Summary
The Drosophila MSL complex associates with active genes specifically on the male X chromosome to acetylate histone H4 at lysine 16, and increase expression approximately two-fold. To date, no DNA sequence has been discovered to explain the specificity of MSL binding. We hypothesized that sequence-specific targeting occurs at “chromatin entry sites”, but the majority of sites are sequence-independent. Here we characterize 150 potential entry sites by ChIP-chip and ChIP-seq and discover a GA-rich MSL recognition element (MRE). The motif is only slightly enriched on the X chromosome (~2 fold), but this is doubled when considering its preferential location within or 3’ to active genes (>4 fold enrichment). When inserted on an autosome, a newly identified site can direct local MSL spreading to flanking active genes. These results provide strong evidence for both sequence-dependent and -independent steps in MSL targeting of dosage compensation to the male X chromosome.
Introduction
The occurrence of sequence motifs and their utilization appear to be well correlated in organisms with small genomes such as bacteria (Wade et al., 2005). However, genomic analyses in eukaryotes have revealed that short sequence-specific DNA binding motifs alone are insufficient to explain the selective occupancy or specificity of regulatory factor function (Lieb et al., 2001; Carroll et al., 2005; Sekinger et al., 2005). Therefore, a very important question in transcriptional regulation is how binding factors identify the motifs at which they will function, selecting them from an excess of sites of equal or greater predicted affinity in the genome.
An additional layer of complexity is introduced when considering the factors that organize chromatin into active and silent domains and play a crucial role in the fidelity of gene expression in development. These chromatin remodeling and modifying enzymes are thought to be directed to their targets initially by sequence-specific factors or the transcriptional machinery, but then recapitulate their functions independently of such factors to maintain the fidelity of essential gene regulation, even after DNA replication and cell division (Goldberg et al., 2007; Li et al., 2007; Schneider and Grosschedl, 2007).
Dosage compensation provides a model system to study how critical chromatin regulatory decisions are initially targeted to genes and local chromatin domains. This is a process in which X chromosomal genes are differentially regulated to equalize expression in males (XY) and females (XX). In the three model systems studied in depth, Drosophila, C. elegans, and mammals, this occurs through sex-specific, X-specific targeting of chromatin regulatory complexes (Csankovszki et al., 2004; Meyer et al., 2004; Lucchesi et al., 2005; Ng et al., 2007). The signal for X chromosome targeting is most clearly understood in mammals, where one of the female X chromosomes is inactivated and acquires characteristics of silenced heterochromatin. In X inactivation, expression of the noncoding Xist RNA appears to be necessary and sufficient to initiate the process of silencing a chromosome in cis (Lee et al., 1996; Herzing et al., 1997; Wutz and Jaenisch, 2000). Xist RNA associates with the chromosome that encodes it (Clemson et al., 1996), and this is linked to the silencing event (Okamoto et al., 2004). Additional X chromosome sequences might play a role as “boosters” for spreading of Xist RNA (Lyon, 2003), but autosomes can be at least partially inactivated if expressing Xist RNA in early development (Lee and Jaenisch, 1997), suggesting that X-specific booster sequences are not obligatory.
In C. elegans, the dosage compensation complex (DCC) down-regulates genes on both hermaphrodite X chromosomes by approximately half to achieve dosage compensation (Lucchesi et al., 2005; Meyer, 2000). In this model system, there is evidence for hundreds of autonomous recognition sites along the length of the X chromosomes that contribute to targeting (Csankovszki et al., 2004). Recently, specific DNA sequences have been correlated with this function (McDonel et al., 2006) and found in locations of high DCC occupancy by ChIP-chip (Ercan et al., 2007). Although these sequences are only enriched ~1.25–1.5 fold on X, non-random clustering may explain how the X chromosome is initially identified (McDonel et al., 2006; Ercan et al., 2007). A second step of sequence-independent spreading has been postulated to explain the full pattern of X chromosome binding (Csankovszki et al., 2004), but characterized recognition sites have not yet been shown to possess this ability when moved to autosomes.
In Drosophila, the MSL complex up-regulates X linked genes in males by approximately two-fold to achieve dosage compensation (Lucchesi et al., 2005). No DNA sequence motif uniquely identifies the X chromosome, although X chromosomes throughout the Drosophila genus show enrichment for simple sequence repeats when compared to autosomes (Stenberg et al., 2005; Gallach et al., 2007). Although the MSL binding pattern on the X chromosome has been mapped at high resolution by ChIP-chip, no sequence within the binding sites has been identified to account for the striking X chromosome specificity (Alekseyenko et al., 2006; Gilfillan et al., 2006; Legube et al., 2006). MSL complex binds broadly within the bodies of active X-linked genes, with a 3’ bias (Alekseyenko et al., 2006; Gilfillan et al., 2006). Reminiscent of mammals, there are at least two noncoding RNAs that participate in dosage compensation in Drosophila, and are encoded by the X chromosome (Amrein and Axel, 1997; Meller et al., 1997). These roX RNAs act like nucleation sites for MSL spreading when ectopically expressed from autosomes, suggesting that they, like Xist, are autonomous targeting elements (Kelley et al., 1999; Park et al., 2002). However, unlike Xist, roX RNAs expressed from other chromosomes can still recognize a roX mutant X chromosome in trans, suggesting that there are additional characteristics that distinguish the X chromosome from the autosomes (Meller et al., 1997; Park et al., 2002).
One model is that the subset of MSL binding sites that remain in the absence of either the MSL3 chromodomain protein, the MLE helicase, or the MOF acetyltransferase, define the set of core MSL1/MSL2 initiation sites, and that the majority of sites are recognized in a second, sequence-independent step in cis (Kelley et al., 1999; Larschan et al., 2007). Cytologically, there are ~30–70 sites seen on polytene chromosomes in msl3 mutant males (Palmer et al., 1993; Demakova et al., 2003). However, the existence of this limited set of sites has not been sufficient to explain the apparently autonomous recognition of many X-linked genes in translocation and transposition stocks (Fagegaltier and Baker, 2004; Oh et al., 2004). This has led to an alternative model in which degenerate sequences define all sites on the X chromosome, with a hierarchical recognition mechanism. However, in this case the sequences have been degenerate enough to confound definition (Dahlsveen et al., 2006; Gilfillan et al., 2006; Gilfillan et al., 2007; Kind and Akhtar, 2007).
Here we use ChIP-chip and ChIP-seq to identify a large set of candidate chromatin entry sites (CES), in order to test the two-step model for MSL binding. These sites are proposed to be recognized in a sequence-dependent manner, and indeed we found a GA- or TC-rich motif common to the majority of sites. Mutagenesis studies reveal the importance of this motif for MSL recognition in a cell culture reporter assay, and when placed on autosomes. Interestingly, a newly identified site can direct local MSL spreading to flanking active genes on an autosome, supporting its proposed activity as a chromatin entry site. These results provide strong evidence for both sequence-dependent and –independent steps in the model for MSL targeting of dosage compensation, and provide a general framework for the identification of initiation sites for other chromatin regulators that function broadly over chromatin domains.
Results
msl3-independent high-affinity sites represent a subset of the wild type binding pattern and are predominantly located within genes
We previously proposed that the incomplete MSL binding pattern seen on polytene chromosomes in the absence of MSL3 might represent specific initiation or “chromatin entry” sites for MSL complex recognition of the X (Kelley et al., 1999). To test this hypothesis, we combined genetic analysis with ChIP-chip. We chose to analyze msl3 mutant embryos to avoid potential complications from working with dying male larvae. Construction of a genetic stock in which all male embryos were msl3 mutant was accomplished by crossing msl3 mutant males carrying an msl3+ transgene on their X chromosome to msl3 mutant females. All sons from such a cross are msl3 mutant, while all daughters receive the msl3+ transgene. Females, however, do not express MSL2 (Kelley et al., 1995; Bashaw and Baker, 1995; Zhou et al., 1995), so MSL2 ChIP of the population results in analysis of only the male embryos. We performed two independent biological replicates of the ChIP-chip experiment (Fig. 1), calculated mean values for each probe on the microarrays, and then determined significant clusters of binding on the X and the autosomes.
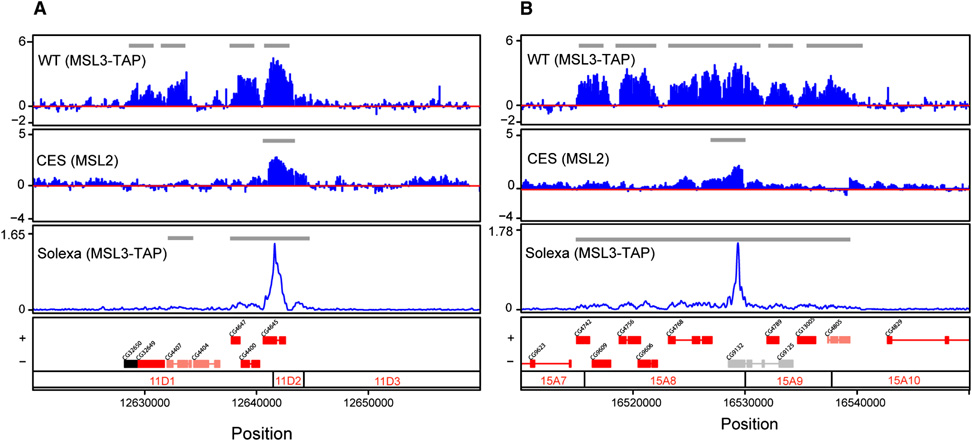
Figure 1. MSL3-independent chromatin entry sites are a subset of the wild-type binding pattern for MSL complex and coincide with the strongest enrichment peaks detected by ChIP-seq.
Two representative chromatin entry sites, CES11D1 (A) and CES15A8 (B) are shown. ChIP-chip profiles were generated from y w; MSL3-TAP; msl3 embryonic chromatin (WT) using IgG to IP the TAP epitope, or msl3 mutant embryos (CES) using anti-MSL2 antibodies. DNA resulting from ChIP was hybridized to custom NimbleGen tiling arrays (Alekseyenko et al., 2006). The y axis shows the log2 ratio of IP/Input signal. The ChIP-seq tag profile (Solexa) was obtained from an MSL3-TAP transformed male cell line, Clone8, using IgG to IP the TAP epitope. The ChIP-seq profile displays broad distribution along WT MSL targets and high peaks that correspond to entry sites. The y axis shows the tag density. Gray lines within ChIP-chip and ChIP-seq panels indicate the regions identified as bound clusters (See Experimental Procedures for details). Genes are color coded based on their transcriptional status (transcribed, red; nontranscribed, black; genes that are differentially transcribed in S2 and Clone 8 cells, salmon; and genes without transcriptional data, gray). Genes on the top row are transcribed left to right, and genes on the bottom row are transcribed from right to left. Numbers along the x axis refer to chromosomal position (bp) (Dm1 release coordinates). Polytene map cytological locations are indicated below.
As expected, our positive clusters were highly enriched on the X chromosome. Using the relative enrichment on the X compared to an autosome, we determined that there were at least 150 sites on the X (and 5 sites on 2L, the autosome represented on the array). There are no absolute criteria for setting the parameters that determine the number of sites; however, this was a reasonable compromise between the enrichment on the X and coverage. 150 is a higher number of MSL3-independent sites than originally predicted based on immunostaining of msl3 mutant polytene chromosomes. However, we recently demonstrated that banding patterns on polytenes do not provide single site resolution and thus can lead to both under-estimation of the number of individual binding sites, as well as an inability to distinguish relatively large changes in binding site occupancy (Larschan et al., 2007). We named the sites CES followed by their cytological position, based on the polytene chromosome map (eg. CES11D1) (Table 1S). Among the 150 sites, we recovered roX1, roX2, and 18D11, previously identified as msl3-independent sites in our lab (Kelley et al., 1999; Oh et al., 2004), as well as DBF3 (CES3A8), DBF7 (CES10F2), DBF9 (CES18D3), and DBF12 (CES1B14), previously characterized as high affinity sites by P. Becker & colleagues (Dahlsveen et al., 2006).
Next we asked whether these 150 candidate CES were a subset of wild-type MSL targets. On average, the clusters observed in our CES ChIP-chip experiments were approximately 1.5 kb in size, and virtually all (98%) were contained within the full set of sites in wild type. Consistent with this observation, we found that CES overlap with genes (135 of 150).
ChIP-seq of the wildtype MSL pattern detects strong peaks of binding within CES candidates
Concurrent with the ChIP-chip analysis of msl3 mutants, we performed Solexa sequencing of MSL3-TAP ChIP from male tissue culture cells. We were testing whether this non-array and non-hybridization-based approach would yield similar or higher resolution results for wild-type MSL3 binding. When we profiled wildtype MSL3-TAP in Clone8 cells, we were able to observe the broad binding patterns covering active genes on the X previously seen in ChIP-chip analysis (Alekseyenko et al., 2006), with 91% of the positive genes in ChIP-seq overlapping with 89% of bound genes in our previous ChIP-chip data set. Using ChIP-seq we found 619 binding sites on X and only 2 on 2L (our negative control), demonstrating the high quality of the experiment. Notably, we also discovered a set of sharp peaks extending far above the broadly bound regions, that were not evident in ChIP-chip analysis. These peaks are of such striking amplitude that when plotted in Fig. 1, the broad and significant signal of overall binding surrounding them is not readily evident at this scale (see gray bars that indicate significant signal). Furthermore, there was a remarkable coincidence of the ChIP-seq peaks from MSL3-TAP in wildtype with our newly identified chromatin entry sites found genetically, with 91% inclusion of the 150 entry sites in the top 309 Solexa peaks (see Fig. 2Sd, Experimental Procedures for details). The set of top peaks covering the entry sites also were distinguished by their magnitude (Fig. 2Se), therefore we used the top 309 peaks for further analysis.
To determine whether or not a bias for recovery of these fragments was intrinsic to the general chromatin composition of those regions rather than being a consequence of the specific ChIP experiments, we sequenced sonicated and amplified input DNA. We found that any intrinsic biases in the input could not account for the sharp peaks observed after MSL pulldown and recovery (data not shown).
To determine whether the peaks were somehow enhanced by the ChIP-seq technique, or quenched in ChIP-chip analysis, we assayed MSL3-TAP chromatin IP samples directly by QPCR for regions encompassing the ChIP-seq peaks. We found an excellent correlation between fold differences seen by QPCR and those measured by ChIP-seq around 3 of 3 peaks analyzed (Fig. 1S). We conclude that the key difference between the ChIP-seq and ChIP-chip platforms appears to be in the dynamic range that can be measured. Several studies have reported a general agreement between the two platforms (Euskirchen et al., 2007; Mikkelsen et al., 2007; Boyle et al., 2008), but those comparisons were mostly based on visual inspection rather than a statistical comparison. Also, if the methods are compared for transcription factor binding, which typically occurs in sharp peaks, the differences would not be so apparent. However, the MSL complex binds quite broadly over transcription units, and before the ChIP-seq analysis, it was less evident that there were strong quantitative differences within the broad binding profiles. For MSL complex binding, ChIP-seq clearly reveals a sub-pattern of high affinity sites within the wildtype binding pattern. The strong coincidence of the peaks of wildtype MSL3-TAP binding detected by ChIP-seq with the subset of sites of MSL2 binding in msl3 mutants, provides strong evidence that this subset of MSL binding sites may have a significant biological role.
Discovery of a sequence motif in entry sites and ChIP-seq peaks that is enriched on the X chromosome
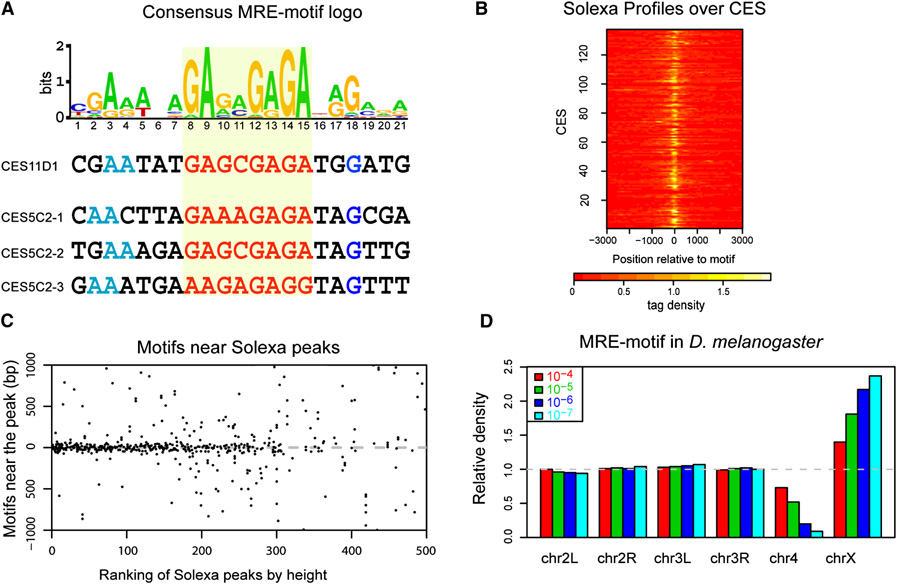
Encouraged by the strong correlation between ChIP-seq and ChIP-chip results, we analyzed the 150 candidate entry sites and the top 309 ChIP-seq peaks for a sequence motif using MEME, a robust searching algorithm (Bailey et al., 2006). We found the same highly significant 21 bp sequence centered around a core GAGA (or TCTC) repeat using both data sets (Fig. 2A, Fig. 2S). This sequence is compatible with the conserved MSL binding motif we previously identified by direct comparison of the roX1 and roX2 genes from multiple species (Park et al., 2003), and the high affinity sequences characterized by Gilfillan et al. (2007). Other motif searching algorithms including WEEDER, and MDscan (Liu et al., 2002; Pavesi et al., 2004), gave a very similar but shorter top motif. We note that while the chromatin entry sites form a subset of the wildtype sites, previous attempts to recover a motif from wild type data failed because of the excess of extraneous DNA sequences included in those regions. For convenience we will refer to our newly discovered motif as the “MSL recognition element” (MRE). Interestingly, the MRE includes the GAGA factor (GAF) consensus sequence GAGAGA, but to date significant coincidence of MSL and GAF binding has not been evident (Sun et al., 2003; van Steensel et al., 2003). Similarly, we wondered whether a CT-rich logo might indicate involvement of CTCF, named initially for its affinity for CT-rich sequences (Lobanenkov et al., 1990). However, the logo discovered for Drosophila CTCF from ChIP-chip studies of the Adh and Bx-C regions bears no resemblance to our GA-motif (Holohan et al., 2007).
Figure 2. A GA-rich motif is a common feature in MSL chromatin entry sites.
(A) Identification of the MRE-motif enriched in CES. The motif logo is shown with representative examples below. This motif occurs once in CES11D1 and three times in CES5C2. The GA-rich core is highlighted in red. We are arbitrarily showing the GA-rather than the TC-rich strand. This motif was identified using the chromatin entry sites as determined by the ChIP-chip data; a nearly identical motif is obtained from the WT Solexa data (Suppl. Fig. 2S). (B) Solexa peaks center on the MRE-motif in chromatin entry sites. Shown is a heatmap of Solexa profiles (tag densities increase from red to white), aligned by the MRE-motif in each of the 137 individual CES that contains the motif. In nearly all cases, the location of the sharp peaks coincides with the motif location. (C) The MRE-motif tends to localize to the center of strong Solexa peaks. The top 500 Solexa peaks were ranked by height on the x axis and then MRE-motifs within a 2 kb window flanking each peak were mapped on the y axis. Some peaks contain multiple motifs; to avoid showing the same region twice, only the strongest peaks in any 3 kb region were considered. Most motifs map near the center, but motif centering decreases as peak intensity weakens. (D) The MRE-motif is enriched on the X chromosome, compared to autosomes. Motif frequency for each chromosome was normalized to its frequency on all autosomes. Colors correspond to different thresholds for defining the occurrence of the motif. At all thresholds, the motif is enriched on the X. As the stringency increases, the enrichment increases but a smaller number of motifs are identified.
In Figure 2B, we display a heatmap of the ChIP-seq profiles around the chromatin entry sites containing MREs. Strikingly, in nearly all cases the sharp wildtype ChIP-seq peaks are centered on the MRE within the chromatin entry site, with a rapid decrease away from the motif. To determine the relationship between the peaks and the occurrence of MREs, we ordered the ChIP-seq peaks by their height, and plotted all the motifs (p-value < 10−5) in the neighborhood (1 kb in each direction) (Fig. 2C). Clearly, MREs are concentrated at the center of the strongest peaks; they are more dispersed as the peaks get weaker. If we are to base the likelihood of a peak being a chromatin entry site on the clustering of MREs within the peak, the estimate of 150 chromatin entry sites appears to be somewhat conservative. But beyond ~300, the alignment degenerates, suggesting that weaker peaks are not dependent on MREs.
A motif utilized for X-specific MSL binding on the X ideally should to be enriched on X. Therefore, we surveyed the D. melanogaster genome for the occurrence of potential MREs on X and autosomes. We found that the motif was approximately two-fold enriched on X versus autosomes for a wide range of thresholds for defining a sequence match, (see Figure 2D). At a p-value of 10−5, there are 3652 and 8770 motifs on the X and autosomes, respectively, for an enrichment of 1.8-fold in terms of motif density (167.7 copies/Mb on X versus 92.3 copies/Mb on autosomes). At this stringency, 137 of the 150 candidate entry sites contain an MRE (91%), with 51 having 2 or more MREs within 500 bp (34%).
The genome sequences of twelve Drosophila species became available recently, and it has been of considerable interest to determine how appropriate dosage compensation evolved following sex chromosome translocation events in the various species. We found that the MRE discovered in D. melanogaster was similarly enriched on the X chromosomes of other sequenced species. Supplementary figure 2S shows results for two species (D. simulans and D. yakuba).
We define chromatin entry sites functionally, as the binding sites for partial MSL complex in the absence of MSL3. In contrast, the MRE is a consensus sequence which is important within CES but clearly is found elsewhere in the genome. The simple presence of an MRE is not sufficient to predict MSL binding, as autosomes carry thousands of matches that are not bound. In addition, among 3652 predicted MREs on X, only ~6% (224/3652) are located within the 150 selected entry sites. At this threshold, ~91% of the entry sites have motifs, but many motifs are ignored. Therefore, we continued our analysis on two fronts, first asking whether the MRE sequence is implicated functionally in MSL binding, and second, identifying additional parameters that would better define the motifs that are actually chosen on X.
Functional analysis of the MRE within candidate entry sites using a luciferase reporter assay
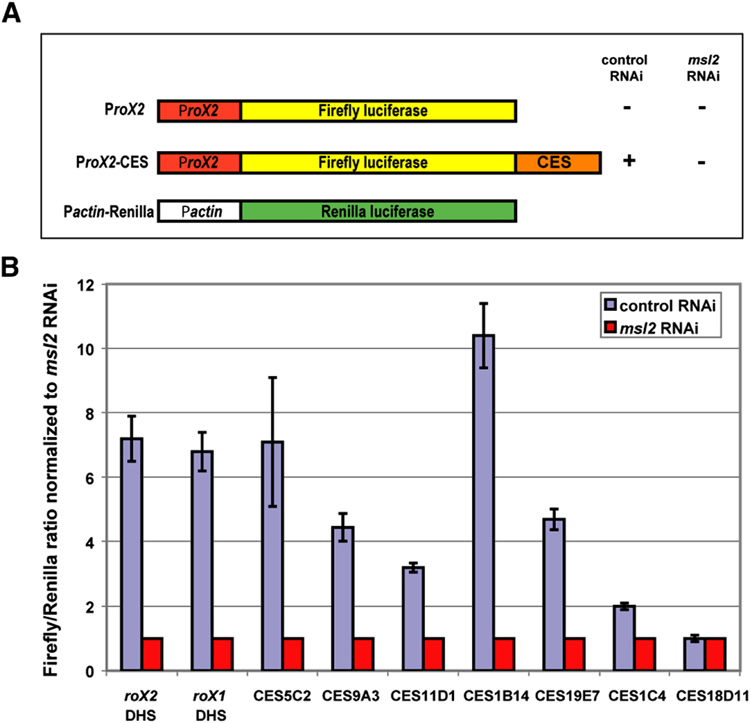
To determine whether the MRE is important for MSL recognition at the newly identified sites, and whether it can function when removed from its normal chromosomal context, we employed a luciferase based reporter assay in male S2 tissue culture cells. The luciferase assay (Fig. 3) is based on the observation that MSL binding downstream of the major roX2 transcription unit positively regulates expression of roX2 (Bai et al., 2004). The MSL binding site in roX2 displays DNase hypersensitivity and thus has been called the roX2 DHS (Kageyama et al., 2001). We constructed a plasmid containing the roX2 promoter region and the roX2 DHS, with the intervening roX2 transcription unit replaced by the firefly luciferase gene (Fig. 3A). Upon transfection into S2 cells, the activity of the firefly luciferase is highly dependent on the presence of MSL complex, as activity is depressed ~7 fold by RNAi treatment for MSL2 (Fig. 3B). The DHS region is required to confer MSL dependence to the reporter gene. To provide accurate quantitation of reporter gene activity, we used a dual luciferase system in which normalization was conducted using a control plasmid that contains the Renilla luciferase gene fused to an MSL-independent promoter. The Firefly and Renilla luciferase reporter genes are quantitatively assayed using independent substrates.
Figure 3. Test for CES function in a luciferase reporter assay.
(A) Experimental design, and structure of the constructs used for a dual Firefly/Renilla luciferase reporter assay. S2 cells co-transfected with the firefly luciferase construct containing each 1.5 kb CES fragment and the control Renilla luciferase plasmid were treated with dsRNA against either GFP (control) or MSL2 (experimental). (B) Examples of Firefly/Renilla luciferase activity conferred by nine chromatin entry sites normalized to the value obtained after msl2 RNAi. CES18D11 is a rare example of a CES that failed to show MSL2-dependent upregulation in this assay.
We found that 1.5 kb wildtype DNA segments containing candidate entry sites gave a clear response in the reporter assay (Fig. 3, Table 2S). We tested 29 of the 150 candidate MSL binding sites in the luciferase assay, and 21 of these responded positively (Table 2S, and data not shown). To exclude roX2 promoter-specific effects, we cloned three different autosomal promoters upstream of the luciferase gene. Derivatives of these constructs carrying various CES also demonstrated dependency on MSL2, similar to the activity observed with the roX2 promoter. Interestingly, the effect inversely correlated with the absolute promoter strength (Table 2S). Next we performed deletion and mutational analysis to test whether or not key residues in the MRE were necessary for the observed MSL-dependent transcriptional response.
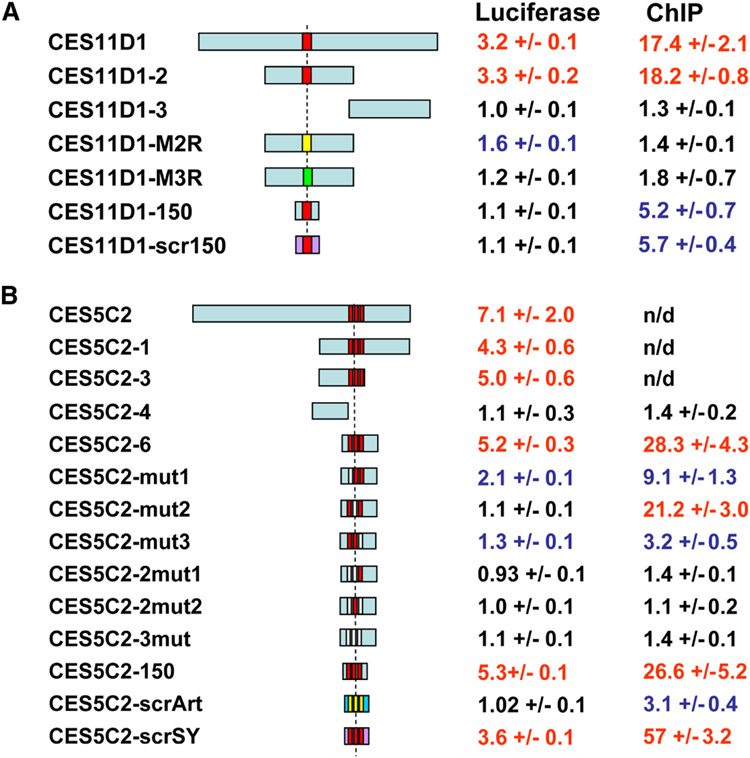
We initially narrowed down the 1.5 kb positive fragments for 5 putative entry sites to 500 bp, and in 4 cases the positive activity tracked unambiguously with the mapped MREs (Fig. 4, Fig. 3S and Table 2S). In subsequent deletion analyses, we were able to identify a 150 bp fragment of CES5C2 with three MREs that retained function, but a 150 bp subclone of CES11D1, with one MRE, did not (Fig. 4, Fig. 3S). We created point mutants in the CES5C2 (150 bp) and CES11D1 (500 bp) fragments to examine the requirement for core residues (Fig. 4, Fig. 3S). We found that the MREs were absolutely required for reporter gene activity in all cases and combinations. We further asked whether the MREs were not only necessary, but might be sufficient in this context, by retaining the motif location but randomizing the rest of the surrounding sequence, keeping the GC content constant. Surprisingly, we found that for CES5C2, such randomized fragments were still functional, demonstrating that the central sequence motif plays the primary role in MSL recognition and function. These results (Fig. 4) provide strong evidence for the importance of key residues in the MRE.
Figure 4. The MRE-motif is required for MSL complex binding to CES.
Structure of the DNA fragments tested in the context of CES11D1 (A) and CES5C2 (B), and the activities they display in the luciferase assay in S2 cells and when inserted in a specific location (37B7) on chromosome 2L in transgenic flies (MSL2 ChIP). Numerical values in red were scored positive, black were negative, and blue were considered intermediate. Positions of the motifs are shown as red boxes; when mutated, MRE-motifs are shown as non-red boxes (please refer to Suppl. Fig. 3S for sequence changes within the motif). Randomized DNA sequences in the shortest subclones not affecting the MRE-motifs are shown in purple or dark blue (with only the motif core remaining intact). Luciferase and ChIP data are based on at least three independent experiments. n/d – not determined.
Transgenic entry sites attract MSL complex to autosomes and are MSL3-independent
Based on the results of the sequence motif analysis and the functional luciferase assays, we proceeded to in vivo cytological and ChIP analysis of five candidates: CES11D1 (1 motif), CES1C4 (1 motif), CES9A3 (2 motifs), CES5C2 (3 motifs) and CES18D11 (1 motif). Of these, all were positive in the luciferase assay except CES18D11, which was chosen for further analysis because of previous cytological and ChIP data supporting its MSL binding capacity (Oh et al., 2004). We inserted these candidate chromatin entry sites and their subclones into two different genomic positions on chromosome 2 by site-specific recombination. Following replacement of a mini-white gene cassette, we determined that each insertion had occurred at the correct position in the genome by PCR. We inserted the roX1 DHS as a positive control, and a GFP gene as a negative control.
We assayed our initial transgenic insertion lines at the level of polytene chromosome staining. We asked whether or not a new MSL3-independent binding site was created in polytene chromosomes by immunostaining for MSL2 in an msl3 mutant background. For this, we used females ectopically expressing MSL2 but homozygous mutant for msl3. The candidates with MREs demonstrated positive signals in all cases (Fig. 5, Fig. 4S, Fig. 5S). On average, the CES candidates integrated at 37B7 displayed better ability to recruit MSL complex than when inserted in 53F8 (Table 3S). This result confirmed that the isolated sites were MSL3-independent, and thus fit our criteria for their initial identification. Our immunostaining results were generally in agreement with the luciferase data. Interestingly though, the CES18D clone that failed to up-regulate the luciferase reporter, displayed consistent binding in our immunostaining assay (Table 3S, Fig. 5S), with improved detection compared to our previous work (Oh et al., 2004).
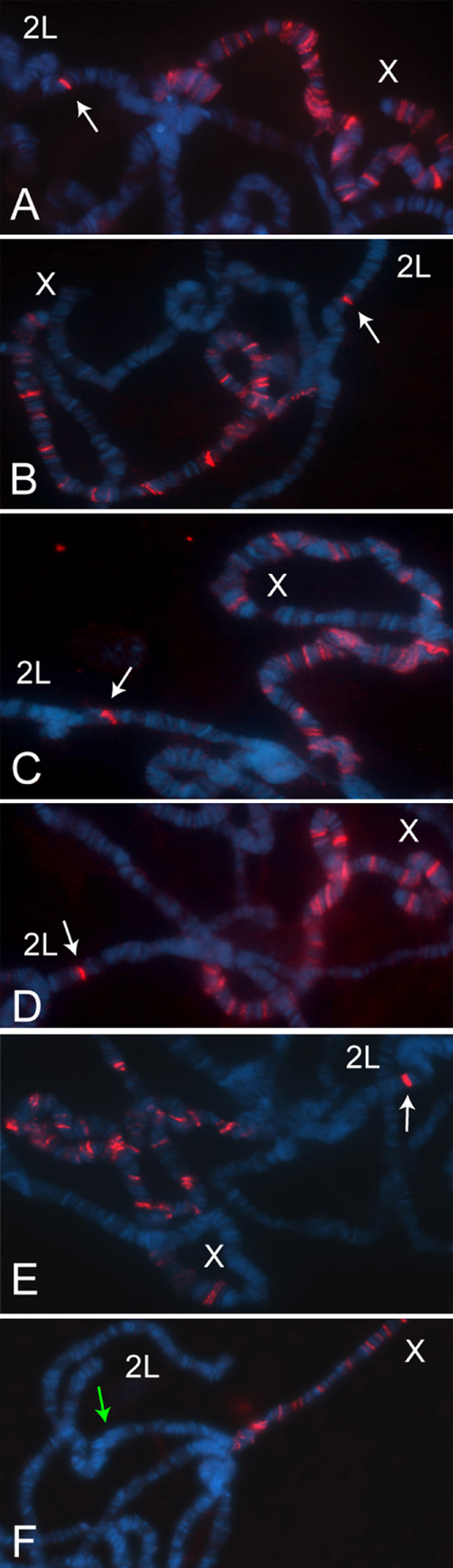
Figure 5. MSL complex is recruited to entry sites placed on autosomes.
Polytene chromosomes from homozygous CES-X{37B7}; H83M2-6I, msl3 females were immunostained with anti-MSL1 antibodies. (A) roX1 DHS (300bp). (B) CES11D1 (1.5 kb). (C) CES9A3 (1.5 kb). (D) CES5C2 (1.5 kb). (E) CES5C2-6 (268 bp, containing three motifs). (F) CES5C2-4 (268 bp, lacking a motif). White arrows point to the positive signal at the integration site, a green arrow points to the cytological location with no visible staining. The X chromosome in each nucleus displays the partial pattern typical of msl3 mutants.
The MRE motif is necessary for MSL complex binding to entry sites in vivo
Does MSL binding within entry sites always follow the MRE? To address this question we used directed ChIP analysis as a high resolution alternative to conventional cytological analysis. We used a mixed population of embryos in which all individuals carried one copy of the transgene insertion, but only 50% (the males) would be expected to form the MSL complex. We designed primers specific for the hybrid attL flanking sites common to all insertions, to create a ChIP assay in which individual strains could be treated identically following chromatin isolation. Following cross-linking, IP with anti-MSL2, isolation of DNA and analysis by QPCR, we found that 0.5–1.5 kb segments of CES5C2 and CES11D1 were positive in all assays, and could be subdivided into positive and negative subclones (eg. CES5C2-6 vs. CES5C2-4) based on the presence or the absence of the MRE (Fig. 3S). Indeed, all the subfragments encompassing MREs recruited MSL complex to the autosomal integration site and were bound by MSL2, as assayed by ChIP (Fig. 4).
Next we tested the same sequences and mutations for CES11D1 and CES5C2 that were tested in the luciferase assay. These fragments contained one (CES11D1) or three (CES5C2) MREs. Three-nucleotide substitution in the core of the single motif in the positive subclone CES11D1-2 completely abolished MSL binding. When we decreased the subclone size to 150 bp (CES11D1-150) and the motif was present, we observed reduced but significant binding. This demonstrates that the ChIP assay was more sensitive than the luciferase assay, possibly because it is directly measuring binding, rather than asking for transcriptional upregulation as a consequence of binding. In this minimized system, we randomized the DNA sequence, keeping the GC-content and position of the MRE constant. Surprisingly, this new altered sequence (CES11D1-scr150) displayed pronounced binding similar to the original DNA indicating that the MRE is sufficient for MSL recruitment at this ectopic location, without multimerization (Fig. 4). Additional confirmation of the importance of the MRE came from subfragments of CES5C2, which carries three motifs. First we found that mutation of individual MREs reduced binding to varying degrees (Fig. 4). Secondly, simultaneous mutation of two MREs completely abolished recruitment of the complex. Interestingly, the smallest fragment tested (CES5C2-150) carrying all three intact MREs displayed binding very similar to larger subclones. We subsequently asked whether it is the whole motif (21 bp) or just its central part (15 bp) that is needed for complex binding. To answer this question we created two different randomized sequences. In one case, scrambling affected all sequences except the three 25bp-segments containing the motifs (CES5C2-scrSY), whereas in CES5C2-scrArt, only the 15 bp core sequences in these motifs remained unaltered. Spacing between the motifs and the GC-content were kept constant. We detected weak residual binding in CES5C2-scrArt, but CES5C2-scrSY showed binding that exceeded the original fragment. These results suggest that integrity of the full MREs in this entry site are essential for binding (Fig. 4) with no evidence for a requirement for local flanking sequences.
If MREs are both necessary and sufficient for binding in the two cases tested, why does the MSL complex bind some MRE-containing sites, but skip many others? This is not a novel problem in genomics, as predicted sites for sequence-specific binding factors often greatly exceed the number of sites actually utilized. Increasing the stringency with which we call a motif a match did not improve our ability to predict entry sites on the X chromosome (Supp Fig. 2S). These results suggest that one or more additional characteristics guide the MSL complex to specific sites on the X. Therefore, we searched for secondary sequence motifs that might contribute to the specificity of the MRE. However, all additional motifs lacked predictive value, as they were no more enriched in CES than they were in the entire X chromosome (data not shown).
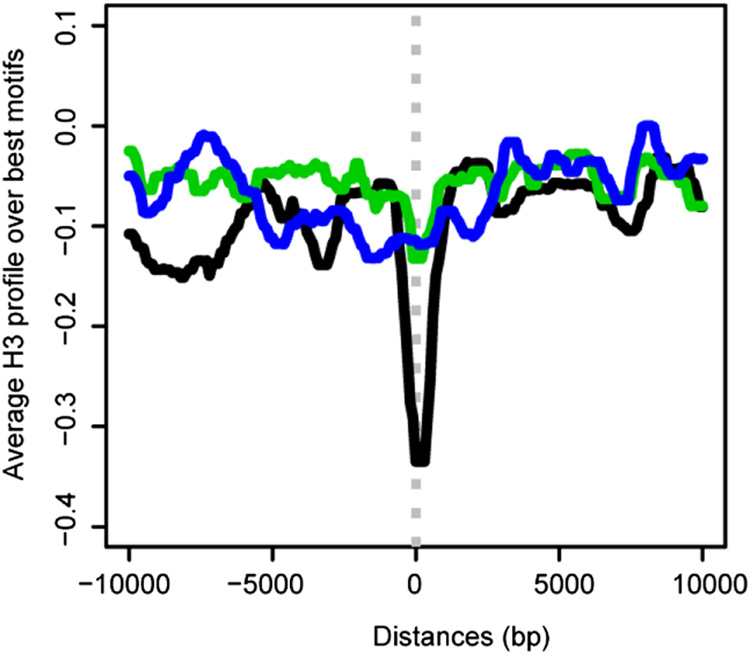
A logical aspect of recognition could be relative accessibility of the MRE within its chromatin environment. For example, functional MREs might be located in a particular position with respect to nucleosomes, or reside in relatively depleted regions. Therefore, we analyzed histone H3 occupancy over the entry sites. We determined the average H3 signal after aligning the 150 entry sites by their best motifs and found a clear H3 depletion centered over these sites (Fig. 6, and data from Larschan et al., 2007). This averaged profile was not observed over the remaining, unselected motifs on X (Fig. 6). These results suggest that nucleosome depletion might be a key defining characteristic of functional chromatin entry sites.
Figure 6. Histone H3 is depleted over MREs in chromatin entry sites.
Average H3 profile centered over the best MRE within 137 CES (black) shows clear depletion. In contrast, the best (green) or a random set (blue) of 137 MRE-motifs outside CES display a relatively flat profile. The histone occupancy data are from S2 and Clone8 cells (Larschan et al., 2007).
We further examined the enrichment of MREs within active genes. Requiring MREs to be coincident with H3K36me3, a chromatin mark in the bodies of active genes, reduces the number of potential sites from 3652 to 449 on X and from 1977 to 105 on 2L. The enrichment on the X compared to 2L therefore increases from 1.8 to 4.3 when H3K36me3 is considered. Furthermore, the proportion of motifs on the X that fall within our data set of 150 CES is ~6% overall (224/3652), but when only the motifs in the H3K36me3 positive regions are considered, the proportion of motifs that belong to CES increases to ~30% (133/449). This is encouraging, as we believe that 150 is likely to be a conservative estimate for the actual number of functional CES Furthermore, 122 of the 150 candidate CES are retained under these criteria, strongly suggesting that additional identifying characteristics correlate with active gene expression. These results suggest that chromatin context could play a key role in selection of potential sites, and that our predictive power will improve as we learn more about the rules that govern the organization of complex genomes.
High affinity sites inserted on autosomes direct MSL spreading to flanking expressed genes
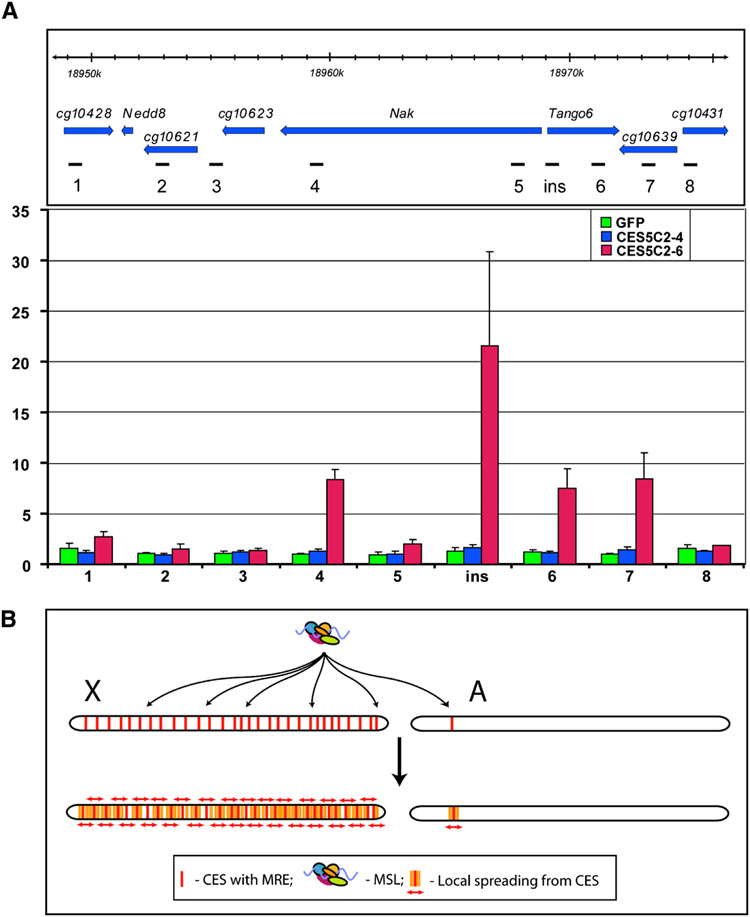
The two-step model for MSL binding to the X chromosome in Drosophila males proposes that after an initial, sequence-specific recognition step, the complex spreads in cis to active genes to achieve its complete pattern. We found that candidate entry sites inserted on chromosome 2L did not show spreading at the level of polytene chromosome staining. However, we recently observed that polytene chromosome analysis cannot distinguish whether or not individual genes within clusters are bound or not. Therefore, to ask whether entry sites might cause MSL binding to occur in flanking autosomal chromatin, we performed ChIP analysis of the local area surrounding the attP insertion site in transgenics carrying the GFP, CES5C2-4, and CES5C2-6 insertions. We probed the region up to 20 kb distant from the insertion site, testing locations in flanking active and inactive genes. We found that GFP and CES5C2-4 (lacking MREs) showed no MSL binding above background at any of the flanking regions tested, but that CES5C2-6 (containing 3 MREs) showed significant binding at specific locations (Fig. 7A, Fig. 6S). Interestingly, these locations correlated with the 3’ ends of active genes. A signal at flanking regions could be due to IP of larger than expected chromatin fragments. However, this was clearly not the case when considering the Nak gene. Probes near the 5’-end of Nak gave no significant signal, even though located between the transgene insertion site and the strongly bound 3’ end of the active gene. Together, these results demonstrate that entry sites removed from the X chromosome have the ability to cause MSL binding in flanking chromatin on autosomes. These results strongly support both sequence-dependent and sequence-independent steps in MSL binding.
Figure 7. MSL complex spreads into neighboring genes from CES5C2 inserted on an autosome.
(A) MSL3-TAP ChIP in the region surrounding the 37B7 integration site where either GFP (1.9 kb), CES5C2-4 (258 bp, no MREs) or CES5C2-6 (268 bp, three MREs) are placed (y w; CES-X{37B7}/ MSL3-TAP; H83M2-6I, msl3 mixed sex embryos). Top panel: genetic map and location of probes, Dm3 release coordinates are shown. Bottom panel: MSL3-TAP data for GFP (green), CES5C2-4 (blue) and CES5C2-6 (red), indicate that only CES5C2-6 effectively targets the MSL complex and directs limited spreading to neighboring genes. (B) Two-step model for MSL targeting of the X chromosome. X, X chromosome, A, autosome. First step: MSL complex targets >150 chromatin entry sites containing MRE motifs on the X chromosome. The autosome is normally ignored, unless a CES from the X is inserted on the autosome. Second step: Local spreading from entry sites leads to MSL binding to the majority of active genes on the X chromosome. In addition, an ectopic CES can lead to targeting of flanking active genes on an autosome, as seen in the results displayed in panel A.
Discussion
We previously proposed that MSL targeting of the X chromosome occurs in at least two steps (Fig. 7B). In the first step, roX genes and other chromatin entry sites attract the complex to the X chromosome (Kelley et al. 1999). In the second step, MSL complex spreads from entry sites to genes in flanking chromatin, by recognizing chromatin marks indicative of active transcription (Larschan et al. 2007). The mechanism governing the first step in MSL recognition of the X chromosome had remained elusive. The isolation of a large set of potential chromatin entry sites by ChIP-chip and ChIP-seq has allowed us for the first time to identify common features of these sites.
Our studies have revealed that they are enriched for a GA (or TC) repeat sequence, and that mutagenesis of this sequence leads to loss of autonomous MSL binding to individual sites. Our findings have several important implications. First, we suggest that there are more MSL entry sites than originally proposed from the analysis of MSL binding to polytene chromosomes in msl3 mutants. Based on ChIP-chip of msl3 mutant embryos using anti-MSL2, there are at least 150 sites. Furthermore, the striking correlation of these sites with strong peaks of wildtype MSL binding using ChIP-seq suggest that the number could approach 300. This is important, as previous studies that appeared to refute the entry site model found apparently autonomous binding of X chromosome genes lacking mapped sites (Fagegaltier and Baker, 2004; Oh et al., 2004). The existence of many more entry sites than were detected at the cytological level could, perhaps, explain this finding.
Interestingly, the X chromosome and autosomes of D. melanogaster and six other species were compared recently for the prevalence of all sequence words up to 13 nt long (Gallach et al., 2007). From this work it is clear that X chromosomes have evolved to be enriched for certain types of simple sequences including GA-rich repeats, which we implicate here in dosage compensation. However, additional parameters must be used to help restrict the recognition of many GA repeats, such as nucleosome depletion to leave these sites relatively open for binding, and co-localization within active gene clusters. We provide correlative evidence for these parameters, and eagerly await additional insights as the Drosophila genome becomes increasingly tractable for analysis at the level of local physical properties and higher order interactions.
Importantly, the question of recognition site specificity of transcription factors in general faces a similar, as yet unsolved problem in metazoans. Of the many sites predicted to be favored by individual factors, how is it that only a small fraction of these are actually occupied? The fidelity of sequence motifs itself is clearly not sufficient to answer this question, and organization relative to genes, other binding factors, positioned nucleosomes and three dimensional location within the nucleus are all factors under investigation.
A second implication of our finding that chromatin entry sites are more numerous than previously expected is that spreading of MSL complex on the X chromosome may normally be quite modest. In particular, it is an appealing idea that entry sites are identified in part because of their location within expressed gene clusters, and may not usually need to spread beyond such clusters (~10–30 kb). This would explain why spreading from candidate entry sites is rarely seen at the cytological level, and requires ChIP for detection. Furthermore, this could explain previous cytological data, in which MSL spreading does not appear to extend into autosomes in X:A translocations (Fagegaltier and Baker, 2004; Oh et al., 2004). Alternatively, it is possible that spreading is normally quite robust on X, but relies on the close proximity and 3D organization of other entry sites and roX genes. These conditions cannot be duplicated when individual chromatin entry sites are placed in ectopic locations for analysis. The next big step in addressing these possibilities will be to transpose large constructs, containing entry sites in combination with roX transgenes, to the autosomes.
Finally, our results suggest that the use of ChIP-seq technology may greatly enhance the ability to identify potential initiation sites for other chromatin organizing complexes that act broadly on large genomes. The dynamic range measurable by ChIP-seq could prove to be a powerful tool for deciphering the currently unknown rules for higher order organization of complex genomes.
Experimental Procedures
Plasmids and cloning
A 359 bp fragment from the roX2 gene promoter (Bai et al., 2004) was amplified with primers roX2promF: 5’-gcgtgctagcggatccctgattgattctaaaaac-3’ and roX2promR: 5’-tttcctcgagctaaacgctcgacttatg-3’ and cloned into the pGL3-Basic vector (Promega) digested with NheI and XhoI to obtain pGL3-roX2prom. Predicted 1.5 kb entry site sequences, centered around the hybridization peaks on the microarray, were amplified by PCR from y w fly stock genomic DNA with primers incorporating SalI sites, and cloned in both orientations into pGL3-roX2prom. Smaller fragments and subclones with mutations were similarily obtained. Three additional derivatives of pGL3-Basic were created and tested with various entry sites. These carried distinct promoter regions from three autosomal genes: CG2937 (367 bp), CG2960 (668 bp), and CG7424 (833 bp), cloned into the SmaI site upstream of the luciferase gene. DNA fragments with multiple mutations were obtained by custom gene synthesis (IDT) and confirmed by sequencing.
The piB-GFP transformation vector (Bateman et al., 2006) was digested with SalI to replace the hs70-GFP cassette with the sequences to be tested, which were cloned as SalI fragments from “pGL3-roX2prom-CES-X” constructs. Integrity of all constructs was verified by sequencing.
Fly genetics and transgenesis
Unless otherwise indicated, all flies were grown at +25°C on standard cornmeal-molasses medium. To obtain msl3 mutant embryos for ChIP-chip experiments, y w; msl3 females were crossed to y w P{w+, msl3+, cos8–1}/Y; msl3 males (a gift of H. Oh). Females do not express MSL2, therefore only msl3 mutant male embryos contributed to the anti-MSL2 ChIP. To generate transgenic flies with a series of sequences placed within the same genetic environment, we used the recombination-mediated cassette exchange approach (Bateman et al., 2006). Two mini-white-marked insertions of pUASTP2.1 at cytological locations 37B7 and 53F8 (upstream of the Nak and GstS1 genes, respectively) were kindly provided by J. Bateman and used as landing sites. Females homozygous for a yellow+, nos-phiC31.NLS transgene on the second chromosome (providing phiC31 integrase, a kind gift of J. Bischof) (Bischof et al., 2007) were crossed with y w; P{UASTP2.1}37B7 or y w; P{UASTP2.1}53F8 males. The resulting embryos were injected with the piB-based constructs at 200–500 ng/µl. Fifty embryos were typically injected per construct. G0 males were crossed with y w; CyO / l(2) KrIf-1; MKRS/ TM6B females, and white−, yellow− progeny were selected and intercrossed to establish homozygous stocks (referred to as y w; CES-X). The msl3 mutant background was introduced in parallel by standard genetic crosses.
To assay for MSL targeting to entry sites in an autosomal context, we used the H83M2-6I, msl3 background where MSL2 is constitutively expressed from the hs83 promoter (Kelley et al., 1995; Lyman et al., 1997). CES-X; H83M2-6I, msl3 females were crossed to MSL3-TAP; msl3 males (Alekseyenko et al., 2006). Both male and female embryos from this cross express the MSL complex, with TAP-tagged MSL3 as the only source of MSL3, making the immunoprecipitation assay particularly robust. Analysis of dosage compensation targeting in embryos under conditions of wild-type, male-specific MSL2 expression was performed using chromatin isolated from mixed sex embryos of the following genotype: y w; CES5C2-6/ MSL3-TAP; msl3.
Dual luciferase reporter assay
A description of the dual luciferase assay is provided in Supplemental Experimental Procedures.
Chromatin IP
Embryo ChIP to initially identify CES
msl3 mutant embryos (1 g) were collected for chromatin preparation as described in Alekseyenko et al. (2006). Immunoprecipitation was performed with anti-MSL2 antibody, as in Larschan et al. (2007). Details on the small scale and large-scale ChIP are provided in the Supplemental Experimental Procedures.
ChIP-chip and ChIP-seq data analysis
Details for ChIP-chip and ChIP-seq data analysis can be found in Supplemental Experimental Procedures.
ChIP DNA Sample Prep for Solexa/Illumina
Immunoprecipitated DNA from Clone 8 cells was isolated as in Alekseyenko et al. (2006). DNA amplification was performed using the Illumina Sample Prep Kit with minor modifications. 1 µl adapter oligo mix was used per 100 ng ChIP DNA. The ligation mix was first amplified as detailed in the protocol and then the PCR products (3–8 µg) were gel-fractionated using Biorad Certified Low Range Ultra Agarose. Different fractions (100–200 bp, 200–300 bp, 300–400 bp) were collected. Purified DNA was run on the Solexa Genome Analyzer. A detailed protocol for Solexa sample preparation is available in Supplemental Experimental Procedures.
Immunocytochemistry
Polytene chromosomes were isolated from female y w; CES-X; H83M2-6I, msl3 larvae and processed for immunostaining as described in Larschan et al. (2007).
Real-time PCR
Quantitation of immunoprecipitated DNA was performed using real time PCR as described in Larschan et al. (2007). Primer sequences used are in Table 4S. Probe design was as follows: at least one of the probes was selected in the center of the Solexa peak, and two probes were 1 or 2 kb distal and proximal to the peak (shoulders). A background control sequence was selected from the baseline Solexa values.
Supplementary Material
Supplemental Data including Supplemental Experimental Procedures, Supplemental References, six figures, and four tables can be found with this article online at.
Acknowledgements
Many thanks to M. Gelbart, T. Sural, P. Yang and M. Tolstorukov for critical comments on the manuscript, H. Oh, J. Bateman, T. Wu, J. Bischof, F. Karch and K. Basler for key fly stocks, S. Elgin for important discussions, Y. Schwartz and T. Kahn for sharing protocols and J. Racine and A. Sarovschii for technical assistance. This work was supported by the National Institutes of Health: GM45744 to M.I.K, GM67825 to P.J.P and HG003079 to E.R.M. (co-PI).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data accessibility
ChIP-chip and ChIP-seq data in this manuscript have been submitted to NCBI GEO public repository with the accession number GSE11485.
References
- Alekseyenko AA, Larschan E, Lai WR, Park PJ, Kuroda MI. High-resolution ChIP-chip analysis reveals that the Drosophila MSL complex selectively identifies active genes on the male X chromosome. Genes Dev. 2006;20:848–857. doi: 10.1101/gad.1400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H, Axel R. Genes expressed in neurons of adult male Drosophila. Cell. 1997;88:459–469. doi: 10.1016/s0092-8674(00)81886-3. [DOI] [PubMed] [Google Scholar]
- Bai X, Alekseyenko AA, Kuroda MI. Sequence-specific targeting of MSL complex regulates transcription of the roX RNA genes. EMBO Journal. 2004;23:2853–2861. doi: 10.1038/sj.emboj.7600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashaw GJ, Baker BS. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development. 1995;121:3245–3258. doi: 10.1242/dev.121.10.3245. [DOI] [PubMed] [Google Scholar]
- Bateman JR, Lee AM, Wu CT. Site-specific transformation of Drosophila via phiC31 integrase-mediated cassette exchange. Genetics. 2006;173:769–777. doi: 10.1534/genetics.106.056945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischof J, Maeda RK, Hediger M, Karch F, Basler K. An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci U S A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, McDonel P, Meyer BJ. Recruitment and spreading of the C. elegans dosage compensation complex along X chromosomes. Science. 2004;303:1182–1185. doi: 10.1126/science.1092938. [DOI] [PubMed] [Google Scholar]
- Dahlsveen IK, Gilfillan GD, Shelest VI, Lamm R, Becker PB. Targeting determinants of dosage compensation in Drosophila. PLoS Genet. 2006;2:e5. doi: 10.1371/journal.pgen.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demakova OV, Kotlikova IV, Gordadze PR, Alekseyenko AA, Kuroda MI, Zhimulev IF. The MSL complex levels are critical for its correct targeting to the chromosomes in Drosophila melanogaster. Chromosoma. 2003;112:103–115. doi: 10.1007/s00412-003-0249-1. [DOI] [PubMed] [Google Scholar]
- Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Rozowsky JS, Wei CL, Lee WH, Zhang ZD, Hartman S, Emanuelsson O, Stolc V, Weissman S, Gerstein MB, et al. Mapping of transcription factor binding regions in mammalian cells by ChIP: comparison of array- and sequencing-based technologies. Genome Research. 2007;17:898–909. doi: 10.1101/gr.5583007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagegaltier D, Baker BS. X chromosome sites autonomously recruit the dosage compensation complex in Drosophila males. PLoS Biol. 2004;2:e341. doi: 10.1371/journal.pbio.0020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallach M, Arnau V, Marin I. Global patterns of sequence evolution in Drosophila. BMC Genomics. 2007;8:408. doi: 10.1186/1471-2164-8-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Konig C, Dahlsveen IK, Prakoura N, Straub T, Lamm R, Fauth T, Becker PB. Cumulative contributions of weak DNA determinants to targeting the Drosophila dosage compensation complex. Nucleic Acids Res. 2007;35:3561–3572. doi: 10.1093/nar/gkm282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan GD, Straub T, de Wit E, Greil F, Lamm R, van Steensel B, Becker PB. Chromosome-wide gene-specific targeting of the Drosophila dosage compensation complex. Genes Dev. 2006;20:858–870. doi: 10.1101/gad.1399406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Herzing LBK, Romer JT, Horn JM, Ashworth A. Xist has properties of the X-chromosome inactivation centre. Nature. 1997;386:272–275. doi: 10.1038/386272a0. [DOI] [PubMed] [Google Scholar]
- Holohan EE, Kwong C, Adryan B, Bartkuhn M, Herold M, Renkawitz R, Russell S, White R. CTCF Genomic Binding Sites in Drosophila and the Organisation of the Bithorax Complex. PLoS Genet. 2007;3:e112. doi: 10.1371/journal.pgen.0030112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Mengus G, Gilfillan G, Kennedy HG, Stuckenholz C, Kelley RL, Becker PB, Kuroda MI. Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J. 2001;20:2236–2245. doi: 10.1093/emboj/20.9.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI. Expression of Msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- Kind J, Akhtar A. Cotranscriptional recruitment of the dosage compensation complex to X-linked target genes. Genes Dev. 2007;21:2030–2040. doi: 10.1101/gad.430807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Alekseyenko AA, Gortchakov AA, Peng S, Li B, Yang P, Workman JL, Park PJ, Kuroda MI. MSL complex is attracted to genes marked by H3K36 trimethylation using a sequence-independent mechanism. Mol Cell. 2007;28:121–133. doi: 10.1016/j.molcel.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Lee JT, Jaenisch R. Long-range cis effects of ectopic X-inactivation centres on a mouse autosome. Nature. 1997;386:275–279. doi: 10.1038/386275a0. [DOI] [PubMed] [Google Scholar]
- Lee JT, Strauss WM, Dausman JA, Jaenisch R. A 450 kb transgene displays properties of the mammalian X-inactivation center. Cell. 1996;86:83–94. doi: 10.1016/s0092-8674(00)80079-3. [DOI] [PubMed] [Google Scholar]
- Legube G, McWeeney SK, Lercher MJ, Akhtar A. X-chromosome-wide profiling of MSL-1 distribution and dosage compensation in Drosophila. Genes Dev. 2006;20:871–883. doi: 10.1101/gad.377506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lieb JD, Liu X, Botstein D, Brown PO. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat Genet. 2001;28:327–334. doi: 10.1038/ng569. [DOI] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS. An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol. 2002;20:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- Lobanenkov VV, Nicolas RH, Adler VV, Paterson H, Klenova EM, Polotskaja AV, Goodwin GH. A novel sequence-specific DNA binding protein which interacts with three regularly spaced direct repeats of the CCCTC-motif in the 5′-flanking sequence of the chicken c-myc gene. Oncogene. 1990;5:1743–1753. [PubMed] [Google Scholar]
- Lucchesi JC, Kelly WG, Panning B. Chromatin remodeling in dosage compensation. Annu Rev Genet. 2005;39:615–651. doi: 10.1146/annurev.genet.39.073003.094210. [DOI] [PubMed] [Google Scholar]
- Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI. Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics. 1997;147:1743–1753. doi: 10.1093/genetics/147.4.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. The Lyon and the LINE hypothesis. Semin Cell Dev Biol. 2003;14:313–318. doi: 10.1016/j.semcdb.2003.09.015. [DOI] [PubMed] [Google Scholar]
- McDonel P, Jans J, Peterson BK, Meyer BJ. Clustered DNA motifs mark X chromosomes for repression by a dosage compensation complex. Nature. 2006;444:614–618. doi: 10.1038/nature05338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL. roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell. 1997;88:445–457. doi: 10.1016/s0092-8674(00)81885-1. [DOI] [PubMed] [Google Scholar]
- Meyer BJ. Sex in the worm counting and compensating X-chromosome dose. Trends Genet. 2000;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- Meyer BJ, McDonel P, Csankovszki G, Ralston E. Sex and X-chromosome-wide repression in Caenorhabditis elegans. Cold Spring Harb Symp Quant Biol. 2004;69:71–79. doi: 10.1101/sqb.2004.69.71. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Pullirsch D, Leeb M, Wutz A. Xist and the order of silencing. EMBO Rep. 2007;8:34–39. doi: 10.1038/sj.embor.7400871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H, Bone JR, Kuroda MI. Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol. 2004;14:481–487. doi: 10.1016/j.cub.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Palmer MJ, Mergner VA, Richman R, Manning JE, Kuroda MI, Lucchesi JC. The male specific lethal-one gene encodes a novel protein that associates with the male X chromosome in Drosophila. Genetics. 1993;134:545–557. doi: 10.1093/genetics/134.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH. Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science. 2002;298:1620–1623. doi: 10.1126/science.1076686. [DOI] [PubMed] [Google Scholar]
- Park Y, Mengus G, Bai X, Kageyama Y, Meller VH, Becker PB, Kuroda MI. Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol Cell. 2003;11:977–986. doi: 10.1016/s1097-2765(03)00147-3. [DOI] [PubMed] [Google Scholar]
- Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–W203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Grosschedl R. Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev. 2007;21:3027–3043. doi: 10.1101/gad.1604607. [DOI] [PubMed] [Google Scholar]
- Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- Stenberg P, Pettersson F, Saura AO, Berglund A, Larsson J. Sequence signature analysis of chromosome identity in three Drosophila species. BMC Bioinformatics. 2005;6:158. doi: 10.1186/1471-2105-6-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun LV, Chen L, Greil F, Negre N, Li TR, Cavalli G, Zhao H, van Steensel B, White KP. Protein-DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc Natl Acad Sci U S A. 2003;100:9428–9433. doi: 10.1073/pnas.1533393100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Bussemaker HJ. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci U S A. 2003;100:2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Reppas NB, Church GM, Struhl K. Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 2005;19:2619–2630. doi: 10.1101/gad.1355605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Zhou S, Yang Y, Scott MJ, Pannuti A, Fehr KC, Eisen A, Koonin EV, Fouts DL, Wrighteman R, Manning JE, et al. Male-specific-lethal 2, a dosage compensation gene of Drosophila that undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cluster. EMBO J. 1995;14:2884–2895. doi: 10.1002/j.1460-2075.1995.tb07288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data including Supplemental Experimental Procedures, Supplemental References, six figures, and four tables can be found with this article online at.