Abstract
How transcription of individual genes is regulated in a single, intact, three-dimensionally organized cell nucleus remains mysterious. Recently, live cell imaging has become an essential tool to dissect the in vivo mechanisms of gene transcription. It not only examines functions of transcription factors at their gene targets within the chromatin context, but it also provides a non-disruptive approach for observing the dynamics of a transcription cycle in real time. However, the identification of any endogenous gene loci and their associated transcription factors remains technically difficult. Here, we describe the method of imaging the transcriptional dynamics of heat shock genes in Drosophila polytene chromosomes in living salivary gland tissues by multiphoton microscopy (MPM) imaging. This method has provided the experimental capability to visualize the assembly and dynamics of individual transcription factors and regulators and to dissect their functions at their endogenous gene targets in living cells.
Keywords: transcription, genes, polytene, chromosomes, chromatin, multiphoton microscopy, DNA, genome, crosslinking, immunostaining
1. Introduction
Examining the spatial and temporal distribution of transcription factors in their target genes is essential for studying transcription regulatory mechanisms in vivo. By protein-DNA crosslinking and chromatin immunoprecipitation (ChIP), one can track the rate of recruitment and movement of a particular transcription factor on a gene [1]. This ChIP assay can also be performed on a genome-wide scale by probing microarrays [2]. In Drosophila, a complementary assay exists: by immunostaining Drosophila polytene chromosome with antibodies against specific proteins, one can identify the transcription factors recruited to DNA both globally (to whole genome) and locally (to a single gene loci) [3].
These methods—protein-DNA crosslinking and polytene chromosome immunostaining—are powerful tools but have common limitations. First, transcription is a physiological process occurring in a three-dimensionally organized cell nucleus; therefore, a complete mechanistic understanding cannot be achieved without studying transcription in the context of native chromatin architecture and native nuclear organization. However, in both of these assays, intact cell nuclei are eventually disrupted and important information is lost. Second, protein-DNA interaction is considered to be highly dynamic in vivo, and requires studies with high time-resolution in individual, living cells. Yet, current methods involve either ensemble averages over a large number of cells, or time averages during sample fixation/crosslinking procedures, or both. A method that is capable of monitoring transcription factor dynamics in real time would be highly valuable. Third, transcription factors interact with each other to execute their roles. However, current immunoprecipitation methods cannot examine direct interactions among transcription factors at individual native genes. All of the above limitations can be overcome by developing a strategy to visualize transcription at specific native loci in living cells.
There are two major challenges to imaging transcription regulation in living diploid cells: 1) the identification of individual native gene loci and 2) the detection of particular transcription factors associated with those genes. To overcome these challenges, imaging must be capable of resolving individual gene loci beyond the diffraction limit (~200 nm) and—more importantly—highlighting a small fraction of transcription factors engaged in gene transcription within a majority of the diffusing molecules that constitute the image background. A further complication is that in many cases these two populations of molecules are under constant exchange at a time scale of seconds to minutes. Therefore, current “super-resolution imaging” methods [4–6] are unlikely to be instrumental in studying the transcription dynamics in the cell nucleus.
Recently, the polytene cells in Drosophila larval salivary glands have proved to be a very good system for live cell imaging of transcription dynamics. During Drosophila larvae development, the interphase chromosomes of salivary gland cells undergo endoreduplication, i.e., DNA replication (approximately 10 rounds) without cell division, resulting in large nuclei with giant polytene chromosome (~1024 DNA copies juxtaposed). Individual gene loci are linearly arrayed on polytene chromosomes and can be distinguished as unique band patterns resolvable under the diffraction-limited optical resolution. The band patterns were categorized and named by Bridges [7] (Figure 1). Transcription at individual euchromatic gene loci leads to the natural local enrichment of transcription factors, which allows one to visualize these chromatin-bound proteins “in action” at these gene loci [8, 9]. In these papers, we described the transcription activation of Drosophila Hsp70 genes. Nevertheless, this method is applicable to other developmental gene loci if the loci can be unambiguously recognized in the polytene nucleus.
Figure 1.
An image of Drosophila polytene chromosomes spread onto a glass slide and stained by Hoechst33342. Salivary glands are dissected, fixed, spread and stained following the described protocol [3]. Each chromosome arm is approximately 150 µm in length and 2 µm in width.
2. Production of transgenic fly lines expressing GFP-tagged proteins in salivary gland
2.1 Protein-tagging strategy
We have assembled a collection of fluorescently labeled transcription factors previously shown to function in Hsp70 gene activation. In order to produce the fly lines that properly express the GFP-tagged transcription factors of interest, we have searched available yeast and Drosophila databases (http://yeastgfp.ucsf.edu/ and http://flybase.bio.indiana.edu/) as well as the published literature to determine 1) whether proteins homologous to those transcription factors in the above list have been successfully tagged in yeast and/or other organisms; 2) if so, at C-terminus or at N-terminus; and 3) whether such tagged proteins are functional in vitro or in vivo. We have designed the tagging scheme individually for each protein following the previous successes found in database and in literature search results.
2.2 DNA construct
We employ the Gateway Cloning System (Invitrogen Inc.) [10] as the primary tool for generating GFP-fusion constructs. We have obtained the Gateway P-element insertion vectors (http://www.ciwemb.edu/labs/murphy/Gatewayvectors.html) containing 1) sequence of EGFP (or ECFP, Venus, mRFP) at 5’ or 3’ of the cloned protein cDNA with an upstream UASGAL promoter (explained in 2.4); 2) P-element transposon sequences; 3) white+ gene as the screening marker. The cDNAs of individual factors of interest were cloned into an “entry” vector and transferred into the Gateway P-element insertion vectors with a simple recombination reaction. We have verified vector sequences before sending purified DNA (~50 µg) to transgenic services (Genetic Services Inc., Cambridge, MA; BestGene Inc., Chino Hills, CA; Rainbow Transgenic Flies, Inc., Newbury Park, CA).
2.3 P-element transformation and subsequent genetic crosses
Transgenic lines were generated by microinjecting the above DNA constructs into the embryos of w1118 fly line which contains the recessive mutation of white gene and shows the phenotype of white eyes (white+ flies have red eyes.). Germline transformants of the injected gene were identified by the red eye phenotype. (Note: Because of the various insertion copies and expression levels of the white+ transgene, the eye color can vary from very pale yellow to dark red, and all should be carried forward as genuine transformants.) Injection of 200 embryos usually yields ~10 transformant lines. Longer cDNAs usually yield fewer transformants and may require injecting more embryos.
We have performed genetic crosses to allocate the chromosome(s) of transgene insertion and to make homozygous alleles of transgenes, although transgenic services can also perform them. Fly transformants were individually crossed to a fly line with balancer chromosomes, which 1) contain multiple inversions of chromosomal segments and suppress homologous recombination; 2) are associated with a dominant phenotype that can be observed and screened; and 3) are homozygous lethal. Therefore, the transgene is preserved over balancer chromosomes and can be made homozygous by selecting against the dominant balancer marker. Transgenes are usually allocated to the X, 2nd or 3rd chromosomes (the 4th chromosome is very small and is very rarely targeted). We have found that about half of the homozygous lines contain a single insertion transgene and can be stably maintained.
2.4 Express the GFP-tagged proteins by Gal4-UAS system
The Gal4-UAS expression system has been the most widely used approach to overexpress proteins of interest in Drosophila [11] (Figure 2A). In this system, GFP-fusion proteins are controlled by UASGAL promoters, which need binding of Gal4 transcription activators in order to be expressed. Gal4 is a yeast transcription activator and is not present as an endogenous gene in Drosophila [12]. The Gal4 enhancer trap lines have Gal4 expressed at particular tissues, which are available from Bloomington Stock and many Drosophila research labs. We have used Gal4 lines (“driver lines”) expressed in larval salivary glands to express the target proteins in polytene cells. For example, Sgs3-Gal4 is expressed under the Sgs3 promoter, which is activated during early third instar larvae [13], and is useful for overexpressing some proteins which might interfere with salivary gland development. Gal4C729 and Gal4C147 lines are expressed before the third instar developmental stage [14] and are useful for expressing some proteins that belong to a multi-component complex. The resulting expressed protein usually shows correct localizations in the cell, and more specifically localizes to the correct chromosome loci [9]. Another useful driver line is the forkhead promoter driven Gal4, which is expressed early in salivary gland development.
Figure 2.
Generating transgenic Drosophila stains expressing GFP-tagged transcription factors. (A) Schematic of the Gal/UAS gene expression system (Brand et al, 1993). (B) Cross scheme to generate fly strains containing Gal4 and UAS-GFP alleles recombined on the same chromosome.
Because more sophisticated applications are being developed, three or more alleles may need to be incorporated into the same fly line. These applications include the simultaneous imaging of multiple transcription factors, the study of protein-protein interaction by Fluorescence Resonance Energy Transfer (FRET) (Förster, 1948; Pollack and Heim, 1999), the imaging of GFP protein after the knock-down of endogenous proteins by RNAi, or the genetic complementation of the GFP-tagged proteins using null alleles or temperature-sensitive alleles. Therefore, it is advantageous to have Gal4 and UAS alleles recombined on the same chromosome to accommodate additional insertions on other chromosomes. We have developed a quick strategy to recombine Gal4 and UAS alleles by sorting living third instar larvae with fluorescent salivary glands (Figure 2B). In short, female heterozygous flies containing Gal4 and UAS alleles were crossed to the male w*; CUX (w*; T(2;3)apXa, apXa/CyO; TM6) flies. Gal4 and UAS alleles recombine during meiosis in females, and a fraction of F1 progenies express fluorescent proteins in the larval salivary gland. These larvae are individually picked under a fluorescence dissection scope and the adults are further screened for those that carry the appropriate balancer chromosome. Note that in Drosophila, meiotic recombination occurs much more frequently in females than in males.
2.5 Functionality test
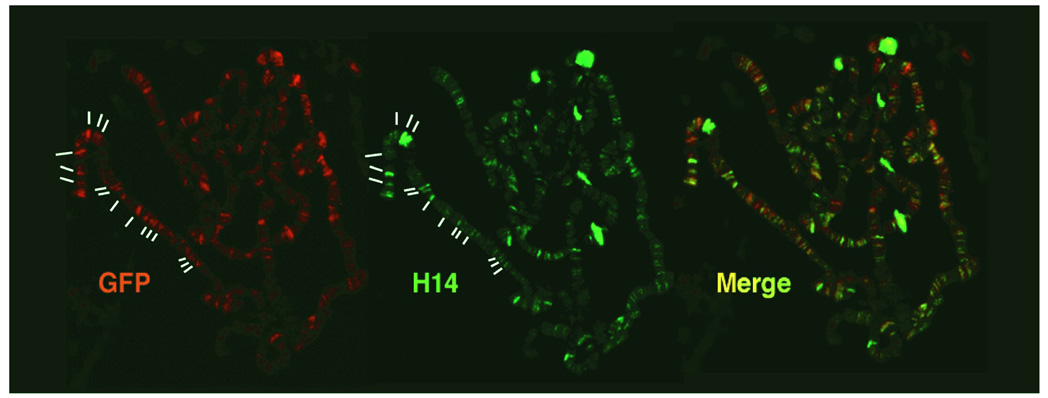
Before performing live-cell imaging experiments, it is critical to assess the functionality of the fluorescently tagged proteins. We have used two such tests. First, we have examined the localization of GFP-tagged proteins and their endogenous partners on polytene chromosomes using corresponding antibodies. For example, we have shown that in the polytene chromosome expressing Rpb3-GFP, the fluorescent staining patterns from anti-GFP and from the antibody against the C-terminal domain of Rpb1 (the largest subunit of RNA Polymerase II) extensively colocalize over 200 chromosomal loci across the polytene chromosomes. This result is highly indicative of the functionality of the Rpb3-GFP fusion protein (Figure 3A). Second, we have performed genetic complementation experiments to test whether the GFP-tagged factors can substitute for the dysfunctional endogenous proteins in vivo. We have examined the functionality of HSF-EGFP using the fly line harboring the temperature-sensitive allele hsf4. The hsf4 allele contains point mutations in the DNA-binding domain of HSF [15]. At heat shock temperature, hsf4 protein is unable to bind DNA and cannot induce Hsp70 transcription activation and heat shock puff formation at 87A&C loci. We have introduced HSF-EGFP and Sgs3-Gal4 alleles into hsf4 homozygous background flies and found that HSF-EGFP restores the heat shock puff formation and Pol II recruitment at 87A&C loci upon HS (Figure 3B).
Figure 3.
Testing the functionality of GFP tagged transcription factors. (A) Immunostaining the GFP-tagged Rpb3 subunit of Pol II with anti-GFP and the largest subunit of Pol II, Rpb1, with antibody H14 (Ser5 phosphorylated C-terminal domain of Rpb1). (B) Genetic complementation for HSF-EGFP. Expressing HSF-EGFP in hsf4 mutant flies can restore the activation of Hsp70 genes, as seen by Pol II recruitment to Hsp70 loci on polytene chromosomes (white arrows).
3. Methods of imaging the transcriptional dynamics of Hsp70 genes in living tissues
3.1 Tissue preparation and culture for MPM imaging
As Drosophila larvae undergo development, they first wander along the culture tube wall, then stop, evert their anterior spiracles, and start puparization. During this time, larval salivary glands grow, secrete glue proteins to help larva and pupa attach to the wall, and eventually undergo cell death upon pupation. Ecdysone-induced transcription activation of a group of genes leads to chromosome puffs at 74EF site at late puff stages [16], which also appear to be a doublet structure similar to heat shock puffs.
We generally dissect salivary glands from wandering third instar larvae and culture them in diluted Grace’s insect medium diluted with sterile water at a volume ratio of 5:1 [17]. A dissected salivary gland is shown in Figure 4A and 4B. Dissected salivary glands examined using live-dead cell stain kit (Molecular Probes) were found to remain viable and heat shock inducible for at least 2 hours (Figure 4C).
Figure 4.
Drosophila salivary glands. (A) Cultured in a Petri dish. (B) An image using transmission light. (C) A salivary gland treated with Live-Dead stain (Molecular Probes) after cultured for 2 hours. (D, E) Optical sections of polytene chromosomes in live salivary glands stained with Hoechst33342. The z-distance is 0.5 µm. Labels identify specific bands on chromosome arm 2L (red) and 3L (blue) are noted. (F, G) Three-dimensional reconstructions of a polytene nucleus. Red and blue arrows indicate the centromeric region (chromocenter) and telomere, respectively. Scale bars 5 µm. Panels D to G are adapted from [9] with permission.
The dissected salivary gland tissues are transferred with media to an FCS2 or FCS3 chamber (Bioptechs) and then examined with transmission light where tissues damaged during dissection are identified and excluded from imaging. When salivary glands are maintained at room temperature, the genes involved in salivary gland development are expressed, but the Hsp70 genes are not activated. A heat shock of the tissue at 36.5°C leads to a turndown in the developmental gene expression and activation of Hsp70 gene transcription.
Polytene cells within one salivary gland are usually identical in shape and show the same level of polytene endoreduplication. In a healthy salivary gland under normal development, a polytene nucleus closest to the coverslip surface is usually at least 30 µm deep in the tissue. Polytene nuclei in the salivary glands can be identified under differential interference contrast (DIC) or transmission light microscopy. They are of spherical shape with diameters ~25–35 µm. Salivary glands dissected from “late” developmental stages (usually dissected from non-wandering (“stopped”) larvae or from prepupae), however, show morphology distinct from those at the early stages: the cell layers are separated from the center of glands, the glands are filled with glue and appear more “transparent” under transmission light compared to those at early developmental stages. Those “late” salivary glands are not the preferred tissue for imaging heat shock gene activation, because 1) the salivary glands at that stage are fragile and difficult to handle; 2) the ecdysone puffs at 74EF are induced in these glands and appear as a doublet that may complicate the identification of heat shock puffs at 87A & 87C.
Expression of GFP-tagged transcription factors by the Gal4/UAS system [12] leads to similar expression level across individual polytene cells in a salivary gland, which is advantageous compared to the significant variation of expression levels among individual transfected mammalian cells. Therefore, choosing cells within the same gland for MPM imaging provides the experimental convenience. In addition, many of the GFP-tagged transcription factors are nuclear-localized or show predominant nuclear-localization (See Figure 5 for an example).
Figure 5.
HSF expression and localization in salivary glands before HS (Upper) and after HS (lower). Shown are the two-photon optical sections of polytene nuclei expressing HSF–EGFP (green) and stained with Hoechst33342 (red), and the three-dimensional reconstructions of the optical sections (rightmost). Adapted from [9] with permission. Scale bars 10 µm.
3.2 Apparatus of imaging polytene nucleus in salivary gland tissue
The structure and function of Drosophila polytene chromosomes have been studied microscopically using various techniques, from optical microscopy to electron microscopy [7, 18]. Dr. Sedat and colleagues have imaged polytene nuclei and their chromosome organization in intact salivary glands by developing widefield deconvolution microscopy [19–22]. However, individual gene loci can be difficult to identify with the image quality provided by widefield deconvolution methods, and studying the dynamics of transcription regulation at individual genes in polytene nucleus is best accomplished with optical sectioning techniques that have high effective resolution in relatively thick tissues.
The most widely used optical sectioning microscopy is confocal laser scanning microscopy [23]. As a variation of confocal laser scanning system, a spinning disk scanning system provides a higher scanning rate at the sacrifice of a loss in penetration [23]. We have tested a variety of widefield deconvolution and laser scanning microscopy systems in imaging polytene nuclei in salivary gland tissue. Images with satisfactory quality have been achieved with confocal scanning units (i.e., Zeiss LSM 510) and with spinning disk scanning on some salivary gland tissues expressing fluorescent proteins, in particular the red fluorescence proteins such as mRFP. The confocal and spinning disc laser scanning systems are commercially available and user-friendly. However, the specific DNA banding pattern of polytene chromosomes as stained by Hoechst dyes is less well imaged in these systems. In addition, photobleaching and photodamage become concerns in confocal LSM when fluorescent proteins are at low concentrations.
Two-photon laser scanning microscopy (TPLSM), usually denoted Multiphoton Microscopy (MPM) has become widely used in biological and biomedical research [24, 25]. Two-photon excitation occurs only at the very proximal region to the focal point [24]. Therefore, the effective “focal volume” is intrinsically confined in both lateral and axial axes, eliminating the autofluorescence background and photobleaching and photodamage from out-of-focus planes [25, 26]. Infrared [27] laser used in TPLSM has deeper tissue penetration (as deep as 0.5 mm) and less scattering [25, 26, 28]. In TPLSM, no pinhole is needed to reject out-of-focus fluorescence as in confocal LSM, which greatly enhances the image signal. Therefore, TPLSM provides an ideal approach to image GFP-tagged transcription factor dynamics in polytene nuclei within Drosophila salivary glands.
A representative set-up for imaging Drosophila salivary gland by TPLSM is illustrated in Figure 6. A Ti:Sapphire laser (80 MHz repetition rate, ~100 fs pulse) is used to excite the tissue sample through a Bio-Rad MRC-1024 scanner. The frame rate/time resolution is about one second per frame (512 × 512 pixels, depending on image size and scan speed). Fluorescence is detected by two bi-alkali PMTs for blue and green fluorescence. In a separate imaging system that is also frequently used, two GaAsP PMTs (H7421-20, Hamamatsu Inc.) are used as detectors. Their high detection quantum efficiency (42% at peak wavelength) is particularly beneficial for imaging red fluorophores [25]. 3-D image series are usually generated by z-series scan of a selected x-y area. Two-color or three-color images can be acquired simultaneously.
Figure 6.
The imaging apparatus. PMT: Photomultiplier tubes.
We use an FCS2 or FCS3 incubation chamber (Bioptechs Inc., shown in Figure 6 as an inverted microscope setup) to control temperature; however, the chamber also allows the perfusion of fresh media or any water-soluble hormones/drugs/inhibitors for treatment of the tissue, as has been demonstrated by inhibiting the transcription elongation factor P-TEFb by perfusion of culture medium containing P-TEFb inhibitor Flavopiridol [29]. It takes about 1 minute for chamber temperature to rise from 24 °C to 36.5 °C, and an objective temperature controller (Bio ptechs) is used to prevent a heat-sink effect. In addition, by applying a preheated objective to contact the coverslip attached to salivary gland tissues, we can instantaneously heat shock these tissues.
Criteria for selecting a good objective are the following: high numerical aperture (>1.2), long working distance (>200 µm), high transmission efficiency of infrared laser power, high fluorescence collection efficiency, minimal perturbation to the sample. We had initially used an oil immersion objective (F-FLUAR, 40X, NA 1.3, Zeiss) to circumvent the problem of water evaporation at 37 °C. Nevertheless, the advantage of a high-NA water-immersion objective is that: 1) It has a long working distance (> 200 µm), and 2) It alleviates the problem of refractive index mismatch between immersion medium and tissues. Indeed, we have obtained images with superior qualities with a water-immersion objective (C-Apochromat, 63X, NA 1.2, Zeiss) deeper into the tissue than with oil-immersion objectives (not shown), and the water-evaporation problem is not a significant issue during the experimental time frame, particularly at an upright microscope.
3.3 Image acquisition and pattern recognition
We use 900 nm as the excitation wavelength, which is near the peak of EGFP two-photon excitation cross-section spectra [25]. By using optimal excitation wavelength and optimizing fluorescence collection and detection efficiencies, we have minimized the excitation laser power delivered to the salivary gland tissue and have alleviated photobleaching and photodamage. The actual laser power, measured at the microscope objective, is usually 5–10 mW for multiphoton imaging.
Sample drifting in the x-y direction during the image time series up to one hour is negligible within the optical resolution and can be minimized by using a high-precision motorized microscope stage and a vibration-free optical table. Position drifting in the z-direction remains a problem during the time course of heat shock, in which the temperature increase causes a small movement between the coverslip and the culture chamber. This movement is often unidirectional and can be estimated in advance and precompensated through taking additional image sections. The sample drifting problem is minimized using a preheated objective.
Although the visualization of individual banding pattern by DNA stain or by GFP-tagged transcription factors is aesthetically satisfying, the recognition of these endogenous gene loci in a 3D-organized polytene nucleus is a significant challenge. Here, we describe the pattern recognition method in two steps. The first step is the “offline” recognition of chromosomal loci; i.e., analyses are performed after images have been taken, usually on another day or time. We have found it useful to recognize chromosome ends by their unique banding patterns [7], followed by tracking each chromosome from the telomere to the centromere in 3D. Nevertheless, we have had only limited success with this approach. Recently, we have studied the localization of GFP-tagged dosage compensation complex and recognized the entire X-chromosome and identified the endogenous loci in many cases (Yao et al, unpublished results). We have found that if one can unambiguously follow one chromosome in 3D, then it becomes practical to identify many gene loci on this chromosome unambiguously. In addition, if one wants to analyze a time series of transcription factor recruitment after induction, the whole 4D series can be taken and the image series can be analyzed offline.
The second step is the recognition of chromosomal loci “online” (i.e., in real time during imaging). If Fluorescence Recovery After Photobleaching (FRAP) (Axelrod, Koppel et al. 1976) is to be performed on a native gene locus, one needs to recognize the genes of interest in real time during imaging. We have found that the accumulation of RNA Polymerase II at heat shock loci at endogenous heat shock gene Hsp70 loci 87A and 87C gives a distinctive doublet pattern throughout a polytene nucleus and can be easily identified in real time during HS. This pattern can be confirmed to be Hsp70 gene loci by two criteria: 1) This doublet is not visible before HS and only appears after HS. 2) This doublet is distinctive and the only pair throughout the entire nuclear section series. We have used this method for studying Pol II dynamics in living cells [8]. Many other factors involved in transcription elongation behave similarly to Pol II, and we have been studying these factors by FRAP as well (Figure 7). However, for imaging other factors present in the cells at less abundance than in Pol II, or for other gene loci that are less highly transcribed than heat shock loci, additional definitive markers are needed. In diploid cells, the locations of transgenes have been identified by including multiple LacO repeats, the binding sequence of the LacI repressor. Upon expression of LacI-GFP, LacI will bind tightly to the LacO sequences, marking the site of interest [30]. This system has been used in Drosophila [31] and can be used to mark endogenous genes. With the recent development of the FC-31 site-specific recombination system in Drosophila [32], it is now possible to integrate the LacO repeats near a specific endogenous locus and visualize its location and dynamics in living tissues.
Figure 7.
Raw images of a salivary gland expressing HSF-EGFP. (Upper) Before heat shock. (Lower) After heat shock. Scale bars 10 µm.
3.4 Considerations for obtaining and interpreting FRAP curves in polytene chromosomes
FRAP uses the irreversible photobleaching of fluorophores to deplete a pool of molecules in a region of interest, and to study the molecular mobility by monitoring the recovery of fluorescence inside the bleached region. FRAP, initially called Fluorescence Photobleaching Recovery (FPR), measures diffusion coefficients of the molecules by rapidly photobleaching the fluorophores within a stationary laser beam with a defined beam shape and intensity profile, and monitors the time course of fluorescence intensity recovery with an attenuated beam. The theory of this method has been developed for one-photon excitation and two-dimensional diffusion [33, 34] and for two-photon excitation and three-dimensional diffusion [35].
Because of the wide use of GFP to tag proteins in vivo, FRAP has found many applications in cell biology as a versatile technique [36]. Rather than photobleaching a diffraction-limited volume, FRAP assay on a specific structure of interest inside living cells (such as an organelle, or a nuclear domain that is visible under an optical microscope) has been extensively used to measure the mobility and the exchange dynamics of protein molecules between various cellular compartments [37, 38]. FRAP routines can now be performed in most confocal LSM units (e.g., Zeiss LSM510 and Leica TCS SP2), which now allow multiple user-defined photobleaching shapes. Inside the cell nucleus, the exchange of transcription factors at the DNA template usually occurs within the timescale of seconds to minutes and is easy to detect using this scanning-based FRAP method. Due to the inability to resolve gene loci inside the diploid cell nucleus, many FRAP studies were performed on an arbitrary region, while the fluorescence recovery was interpreted as “genome-wide” rapid exchange of protein molecules [27]. On the other hand, individual gene loci are resolvable and highlighted in Drosophila polytene nuclei, and the dynamic exchange of transcription factors can be measured by FRAP at specific genes.
To perform FRAP with the MRC-600 scanner, we used a high-scan zoom (40 ×) to achieve bleaching followed by time series imaging (Figure 8A). We also used the standard bleaching procedures on the commercially available Zeiss LSM510 system and have found identical results on both systems. We have found that it is crucial to limit the bleaching depth for performing FRAP at endogenous gene loci in polytene nuclei for several reasons. First, deeper bleaching usually requires high power or more bleach repeats, which might yield unwanted bleaching of molecules surrounding the targeted volume and complicate further analysis. Second, high bleaching power always raises concerns of potential photodamage to the cell. Third, unbound diffusing transcription factors, although being partially excluded from gene loci in polytene chromosomes, remain as a significant fraction at the photobleached volume; fluorescence recovery from deeper bleaching introduces a “rapid recovery” phase, which likely reflects the rapid equilibration among these unbound diffusing molecules inside and outside of the targeted volume after photobleaching. This requires monitoring but needs to be distinguished from the bona fide protein-DNA interaction kinetics. This effect is particularly pronounced in the current experimental setup, because the time delay between photobleaching and acquisition of the first image is not negligible. Last, a non-uniform bleach profile makes the calculation of diffusion constant dependent on the actual bleach depth [33]. In practice, different bleach depths should be performed to test the reproducibility and computational model for fitting the recovery process (Figure 8B).
Figure 8.
FRAP of HSF-EGFP at endogenous gene loci in salivary glands. (A) Intensity images of various sections in a nucleus of a heat shocked salivary gland in pseudocolor. Scale bars 10 µm. (B) FRAP recovery curves.
FRAP on active transcription sites is practical to perform but difficult for quantitative interpretation because of: 1) the uncertainty of the completeness or the validity of the transcription kinetics model, and 2) the complex mathematical nature of FRAP recovery kinetics of diffusing molecules. Instead, we have taken an approach to dissect the kinetic steps by qualitatively defining these steps with FRAP and perturbing these steps with specific inhibitors and RNAi. For example, by treating the living salivary glands under HS with Flavopiridol, a specific inhibitor to P-TEFb, Pol II are unable to enter productive elongation, and leave a promoter-proximally engaged Pol II population, and we have demonstrated that this subpopulation is stably associated with Hsp70 genes [29].
4. Discussion and potential improvements
4.1 Additional approaches to tag proteins with GFPs in Drosophila
In addition to the Gal4/UAS approach to express GFP-fusion proteins, other methods have been developed to introduce GFPs of various fluorescence colors into endogenous genes with their native promoters. FlyTrap is a method using the P-element insertion into the introns of many native genes, with GFP being expressed as an additional exon [39]. The chimera proteins are expressed under their native promoters and are assumed to behave like native proteins because many of those tagged lines are not lethal and do not show defects in development. Therefore, the FlyTrap method may provide an alternative to the standard transgene approach. Many trap lines have been made and can be found at the FlyTrap database (http://flytrap.med.yale.edu/) as well as other labs [40, 41]. Nevertheless, the P-elements’ transposons have “hot-spots” for integration, and such “hot-spots” have almost been saturated by now. A different transposon, piggyBac, has less sequence specificity for insertion, and is now being developed to generate new fly trap lines [42]. Additionally, it is possible to make homologous recombination into endogenous genes in Drosophila [43, 44]. “Ends-in” and “ends-out” recombination approaches are being used for a number of genes, including the Hsp70 genes [45]. This approach could also be useful, especially for the long genes, which do not have cDNA available and may be difficult to clone.
4.2 GFP variants and their applications
The imaging of transcription dynamics in live cells will benefit from advances in developing novel genetically-encodable fluorophores. As new fluorescent proteins are created, they can be incorporated into the toolbox for dissecting transcription factor dynamics in living Drosophila tissues. Of particular interest is the development of photoactivatable and photoswitchable proteins [46, 47]. With photoactivatable GFP (paGFP) and photoswitchable GFPs it is possible to activate a subset of molecules in order to track the localization of the protein and measure the dynamics at which it leaves the locus. Photoswitchable GFPs emit fluorescence prior to activation; upon excitation at a short wavelength their emission spectra are shifted to different colors. This allows one to localize specific regions of interest before activating and allows for FRAP measurements as well as photoactivation measurements. Finally, another application that can be applied to GFP and its variants is FRET. With the battery of live cell imaging techniques, we hope to achieve a comprehensive understanding regarding the dynamics and specific interactions of transcription factors at specific genes in live cells.
5. Concluding remarks
Direct visualization of transcription while resolving individual components in real time will fundamentally address the mechanisms of eukaryotic transcription regulation. Achieving this goal has been hampered by the inability to resolve endogenous gene loci in diploid cells. Therefore, two-photon imaging of Drosophila polytene nuclei provides an attractive alternative to tackle this difficult problem by using naturally amplified chromosomes. We have demonstrated the generation, maintenance and imaging of transgenic Drosophila containing GFP-tagged proteins in larval salivary glands. For the future, we envision that the dynamic assembly of individual positive and negative transcription regulatory factors and RNA processing factors can be examined, and a chronological and integrative view of gene activation can be documented at individual native genes within living cells.
Acknowledgements
We thank K. Munson for generating the DNA constructs, and J. Werner for preparing spread polytene chromosomes and immunostaining, C. Fecko for experimental contributions and discussions, B. Ardehali for experimental contribution of RNAi fly generation. We thank Drs. M. Kuroda and M. Gelbart for collaborating on imaging Drosophila X chromosome in polytene cells. We thank M. Williams for editorial assistance. J.Y. thanks Dr. Robert Tjian for his support. This research was partly performed in the Developmental Resource for Biophysical Imaging Opto-Electronics and was supported by an NSF grant CHE-0242328 to W.W.W. and J.T.L., an NIH grant GM25232 to J.T.L., and an NIH-NIBIB grant 9 P41 EB001976-17 to W.W.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Boehm AK, Saunders A, Werner J, Lis JT. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, Volkert TL, Wilson CJ, Bell SP, Young RA. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 3.Schwartz BE, Werner JK, Lis JT. Methods Enzymol. 2004;376:393–404. doi: 10.1016/S0076-6879(03)76026-1. [DOI] [PubMed] [Google Scholar]
- 4.Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 5.Hess ST, Gould TJ, Gudheti MV, Maas SA, Mills KD, Zimmerberg J. Proc Natl Acad Sci U S A. 2007;104:17370–17375. doi: 10.1073/pnas.0708066104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willig KI, Kellner RR, Medda R, Hein B, Jakobs S, Hell SW. Nat Methods. 2006;3:721–723. doi: 10.1038/nmeth922. [DOI] [PubMed] [Google Scholar]
- 7.Bridges CB. J. Hered. 1935;26:60–64. [Google Scholar]
- 8.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Mol Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 9.Yao J, Munson KM, Webb WW, Lis JT. Nature. 2006;442:1050–1053. doi: 10.1038/nature05025. [DOI] [PubMed] [Google Scholar]
- 10.Hartley JL, Temple GF, Brasch MA. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand AH, Perrimon N. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 12.Duffy JB. Genesis. 2002;34:1–15. doi: 10.1002/gene.10150. [DOI] [PubMed] [Google Scholar]
- 13.Cherbas L, Hu X, Zhimulev I, Belyaeva E, Cherbas P. Development. 2003;130:271–284. doi: 10.1242/dev.00205. [DOI] [PubMed] [Google Scholar]
- 14.Hrdlicka L, Gibson M, Kiger A, Micchelli C, Schober M, Schock F, Perrimon N. Genesis. 2002;34:51–57. doi: 10.1002/gene.10125. [DOI] [PubMed] [Google Scholar]
- 15.Jedlicka P, Mortin MA, Wu C. Embo J. 1997;16:2452–2462. doi: 10.1093/emboj/16.9.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andres AJ, Thummel CS. Trends Genet. 1992;8:132–138. doi: 10.1016/0168-9525(92)90371-A. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M. Drosophila: A Laboratory Manual. PlainView, NY: Cold Spring Harbor Laboratory Press; 1989. p. 245. [Google Scholar]
- 18.Semeshin VF, Belyaeva ES, Shloma VV, Zhimulev IF. Methods Mol Biol. 2004;247:305–324. doi: 10.1385/1-59259-665-7:305. [DOI] [PubMed] [Google Scholar]
- 19.Mathog D, Hochstrasser M, Gruenbaum Y, Saumweber H, Sedat J. Nature. 1984;308:414–421. doi: 10.1038/308414a0. [DOI] [PubMed] [Google Scholar]
- 20.Agard DA, Sedat JW. Nature. 1983;302:676–681. doi: 10.1038/302676a0. [DOI] [PubMed] [Google Scholar]
- 21.Hochstrasser M, Mathog D, Gruenbaum Y, Saumweber H, Sedat JW. J Cell Biol. 1986;102:112–123. doi: 10.1083/jcb.102.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochstrasser M, Sedat JW. J Cell Biol. 1987;104:1471–1483. doi: 10.1083/jcb.104.6.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conchello JA, Lichtman JW. Nat Methods. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- 24.Denk W, Strickler JH, Webb WW. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 25.Zipfel WR, Williams RM, Webb WW. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 26.Williams RM, Zipfel WR, Webb WW. Curr Opin Chem Biol. 2001;5:603–608. doi: 10.1016/s1367-5931(00)00241-6. [DOI] [PubMed] [Google Scholar]
- 27.Phair RD, Scaffidi P, Elbi C, Vecerova J, Dey A, Ozato K, Brown DT, Hager G, Bustin M, Misteli T. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gratton E, Barry NP, Beretta S, Celli A. Methods. 2001;25:103–110. doi: 10.1006/meth.2001.1219. [DOI] [PubMed] [Google Scholar]
- 29.Ni Z, Saunders A, Fuda NJ, Yao J, Suarez JR, Webb WW, Lis JT. Mol Cell Biol. 2008;28:1161–1170. doi: 10.1128/MCB.01859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinett CC, Straight A, Li G, Willhelm C, Sudlow G, Murray A, Belmont AS. J Cell Biol. 1996;135:1685–1700. doi: 10.1083/jcb.135.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez J, Belmont AS, Sedat JW. Curr Biol. 2001;11:1227–1239. doi: 10.1016/s0960-9822(01)00390-6. [DOI] [PubMed] [Google Scholar]
- 32.Bischof J, Maeda RK, Hediger M, Karch v, Basler K. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Axelrod D, Koppel DE, Schlessinger J, Elson E, Webb WW. Biophys J. 1976;16:1055–1069. doi: 10.1016/S0006-3495(76)85755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Periasamy N, Verkman AS. Biophys J. 1998;75:557–567. doi: 10.1016/S0006-3495(98)77545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown EB, Wu ES, Zipfel W, Webb WW. Biophys J. 1999;77:2837–2849. doi: 10.1016/S0006-3495(99)77115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lippincott-Schwartz J, Snapp E, Kenworthy A. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- 37.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. Science. 2000;287:1262–1265. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 38.Cole NB, Smith CL, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 39.Morin X, Daneman R, Zavortink M, Chia W. Proc Natl Acad Sci U S A. 2001;98:15050–15055. doi: 10.1073/pnas.261408198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelso RJ, Buszczak M, Quinones AT, Castiblanco C, Mazzalupo S, Cooley L. Nucleic Acids Res. 2004;32:D418–D420. doi: 10.1093/nar/gkh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clyne PJ, Brotman JS, Sweeney ST, Davis G. Genetics. 2003;165:1433–1441. doi: 10.1093/genetics/165.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buszczak M, Paterno S, Lighthouse D, Bachman J, Planck J, Owen S, Skora AD, Nystul TG, Ohlstein B, Allen A, Wilhelm JE, Murphy TD, Levis RW, Matunis E, Srivali N, Hoskins RA, Spradling AC. Genetics. 2007;175:1505–1531. doi: 10.1534/genetics.106.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong WJ, Golic KG. Proc Natl Acad Sci U S A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rong YS, Golic KG. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 45.Gong WJ, Golic KG. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Nat Rev Mol Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 47.Patterson GH, Lippincott-Schwartz J. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]