UCSC Genome Browser, Human, March 2006 assembly, was used for homology searches.
Quantitative analysis of DNA methylation using base-specific cleavage and Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
Sequenom MassARRAY quantitative methylation analysis [16] using the MassARRAY Compact System (www.sequenom.com) was employed for the quantitative DNA methylation analysis at CpG dinucleotides. This system utilizes mass spectrometry (MS) for the detection and quantitative analysis of DNA methylation using Homogeneous MassCLEAVE (hMC) base-specific cleavage and matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS [16]. Genomic DNA was isolated, as described above. DNA (1 ug) was converted with sodium bisulfite using the EZ DNA methylation kit (Zymo Research, Orange, CA) described above. The primers were designed using Methprimer [17] for the target human orthologous region of mouse tDMRs. Each reverse primer has a T7-promotor tag for in vitro transcription (5′-cagtaatacgactcactatagggagaaggct-3′), and the forward primer is tagged with a 10mer to balance TM (5′aggaagagag-3′). The primer pairs were designed to span the tDMRs or closely adjacent region or CG rich region, as indicated. PCR amplification of 1 ul bisulfite treated DNA (~20 ng/ml) was performed using HotStar Taq Polymerase (Qiagen) in a 5 ul reaction volume using PCR primers at a 200 nM final concentration. After Shrimp Alkaline Phosphatase treatment, 2 ul of the PCR products were used as a template for in vitro transcription and RNase A Cleavage for the T-reverse reaction, as per manufacturer’s instructions (Sequenom hMC). The samples were desalted and spotted on a 384-pad SpectroCHIP (Sequenom) using a MassARRAY nanodispenser (Samsung), followed by spectral acquisition on a MassARRAY Analyzer Compact MALDI-TOF MS (Sequenom). The resultant methylation calls were performed by EpiTyper software v1.0 (Sequenom) to generate quantitative results for each CpG site or an aggregate of multiple CpG sites. Since Maldi-TOF mass methylated peaks does not denote a particular CpG site, but rather corresponds to the number of CpG sites methylated within the cleavage fragment, we decided to present average percent methylation of all CpG sites in the bisulfite PCR fragment with the standard deviation. Small standard deviation means low variation at each CpG site in analyses. The experiment was performed in triplicate. The non-applicable reading and its corresponding site were eliminated in calculation. More detailed methodology may be found in [18].
Histological evaluation
A section of normal tissue was subjected to paraffin slides and stained by routine hematoxylin and eosin (H&E) staining to observe pathological features. The number of spermatocytes was counted from 30 seminiferous tubules of each testis in triplicate by the pathologist. The average number of spermatocytes per seminiferous tube was used to evaluate spermatogenesis.
Results and Discussion
Mass array evaluation using mouse tDMRs
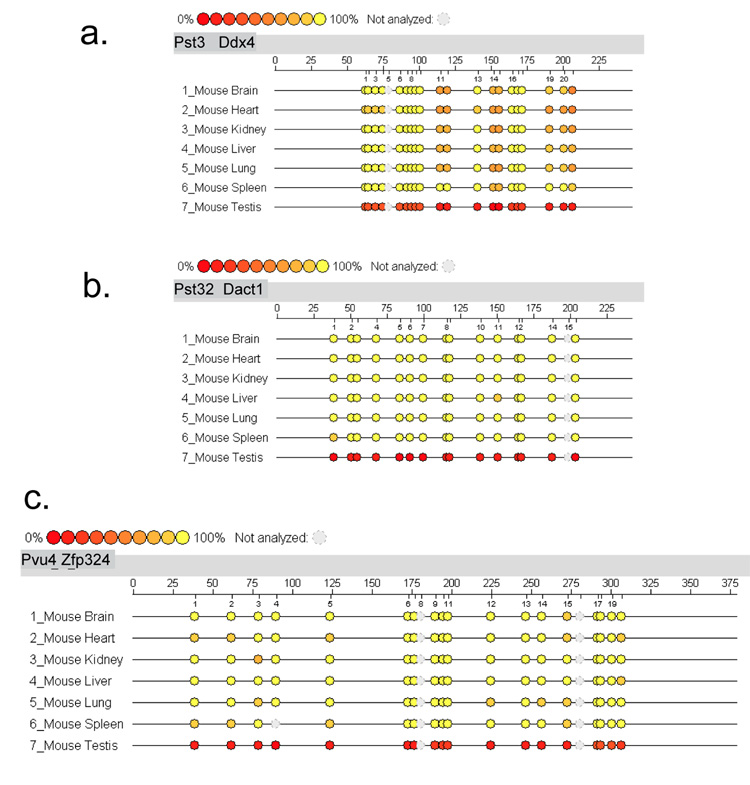
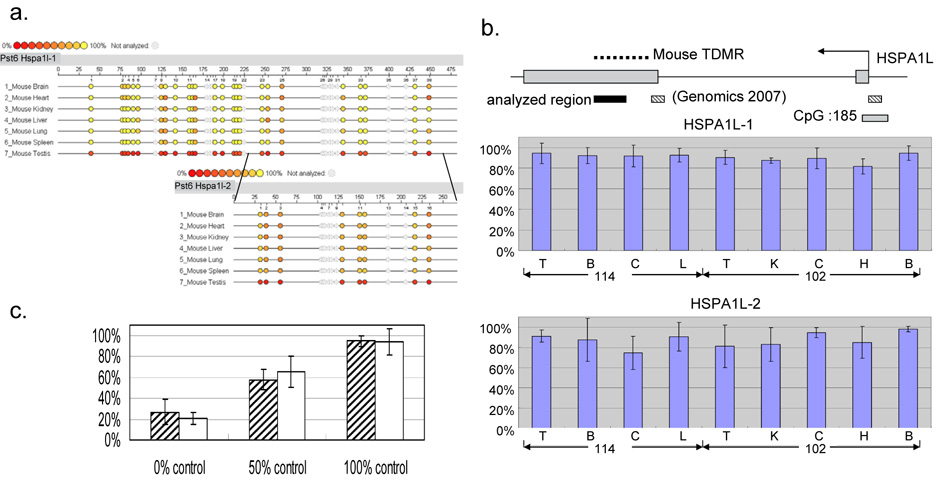
In order to evaluate the Sequenom MassARRAY quantitative methylation analysis in the mammalian genome, we first applied this methodology to the previously confirmed three mouse tDMRs of Pst3, Pst32 and Pvu4, in the Ddx4, Dact1 and Zfp324 genes, respectively [6; 14]. The Sequenom MassARRAY data were presented as epigrams in figures 1. The MassArray results showed testis-specific tDMRs in a good quantitative corresponding to previous publication. We also examined a human orthologous region of mouse tDMR Pst6, which was located in the exon 2 CpG island (CpG:33) for Hspa1l (heat shock protein a1-like) that is expressed virtually exclusively in testis (http://symatlas.gnf.org/SymAtlas/)[19]. This region is recently reassigned for Pst6 and was not analyzed in previous study, in which only 5’ exon 2 and promoter CpG :185 region were analyzed as illustrated in Figure 2b. We initially analyzed the mouse Pst6 tDMR and then analyzed the orthologous region in humans using two distinct primer sets. The CpG:33 was indeed the testis-specific hypomethylation in mice by both the primer sets, as described in the epigrams (Figure 2-a). In humans no CpG island was defined in the region homologous to the mouse tDMR at the HSPA1L gene, which he testis-specific expression is see, and no tissue-specific difference in methylation by either primer set were detected (Figure 2b). This suggested that the mouse tDMR at the Hspa1L gene may not be conserved in humans, although more extensive study may be needed to confirm this. Prior to applying the Sequenom MassArray technology, the test PCR experiments are important using 0% and 100% methylated control DNAs to make sure of equal amplification. Figure 2-c showed an average percent methylation analysis using 0%, 100% and its equal amount mixture of (50%) control DNA for two primer sets for BC048318 transcript. The Sequenom MassArray analysis using two distinct but overlapping primer-sets showed good agreement, suggesting the accuracy of methodology under our protocol.
Figure 1.
Sequenom MassARRAY Analysis of percent DNA methylation at mouse testis-specific differentially methylated regions. The EpiTYPER program from Sequenom MassARRAY Analysis provides the results of percent methylation as an epigram. The epigram shows the percent DNA methylation level of each CpG site of the target region. The aligned epigram shows methylation signature differences in the target genomic region. Different colors display relative methylation changes in 10% increments. The yellow circle indicates 100% methylation, and the red circle is 0% methylation at each CpG site. The number of CpG sites, target sequence length and sample names are included in each epigram. Seven DNA samples from various mouse tissues (brain, heart, kidney, liver, lung, spleen and testes) were used in this analysis. The epigrams for a.Ddx4, b.Dact1 and c. Znf324 were shown.
Figure 2.
Mouse and human DNA methylation levels at the Hspa1l tDMR. a. Percent methylation levels are indicated by epigrams for the mouse Hspa1l tDMR using two primer sets. Top is Hspa1l-1 set and bottom is Hspa1l-2 set within the Hspa1l-1 fragment. The aligned epigram shows a methylation signature difference in the target genomic region as shown in Figure 1. b. Genomic structure of human HSPA1L (top). Mouse tDMR, previously analyzed regions [14] and the region analyzed in this paper are indicated as dotted lines, shaded boxes and a black box, respectively. Two independent primer sets are designed to cover the region. The average percent methylation level and the standard deviation are indicated as bar graphs for HSPA1L-1 (middle) and HSPA1L-2 (bottom). c. 0%, 50% and 100% methylation control analysis. A bar graph shows an average percent methylation of two primer sets (BC048318-A and BC048318-B). Average percent methylation levels with standard deviations were depicted for 0%, 50% and 100% controls in each primer set.
Human tissue-specific difference in methylation
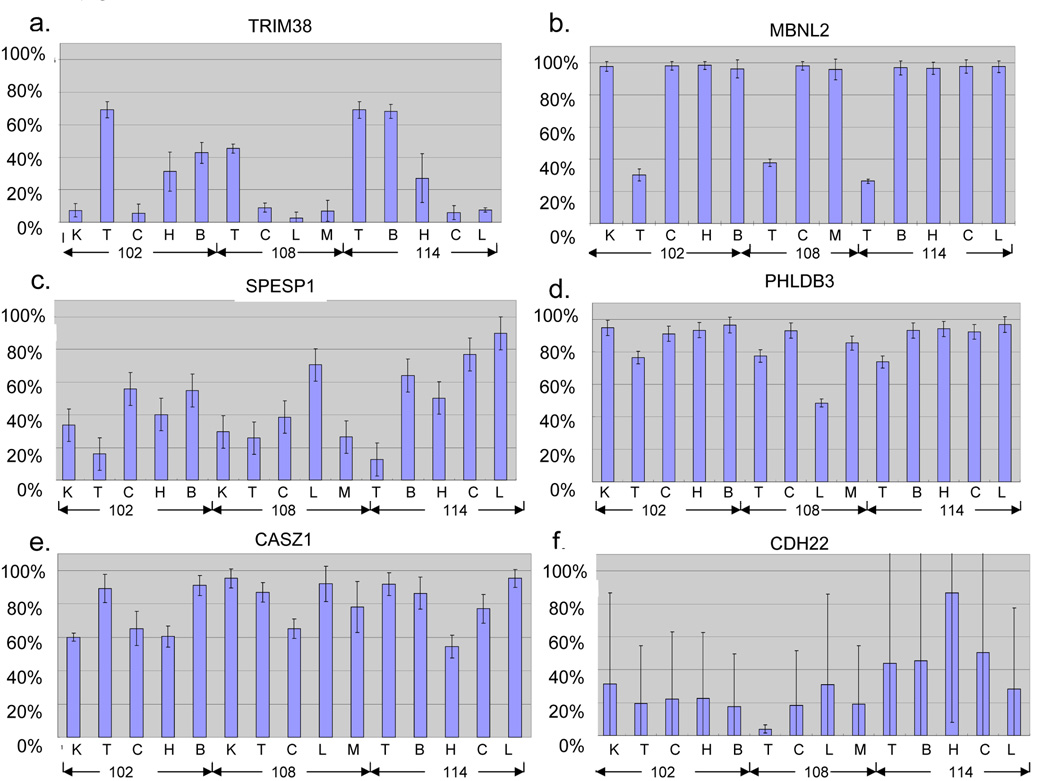
Thirteen additional human orthologous regions of mouse tDMRs were analyzed by the Sequenom MassARRAY method in this study and summarized in Table 1. These 13 regions were identified in the mouse by using Restriction Landmark Genomic Scanning (RLGS) in conjunction with in silico RLGS analysis [6] and the most were confirmed by Sequenom MassARRAY analysis (Songet al, submitted.). The results of a Sequenom MassARRAY analysis of the human orthologous genomic regions were summarized in Supplemental Table 2. Eleven human regions showed at least one significant difference in methylation among tissues. At least eight out of 11 tDMRs showed multiple significant tissue differences in methylation separating two low- and high-methylation groups. Six out of 8 showed DNA methylation in testis belonging at the low-methylation group and two belonged in a high-percentage group. More analysis is necessary to confirm the mouse and human corresponding at tDMRs. However, we found interesting distinct patterns of human tDMRs and presented six examples in Figure 3.
Table 1.
| mouse chr location | mouseCpG island | Human | human chr location | Human CpG island | description | function | |
|---|---|---|---|---|---|---|---|
| Pst2 | chr13:23870412-23870679 | no | TRIM38 | chr6:26070797-26071050 | no | tripartite motif-containing 38 | metal ion binding (GO:0046872) |
| Pst31 | chr7:25411496-25411672 | no | PHLDB3 | chr19:48675345-48675649 | yes | PHLDB3 protein (Fragment). | unknown |
| Pst4 | chr9:62130029-62130377 | yes | SPESP1 | chr15:67009622-67009949 | yes | sperm equatorial segment protein 1 | development (GO:0007275) |
| Pst44 | chr2:72650905-72651173 | no | SP3 | chr2:174366228-174366453 | yes | Sp3 transcription fuctor | DNA binding (GO:0003677) |
| Pst49 | chr7:28669202-28669492 | no | PRKCZ | chr1:1971083-1971328 | yes | protein kinase C, zeta isoform 1 | anti-apoptosis(GO:0006916) |
| Pvu16 | chr1:167973707-167974203 | no | APLP2 | chr11:129407978-129408295 | yes | amyloid beta (A4) precursor-like protein 2 | binding(GO:0005488) |
| Pvu35 | chr4:148294717-148294921 | no | CASZ1 | chr1:10656183-10656406 | yes | castor homolog 1, zinc finger isoform b | DNA binding (GO:0003677) |
| Pvu42 | chr8:124860388-124860558 | yes | ZFPM1 | chr16:87127203-87127507 | yes | zinc finger protein, multitype 1 | DNA binding (GO:0003677) |
| Pvu5 | chr6:48570188-48570357 | yes | ZNF755 | chr7:149725356-149725564 | yes | zinc finger protein 775 | DNA binding (GO:0003677) |
| Pvu57 | chr14:120788365-120788475 | no | MBNL2 | chr13:96797225-96797468 | yes | muscleblind-like 2 | nucleic acid binding(GO:0003676) |
| Pvu74 | chr2:165006494-165006766 | no | CDH22 | chr20:44313358-44313635 | yes | cadherin 22 precursor | calcium ion binding(GO:0005509) |
| Pvu75 | chr1:156572334-156572941 | no | CACNA1E | chr1:179719485-179719749 | yes | calcium channel, voltage-dependent, R type | calcium ion binding(GO:0005509) |
| Pst10 | chr17:23745919-23746668 | yes | ZNF206 | chr16:3080152-3080446 | yes | ZNF206 protein (Fragment) | metal ion binding (GO:0046872) |
Figure 3.
Average percent methylation level and standard deviation are indicated as bar graphs for the a. TRIM38, b. MBNL2, c. SPESP1, d. PHLDB3, e. CASZ1 and f. CDH22. Three individuals ID numbers (102, 108 and 114) and tissue samples (K: kidney, T: testis, C: colon, H: heart, B: brain, L: liver and M: muscle) are indicated in each bar.
Testis-specific difference in methylation
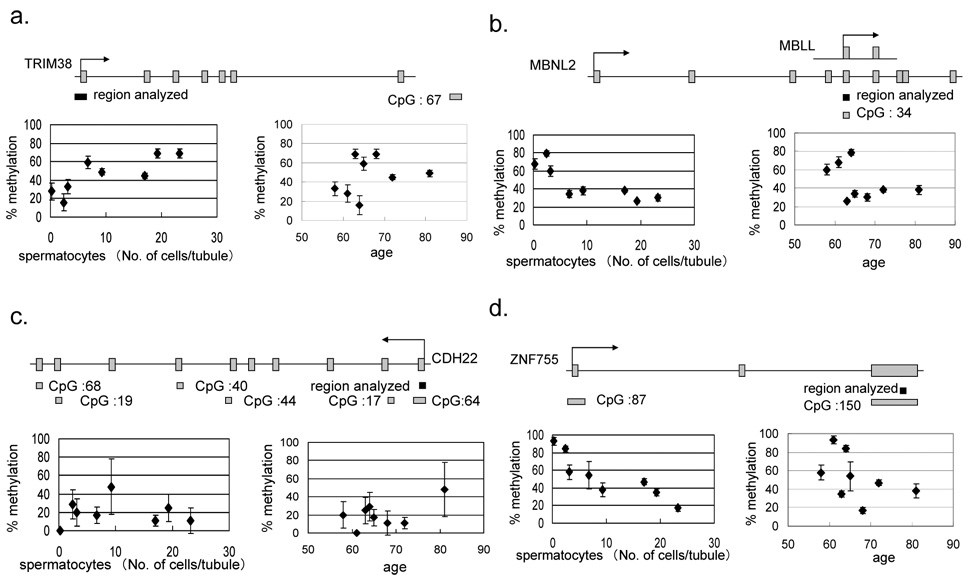
The testes are the unique organ that produces sperm, the male reproductive cell. The testes include various levels of cells from spermatocytes to sperm as well as the surrounding mesenchymal cell population. A sample set from eight testes consists of three healthy sperm-producing testes, two intermediate and three non-sperm producing testes from people of various ages (Supplementary Figure 1-b and Table 2). Sequenom Massarray data from the testes sample-set of each human orthologous region was performed, and the result was summarized in Supplementary Table 2. Ten out of 13 regions analyzed have shown at least one significant difference in the average percentage of DNA methylation among eight testes DNA samples. Seven of 10 showed multiple significant differences. We plotted scatter-graphs (Figure 4) for percent average methylation, comparing the number of spermatocytes and the age of the individual in order to analyze the linear correlation or inverse correlation at each genomic region. An inverse correlation between methylation and the number of spermatocytes was identified in five out of eight tDMRs; one showed linear correlation, one showed a pattern of two distinct groups and another was methylated in all testes tissues tested. There were no significant correlation between methylation and age in this study.
Table 2.
| case No. | age | spermatocyte/tubule | SD |
|---|---|---|---|
| 102 | 68 | 23.20 | 8.92 |
| 114 | 63 | 19.30 | 9.33 |
| 108 | 72 | 17.00 | 8.76 |
| 128 | 81 | 9.23 | 5.96 |
| 126 | 65 | 6.73 | 3.39 |
| 106 | 58 | 3.13 | 2.42 |
| 127 | 64 | 2.43 | 3.19 |
| 101 | 61 | 0.27 | 0.64 |
Figure 4.
Genomic structure of a. TRIM38, b. MBNL2, c. CDH22 and d. NF755. The area indicated by a black rectangle is the region examined as a T-DMR. The left scatter-graph shows promoter DNA methylation and spermatocytes. The right scatter-graph shows promoter DNA methylation and age. Eight human autopsy cases were analyzed and their number of spermatocytes per seminiferous tubule was examined by the pathologist.
Various characteristics of differentially methylated regions
The tripartite motif-containing 38 (TRIM38) gene belongs to the TRIM family, which is implicated in a variety of processes, such as development and cell growth [20]. The TRIM family is composed of three zinc-binding domains, a RING (R), a B-box type 1 (B1) and a B-box type 2 (B2), followed by a coiled-coil (CC) region [21]. TRIM38 is characterized by the presence of a C-terminal SPRY domain, which is a group of the TRIM family absent in invertebrates [22]. The 5’ non-CpG promoter of TMRI38 was subjected to quantitative DNA methylation analysis and showed testis, brain and heart specific hyper methylation and hypomethylation in the kidney, colon, liver and muscle. Expression is mainly in hematopoietic cells; otherwise, it shows ubiquitously low expression (http://symatlas.gnf.org/SymAtlas/Geneatlas) [19]. The tDMR for muscle blind-like 2 (MBNL2) is located in the middle of the gene flanking 5’ exon-intone junction region, where a CpG:34 is harbored and starts alternative transcript MBLL. The CpG:34 was hypomethylated in testis tissues from three independent individuals (figure 3b and figure 4b). Figure 4b showed no correlation between the number of spermatocytes and methylation, but all three individuals with no sperm production had hypermethylation and the rest of the five individuals showed a similar level of hypomethylation. It is speculated that the tDMR for MBNL2 might control that small transcript demethylating the CpG island only in the cell populations of testis tissue, such as cells related to spermatogenesis. MBNL2 is RNA-binding protein that binds to a 5′ACACCC-3′ core sequence and co-localizes with nuclear foci of expanded-repeat transcripts in myotonic dystrophy muscle cells, presumably sequestering the target RNAs [23], but the function of the short alternative transcript is not known. The tDMR for sperm equatorial segment protein 1(SPESP1) is located in the 5’ CpG islands promoter. The region was highly methylated in the liver, had low methylation in the testis and was intermediate in the other tissues (Figure 3c). SPESP demethylation also showed a significant correlation with the number of spermatocytes (Supplemental Table 2), but no testis-specific expression was reported in the database. The tDMR for Pleckstrin homology-like domain family B member 3 (PHLDB3) is located within the non-CpG 3’ last third exon. The PHLDB3 gene showed high expression exclusively in testis but its function is not known. We found the region was highly methylated, most of the tissues were examined and around 25% reduction was seen in all three testis samples (Figure 3d), although the mouse sequenome analysis did not agree this reduction. The reduction is well correlated with an increase in the number of spermatocytes. Interestingly, only in the liver of Case 108 was almost 50% DNA methylation seen. The similarity of the histological findings from livers was seen in Supplemental Figure 1, and a substantial portion of the cell population was hepatocellular cells. Therefore, this 50% methylation may be an individual variation of DNA methylation. We also investigate any polymorphisms of this genomic region in this individual. There were no polymorphisms distinguish paternal and maternal alleles to investigate allele specific methylation. The CASZ1 (castor zinc finger 1) gene is located on 1p36.2, where frequent chromosomal losses are detected, and there is decreased expression in neuroblastomas [24]. tDMRs for CASZ1 were detected in humans. A 30–40% reduction in methylation was observed in the heart, colon and kidney from Case 102 (Figure 3e). This is another example of complete methylation in the testes but partial methylation in other tissue types, such as the tDMR for TRIM38. The tDMR for the CDH22 gene, which encodes cadherin-like 22 protein, showed a different methylation level among samples examined without any tissue specificity. Patchy methylation changes were also seen in their epigrams, reflecting the high standard deviations shown in Figure 3f. Although it is not significant (p=0.1394), there is a tendency toward a linear correlation between age and the DNA methylation level in testes tissues examined. Age-related DNA methylation may affect the scattered CpG methylation pattern at this region and may affect late-onset neural diseases, since the CDH22 gene may have a role in the morphological organization of pituitary glands and brain tissues. A recent publication suggested age-specific epigenetic drift was likely to be a substantial mechanism predisposing individuals to late-onset Alzheimer’s disease [25]. Further analysis is necessary to confirm this important hypothesis of the existence of age-specific differentially methylated regions for the epigenetic drift. The tDMR for ZNF755 is located at the 3’, the last exon with a CpG island. The region showed low methylation only in DNAs from testis tissues and significant inverse correlation between the number of spermatocytes and DNA methylation in eight testis samples (Figure 4-d). Testis-specific hypomethylation was one of the most popular DNA methylation changes identified through RLGS analysis. This is because this pattern of tDMR shows almost complete demethylation in testes but was highly methylated in most of the other somatic tissues. Including four previous published testis-specific DMRs (DDX4, DACT1, ZNF324 and USP49) and five in this publication (PHLDB3, SPESP1, ZFPM1, ZNF755 and MBNL2), nine tDMRs so far are in this pattern. In the mouse study, eight testis-specific DMRs were examined in neonatals to adult development, and a gradual progression of loss of methylation from almost fully methylated in neonatal testis to almost completely unmethylated in 20-day-old adult testes was found (Song et al, submitted). Oakes et al found both de novo methylation and demethylation in spermatogonia and spermatocytes in the early meioic prophase [26]. Recently, a majority of described germ-line-specific genes were silenced and showed hypermethylation in mature sperm, associated with testis-specific histone variants, such as HIST1H2BA and HIST1H1T [5]. Demethylation at the testis-specific DMRs also shared the above mentioned similarities and may also have a common mechanism for demethylation in mammalian testis tissues. Further analyses are necessary to answer this question.
Interestingly, the patterns of percent DNA methylation between TRIM38 and SPESP1 showed the opposite direction. The same thing was seen between PHLDB3 and CASZ1 (Figure 3 and Supplemental Table 2). Distinct cell populations of each tissue may behave in the opposite DNA methylation manner at those regions.
In summary we have identified 15 tDMRs in humans from the quantitative methylation study of orthologous regions of 28 mouse tDMRs including our previous publication [14]. Our results indicate dynamic changes in DNA methylation that may involve both methylation and de-methylation in humans. In addition, there may be many changes in methylation that occur among the human population. Additional studies will be necessary to determine whether DNA methylation influences normal or disease functional phenotypes, or maybe an age-related phenotype of the human cell population in each tissue type.
Supplementary Material
Figure S1. Pathological HE sections of tissues (colon, kidney, brain, testes, heart and liver × 100) from autopsy cases.
Figure S2. Six pathological HE sections of tissues (× 400) from autopsy cases are shown.
Acknowledgements
The authors wish to thank Yukari Obana and Kunio Goto for their excellent technical assistance. This work is supported by a Nihon University Research Grant (2006–2007), an academic frontier project in 2006 (to H.N.) and National Cancer Institute Grant CA102423 (to W.A.H. and H.N.).
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 3.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–37. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 4.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat.Genet. 2003;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 5.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, Schubeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat.Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 6.Song F, Smith JF, Kimura MT, Morrow AD, Matsuyama T, Nagase H, Held WA. Association of tissue-specific differentially methylated regions (TDMs) with differential gene expression. Proc.Natl.Acad.Sci.U.S.A. 2005;102:3336–3341. doi: 10.1073/pnas.0408436102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eckhardt F, Lewin J, Cortese R, Rakyan VK, Attwood J, Burger M, Burton J, Cox TV, Davies R, Down TA, Haefliger C, Horton R, Howe K, Jackson DK, Kunde J, Koenig C, Liddle J, Niblett D, Otto T, Pettett R, Seemann S, Thompson C, West T, Rogers J, Olek A, Berlin K, Beck S. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat.Genet. 2006;38:1378–1385. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schilling E, Rehli M. Global, comparative analysis of tissue-specific promoter CpG methylation. Genomics. 2007;90:314–323. doi: 10.1016/j.ygeno.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 10.Ushijima T, Okochi-Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci. 2005;96:206–211. doi: 10.1111/j.1349-7006.2005.00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 12.Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat.Rev.Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- 13.Laird PW. Cancer epigenetics. Hum.Mol.Genet. 2005;14:R65–R76. doi: 10.1093/hmg/ddi113. Spec No 1. [DOI] [PubMed] [Google Scholar]
- 14.Kitamura E, Igarashi J, Morohashi A, Hida N, Oinuma T, Nemoto N, Song F, Ghosh S, Held WA, Yoshida-Noro C, Nagase H. Analysis of tissue-specific differentially methylated regions (TDMs) in humans. Genomics. 2007;89:326–337. doi: 10.1016/j.ygeno.2006.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 16.Ehrich M, Nelson MR, Stanssens P, Zabeau M, Liloglou T, Xinarianos G, Cantor CR, Field JK, van den BD. Quantitative high-throughput analysis of DNA methylation patterns by base-specific cleavage and mass spectrometry. Proc.Natl.Acad.Sci.U.S.A. 2005;102:15785–15790. doi: 10.1073/pnas.0507816102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–1431. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 18.Coolen MW, Statham AL, Gardiner-Garden M, Clark SJ. Genomic profiling of CpG methylation and allelic specificity using quantitative high-throughput mass spectrometry: critical evaluation and improvements. Nucleic Acids Res. 2007;35:e119. doi: 10.1093/nar/gkm662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc.Natl.Acad.Sci.U.S.A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reymond A, Meroni G, Fantozzi A, Merla G, Cairo S, Luzi L, Riganelli D, Zanaria E, Messali S, Cainarca S, Guffanti A, Minucci S, Pelicci PG, Ballabio A. The tripartite motif family identifies cell compartments. EMBO J. 2001;20:2140–2151. doi: 10.1093/emboj/20.9.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borden KL. RING fingers and B-boxes: zinc-binding protein-protein interaction domains. Biochem.Cell Biol. 1998;76:351–358. doi: 10.1139/bcb-76-2-3-351. [DOI] [PubMed] [Google Scholar]
- 22.Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC.Evol.Biol. 2008;8:225. doi: 10.1186/1471-2148-8-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum.Mol.Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 24.Fransson S, Martinsson T, Ejeskar K. Neuroblastoma tumors with favorable and unfavorable outcomes: Significant differences in mRNA expression of genes mapped at 1p36.2 Genes Chromosomes. Cancer. 2007;46:45–52. doi: 10.1002/gcc.20387. [DOI] [PubMed] [Google Scholar]
- 25.Wang S-C, Oelze B, Schumacher A. Age-Specific Epigenetic Drift in Late-Onset Alzheimer's Disease. Anonymous. 2008. p. e2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oakes CC, La SS, Smiraglia DJ, Robaire B, Trasler JM. A unique configuration of genome-wide DNA methylation patterns in the testis. Proc.Natl.Acad.Sci.U.S.A. 2007;104:228–233. doi: 10.1073/pnas.0607521104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Pathological HE sections of tissues (colon, kidney, brain, testes, heart and liver × 100) from autopsy cases.
Figure S2. Six pathological HE sections of tissues (× 400) from autopsy cases are shown.