Abstract
During the reproductive cycle, fluctuations in circulating estrogens affect multiple homeostatic systems controlled by hypothalamic neurons. Two of these neuronal populations are arcuate proopiomelanocortin and neuropeptide Y neurons, which control energy homeostasis and feeding. Estradiol modulates these neurons either through the classical estrogen receptors (ERs) to control gene transcription or through a G protein-coupled receptor (mER) activating multiple signaling pathways. To differentiate between these two divergent ER-mediated mechanisms and their effects on homeostasis, female guinea pigs were ovariectomized and treated systemically with vehicle, estradiol benzoate (EB) or STX, a selective mER agonist, for 4 wk, starting 7 d after ovariectomy. Individual body weights were measured after each injection day for 28 d, at which time the animals were euthanized, and the arcuate nucleus was microdissected. As predicted, the body weight gain was significantly lower for EB-treated females after d 5 and for STX-treated females after d 12 compared with vehicle-treated females. Total arcuate RNA was extracted from all groups, but only the vehicle and STX-treated samples were prepared for gene microarray analysis using a custom guinea pig gene microarray. In the arcuate nucleus, 241 identified genes were significantly regulated by STX, several of which were confirmed by quantitative real-time PCR and compared with EB-treated groups. The lower weight gain of EB-treated and STX-treated females suggests that estradiol controls energy homeostasis through both ERα and mER-mediated mechanisms. Genes regulated by STX indicate that not only does it control neuronal excitability but also alters gene transcription via signal transduction cascades initiated from mER activation.
THE HYPOTHALAMUS is a key central nervous system center for controlling many homeostatic processes, including reproduction, stress responses, fluid balance, temperature, and appetite. However, the cellular mechanisms and hypothalamic circuits involved in the control of energy homeostasis are only partially understood. Estradiol is known to affect many different types of neurons involved in the control of energy homeostasis, including the hypothalamic arcuate neurons, proopiomelanocortin (POMC), and neuropeptide Y (NPY), through multiple receptor-mediated mechanisms (1,2,3,4). Estradiol is known to attenuate weight gain after ovariectomy in multiple rodent models primarily through classical estrogen receptor (ER)α-mediated mechanisms and potentially through a novel G protein-coupled receptor (mER) (3,5). Furthermore, physiological levels of estradiol can rapidly attenuate food intake within hours of third ventricular intracerebroventricular administration (6).
Previously, we have identified a putative mER that is Gαq coupled to a phospholipase C (PLC)-protein kinase C (PKC) C-PKA pathway (see Fig. 5) (2). Through this receptor, estradiol reduces the efficacy of the γ-amino-butyric acid (GABA)B receptor agonist baclofen to activate G protein-coupled inwardly rectifying K+ channels in hypothalamic neurons (3). The inhibition of the G protein-coupled inwardly rectifying potassium channel activity will depolarize POMC neurons and increase neuronal activity potentially adding another mechanism for estradiol to control POMC-associated functions. Although the mER gene or proteins has not been identified, the putative mER has been functionally identified in at least three types of arcuate neurons, including POMC, dopamine, and GABA neurons. The mER was initially characterized in female guinea pigs but has also been functionally examined in ERα, ERβ, and ERαβ knockout (KO) mice (3). To elucidate this estradiol-induced signaling pathway, a selective agonist for the mER was discovered that has no binding capacity to classical ERs (7). This compound, called STX, is more potent than estradiol in attenuating the activation of G protein-coupled inwardly rectifying K+ channels by GABAB receptors in POMC neurons (2,3). Because STX can increase the neuronal excitability of POMC neurons, we hypothesized that the putative mER has a role in hypothalamic functions controlled by POMC neurons such as energy homeostasis. Indeed, we have previously reported that chronic, systemic injections of STX lowered post-ovariectomy body weight gain in guinea pigs similar to estradiol treatment (3). In the present study, we repeated the previous whole animal studies using a higher dose of the mER selective ligand, STX, to demonstrate a dose effect of the compound on body weight gain.
Figure 5.
A, Classical ER membrane-associated and nuclear-associated signaling. B, Signaling pathways for the novel membrane ER (mER). 1) Estrogen can activate the classical ER-mediated (ERα/β) pathways leading to changes in mRNA transcription of relevant genes via ERE. 2) Both ERs are regulated by STX, and may be a mechanism for the interaction of classical ER and mER signaling. 3) Estrogen can bind to membrane-associated classical ER leading to the activation of signal transduction pathways to control gene transcription and cellular functions. The putative mER is a G protein-coupled receptor (GPCR) linked to Gq/11 protein that activates PLC-PKC-PKA pathway may control gene transcription via pCREB-CRE. The activation of PKC can also lead to changes in gene transcription via the phosphorylation of other signaling proteins (MAPK/ERK). 4) The hydrolysis of phosphatidylinositol (4,5)-bisphosphate (PIP2) into 1,4,5-trisphosphate (IP3) may also initiate calcium-dependent signaling molecules (CaM/CaMK) to control transcription. Several of these genes are associated with cation and anion channels (Cav3.1, GLR) or proteins that impinge on channel functions (AKAP). 5) Changes in neuronal excitability either by channel expression or channel modulation may also control gene expression. A few of the genes analyzed by qPCR are involved in calcium signaling (CaMK II) and protein trafficking (GEC1).
Furthermore, we hypothesized that STX like estradiol acting through a mER can also activate signaling cascades that could modulate gene transcription. To measure STX-induced gene regulation in the arcuate nucleus, we used a brain-specific cDNA library, produced by suppression subtractive hybridization (SSH), of estradiol-regulated genes from the guinea pig (8) coupled with cDNA gene microarray production. We have previously published a description of the genes in the cDNA library (9) and the initial microarray results of estradiol-treated hypothalamic tissue (8). Presently, we found that STX altered gene transcription in the arcuate nucleus, including genes that are relevant to neuronal excitability, signaling pathways, and energy homeostasis.
Materials and Methods
Animals
Our chosen model for the human reproductive cycle is the female guinea pig because the guinea pig has a long ovulatory cycle (16–18 d), and the regulation of reproductive functions by estrogens is similar to that of primates (10,11,12,13), with a suppression of LH secretion 24 h after estradiol treatment (negative feedback), followed by the LH surge at 42 h after estradiol treatment (positive feedback) (14). All animal procedures described in this study are in accordance with institutional guidelines based on National Institutes of Health standards, and were performed with institutional Animal Care and Use Committee approval at the Oregon Health & Science University. Female multicolor guinea pigs (400–500 g; Elm Hill Breeding Labs, Chelmsford, MA) were used in these experiments. The guinea pigs were maintained under constant temperature (22 C), and the lights were on between 0630 and 2030 h. Animals were housed in social groups with food and water provided ad libitum.
Measurement of the effects of estradiol and STX on post-ovariectomy body weight gain
Adult female guinea pigs weighing approximately 400 g were ovariectomized under ketamine-xylazine anesthesia (33 and 6 mg/kg, respectively, sc) and allowed to recover for 1 wk before initiation of injection treatments. The animals were injected three times per week (Monday, Wednesday, and Friday) for 4 wk with propylene glycol (vehicle) (n = 5), estradiol benzoate (EB) (8 μg/kg; n = 5), or STX (6 mg/kg; n = 7). The body weight was recorded before each injection. The dosage of EB used in this experiment produced serum estradiol levels well below the preovulatory surge of estradiol. Using RIA, the average level of estradiol from the EB-treated females was 25.2 ± 3.2 pg/ml, with the vehicle-treated serum estradiol levels less than 10 pg/ml, which is near or below the detection limit of the assay (Oregon National Primate Research Center Hormone Assay Core, Beaverton, OR). Previously, we have reported the biochemical recipe for the production of a nonsteroidal diphenylacrylamide compound, STX, which does not bind to ERα or ERβ (2,7). The STX was produced in the laboratory of T.S.S. with a purity of more than 95%. A two-way ANOVA (repeated measures) followed by a Newman-Keuls post hoc test was used to determine statistical significance for body weight.
Tissue dissection
Vehicle-, STX-, and EB-treated ovariectomized females were decapitated after sedation with ketamine (33 mg/kg, sc) 24 h after the final treatment injection. Hypothalamic nuclei were microdissected for RNA extraction and microarray analysis using our customized guinea pig microarray gene chip created from a cDNA library of estradiol-regulated, brain-specific genes (9). The basal hypothalamus was cut using a brain slicer (EM Corp., Chestnut Hill, MA) into 1-mm thick coronal rostral and caudal blocks corresponding to certain figures in the Bleier (15) guinea pig atlas of the hypothalamus (Figs. 18–22 and 23–26). The tissue blocks were placed in RNAlater (Ambion, Inc., Austin, TX), and the rostral and caudal parts of the arcuate nucleus were dissected from the rostral and caudal blocks, respectively, using a dissecting microscope. Dissected tissue was stored at −80 C. Total RNA was extracted from the combined nuclei (rostral and caudal arcuate) using Ambion RNAqueous Micro Kits according to the manufacturer’s protocol, and tested for RNA quantity using the NanoDrop ND-100 spectrophotometer (NanoDrop Technologies, Wilmington, DE). RNA quality was assessed by applying 200 ng of each RNA sample to a channel of an Agilent Nanochip and analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA).
Microarray chip production and printing
Two microliters of bacterial culture containing the cloned DNA fragments were used for amplification. Amplifications were performed using DynaZyme EXT (New England Biolabs, Ipswich, MA) with universal primers. Reactions were run in PTC-225 thermocyclers (Bio-Rad Laboratories, Inc., Hercules, CA) for 30 cycles. Reaction products were separated from unincorporated nucleotides using MultiScreen PCR plates (Millipore Corp., Billerica, MA) and verified by visualization by agarose gel electrophoresis. Reaction products were reduced to dryness, then resuspended in 30 μl printing solution [50% dimethylsulfoxide/TE (10 mM Tris, 10 mM EDTA)] and stored for use in 96-well microtiter plates at −20 C. Printing plates were prepared using 3 μl of each reaction product in 384-well microtiter plates (Whatman, Maidstone, Kent, UK). Arrays were printed onto UltraGAPS slides (Corning, Corning, NY) on a PixSys 5500XL microarray printer (Genomic Solutions, Ann Arbor, MI) using CMP3 printing pins (TeleChem, Sunnyvale, CA). Arrays were printed in duplicate on each slide. After printing, slides were baked at 80 C for 3.5 h and stored in a desiccator until use.
RNA amplification, labeling, and cDNA hybridization
Amplified RNA was produced from total RNA with a single round of amplification using MessageAmp kits (Ambion). Labeled target was then made from 2.5 μg amplified RNA using the SuperScript Indirect cDNA labeling kit (Invitrogen Corp., Carlsbad, CA) with either single-use cyanine-5 (Cy5) or cyanine-3 (Cy3) dye packs (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). Before hybridization, slides were blocked for 1 h in 2% BSA (Fisher Scientific, Waltham, MA). Hybridizations were done with two slides per sample pair, arranged facing each other with the barcode labels functioning as the slide separators. Cy5-labeled sample and Cy3-labeled control were combined in 5× standard saline solution (SSC)/0.1% sodium dodecyl sulfate (SDS) and hybridized at 65 C overnight in InSlideOut slide hybridization chambers (Boekel, Feasterville, PA). After hybridizations, slide “sandwiches” were placed in 0.5× SSC/0.1% SDS and separated, then washed sequentially in 0.5× SSC/0.1% SDS, 0.06× SSC/0.1% SDS, and 0.06× SSC. Slides were then spun dry and placed in dark slide boxes until scanned.
Microarray scanning and analysis
Slides were scanned on a ScanArray 4000XL microarray scanner (PerkinElmer, Waltham, MA) using the proprietary ScanArray software. Scans were done at 5-μm resolution. Images were stored as 16-bit tiff files. Images were quantified using ImaGene image analysis software (BioDiscovery, El Segundo, CA). Preprocessing for quality control was done using custom R scripts based on BioConductor 1 (16).
Microarray data preprocessing and normalization
We computed the background-adjusted signal intensities by subtracting the mean local background intensity from the mean signal intensity. Negatively adjusted signals were flagged and removed from subsequent analysis. The data were filtered using a threshold value based on the 95th percentile of blank spots. For each gene there were four technical replicates. Any genes with expression values below this threshold for any of the four technical replicates were considered absent or not expressed, and were not included for further analysis. Data were transformed into a ratio by dividing the signal of the Cy3 channel by Cy5 channel. A ratio of one is considered normal expression, and a ratio of more than one or less than one is considered overexpression and underexpression, respectively. In general, when the distribution of the ratios is positively skewed, the ratios are Log2 transformed. Normalization was performed using the within-slide, intensity-dependent normalization method of Yang et al. (17). The normalized log ratio is the main measurement used to analyze the data set. To address the issue of outlier detection, any gene with an absolute value of studentized residual 2 or Cook’s distance 2 for any of the four technical replicates was marked as an outlier and filtered out.
Discovery of differentially expressed genes
One-way ANOVA was used to determine whether the STX treatment group is significantly expressed or not, and whether genes from STX treatment are significantly different compared with vehicle. The false discovery rate (FDR) (18) adjusted P value had been used to identify the differentially expressed genes. The FDR of a set of predictions is the expected percentage of false predictions in the set of predictions. For example, if the algorithm returns 100 genes with a FDR of 0.3, then we expect 70% of them to be correct. A fold-change (FC) cutoff of more than ±1.25 (FDR P < 0.05) was used as an initial criterion for the selection of STX-regulated genes. All these genes were sorted into functional categories.
Quantitative real-time PCR (qPCR)
To confirm the microarray results of selected genes known to be regulated by estradiol, we designed guinea pig-specific primers using areas of high homology between multiple species by aligning cDNA sequences from our guinea pig cDNA library (8) or known guinea pig sequences with sequences from human and rodents. All primers were designed to span introns and synthesized by Invitrogen using Clone Manager 5 software (Scientific & Educational Software, Cary, NC). See Table 1 for a listing of all the primer sets used for qPCR. Because of the loss of one of the EB-treated RNA samples leaving us with a number of four EB-treated samples, we decided to also compare selected STX-regulated genes with another set of long-term EB-treated arcuate RNA samples (n = 6) from a previous body weight experiment (3). The vehicle for this earlier set of RNA samples was sesame oil, therefore, the oil samples (n = 7) from that experiment were used as the control for the EB-treated qPCR analysis. Both EB-treated groups were analyzed by qPCR for some of the selected genes.
Table 1.
Primer sequences used for qPCR
| Name | Product length | Primer efficiency | Primer sequence | Base pair no. | Accession no. |
|---|---|---|---|---|---|
| KCNQ5 | 225 bp | 100 | CTC ACC CCA CCA CTT AAA | 1623–1640 | NM_019842 |
| CTC TCG GCT CTT CTT ATC TG | 1847–1828 | ||||
| SUR1 | 145 bp | 91 | TCG CCG GGA CAG AAG ATT GG | 911–930 | AF183921 |
| GGG TGT GCA GTG GCA GTT TG | 1055–1036 | ||||
| GEC-1 | 234 bp | 100 | GCG CGA GTC TTC ATC AAG GG | 244–263 | AF012920.2 |
| TGA GGT CAG AGG GCA CAA GG | 477–458 | ||||
| ERα | 56 | 97 | CTG CGC AGT GTG CAA TGA C | 19–37 | DQ218311 |
| TCA CAG GAC CAG ACC CCA TAA | 75–55 | ||||
| ERβ | 58 | 99 | AGA ACC GGC GGA AAA GCT | 96–113 | DQ218312 |
| CAT TCC TAC TCC ATA GCA CTT TCG | 154–131 | ||||
| Cav3.1 | 133 bp | 96 | CTG GAG CTG GAG ATG AAG AC | 162–181 | DQ992353.1 |
| AGT GGG CTG CCT TGT ATG | 294–277 | ||||
| CaM-1 | 176 bp | 92 | TCT GCA CAT CAT CCT GAG | 164–181 | NM_009790 |
| CAT GCA GCT TGA CGA TTG | 339–322 | ||||
| AKAP11 | 177 bp | 94 | GTC CAA TGA TGG CAA GTC AG | 217–236 | NM_144490 |
| CGT GCA ATC CAT CTA CGC AG | 393–374 | ||||
| PKAα1 | 162 bp | 95 | GAG CAG CCA CTG TCA AAG C | 827–845 | NM_212471 |
| CGT TCC CAC TTG TCC AGA G | 988–970 | ||||
| PI3K p85α | 190 bp | 93 | GCC AGA CCT CAT TCA GCT AAG | 1701–1721 | NM_174575 |
| GTT GCT GCT GCC AAC ATT C | 1890–1872 | ||||
| GLRβ | 246 bp | 98 | CAT GCC CTT TGG ACT TGA C | 621–639 | NM_000824 |
| TCC TCA GGG TGA AGA TGA C | 866–848 | ||||
| CaMKIIα | 101 bp | 93 | GAA GAC ACC AAA GTG CGG AAA C | 1151–1172 | BC040457 |
| TCG CAC ATC TTC GTG TAG GAC | 1251–1231 | ||||
| PITPβ | 114 bp | 93 | GTTG TGC CTA TAA GCT GGT GAC | 602–622 | BC031427 |
| AGC TGG CGA TGG AAG TTT G | 715–697 | ||||
| TH | 223 bp | 93 | TCC ACG TTA TAC TGG TTC AC | 64–83 | DQ992354 |
| TTG CAT CAC TGA AGC TCT C | 286–268 | ||||
| GAPDH | 212 bp | 100 | CAT CCA CTG GTG CTG CCA | 123–142 | U51572 |
| GTC CTC GGT GTA GCC CAA G | 334–315 | ||||
| NPY | 91 | 94 | CCC GAG ACA CTG ATT TCA GAT CT | 294–316 | NM_000905 |
| Probe | CCC ATC ACC ACA TAG AAG GAT CTT | 384–361 | |||
| AAA GCA CAG AAA ATG TCC CCA GAA CTA GGC | 328–357 | ||||
| POMC | 63 | 96 | CGC TGG TGA CGC TGT TCA | 713–730 | S78260.1 |
| Probe | CGC CTC ACT GGC CCT TCT | 775–758 | |||
| AAC GCC ATC GTC AAG A | 733–748 | ||||
| β -Actin | 60 | 100 | AAG CCC AGA GCA AAA GAG GTA TC | 177–199 | AF508792 |
| Probe | TAA CGA TGC CGT GCT CAA TG | 236–217 | |||
| TGACCCTGAAATACC | 201–215 |
The sense primer is listed first with the antisense primer below.
In preparation for qPCR, the remaining total RNA from the microdissected arcuate nucleus samples was deoxyribonuclease I treated (DNAfree; Ambion) at 37 C for 30 min to minimize any genomic DNA contamination. cDNA was synthesized from 200 ng total RNA using 50 U murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA), 4 μl 5 × buffer, 25 mm MgCl2, 10 mm deoxynucleotide triphosphate, 100 ng random hexamer primers (Promega Corp., Madison, WI), 40 U/μl Rnasin (Promega), and 100 mm dithiothreitol in diethylpyrocarbonate-treated water (Ambion) in a total volume of 20 μl. Reverse transcription was conducted using the following protocol: 60 min at 42 C, 5 min at 95 C, and 5 min at 4 C. The cDNA was diluted to 1:20 with Nuclease-free water (Ambion) for a final cDNA concentration of 0.5 ng/μl and stored at −20 C. Basal hypothalamic test tissue RNA was used for positive and negative controls (no reverse transcriptase) and processed simultaneously with the experimental samples.
For qPCR, 4 μl cDNA template (an equivalent of 2 ng total RNA) was amplified using PowerSyber Green master mix (Applied Biosystems) on an ABI 7500 Fast Real-time PCR instrument. Standard curves for each primer pair were prepared using serial dilutions of basal hypothalamus cDNA in triplicate to determine the efficiency [E = 10(−1/m) − 1, m = slope] of each primer pair. All efficiencies expressed as percent efficiency were approximately equal (one doubling per cycle, 90–100%; Table 1), therefore, the relative mRNA expression data were analyzed using the ΔΔCT (cycle threshold) method (19,20). The amplification protocol for all the genes was as follows: 95 C for 10 min (initial denaturing), followed by 45 cycles of amplification at 94 C for 15 sec (denaturing); 60 C for 30 sec (annealing); and completed with a dissociation step for melting point analysis with 35 cycles of 95 C for 15 sec, 60–95 C (in increments of 1 C) for 1 min, and 95 C for 15 sec. However, primers for M-current potassium channel subunit 5 (KCNQ5) and protein kinase A α1 subunit (PKAα1) were optimized with an annealing temperature of 57 C and 62 C for 30 sec, respectively. For NPY and POMC, TaqMan primers and probes were used with β-actin as the reference gene. Positive and negative controls were added to each amplification run, including a water blank. Quantification values were generated only from samples showing a single product at the expected melting point.
Final relative quantitation was done using the comparative CT method (19,20) using a calibrator of pooled, diluted cDNAs from each oil-treated arcuate sample (50 μl/sample). The data are reported as relative mRNA expression. To determine the CT for each transcript, the threshold was consistently set at the lowest point of the exponential curve where the slope of the curve was the steepest and above the baseline of the first 15 cycles. The CT method normalizes the CT from each sample for each target gene by subtracting the CT of the reference gene [glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin], which is unresponsive to estradiol-treatment (ΔCT). The ΔΔCT values were calculated using the pooled oil-cDNA calibrator ΔCT: ΔΔCT = (CT target gene − CT reference gene) − ΔCT of calibrator. The relative linear quantity of target molecules was calculated using the formula 2−ΔΔCT. Therefore, all transcription data are expressed as an n-fold difference relative to the calibrator. The n-fold difference was averaged for each treatment and analyzed statistically using a two-tailed Student’s t test (P < 0.05).
Results
Body weight gain is attenuated in ovariectomized females by EB and STX
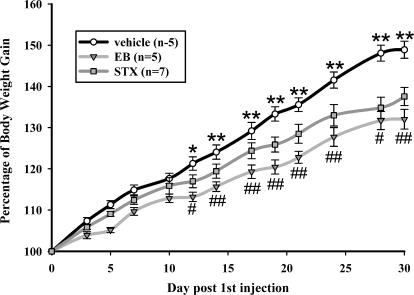
We have previously shown that systemic treatment with STX (2 mg/kg) in guinea pigs will lower the body weight gain after ovariectomy similar to systemic treatment with EB (3). We repeated the experiments with more animals and with a higher dose (6 mg/kg) of STX (Fig. 1). The present study not only reproduced the same effect on post-ovariectomy body weight gain but even showed a significantly greater reduction in body weight gain than in the former STX study. Therefore, when both studies were analyzed for percent body weight gain at the end of each experiment normalized to body weight at ovariectomy, the lower dose of STX has significantly higher weight gain (149 ± 3.0%) compared with the higher dose used in the present study (139 ± 2.0%; P < 0.05). To confirm further the reproducibility of the former study, we also measured uterine weight and found that STX did not have a significant effect on uterine weight (STX: 0.3 ± 0.02 g vs. vehicle: 0.24 ± 0.02), whereas EB did significantly increase uterine weight by nearly 10-fold (EB: 2.24 ± 0.17).
Figure 1.
STX and estradiol significantly reduced body weight gain in ovariectomized female guinea pigs. Estradiol significantly reduced body weight gain by the fifth day after first injection. STX significantly reduced body weight gain compared with vehicle by d 12 (*, P < 0.05; **, P < 0.01). The body weights of STX-treated females were also significantly different from EB-treated females (#, P < 0.05; ##, P < 0.01). Data points represent the mean ± sem of five, five, and seven animals per group for vehicle (circles), EB (triangles), and STX (squares) treatments, respectively. Comparisons between groups were made using a two-way ANOVA (repeated measures) with post hoc Newman-Keuls paired analysis.
STX regulates gene expression in the arcuate nucleus
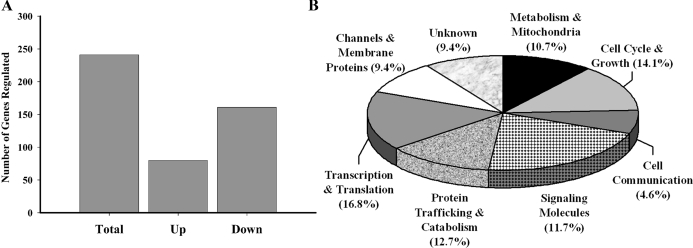
Pairwise comparisons of the mean difference between the vehicle- and STX-treated samples (n = 5 and 7, respectively) for each gene determined that 241 identified genes were significantly different (>±1.25 FC; P < 0.05) between the treatments. We used this FC and P value cutoff because these genes were previously identified as estradiol-regulated during the suppressive subtractive hybridization to produce the cDNA library. Moreover, we found that several of our known estradiol-regulated genes, including tyrosine hydroxylase (TH), GABA-A receptor associated protein-like 1 (GEC-1), and NPY, were regulated by STX according to this criterion. Of these 241 genes, 80 genes were up-regulated, and 161 genes were down-regulated (Fig. 2A). On the microarray gene chip, over 400 cDNA transcripts were not identified by Basic Local Alignment Search Tool searching, and of these unidentified transcripts, 196 were significantly regulated by STX. Thus, a combined total of 437 transcripts in the cDNA library were regulated by STX according to the microarray. Of the identified genes, 10.7% of them were functionally categorized to be involved in metabolism and mitochondrial function, and 9.4% were channels or other membrane proteins (Fig. 2B). Signaling molecules made up 11.7% of the total, whereas 12.7% of the genes were involved in protein trafficking and catabolism. Genes involved in transcription and translation made up 16.8% of the total with genes involved in cell communication or cell cycle and growth made up 4.6 and 14.1%, respectively.
Figure 2.
A, STX (gray) significantly (FDR P < 0.05; FC > ±1.25) regulates a number of genes compared with vehicle in the arcuate. “Up” are up-regulated genes, and “Down” are down-regulated genes. B, The percent distribution of the STX-regulated genes from the arcuate total RNA samples analyzed by microarray and divided into eight functional categories: Translation & Transcription; Channels & and Membrane Proteins; Signaling Molecules; Cell Cycle & Growth; Metabolism and Mitochondria; Protein Trafficking & Catabolism; Cell Communication; and Unknown. The cDNA microarray chip was created from a brain-specific, estrogen-regulated guinea pig cDNA library produced using SSH.
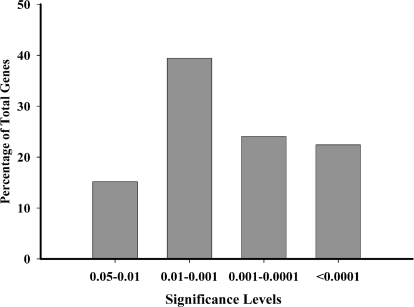
Another cutoff commonly used in microarray analysis is the statistical P values produced during the ANOVA comparison between the treatments. In our study the genes more than ±1.25 FC were divided into four groups of P value ranges. Of the 241 genes, 34 had a P value between 0.05 and 0.01, and 95 had a P value between 0.01 and 0.001 (Fig. 3). Another 58 genes have a P value less than 0.001 but above 0.0001, with the remaining 54 less than 0.0001. A P value cutoff of P < 0.01 for eliminating false positives is commonly used in the analysis of microarray data (21,22,23). However, we chose a modification of the FDR to estimate the percentage of observed total positives expected to be false. With our microarray of 954 genes, a P < 0.05 would result in 48 false positives (954 × 0.05). The FDR would be 48/241 (the number of genes with a FC ± 1.25) or 0.199. This FDR indicates that 20% of the 241 genes should be positive by chance alone and that any single positive has a 20% chance of being a false positive (24). Because the genes are from an estradiol-regulated cDNA library and many of them are known to be involved in homeostatic and neuroendocrine functions, we used the P < 0.05 cutoff and present selected genes from each P value range in Tables 2–5.
Figure 3.
A representation of the percentage of the STX-regulated genes within each of the four P value ranges. All 241 genes with a FC more than ±1.25 were divided into four categories based on their associated P value for the FDR. These categories are: P < 0.05 but more than 0.01; P < 0.01 but more than 0.001; P < 0.001 but more than 0.0001; and P < 0.0001.
Table 2.
A list of genes regulated by long-term STX treatment (0.01 < P < 0.05)
| Accession no. | Gene name | Function | FC | se | P value |
|---|---|---|---|---|---|
| AF183921 | SUR1a | Channels | 1.33 | 0.08 | 0.013 |
| NM_174324 | Adenylate cyclase-inhibiting G protein αI1 | G proteins | −1.38 | 0.12 | 0.022 |
| AB038514 | Na+/K+-ATPase β -1 | Membrane transport | 2.30 | 0.36 | 0.029 |
| NM_004734 | Double cortin and CaM kinase-like 1 | PKs | 1.49 | 0.14 | 0.015 |
| NM_002576 | p21/Cdc42/Rac1-activated kinase 1 | −1.28 | 0.08 | 0.011 | |
| AY257469 | ρ /rac-interacting citron kinase | −1.44 | 0.12 | 0.010 | |
| AF012920.2 | GEC-1a | Protein trafficking | 1.35 | 0.10 | 0.012 |
| DQ166628 | Glucose-regulated protein GRP78 | −1.51 | 0.18 | 0.037 | |
| DQ218312 | ERβa | Receptors | 1.50 | 0.07 | 0.042 |
| NM_144490 | AKAP11a | Signal transduction | 1.25 | 0.09 | 0.016 |
| AF250396 | Dopamine receptor interacting protein 2 | −1.26 | 0.09 | 0.024 | |
| NR_003252.1 | Inositol polyphosphate-5-phosphatase F | −1.38 | 0.11 | 0.011 | |
| NM_001005473 | Phosphatidylinositol-specific PLC, X domain containing 3 | −1.52 | 0.14 | 0.014 | |
| NM_000905 | NPYa | Synaptic transmission | −1.36 | 0.11 | 0.019 |
| S78260.1 | POMCa | 3.28 | 0.46 | 0.030 | |
| DQ992354 | THa | 1.40 | 0.11 | 0.028 |
Transcript tested with qPCR.
Table 3.
A list of genes regulated by long-term STX treatment (0.001 < P < 0.01)
| Accession no. | Gene name | Function | FC | se | P value |
|---|---|---|---|---|---|
| XM_509005.2 | Contactin 1 | Cell adhesion | −1.28 | 0.07 | 0.0028 |
| XM_535617 | Cyclin I | Cell cycle | −1.27 | 0.07 | 0.0037 |
| NM_205842 | NCK-associated protein 1 | Cell proliferation | −1.26 | 0.07 | 0.0022 |
| DQ992353.1 | Cav3.1a | Channels | 1.37 | 0.08 | 0.0086 |
| AF467766 | Guanine nucleotide exchange factor | G proteins | −1.47 | 0.10 | 0.0016 |
| NM_002872.3 | Ras-related C3 botulinum toxin substrate 2 | −1.42 | 0.09 | 0.0035 | |
| BC047214 | F-box and leucine-rich repeat protein 5 | Protein catabolism | −1.50 | 0.11 | 0.0030 |
| BC053537 | F-box protein 33 | −1.27 | 0.07 | 0.0062 | |
| NM_007106 | Ubiquitin-like 3 | −1.30 | 0.08 | 0.0062 | |
| NM_212471 | PKAα 1a | PKs | −1.41 | 0.09 | 0.0030 |
| NM_009790.4 | CaM-1a | −1.27 | 0.07 | 0.0064 | |
| NM_080885 | Cyclin-dependent kinase 5 | 2.38 | 0.18 | 0.0014 | |
| M29550 | Calcineurin A1 | Protein phosphatase | −1.47 | 0.11 | 0.0030 |
| AF494296 | Syntaxin-interacting protein | Protein trafficking | −1.27 | 0.07 | 0.0021 |
| AJ012185 | GABA(B)-R1 | Receptors | 2.04 | 0.18 | 0.0041 |
| NM_174575 | PI3K p85αa | Signal transduction | −1.61 | 0.14 | 0.0083 |
| NM_080555.2 | Phosphatidic acid phosphatase type 2B | 1.43 | 0.11 | 0.0082 | |
| NM_000817 | Glutamate decarboxylase 67 | Synaptic transmission | −2.03 | 0.20 | 0.0060 |
Transcript tested with qPCR.
Table 4.
A list of genes regulated by long-term STX treatment (0.0001 < P < 0.001)
| Accession no. | Gene name | Function | FC | se | P value |
|---|---|---|---|---|---|
| NM_019842 | KCNQ5a | Channels | −1.81 | 0.14 | 0.0009 |
| NM_171830 | Maxi Kβ 1 | 1.54 | 0.08 | 0.0005 | |
| NM_011695 | Voltage-dependent anion channel 2 | −1.58 | 0.09 | 0.0003 | |
| AF508792 | β -Actina | Cytoskeleton | 2.01 | 0.13 | 0.0001 |
| NM_006098.4 | G protein, β -polypeptide 2-like 1 | G proteins | 1.36 | 0.07 | 0.0010 |
| NM_194301 | GTPase activating Rap/RanGAP domain-like 1 | −1.45 | 0.08 | 0.0003 | |
| NM_020988.2 | Gα, ο−1 activating activity polypeptide | −1.40 | 0.07 | 0.0003 | |
| NM_016131.3 | RAB10, member RAS oncogene family | −1.38 | 0.07 | 0.0009 | |
| AF480466 | ρ -GTPase activating protein 10 | −1.87 | 0.12 | 0.0002 | |
| BC040457 | CaMKII αa | PKs | 2.07 | 0.17 | 0.0009 |
| NM_006869 | Centaurin, α 1 | Receptors | 1.81 | 0.14 | 0.0006 |
| NM_000824 | GLRβa | −1.72 | 0.10 | 0.0001 | |
| NM_016783.3 | Progesterone receptor membrane component 1 | −1.27 | 0.06 | 0.0009 | |
| BC031427 | PITPβa | Signal transduction | −1.31 | 0.06 | 0.0004 |
| NM_006226 | PLC-like 1 | −1.60 | 0.11 | 0.0008 |
Transcript tested with qPCR.
Table 5.
A list of genes regulated by long-term STX treatment (P < 0.0001)
| Accession no. | Gene name | Function | FC | se | P value |
|---|---|---|---|---|---|
| AY339885 | Cytochrome c oxidase subunit VIIβ | Electron transport | −1.32 | 0.05 | 0.0000 |
| BC000484 | Ubiquinol-cytochrome c reductase core protein II | −1.60 | 0.08 | 0.0000 | |
| NM_003716 | Ca2+-dependent secretion activator | Exocytosis | −1.57 | 0.08 | 0.0000 |
| BT020037 | RAB, RAS oncogene family-like 2B | G proteins | 17.44 | 0.34 | 0.0000 |
| NM_138647 | Longevity assurance gene homolog 1 | Lipid biosynthesis | 1.90 | 0.08 | 0.0000 |
| NM_004171.3 | Glutamate transporter A2 | Membrane transport | −1.50 | 0.05 | 0.0000 |
| X75252.1 | Phosphatidylethanolamine binding protein | Metabolism | 1.86 | 0.10 | 0.0000 |
| NM_006313 | Ubiquitin specific peptidase 15 | Protein catabolism | −1.60 | 0.07 | 0.0000 |
| NM_008913 | Protein phosphatase 3, catalytic subunit, α | Protein phosphatase | 1.78 | 0.08 | 0.0000 |
| BC026733 | Protein phosphatase 2, regulatory subunit A, α | 3.13 | 0.16 | 0.0000 | |
| NM_001658 | ADP-ribosylation factor 1 | Protein trafficking | 1.82 | 0.08 | 0.0000 |
| BC01083 | Ribophorin I | 1.55 | 0.09 | 0.0000 | |
| NM_053329 | IGF binding protein, acid labile | Signal transduction | 5.63 | 0.31 | 0.0000 |
| NM_012324 | MAPK 8 interacting protein 2 | −1.38 | 0.05 | 0.0001 | |
| AF033850 | Phospholipase D2 | 1.75 | 0.09 | 0.0000 | |
| AY305871 | Migration-inducing protein 6 | Stress response | 1.97 | 0.12 | 0.0000 |
| BC013850 | Amyloid β (A4) precursor-like protein 1 | Synaptic transmission | −1.38 | 0.05 | 0.0000 |
Confirmation of selected genes by qPCR
The genes listed in Tables 2–5 demonstrate the wide variety of genes that are regulated by systemic STX in the arcuate nucleus. These identified genes have a multitude of functions in neurons but collectively suggest that estradiol through the mER may affect a wide variety of cellular mechanisms through the control of gene transcription. However, because we are primarily interested in the signaling pathways involved in energy homeostasis, we selected nine genes isolated during the cDNA library production that were regulated by STX plus eight genes added to the microarray as controls to be analyzed by qPCR. These 17 genes include receptors, cation channel subunits, calcium and phosphatidylinositol signaling molecules, and genes involved in the control of energy homeostasis and feeding behavior. Estradiol has previously regulated a number of these genes in the arcuate nucleus (9,25,26,27).
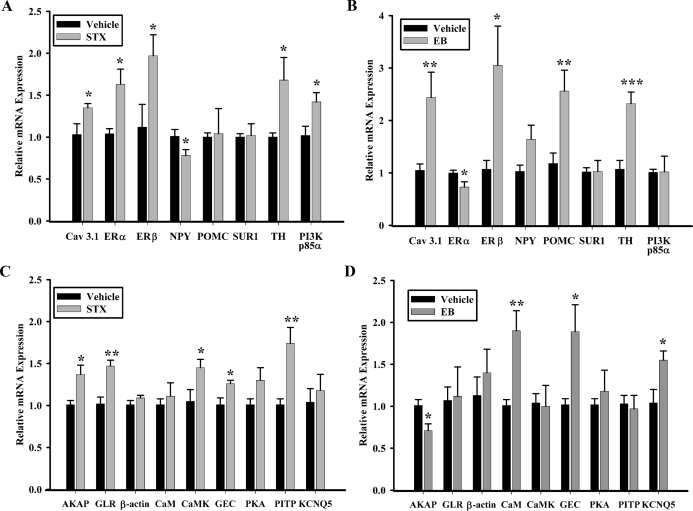
Of these 17 genes, 16 of them had significantly different expression levels in STX-treated females compared with vehicle-treated females, with ERα the only gene that did not reach significance in the microarray according to our criteria but was significantly up-regulated by STX treatment using qPCR (Fig. 4A). In Table 2 (P < 0.05 to > 0.01), we confirmed STX-induced expression differences by qPCR for five of the seven genes in this table (Fig. 4, A and C). In Table 3 (P < 0.01 to P > 0.001), we examined four genes with qPCR and confirmed directional regulation in only one gene [T-type calcium channel 3.1 (Cav3.1)]. Another gene, phosphatidylinositol 3-kinase (PI3K), p85α subunit (PI3K p85α), was suppressed by STX according tomicroarray analysis but was significantly up-regulated by STX using qPCR. This confirmation of gene regulation in the opposite direction occurred with two more genes, and in all three instances, gene expression was suppressed based on the microarray but enhanced using qPCR. The other two genes were this occurred are presented in Table 4 (P < 0.001 to P > 0.0001) along with three other genes that were analyzed with qPCR. In this P value category, three of the five genes were regulated by STX treatment in the arcuate nucleus.
Figure 4.
qPCR confirms regulation of multiple genes by STX in the arcuate nucleus. The relative mRNA expression for selected control genes (A and B) and selected genes of interest (C and D) from microdissected arcuate tissue from vehicle-treated [propylene glycol (n = 5) or oil (n = 7); black bars] females and STX- (n = 7) or EB-treated (n = 6) females (gray bars). The expression values were calculated using the ΔΔCT method where the calibrator was the average ΔCT of the vehicle-treated samples. Bar graphs represent the mean ± sem. Statistics: two-tailed Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001 compared with vehicle).
Comparison to estradiol treatment using qPCR
For comparison to chronic STX treatment, we also analyzed the expression of the 17 selected genes in the arcuate nucleus from long-term EB- and vehicle-treated females (an independent set of experiments in which body weight was significantly reduced by EB) using qPCR (Fig. 4, B and D). Among the control genes that are known to be regulated by estradiol, EB up-regulated Cav3.1 (α1 G) (2.44 ± 0.48 vs. 1.05 ± 0.12, vehicle; all genes were normalized to vehicle expression, i.e. ∼1.00), ERβ (3.05 ± 0.75), POMC (2.56 ± 0.4), and TH (2.32 ± 0.22), while down-regulating ERα (0.73 ± 0.1). STX up-regulated Cav3.1 (1.35 ± 0.05), ERβ (1.97 ± 0.25), and TH (1.68 ± 0.27), and also up-regulated PI3K p85α (1.42 ± 0.11), which EB did not affect. Unlike EB, STX did not have an effect on POMC relative mRNA expression but did significantly decrease NPY expression in the arcuate nucleus (0.78 ± 0.07). Among the genes isolated during the SSH, chronic EB treatment up-regulated calmodulin 1 (CaM-1) (1.9 ± 0.24), GEC-1 (1.89 ± 0.32), and KCNQ5 (1.55 ± 0.11), while down-regulating A-kinase anchoring protein 11 (AKAP11) (0.71 ± 0.08). Conversely, STX up-regulated AKAP11 (1.37 ± 0.11), calcium/calmodulin-dependent kinase II α subunit (CaMKIIα) (1.45 ± 0.1), GEC-1 (1.26 ± 0.04), glycine receptor β-subunit (GLRβ) (1.47 ± 0.07), and phosphatidylinositol transfer protein β (PITPβ) (1.74 ± 0.19). Although the microarray indicated a 2-fold increase in the expression of β-actin, a commonly used qPCR reference gene, neither EB- nor STX-treatment changed the relative expression of β-actin using qPCR and GAPDH as the reference gene.
To compare between the two sets of EB-treated samples [long-term treated EB samples from the earlier study (n = 6) or the current study (n = 4)], we analyzed gene expression of 11 of the 17 selected genes using the current EB-treated samples (n = 4) and found similar levels of relative mRNA expression in nine of the 11 genes. In the current EB-treated group (n = 4), AKAP11 and ERα mRNA expression was suppressed (0.64 ± 0.11** and 0.8 ± 0.03*, respectively; *, P < 0.05; **, P < 0.01 compared with oil-treated samples). Also in this group, EB treatment increased the relative mRNA expression of CaM-1 (1.9 ± 0.24**) Cav3.1 (1.76 ± 0.2*), ERβ (2.01 ± 0.3*), and KCNQ5 (1.55 ± 0.11*). Similar to the earlier experiment, EB treatment did not have an effect on NPY, PKAα1, and PI3K p85α expression. However, in the current (n = 4) samples, EB treatment did not affect GEC-1 expression but did increase sulfonylurea receptor 1 (SUR1) expression (1.34 ± 0.09**). It should be noted that in the current group with a lower sample number, the variation was greater compared with the n = 6 group, but the majority of genes were similarly affected by EB.
Discussion
In this study we examined the effects of STX, a selective agonist for the putative mER, on body weight gain in ovariectomized guinea pigs. We then determined whether systemic STX regulates gene transcription in the arcuate nucleus using microarray analysis in conjunction with qPCR in the same group of animals. The potential signaling pathways leading to gene expression are illustrated in Fig. 5. We have shown that a unique gene regulation signature occurs in the arcuate nucleus in the STX-treated females. The genes regulated have a variety of functions in the cell, including signal transduction, transcription, translation, protein trafficking, and other vital aspects of neuronal activity and survival. Because microarray analysis is a crude indicator of individual gene expression, relevant genes should be verified by other techniques of mRNA analysis such as qPCR. Of the 16 selected genes that had significant differences in expression between the vehicle and STX after microarray analysis, 10 of these selected genes were regulated by STX using qPCR. Therefore, the confirmation of the microarray results suggests that our novel array is useful in obtaining data for a global picture of gene regulation in the context of STX treatment and an adequate indicator of the genes regulated by STX.
The role of a mER and membrane-initiated gene transcription in energy homeostasis
The central, hypothalamic effects of estradiol on energy homeostasis are primarily thought to function through ERα-mediated mechanisms. This hypothesis is partially due to early observations of an obesity phenotype in αERKO mice. The αERKO mice exhibited an obesity phenotype, whereas the βERKO mice did not (28,29). ERα is also considered important for the attenuation of post-ovariectomy body weight gain by estradiol in mice because estradiol did not have a significant effect in αERKO mice on weight gain (5).
An important nucleus in the hypothalamic control of energy homeostasis besides the arcuate nucleus is the ventromedial nucleus of the hypothalamus (VMH). Estradiol has many actions in this nucleus, and, in fact, the suppression of ERα by RNA interference in the VMH of both mice and rats produced a phenotype similar to metabolic syndrome with an increase in body weight, food intake, and reduced metabolism (30). There are synaptic interactions between the neurons of the VMH and arcuate POMC neurons, and VMH neurons do express the melanocortin-4 receptors (31,32). Because these neurons are also activated by STX and estradiol increasing their neuronal excitability, this suggests that estradiol may have more than one pathway to effect VMH-mediated control of energy homeostasis.
The effects of estradiol in the transgenic mouse model are also thought to be due to the loss of estradiol-mediated actions at extrahypothalamic sites in the brain such as the nucleus tractus solitarius. The nucleus tractus solitarius controls feeding behaviors and is prominently involved in the effects of the gut peptide, cholecystokinin, on the control of food intake, which is potentiated by estradiol (5,33,34). In peripheral tissues, another study suggested a role for ERβ in adipose tissue accumulation based on an increase in weight gain and fat accumulation during estradiol treatment in αERKO mice (29). These findings collectively suggest that ERα is the prominent nuclear receptor involved in the estrogenic effects on the neural control of energy homeostasis, although ERβ-mediated signaling may have a part in the periphery.
Another possible pathway for estradiol to control certain aspects of energy homeostasis is through the activation of the putative mER in arcuate neurons. Our previous data and the current data suggest a role for the mER (via STX-induced activation) in the effects of estradiol on energy homeostasis. At this time, we are not certain what part of energy homeostasis (metabolism, feeding behavior, etc.) is directly affected by STX (mER activation). Because estradiol and STX are known to activate POMC neurons, we can hypothesize that part of the STX-mediated effects involves those functions that are controlled by POMC activation and gene expression, i.e. feeding behaviors (35). These effects on POMC neuronal excitability occur before the activation of the melanocortin 3/4 receptors in other hypothalamic nuclei that control energy homeostasis. Indeed, injections of a melanocortin 3/4 receptor antagonist in the lateral ventricle, which blocked the downstream effects of melanocortin signaling, ablated any estrogenic effects on food intake in ovariectomized rats (36).
In recent years, substantial evidence indicates that estradiol can activate a host of rapid signaling cascades to induce gene transcription (membrane-initiated steroid signaling) (37,38,39,40). Through membrane-associated ER, estradiol activates multiple signaling pathways, including PI3K, PLC, and PKA/phosphorylated cAMP response element-binding protein (pCREB). In fact, estrogenic activation of pCREB can induce transcription of genes for important neurotransmitters such as dopamine, enkephalin, dynorphin, and neurotensin (41,42), and regulate CRH gene expression through ERα- and/or ERβ-CRE pathways (43). Although there is little to no direct evidence suggesting that membrane-initiated signaling via classical ER controls energy homeostasis, the rapid signaling initiated by the putative mER (PLC-PKC-PKA) has now been shown to generate nascent transcription potentially through multiple signaling pathways or even due to an increase in neuronal excitability (44,45) (Fig. 5).
Interestingly, STX up-regulated the mRNA expression of both types of ERs in the arcuate nucleus. Although both ERα and ERβ are expressed in the guinea pig arcuate, ERα expression is 4-fold higher than ERβ (9), and estradiol-induced down-regulation of ERα has been previously found to be an estrogen response element (ERE)-dependent transcriptional event (46). In our study, long-term EB treatment also down-regulated ERα; however, in STX-treated females, ERα is up-regulated, which is not surprising because ERα down-regulation is ERE dependent. Expression of the other nuclear ER, ERβ, is increased by both EB and STX treatment. An increase in ERβ expression in the hypothalamus of females during long-term estradiol treatment has not been previously reported. In the pituitary, estradiol treatment suppressed ERβ expression after ovariectomy in rats again through an ERα-dependent mechanism (47). In peripheral cells, PKC- and Ras-mediated pathways regulate the expression of ERα/β genes (48,49), and potentially, similar mechanisms may be involved in the STX-induced increase in classical ER gene expression. The up-regulation of ERα/β by STX in the arcuate nucleus is a potential mechanism whereby the activity of the mER and the classical ERs intersect to control arcuate neuronal functions.
We found that chronic treatment with estradiol increased POMC mRNA expression, whereas STX decreased NPY expression according to qPCR. These two types of arcuate neurons (POMC and NPY) are relevant to the estrogenic effects on energy homeostasis (2,3,4,50,51). POMC neurons decrease food intake (anorectic), whereas NPY neurons increase food intake (orexigenic) (35). Previous studies have shown that estradiol regulates the expression of both of these genes depending upon the treatment paradigm and experimental model (1,4,27,50,51,52,53,54,55). In our study, estradiol did not affect NPY expression, whereas STX suppressed NPY expression. Potentially, STX, the mER selective ligand, exerts some of its effects by suppressing NPY while not directly effecting POMC mRNA expression. The suppression of NPY or the increase in POMC expression would both lead to a similar physiological effect: a decrease in food intake. The changes in POMC or NPY mRNA expression may be a response to changes in caloric intake, and not directly from STX or EB treatments. However, in the mouse the increase in POMC gene expression by estradiol treatment occurs within 12 h (4), which is before there is a measurable effect on body weight in the guinea pigs under the current experimental design. Other genes regulated by the long-term treatments by EB and/or STX are also regulated by 24-h treatment, including AKAP11, Cav3.1, CaM-1, KCNQ5, etc. (9,26), which suggests that some of the actions of STX on gene expression are directly on arcuate neurons. Although we cannot exclude the long-term impact of ovariectomy on NPY expression as reported by Clegg et al. (56), further studies are needed to elucidate central and peripheral effects of STX treatment on homeostatic functions. Regardless of the different responses to EB or STX treatment, it is apparent that the effects of STX are mediated, in part, by direct effects on POMC and NPY neurons in the arcuate nucleus.
Another arcuate neuronal cell type is dopaminergic neurons that are marked by the expression of TH, the rate-limiting enzyme in the catecholamine synthesis pathway. Our data suggest that long-term estradiol treatment up-regulates TH expression in the arcuate nucleus partially through activation of the mER, which has been functionally identified in arcuate dopamine neurons (2). The mER when activated by STX initiates a PLC-PKC signaling pathway that activates PKA. The activation of PKA will initiate pCREB gene regulation, which has already been suggested as a transcriptional pathway for estradiol-induced regulation of TH mRNA expression (41,57). Previous studies have shown that estradiol treatment will increase the expression of both TH mRNA and protein in other dopaminergic neurons (58,59). Although dopamine-deficient mice exhibit a lack of feeding through the loss of motivation that is controlled by the midbrain region (60), it is unknown at this time if the activation of dopamine neurons in the arcuate nucleus by estradiol is a part of estradiol’s control of energy homeostasis.
The regulation of channels and signaling molecules
Estradiol induces gene expression of a Ca2+ channel (Cav3.1) subunit in the arcuate nucleus after 24 h treatment (26) and after long-term estradiol treatment. STX also significantly up-regulates the Cav3.1 in the arcuate nucleus after long-term treatment. Because the mER is found in POMC neurons, one can hypothesize that the up-regulation after STX treatment is occurring in these neurons. The estradiol-induced increase in Cav3.1 expression has previously been demonstrated to increase the peak T-type Ca2+ current by 2-fold in arcuate neurons (26), which includes POMC neurons. The increase in the T-type Ca2+ current would augment burst firing, and cause increases in neurotransmitter and secretory protein (α-MSH) release.
STX, but not estradiol, altered the expression of another type of channel subunit in the arcuate nucleus, the GLRβ, which is the structural subunit for this ligand-gated Cl− channel. Glycine is a major inhibitory neurotransmitter and initiates a postsynaptic increase in chloride conductance through the glycine receptor (61). The inhibitory nature of the glycine receptor may have a role in rapid leptin signaling in the arcuate nucleus because the α1 subunit of the glycine receptor associates with leptin receptors in rat brainstem and may do so in the arcuate nucleus where the effects of leptin on energy homeostasis is centered (62). Therefore, any alteration in the expression of the glycine receptor by the mER may be relevant to the anorectic effects of leptin that occurs through hypothalamic neurons.
There are two signaling pathways that STX controls through modulation of gene expression. The first such pathway is the PI3K pathway. STX up-regulates the p85α subunit of PI3K and PITPβ, whereas estradiol did not significantly regulate either of these genes. Estradiol has recently up-regulated PI3K p85α in the dorsomedial portion of the ventromedial hypothalamic nuclei (adjacent to the arcuate nucleus) after 24 h treatment but did not regulate p85α expression in the arcuate nucleus (25). The other PI3K-associated gene, PITPβ, transports lipids (phosphatidylinositols) from their site of synthesis (endoplasmic reticulum) to the cellular membrane where they are the preferred substrates for the lipid kinases (PI3K, PI4K, etc.) (63). Not only is PITPβ activity required for PI3K, this protein is also necessary for PLC-mediated signaling (64). Because PI3K- and PLC-mediated signaling is implicated in the membrane-mediated effects of estradiol and other factors that control energy homeostasis (leptin, insulin, etc.), any changes in activity or expression of the transfer proteins may be another indirect mechanism for estradiol to potentiate the effects of these peripheral signals on energy homeostasis.
Another signaling pathway affected transcriptionally by STX is the calcium signaling pathways specifically calmodulin-dependent kinase, CaM kinase II. CaMK II is a modulator of ion channels (Ca2+, K+, Na+) (65) and is required for the Ca2+-sensitive production of long-term potentiation in neurons from the hypothalamus (66) and the hippocampus, where leptin enhances long-term potentiation through the Ca2+-independent activation of CaMK II (67). STX-induced gene expression of CaMK II is one more indirect mechanism for estradiol to potentiate the effects of peripheral signals and neuronal functions necessary for the control of energy homeostasis. Furthermore, calmodulin is up-regulated by estradiol both at 24 h (9) and after long-term treatment, as presently illustrated, and an increase in calmodulin expression would also affect the activity of the CaM kinase II.
The regulation of trafficking and scaffold protein genes
In our study we show that long-term EB and STX treatment increased the expression of the GABAA receptor trafficking gene, gec1, in the arcuate nucleus, confirming and expanding previous findings that estradiol increases gec1 mRNA expression (8). GABA neurotransmission relies on appropriate clustering of GABA receptors to the postsynaptic neuronal membrane. GABAA receptor targeting to the membrane is facilitated by the GEC-1 protein, which connects the receptors to the cytoskeleton (68). Because there is an ERE promoter sequence associated with the gec1 gene (69), the up-regulation of gec1 by both treatments suggests that estradiol has at least two pathways to initiate gec1 gene transcription. Because GABA is an inhibitory amino acid transmitter that is critical to hypothalamic functions, an increase in gec1 gene expression could potentiate the GABAA-mediated activity in the arcuate nucleus and may be a mediator of estradiol-regulated inhibitory tone for particular hypothalamic functions.
Previously, we have shown that 24 h EB treatment, down-regulated AKAP11 in the arcuate nucleus (9). In the present study, long-term estradiol treatment also down-regulated AKAP11, whereas STX treatment increases the expression of the AKAP11 mRNA using qPCR. AKAP11 (also known as AKAP220) is a scaffolding protein for kinases and phosphatases, and has binding sites for PKA, protein phosphatase 1, and glycogen synthase-3β (70). Glycogen synthase-3β is a serine/threonine kinase involved in metabolic pathways for glycogen synthesis, and also controls numerous other cellular functions like protein synthesis and gene transcription, including ERα-mediated transcription (71). Glycogen synthase kinase-3β is inhibited by PKA phosphorylation through its common association with AKAP11 (72). A reduction in AKAP11 availability by gene suppression from estradiol treatment in arcuate neurons may be a disinhibitory mechanism for glycogen synthase kinase-3β activities associated with cellular functions.
Conclusions
In this study we have demonstrated that estradiol has a novel hypothalamic signal transduction pathway for the control of gene transcription that is initiated by an estradiol-responsive Gq-coupled membrane receptor. This novel pathway is in addition to the classical ERE-mediated and non-ERE-mediated transcriptional pathways that have been extensively investigated in central and peripheral tissues. The putative hypothalamic Gq-mER via STX is associated not only with new gene transcription but with attenuation in post-ovariectomy body weight gain. The selective mER ligand, STX, attenuates body weight gain in a dose-dependent manner. STX is currently being examined in other homeostatic functions and may be an excellent tool to delineate the effects of the mER on these hypothalamic functions. The mER clearly is a coupling mechanism between the control of gene transcription and rapid signaling events (control of neuronal excitability) that will ultimately determine the effects of estradiol on hypothalamic functions.
Acknowledgments
We thank Dr. Robert Searles at Oregon Health & Science University’s Spotted Microarray Core for conducting the gene microarray analysis and Dr. Byung Park at Oregon Health & Science University’s Center for Biostatistics, Computing and Informatics in Biology and Medicine for statistical analysis of the microarray data. We also thank Beth Rick for her technical expertise in the guinea pig experiments.
Footnotes
This work was supported by Public Health Service Grants NS 38809, NS 43330, DK 68098, and DK 57574. T.A.R. was supported by Public Health Service training Grants 5T32 DA07262 and 1F32 DK079508.
Disclosure Statement: The authors have nothing to disclose.
First Published Online August 28, 2008
Abbreviations: AKAP11, A-kinase anchoring protein 11; CaM-1, calmodulin 1; CaMKIIα, calcium/calmodulin-dependent kinase II α subunit; Cav3.1, T-type calcium channel 3.1 subunit; Cy3, cyanine 3; Cy5, cyanine 5; EB, estradiol benzoate; ER, estrogen receptor; ERE, estrogen response element; FC, fold change; FDR, false discovery rate; GABA, γ-amino-butyric acid; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GEC-1, γ-amino-butyric acid-A receptor associated protein-like 1; GLRβ, glycine receptor β-subunit; KCNQ5, M-current potassium channel subunit 5; KO, knockout; mER, putative G protein-coupled ER; NPY, neuropeptide Y; pCREB, phosphorylated cAMP response element-binding protein; PI3K, phosphatidylinositol 3-kinase; PI3K p85, phosphatidylinositol 3-kinase, p85α subunit; PLC, phospholipase C; PKC, protein kinase C; PITPβ, phosphatidylinositol transfer protein β; PKAα1, protein kinase A α1 subunit; POMC, proopiomelanocortin; qPCR, quantitative real-time PCR; SDS, sodium dodecyl sulfate; SSC, standard saline solution; SSH, suppression subtractive hybridization; SUR1, sulfonylurea receptor 1; TH, tyrosine hydroxylase; VMH, ventromedial nucleus of the hypothalamus.
References
- Priest CA, Roberts JL 2000 Estrogen and tamoxifen differentially regulate β-endorphin and cFos expression and neuronal colocalization in the arcuate nucleus of the rat. Neuroendocrinology 72:293–305 [DOI] [PubMed] [Google Scholar]
- Qiu J, Bosch MA, Tobias SC, Grandy DK, Scanlan TS, Rønnekleiv OK, Kelly MJ 2003 Rapid signaling of estrogen in hypothalamic neurons involves a novel G protein-coupled estrogen receptor that activates protein kinase C. J Neurosci 23:9529–9540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Bosch MA, Tobias SC, Krust A, Graham S, Murphy S, Korach KS, Chambon P, Scanlan TS, Rønnekleiv OK, Kelly MJ 2006 A G protein-coupled estrogen receptor is involved in hypothalamic control of energy homeostasis. J Neurosci 26:5649–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier G, Li S, Luu-The V, Labrie F 2007 Oestrogenic regulation of pro-opiomelanocortin, neuropeptide Y and corticotrophin-releasing hormone mRNAs in mouse hypothalamus. J Neuroendocrinol 19:426–431 [DOI] [PubMed] [Google Scholar]
- Geary N, Asarian L, Korach KS, Pfaff DW, Ogawa S 2001 Deficits in E2-dependent control of feeding, weight gain, and cholecystokinin satiation in ER-α null mice. Endocrinology 142:4751–4757 [DOI] [PubMed] [Google Scholar]
- Qiu J, Xue C, Bosch MA, Murphy JG, Fan W, Rønnekleiv OK, Kelly MJ 2007 Serotonin 5-hydroxytryptamine2C receptor signaling in hypothalamic proopiomelanocortin neurons: role in energy homeostasis in females. Mol Pharmacol 72:885–896 [DOI] [PubMed] [Google Scholar]
- Tobias SC, Qiu J, Kelly MJ, Scanlan TS 2006 Synthesis and biological evaluation of SERMs with potent nongenomic estrogenic activity. ChemMedChem 1:565–571 [DOI] [PubMed] [Google Scholar]
- Malyala A, Pattee P, Nagalla SR, Kelly MJ, Rønnekleiv OK 2004 Suppression subtractive hybridization and microarray identification of estrogen regulated hypothalamic genes. Neurochem Res 29:1189–1200 [DOI] [PubMed] [Google Scholar]
- Roepke TA, Malyala A, Bosch MA, Kelly MJ, Rønnekleiv OK 2007 Estrogen regulation of genes important for K+ channel signaling in the arcuate nucleus. Endocrinology 148:4937–4951 [DOI] [PubMed] [Google Scholar]
- Bethea CL, Hess DL, Widmann AA, Henningfeld JM 1995 Effects of progesterone on prolactin, hypothalamic β-endorphin, hypothalamic substance P, and midbrain serotonin in guinea pigs. Neuroendocrinology 61:695–703 [DOI] [PubMed] [Google Scholar]
- King JC, Ronsheim PM, Liu E, Powers L, Slonimski M, Rubin BS 1998 Fos expression in luteinizing hormone-releasing hormone neurons of guinea pigs, with knife cuts separating the preoptic area and the hypothalamus, demonstrating luteinizing hormone surges. Biol Reprod 58:323–329 [DOI] [PubMed] [Google Scholar]
- Terasawa E, Rodriguez JS, Bridson WE, Wiegand SJ 1979 Factors influencing the positive feedback action of estrogen upon luteinizing hormone surge in the ovariectomized guinea pig. Endocrinology 104:680–686 [DOI] [PubMed] [Google Scholar]
- Terasawa E, Yeoman RR, Schultz NJ 1984 Factors influencing the progesterone-induced luteinizing hormone surge in rhesus monkeys: diurnal influence and time interval after estrogen. Biol Reprod 31:732–741 [DOI] [PubMed] [Google Scholar]
- Wagner EJ, Rønnekleiv OK, Bosch MA, Kelly MJ 2001 Estrogen biphasically modifies hypothalamic GABAergic function concomitantly with negative and positive control of luteinizing hormone release. J Neurosci 21:2085–2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleier R 1983 The hypothalamus of the guinea pig: a cytoarchitectonic atlas. Madison, WI: University of Wisconsin Press [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J 2004 Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP 2002 Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30:e15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Stat Method 57:289–300 [Google Scholar]
- Livak KJ, Schmittgen TD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ-Δ C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW 2001 A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu TT, Fink MY, Mong JA, John G, Auger AP, Ge Y, Sealfon SC 2007 Effective use of microarrays in neuroendocrine research. J Neuroendocrinol 19:145–161 [DOI] [PubMed] [Google Scholar]
- Koza RA, Nikonova L, Hogan J, Rim JS, Mendoza T, Faulk C, Skaf J, Kozak LP 2006 Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PLoS Genet 2:e81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mobbs CV, Yen K, Mastaitis J, Nguyen H, Watson E, Wurmbach E, Sealfon SC, Brooks A, Salton SR 2004 Mining microarrays for metabolic meaning: nutritional regulation of hypothalamic gene expression. Neurochem Res 29:1093–1103 [DOI] [PubMed] [Google Scholar]
- Blalock EM, Chen KC, Sharrow K, Herman JP, Porter NM, Foster TC, Landfield PW 2003 Gene microarrays in hippocampal aging: statistical profiling identifies novel processes correlated with cognitive impairment. J Neurosci 23:3807–3819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyala A, Zhang C, Bryant D, Kelly MJ, Rønnekleiv OK 2008 PI3K signaling effects in hypothalamic neurons mediated by estrogen. J Comp Neurol 506:895–911 [DOI] [PubMed] [Google Scholar]
- Qiu J, Bosch MA, Jamali K, Xue C, Kelly MJ, Rønnekleiv OK 2006 Estrogen upregulates T-type calcium channels in the hypothalamus and pituitary. J Neurosci 26:11072–11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton JE, Loose MD, Kelly MJ, Rønnekleiv OK 1994 Effects of estrogen on the number of neurons expressing β-endorphin in the medial basal hypothalamus of the female guinea pig. J Comp Neurol 341:68–77 [DOI] [PubMed] [Google Scholar]
- Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, Cooke PS 2000 Increased adipose tissue in male and female estrogen receptor-α knockout mice. Proc Natl Acad Sci USA 97:12729–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naaz A, Zakroczymski M, Heine PA, Taylor J, Saunders PT, Lubahn DB, Cooke PS 2002 Effect of Ovariectomy on adipose tissue of mice in the absence of estrogen receptor α (ERα): a potential role for estrogen receptor β (ERβ). Horm Metab Res 34:758–763 [DOI] [PubMed] [Google Scholar]
- Musatov S, Chen W, Pfaff DW, Mobbs CV, Yang XJ, Clegg DJ, Kaplitt MG, Ogawa S 2007 Silencing of estrogen receptor α in the ventromedial nucleus of hypothalamus leads to metabolic syndrome. Proc Natl Acad Sci USA 104:2501–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrold JA, Widdowson PS, Williams G 2003 β-MISH: a functional ligand that regulated energy homeostasis via hypothalamic MC4-R? Peptides 24:397–405 [DOI] [PubMed] [Google Scholar]
- Harrold JA, Williams G, Widdowson PS 2000 Early leptin response to a palatable diet predicts dietary obesity in rats: key role of melancortin-4 receptors in the ventromedial hypothalamic nucleus. J Neurochem 74:1224–1228 [DOI] [PubMed] [Google Scholar]
- Thammacharoen S, Lutz TA, Geary N, Asarian L 2008 Hindbrain administration of estradiol inhibits feeding and activates estrogen receptor-α-expressing cells in the nucleus tractus solitarius of ovariectomized rats. Endocrinology 149:1609–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asarian L, Geary N 2007 Estradiol enhances cholecystokinin-dependent lipid-induced satiation and activates estrogen receptor-α-expressing cells in the nucleus tractus solitarius of ovariectomized rats. Endocrinology 148:5656–5666 [DOI] [PubMed] [Google Scholar]
- Cone RD 2005 Anatomy and regulation of the central melanocortin system. Nat Neurosci 8:571–578 [DOI] [PubMed] [Google Scholar]
- Polidori C, Geary N 2002 Estradiol treatment fails to affect the feeding responses to melanocortin-3/4 receptor agonism or antagonism in ovariectomized rats. Peptides 23:1697–1700 [DOI] [PubMed] [Google Scholar]
- Hammes SR, Levin ER 2007 Extranuclear steroid receptors: nature and actions. Endocr Rev 28:726–741 [DOI] [PubMed] [Google Scholar]
- Kelly MJ, Qiu J, Rønnekleiv OK 2005 Estrogen signaling in the hypothalamus. Vitam Horm 71:123–145 [DOI] [PubMed] [Google Scholar]
- Rønnekleiv OK, Malyala A, Kelly MJ 2007 Membrane-initiated signaling of estrogen in the brain. Semin Reprod Med 25:165–176 [DOI] [PubMed] [Google Scholar]
- Mhyre AJ, Dorsa DM 2006 Estrogen activates rapid signaling in the brain: role of estrogen receptor α and estrogen receptor β in neurons and glia. Neuroscience 138:851–858 [DOI] [PubMed] [Google Scholar]
- Gu G, Rojo AA, Zee MC, Yu J, Simerly RB 1996 Hormonal regulation of CREB phosphorylation in the anteroventral periventricular nucleus. J Neurosci 16:3035–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watters JJ, Dorsa DM 1998 Transcriptional effects of estrogen on neuronal neurotensin gene expression involve cAMP/protein kinase A-dependent signaling mechanisms. J Neurosci 18:6672–6680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalmansingh AS, Uht RM 2008 Estradiol regulates corticotropin-releasing hormone gene (crh) expression in a rapid and phasic manner that parallels estrogen receptor-α and -β recruitment to a 3′,5′-cyclic adenosine 5′-monophosphate regulatory region of the proximal crh promoter. Endocrinology 149:346–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME 2002 Regulation of transcription factors by neuronal activity. Nat Rev Neurosci 3:921–931 [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Mermelstein PG, Xia H, Tsien RW 2003 Signaling from synapse to nucleus: the logic behind the mechanisms. Curr Opin Neurobiol 13:354–365 [DOI] [PubMed] [Google Scholar]
- Lagrange AH, Rønnekleiv OK, Kelly MJ 1997 Modulation of G protein-coupled receptors by an estrogen receptor that activates protein kinase A. Mol Pharmacol 51:605–612 [DOI] [PubMed] [Google Scholar]
- Tena-Sempere M, Navarro VM, Mayer A, Bellido C, Sánchez-Criado JE 2003 Regulation of estrogen receptor (ER) isoform messenger RNA expression by different ER ligands in female rat pituitary. Biol Reprod 70:671–678 [DOI] [PubMed] [Google Scholar]
- Longo M, Peruzzi B, Fortunati D, De Luca V, Denger S, Caselli G, Migliaccio S, Teti A 2006 Modulation of human estrogen receptor a F promoter by a protein kinase C/c-Src-dependent mechanism in osteoblast-like cells. J Mol Endocrinol 37:489–502 [DOI] [PubMed] [Google Scholar]
- Tu Z, Gui L, Wang J, Li X, Sun P, Wei L 2006 Tumorigenesis of K-rats mutation in human endometrial carcinoma via upregulation of estrogen receptor. Gynecol Endocrinol 101:274–279 [DOI] [PubMed] [Google Scholar]
- Bonavera JJ, Dube MG, Kalra PS, Kalra SP 1994 Anorectic effects of estrogen may be mediated by decreased neuropeptide-Y release in the hypothalamic paraventricular nucleus. Endocrinology 134:2367–2370 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Ohtani K, Kato Y, Tanaka Y, Mori M 1996 Estrogen increases hypothalamic neuropeptide Y (NPY) mRNA expression in ovariectomized obese rat. Neurosci Lett 204:81–84 [DOI] [PubMed] [Google Scholar]
- Broad KD, Kendrick KM, Sirinathsinghji DJ, Keverne EB 1993 Changes in pro-opiomelanocortin and pre-proenkephalin mRNA levels in the ovine brain during pregnancy, parturition and lactation and in response to oestrogen and progesterone. J Neuroendocrinol 5:711–719 [DOI] [PubMed] [Google Scholar]
- Crowley WR, Tessel RE, O'Donohue TL, Adler BA, Kalra SP 1985 Effects of ovarian hormones on the concentrations of immunoreactive neuropeptide Y in discrete brain regions of the female rat: correlation with serum luteinizing hormone (LH) and median eminence LH-releasing hormone. Endocrinology 117:1151–1155 [DOI] [PubMed] [Google Scholar]
- Taylor JA, Goubillon M-L, Broad KD, Robinson JE 2007 Steroid control of gonadotropin-releasing hormone secretion: associated changes in pro-opiomelanocortin and preproenkephalin messenger RNA expression in the ovine hypothalamus. Biol Reprod 76:524–531 [DOI] [PubMed] [Google Scholar]
- Treiser SL, Wardlaw SL 1992 Estradiol regulation of proopiomelanocortin gene expression and peptide content in the hypothalamus. Neuroendocrinology 55:167–173 [DOI] [PubMed] [Google Scholar]
- Clegg DJ, Brown LM, Zigman JM, Kemp CJ, Strader AD, Benoit SC, Woods SC, Mangiaracina M, Geary N 2007 Estradiol-dependent decrease in the orexigenic potency of ghrelin in female rats. Diabetes 56:1051–1058 [DOI] [PubMed] [Google Scholar]
- Maharjan S, Serova L, Sabban EL 2005 Transcriptional regulation of tyrosine hydroxylase by estrogen: opposite effects with estrogen receptors α and β and interactions with cyclic AMP. J Neurosci 93:1502–1514 [DOI] [PubMed] [Google Scholar]
- Ivanova T, Beyer C 2003 Estrogen regulates tyrosine hydroxylase expression in the neonate mouse midbrain. J Neurobiol 54:638–647 [DOI] [PubMed] [Google Scholar]
- Serova LI, Maharjan S, Huang A, Sun D, Kaley G, Sabban EL 2004 Response of tyrosine hydroxylase and GTP cyclohydrolase I gene expression to estrogen in brain catecholaminergic regions varies with mode of administration. Brain Res 1015:1–8 [DOI] [PubMed] [Google Scholar]
- Sotak BN, Hnasko TS, Robinson S, Kremer EJ, Palmiter RD 2005 Dysregulation of dopamine signaling in the dorsal striatum inhibits feeding. Brain Res 1061:88–96 [DOI] [PubMed] [Google Scholar]
- Kirsch J 2006 Glycinergic transmission. Cell Tissue Res 326:535–540 [DOI] [PubMed] [Google Scholar]
- Leite JF, Gribble B, Randolph N, Cascio M 2002 In vitro interaction of the glycine receptor with the leptin receptor. Physiol Behav 77:565–569 [DOI] [PubMed] [Google Scholar]
- Cockcroft S, Carvou N 2007 Biochemical and biological functions of class I phosphatidylinositol transfer proteins. Biochim Biophys Acta 1771:677–691 [DOI] [PubMed] [Google Scholar]
- Thomas GM, Cunningham E, Fensome A, Ball A, Totty NF, Truong O, Hsuan JJ, Cockcroft S 1993 An essential role for phosphatidylinositol transfer protein in phospholipase C-mediated inositol lipid signaling. Cell 74:919–928 [DOI] [PubMed] [Google Scholar]
- Pitt GS 2007 Calmodulin and CaMKII as molecular switches for cardiac ion channels. Cardiovasc Res 73:641–647 [DOI] [PubMed] [Google Scholar]
- Fukunaga K, Horikawa K, Shibata S, Takeuchi Y, Miyamoto E 2002 Ca2+/calmodulin-dependent protein kinase II-dependent long-term potentiation in the rat suprachiasmatic nucleus and its inhibition by melatonin. J Neurosci Res 70:799–807 [DOI] [PubMed] [Google Scholar]
- Oomura Y, Hori N, Shirasahi T, Fukunaga K, Takeda H, Tsuji M, Matsumiya T, Ishibashi M, Aou S, Li XL, Kohno D, Uramura K, Sougawa H, Yada T, Wayner MJ, Sasaki K 2006 Leptin facilitates learning and memory performance and enhances hippocampal CA1 long-term potentiation and CaMK II phosphorylation in rats. Peptides 27:2738–2749 [DOI] [PubMed] [Google Scholar]
- Wang H, Bedford FK, Brandon NJ, Moss SJ, Olsen RW 1999 GABA(A)-receptor-associated protein links GABA(A) receptors and the cytoskeleton. Nature 397:69–72 [DOI] [PubMed] [Google Scholar]
- Vernier-Magnin S, Nemos C, Mansuy V, Tolle F, Guichard L, Delage-Mourroux R, Jouvenot M, Fraichard A 2005 Analysis of the guinea-pig estrogen-regulated gec1/GABARAPL1 gene promoter and identification of a functional ERE in the first exon. Biochim Biophys Acta 1731:23–31 [DOI] [PubMed] [Google Scholar]
- Wong W, Scott JD 2004 AKAP signalling complexes focal points in space and time. Nat Rev Mol Cell Biol 5:959–970 [DOI] [PubMed] [Google Scholar]
- Mendez P, Garcia-Segura LM 2006 Phosphatidylinositol 3-kinase and glycogen synthase kinase 3 regulate estrogen receptor-mediated transcription in neuronal cells. Endocrinology 147:3027–3039 [DOI] [PubMed] [Google Scholar]
- Tanji C, Yamamoto H, Yurioka N, Kohno N, Kikuchi K, Kikuchi A 2002 A-kinase anchoring protein AKAP220 binds to glycogen synthase kinase-3β (GSK-3β) and mediates protein kinase A-dependent inhibition of GSK-3β. J Biol Chem 277:36955–36961 [DOI] [PubMed] [Google Scholar]